Abstract
The telomerase ribonucleoprotein complex ensures complete replication of eukaryotic chromosomes. Telomerase RNA, TER, provides the template for replicating the G-rich strand of telomeric DNA, provides an anchor site for telomerase-associated proteins, and participates in catalysis through several incompletely characterized mechanisms. A major impediment towards understanding its non-templating roles is the absence of high content structural information for TER within the telomerase complex. Here, we used selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) to examine the structure of Tetrahymena TER free in solution and bound to tTERT in the minimal telomerase RNP. We discovered a striking difference in the two conformations and established direct evidence for base pair triples in the tTER pseudoknot. We then used SHAPE data, previously published FRET data, and biochemical inference to model the structure of tTER using discrete molecular dynamics simulations. The resulting tTER structure was docked with a homology model of tTERT to characterize the conformational changes of tTER that attend binding to tTERT. Free in solution, tTER appears to contain four pairing regions: stems I, II, and IV, which are present in the commonly accepted structure, and stem III, a large paired region that encompasses the template and pseudoknot domains. Our interpretation of the data and subsequent modeling affords a molecular model for telomerase assemblage in which a large stem III of tTER unwinds to allow proper association of the template with the tTERT active site and formation of the pseudoknot. Additionally, analysis of our SHAPE data and previous enzymatic footpinting allows us to propose a model for stem-loop IV function in which tTERT is activated by binding stem IV in the major grove of the helix-capping loop.
Keywords: Telomerase, ribonucleoprotein complex, RNA footprinting, pseudoknot
INTRODUCTION
Ribonucleic acid has vast functions beyond its canonical roles in the transcription and translation of genetic information. 1 Many of these functions require specific RNA folding, and like proteins, many RNAs fold into complex three-dimensional structures that are essential for their function.1–2 Generally, RNAs are considered more conformationally dynamic than proteins, in part because RNAs possess six backbone torsion angles rather than three backbone torsion angles present in peptides.3 A detailed understanding of RNA function therefore requires a description of both the RNA tertiary structure as well as major available alternative conformations. However, many larger RNAs, particularly those in ribonucleoprotein complexes (RNPs), are challenging to study by X-ray crystallography or NMR. To overcome this problem, computational methods using experimental constraints afford an approach towards obtaining high-resolution structural models as well as assessing conformational flexibility of RNAs.
Telomerase is an important RNP for which high-resolution structural data of the RNA within the RNP remains incomplete. 4 In fact, no structural data of an intact, minimally functional telomerase complex has been reported except for low-resolution electron microscopic analysis of telomerase isolated from Euplotes aediculatus.5 Telomerase elongates the linear chromosomes of most eukaryotes with repeating sequences of guanosine-rich DNA to solve the end replication problem faced during DNA replication.6 Telomerase is critical for the genomic integrity of dividing cells because of its central role in maintaining the chromosome ends. Mutations that disrupt telomerase function have been linked to several genetic disorders such as dyskeratosis congenita and idiopathic pulmonary fibrosis,7 and telomerase activity is elevated in cancer cells.8 The important role of telomerase activity in many human disease states suggests that the detection and control of telomerase may prove to be effective diagnosis and treatment strategies. 9 However, the incomplete understanding of telomerase structure and catalytic mechanism hinders the rational design of effective telomerase-based therapies.
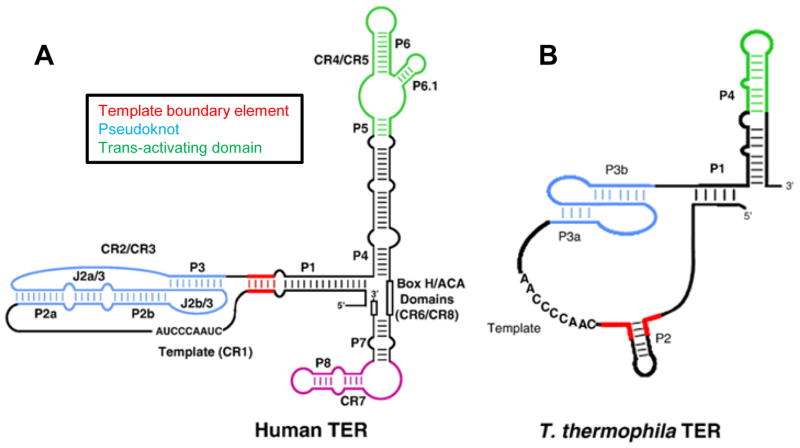
Telomerase RNPs demonstrate rapid evolutionary divergence, but all minimally contain a catalytic subunit, telomerase reverse transcriptase (TERT) and a RNA subunit, telomerase RNA (TER).10 TERT is conserved in telomerase containing species and contains several highly conserved domains including RNA-binding and reverse transcriptase domains. TER’s are not well conserved but do share functionally related domains: a template, a pseudoknot adjacent to the template, and a trans-activating domain that enhances catalytic activity in what appears to be an allosteric fashion.11 Telomerase from the ciliate Tetrahymena thermophila has served as an important model since its activity was first detected,12 and it can be reconstituted in vitro using rabbit reticulocyte lysates and recombinant Tetrahymena TERT and TER.13 Tetrahymena TER (tTER) is 159 nucleotides long and contains the functionally conserved TER domains.14 In addition to these, tTER has several well-characterized domains that contribute to RNP assemblage and biochemical activity (Figure 1). Endogenous telomerase RNPs generally contain an RNA binding protein required for biogenesis and stability of the complex.15 In humans this activity is supplied by the box H/ACA binding protein dyskerin.16 In Tetrahymena, the core telomerase RNP contains the specific tTER-binding protein p65.17 In vitro, the efficiency of telomerase assemblage is enhanced by p65 because of its apparent ability to facilitate conformational changes in tTER and stabilize the active conformation.18
Figure 1.
Cartoon of human and Tetrahymena telomerase RNA.
Telomerase exhibits several unique structural and biochemical features. Unlike most reverse transcriptases, telomerase appears remarkably specific for the template embedded in its RNA subunit, and the RNA subunit also appears to activate telomerase activity through poorly understood mechanisms.14c Although there is some evidence that TERT can utilize alternative templates, this alternative activity appears much less efficient than its canonical acticty.19 Like all reverse transcriptases, telomerase catalyzes processive nucleotide addition to its primer. Uniquely, telomerase also efficiently conducts repeat addition processivity to generate long copies of its repetitive DNA product.20 TER therefore must exist in multiple conformations throughout the catalytic cycle, and these conformations are constrained by RNA-RNA, RNA-DNA, and RNA-protein interactions. Accurate descriptions of these interactions in the minimal telomerase complex and at discrete steps of catalysis remain elusive. To date, the structures of ciliate and vertebrate TERs within the telomerase complex have been suggested based on phylogenetic comparative analysis 21 and many aspects of these models have been validated experimentally.14a
We sought to better understand the three-dimensional structure and conformational changes associated with tTER function within the telomerase ribonucleoprotein. We combined secondary structural constraints of tTER obtained using the high resolution footprinting technique selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE),22 distance constraints obtained from single molecule FRET data,18b and biochemical inference gleaned from previous biochemical experiments to generate constraints. We then modeled the structure of tTER in the minimal complex using discrete molecular dynamics (DMD) that allows facile incorporation of experimental information.23 In addition, we docked the resulting model with a homology model of tTERT based on crystal structure of the T. Castaneum TERT24 and the tTERT RNA binding domain25 to generate a three dimensional model of tTER in the minimal telomerase complex. The results reveal conformational changes that occur during telomerase assembly and suggest a model for stem IV binding to tTERT.
RESULTS
A recombinant telomerase complex for chemical probing experiments
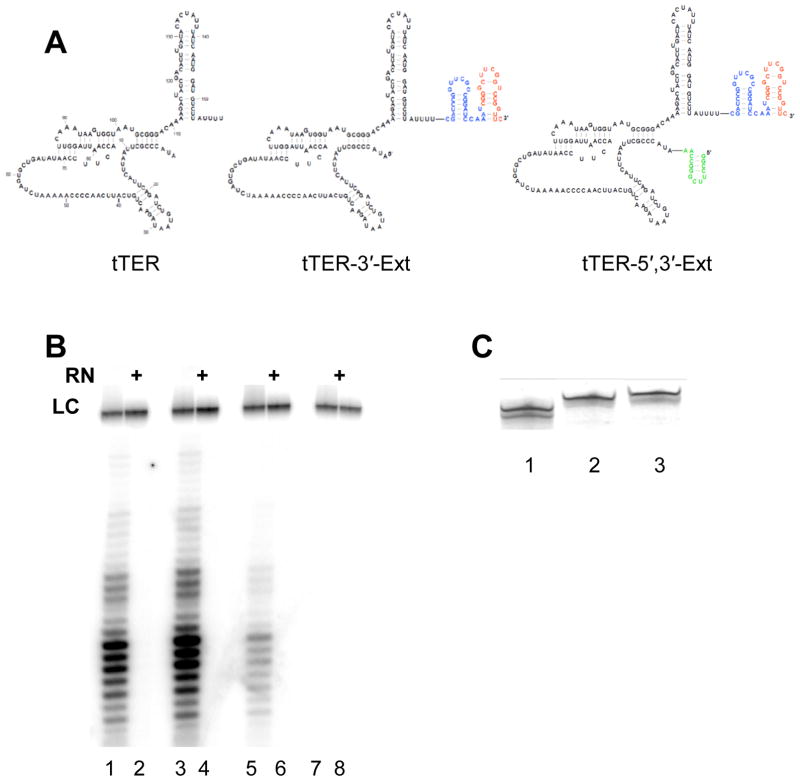
Because accurate structural modeling requires robust experimental constraints, we generated quantifiable data reporting on individual tTER nucleotides using SHAPE chemistry.22 SHAPE chemistry measures the reactivity of RNA 2′-hydroxyl groups with isatoic anhydride derivatives. Reactivity is primarily governed by nucleotide flexibility with more flexible nucleotides exhibiting greater reactivity.26 Nucleotide reactivity was mapped as reverse transcription stops. To maximize coverage of tTER in SHAPE experiments, we added a 3′-extension with a primer binding site for reverse transcription and a linker that separated the primer binding site from tTER to generate tTER-3′-Ext (Figure 2). We confirmed that the extension allowed reconstitution of active telomerase and conducted SHAPE experiments on tTER-3′-Ext in the presence and absence of tTERT.
Figure 2.
A tTER construct for SHAPE experiments. (A) Wildtype tTER is shown together with tTER-3′-Ext: tTER with a 3′-extension containing a linker and a primer binding site for reverse transcription, and tTER-5′,3′-Ext: tTER with a 3′-extension containing a linker (blue) and a primer binding site (red) for reverse transcription and a 5′-extension. The extensions facilitate analysis of the entire RNA by reverse transcription. (B) Activity of telomerase reconstituted with tTERT and wildtype tTER (lanes 1 and 2), tTER-3′-Ext (lanes 3 and 4), tTER-5′,3′-Ext (lanes 5 and 6), or no RNA (lanes 7 and 8). RN indicates treatment with RNase A prior to conducting telomerase assays. LC indicates a 32P-labeled, 100 nucleotide loading control. Telomerase was assayed by primer extension as described in methods. (C) Analysis of in vitro transcribed tTER constructs by denaturing gel electrophoresis. Lane 1, wildtype; lane 2, tTER-3′-Ext; Lane 3, tTER-5′,3′-Ext.
In vitro transcribed Tetrahymena telomerase RNA forms an extended stem III instead of a stem IIIa/IIIb pseudoknot
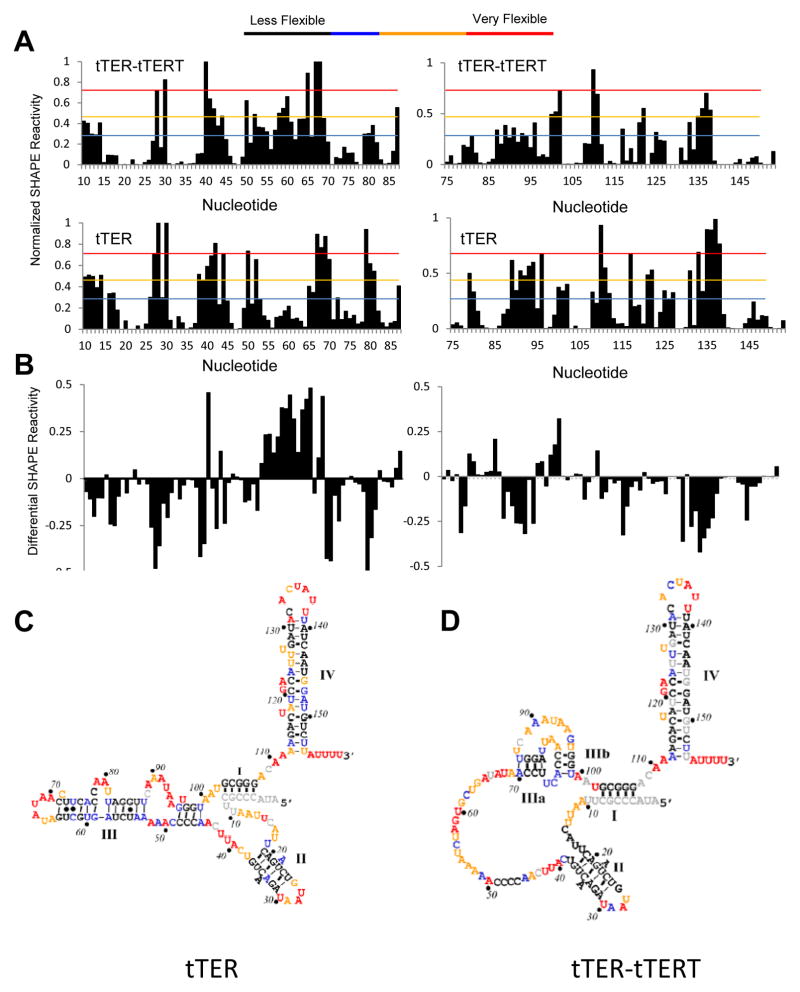
We used N-Methylisatoic anhydride (NMIA) to generate the SHAPE profile of tTER-3′-Ext in the absence of tTERT (Figure 3 and S1). We quantified SHAPE reactivities for 149 of the 159 tTER nucleotides and the data were used to constrain predictions using the program RNAstructure.27 Remarkably, the secondary structure of tTER using SHAPE data contained all but four of the basepairs predicted by RNAstructure using only the primary sequence.27 Both the SHAPE constrained and unconstrained models of protein-free tTER contained many features present in the currently accepted secondary structure model of tTER including stems I, II and IV. The most prominent difference was in the template and pseudoknot domains, which included a large stem III that encompassed several template residues instead of the stem IIIa/IIIb pseudoknot (Figure 3). To ensure that the 3′-extension did not perturb the native tTER structure leading to the formation of the large stem III confirmation, we compared both the SHAPE and RNase V1 profiles of wildtype tTER to tTER-3′-Ext. We detected no difference in the footprinting profiles of the RNAs and interpretation of the SHAPE data by RNAstructure generated the same secondary structures for tTER and tTER-3′-Ext (Figure S2 and data not shown).
Figure 3.
SHAPE analysis of tTER in solution and bound to tTERT. Quantified data from SHAPE experiments were plotted versus nucleotide position. Data are for: (A) tTER bound to tTERT. (B) tTER in the absence of proteins. (C) Differential plot of SHAPE reactivities: tTER-tTERT minus free tTER reactivities. (D) Secondary structure of free tTER color coded for SHAPE reactivity. The structure was generated using RNAstructure. (E) Secondary structure of tTER bound to tTERT color coded for SHAPE reactivity. Stems I, II, and IV were generated using RNAstructure. The base-paring of the pseudoknot region was set manually. See Figure S1 of the supporting information for representative raw data.
We compared the SHAPE data to reported NMR structures and found excellent agreement (Figure S3).28 The SHAPE profile for stem IV correlates with the generalized order parameter S2 consistent with previously reported data for a small stem IV model,29 and is consistent with the stem IV solution structure, as we previously reported.28a SHAPE reactivity of stem II also correlated with its solution structure. Each nucleotide forming the predicted stem II helix was unreactive to NMIA, including A22 and A34. This suggests that A22 and A34 are stacked within the helix and not bulged as is typically drawn, consistent with NMR data.28b Interestingly, the loop residues of stem II exhibited mixed levels of reactivity. G26 and A29 were less reactive than A28 and U30 suggesting that G26 and A29 are structured despite residing in a single-stranded loop. Indeed, the solution structure indicates that stem II is capped by a structured pentaloop with G26 stacked on top of the terminal U25-A31 base pair and A29 is tucked into the pentaloop structure. We conclude that the previously reported solution structure of stems II and IV accurately represent these domains in full-length tTER.
Surprisingly, the SHAPE profiles of the template and pseudoknot nucleotides (nts 45-99) are inconsistent with the accepted secondary structure of tTER. Instead, the data suggest with high probability that these nucleotides are involved in a large and stable stem-loop structure. This model is remarkably consistent with previous footprinting data,11b, 18a, 30 but the SHAPE experiment revealed sufficient constraints to confidently make this conclusion (Table S2). Moreover, recently reported FRET data also suggest that a pseudoknot does not form in protein-free tTER owing to disruptive interactions with other parts of the RNA.31 These interactions now appear defined. Instead of a pseudoknot and single-stranded template region, the pseudoknot and template nucleotides participate in extensive base-pairing to form an extended stem III (Figure 3C).
TERT induces a conformational change in tTER
The structure of in vitro transcribed tTER we determined is incompatible with a functional telomerase RNP as it would prevent association of the template with the active site. We predicted, therefore, that binding to tTERT would result in significant conformational changes in the template and pseudoknot domains of tTER. To test this hypothesis, we assembled telomerase in rabbit reticulocyte lysates, immunopurified the complex, and analyzed the structure of tTER by SHAPE. We posited that acylation by a SHAPE reagent might destabilize the telomerase complex and compromise the experiments. This concern was validated by demonstrating that acylation with NMIA was destabilizing to telomerase complex (Figure S4). For our studies, we therefore performed SHAPE on the telomerase complex for 1 reagent half-life to maximize signal to noise and avoid potential contribution of tTER that has dissociated from tTERT. We also confirmed that the SHAPE reactivity profile of tTER in the tTERT complex was not time dependent over the course of the experiments (Figure S5).
A histogram of all SHAPE reactivities for free tTER compared to reactivities of tTER bound to TERT shows no appreciable difference in the overall distribution of specific SHAPE values between the two conditions: an equal number of nucleotides became more reactive to the SHAPE reagent as became less reactive when tTER was bound to TERT. However, we observed significant localized SHAPE-reactivity changes in TERT bound tTER when compared to protein-free tTER (Figure 3 and S1). Remarkably, nucleotides exhibiting increased reactivity were concentrated from A53-G65: the template recognition element and proposed tTERT binding site 3′ of the template. In the absence of tTERT, these nucleotides are resistant to NMIA. However, upon assembly they become SHAPE reactive suggesting they become single stranded upon binding to tTERT.
Nucleotides with decreased SHAPE reactivity were present in provocative locations: the residues flanking the base of stem II, the loops of stems II and IV, and nucleotides that constitute the presumed pseudoknot stems IIIa and IIIb: A69-C72, A79-A80, and A89-U96. The SHAPE profiles of stems II and IV are consistent with reported solution structures, including correlation of S2 for stem IV nucleotides.29 Nucleotides predicted to be base paired demonstrated low SHAPE reactivity while loop nucleotides displayed mixed SHAPE reactivity. Notably, nucleotides predicted to be ordered by NMR displayed low SHAPE reactivity.28 These observations further validate the solution structures as accurate models of these domains in the functional telomerase RNP.
Predictions of tTER basepairing
We utilized the folding algorithm RNAstructure to predict basepairing probability for tTER nucleotides and compared these predictions to the tTER model based on comparative sequence analysis (CSA model).27 The currently accepted tTER model contains 40 base pairs, including 13 in the pseudoknot region.30, 32 Stems I, II, and IV contribute 27 of these 40 base pairs. RNAstructure predicted the 27 base pairs in stems I, II, and IV without experimental constraints. The five base pairs in stem I are predicted without the aid of SHAPE constraints to exist with probabilities exceeding 99%. The six base pairs in stem II are all predicted with probabilities exceeding 95%. Fourteen of the sixteen base pairs in stem 4 are predicted with probabilities exceeding 80%. Both MaxExpect33 and ProbKnot34 predict U126 and U127 to base pair with A144 and A143 respectively instead of U126 and U125. The U126-A144 base pair is predicted with 45% probability while the U127-A143 pair is predicted with 86% (Figure S6.).
We incorporated the SHAPE intensities of tTER in complex with tTERT into RNAstructure predictions. RNAstructure predicted 32 of the 40 tTER base pairs present in the CSA model (sensitivity of 80%) and 32 of the 38 base pairs in RNAstructure model are found in the CSA model (positive predictive value of 84%). All of the 27 base pairs in stems I, II, and IV of the CSA model were predicted. Additionally, the five base pairs in stem IIIA of the putative pseudoknot in the CSA model were also correctly predicted. However, stem IIIb of the pseudoknot in the CSA model was not present in predictions. Instead, structures that included long range base-pairs between nucleotides 94-98 and 14-18. This long range interaction is not consistent with the biochemical understanding of telomerase, and likely results from the inability of RNAstructure to accurately predict the tTER pseudoknot (Figure S7). Since the pseudoknot domain was not accurately predicted, we compared the SHAPE profiles to several possible pseudoknot structures including the CSA model,30, 32 a model predicted by the Tzaffati lab,35 and models predicted by several heuristic algorithms (Figure S8).36 In no case was the SHAPE profile completely consistent with the predicted base-pairing pattern. Overall, only two sets of base-pairing interactions are consistently supported by predictions and the SHAPE data: 70-ACCU/83-AGGU and 76-ACC/97-GGU.
Test of predicted stemIIIa base pairs by SHAPE and activity analysis of tTER mutants
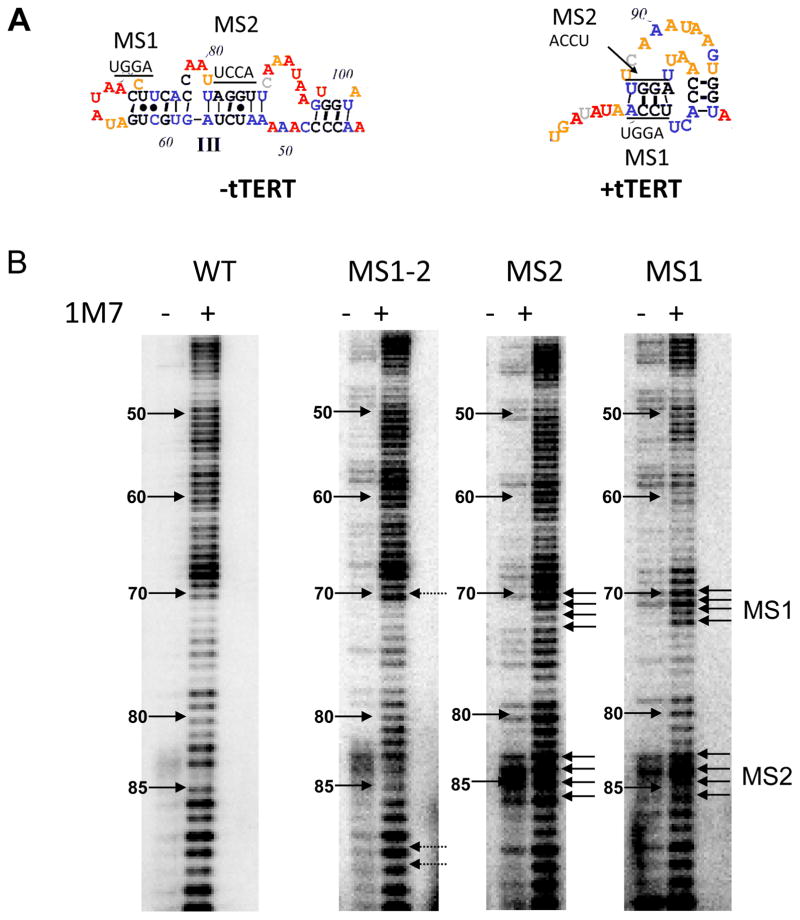
We designed three mutants to test the SHAPE-informed protein-free and tTERT-bound tTER models. Mutants were designed to deferentially affect the stability of stem III in the protein-free model and stem IIIa in the tTERT-bound model (Figure S9). The models predict that two mutants, MS1 (70-ACCU → 70-UGGA) and MS2 (83-AGGU → 83-UCCA) would alter several base-pairing interactions in both protein-free and tTERT-bound tTER, whereas double mutant MS1/MS2 is predicted to dramatically destabilize the protein-free structure but allow base-pairing in the tTER-complex owing to the compensatory mutations. SHAPE profiles of protein-free MS1, MS2 and MS1/MS2 suggest disruption of the wild type tTER stem III structure and new but poorly formed structures or mixtures of several structures (Figure S9). Importantly, the SHAPE profile of protein-free MS1/MS2 was distinct from wild-type, protein-free tTER consistent with the hypothesis that these residues are not associated by base-pairing.
When bound to tTERT, MS1 exhibited a shift in SHAPE reactivity making it appear to have formed a new structure (Figure 4). We examined possible secondary structures of the MS1 pseudoknot using heuristic modeling algorithms and found that several stable pseudoknot structures are compatible with the MS1 sequence and SHAPE reactivity of MS1 bound to tTERT. MS2 exhibited a much greater increase in SHAPE reactivity of both the mutated residues as well as their predicted base-pairing partners. Unlike MS1, none of the algorithms we tested predicted a stable structure for an MS2 pseudoknot. In striking contrast to protein free tTER, the SHAPE profile of MS1/MS2 is nearly indistinguishable from the profile of wild-type tTER. Notable exceptions include A70 and A90, which exhibited increased reactivity in the MS1/MS2 mutant when compared to wild type tTER. We examined the effect of disrupting predicted base-pairs in stem IIIa on telomerase activity and found that both MS1 and MS2 exhibited severely reduced telomerase activity while MS1/MS2 retained wild type activity (Figure 5). It should be pointed out that similar mutations: 71-CC → 71-GG and 84-GG → 84-CC were reported to show decreased telomerase activity that can be rescued by p65.18a
Figure 4.
Mutational analysis of tTER provides evidence for base pairing interactions in the stem III pseudoknot. (A) Positions of mutations in tTER are indicated. (B) SHAPE analysis of tTER mutants in complex with tTERT. Arrows indicate positions of the MS1 and MS2 mutations.
Figure 5.

tTER mutants that disrupt base pairing in the pseudoknot prevent reconstitution of robust telomerase activity. Telomerase activity of the tTERT-tTER minimal complex was determined by direct primer extension. LC indicates a loading control used form normalization.
DMD Analysis of tTER
DMD simulations have been successfully used to model the three dimensional structures of RNAs, and the accuracy of modeling can be greatly enhanced by experimental constraints.23, 37 We performed DMD simulations to generate structural models of protein free and tTERT-bound tTER using both SHAPE-derived secondary structure and FRET-derived distance constraints (Figure S10).23 Protein free tTER formed the predicted pairing regions stems I, II, and IV present in the CSA as well as the large stem III predicted by SHAPE constrained RNAstructure. In exploratory studies, initial models of tTERT-bound tTER generated with SHAPE and FRET data alone exhibited long-range base pairing like that found using RNAstructure (see Figure S7) that would block a proposed tTERT binding site as well as seemingly preclude proper association of the template with the tTERT active site. We therefore included several constraints based on biochemically inference (Figure S11). First, we introduced a nine-nucleotide RNA sequence that was complimentary to tTER template nucleotides 43-51 to mimic association of tTER with its primer and provide a steric block of the template from other tTER domains. Because nucleotides 15-18 are predicted to function as a protein binding site,38 we also restricted the distances between tTER nucleotides 10-18 and the rest of the RNA to no less than 10 Å in order to ensure these nucleotides remain single-stranded. The resulting models recapitulate all of the established base pairs in stems I, II, and IV. The models also predicted the stacked adenosines 22 and 34 of stem II, consistent with models from NMR data.28b Cluster analysis of tTER folding trajectories, which identifies distinct conformational states sampled in simulations, revealed three stable domains that are internally stable: a region encompassing stem IV (nucleotides 112-159), a region encompassing the template (nucleotides 1-107), and a flexible linker between stem IV and stem I (108-111). In the simulations, the flexible linker allows stem IV and the template domains of tTER to change coordinates with respect to each other. Examination of the representative structures from cluster analysis indicates that movement in the flexible linker region enables the template nucleotides 43-51 to rotate approximately 90° in relation to stationary stem IV, which suggests that the template nucleotides 43-51 can exist in several discrete positions with respect to stem IV.
One aspect of the DMD-generated models that did not appear to allow tTER function in the telomerase complex was the close association of the template with the body of the RNA (Figure 6B and C). We therefore modeled tTER bound to a homology model of tTERT. To constrain tTER binding, we aligned the template to the coordinates of a DNA primer available from the T. castaneum TERT crystal structure, which contains a model of the predicted T. castaneum telomerase template RNA residues base paired to the complementary DNA contained in a chimeric hairpin.24 We performed DMD simulations to relax tTER while maintaining the secondary and tertiary structures of tTER. As we expected, docking to the tTERT model as a constraint altered the tTER structure (Figure 6D). The major change was a twist in the template containing strand away from the remainder of tTER commensurate with extending stem II away from the main body of the RNA. The tTER model contains stems I, II, IIIa/IIIb and IV with stem IV pointed towards the IIIa/IIIb pseudoknot. The template recognition element and the template are positioned away from the main body of tTER to accommodate association with the tTERT active site. The stem II model aligns within 1.5 Å root mean square deviation (RMSD) of the published NMR structure,28b and stem IV aligns to within 5.4 Å RMSD of the published NMR structure (Figure S12). 28a, 28c
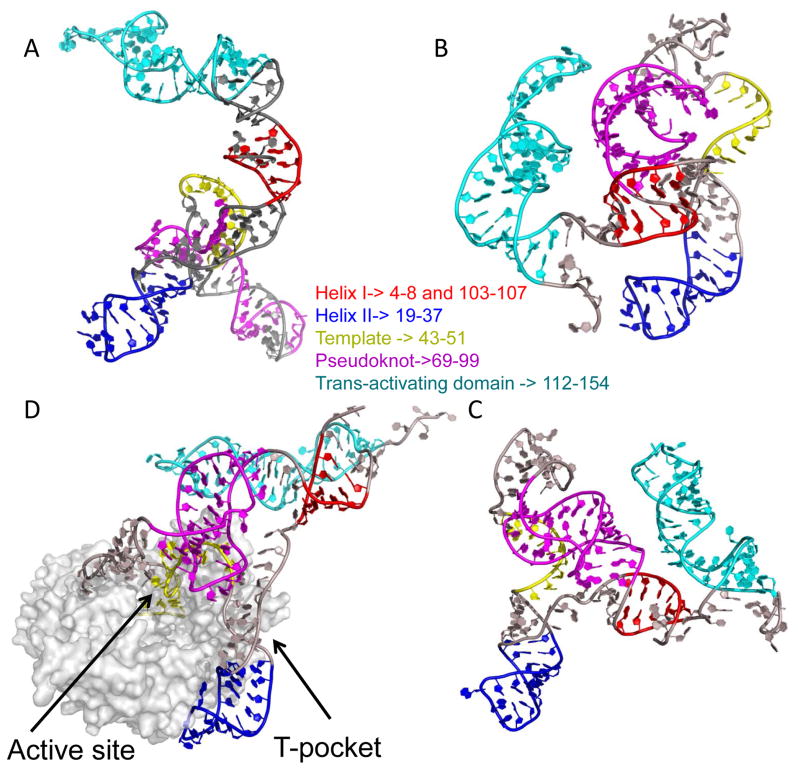
Figure 6.
Conformations of tTER free in solution and bound to tTERT. tTER was modeled using DMD simulations using constraints describe in the Methods section. (A) Unbound tTER. (B) and (C) The two most extreme populations of tTER bund to tTERT predicted by DMD. (C) Was the most populated state. (D) The structure of tTER predicted by DMD when docked to a homology model of tTERT. The TEN domain and flexible linker to the reverse transcriptase domain of TERT was not modeled. Three DNA substrate nucleotides buried within the active site in very close proximity to the catalytic triad of aspartates are colored yellow. The T-pocket of the RNA binding domain of tTERT is indicated. All tTER models are aligned along stem IV nucleotides. Telomerase RNA is shown with stem I nucleotides colored red, stem II nucleotides colored blue, template nucleotides colored yellow, pseudoknot nucleotides colored magenta, and stem IV nucleotides colored cyan. All remaining nucleotides are colored gray.
DISCUSSION
Binding to tTER Causes a Major Conformational Change in tTER
We combined SHAPE chemistry, published FRET, and molecular modeling to examine the three-dimensional structure of tTER. By comparing unbound tTER to tTER in the minimal telomerase complex, we revealed a dramatic conformational change in tTER that attends assemblage in the minimum telomerase complex. The most noteworthy evidence for the conformational change is indicated by the SHAPE profile for nucleotides A53 through A100, which includes the template recognition element and presumed pseudoknot domain (Figure 3). We interpret this change as evidence for the absence of a pseudoknot in protein-free tTER and formation of a pseudoknot in tTERT-bound tTER.
The SHAPE inferred-secondary structure of tTER in solution is remarkably different than the accepted secondary structure associated with tTER function in that the template and pseudoknot are enveloped by a large paired region, which we refer to a stem III. Importantly, this model is consistent with previous enzymatic and chemical footprinting of tTER (Table S1). It is notable that previous tTER structure probing experiments did not lead to the protein-free structure of tTER we predict. In part, this is a result of insufficient data available to accurately assign the structure. Recent experiments using RNase One does provide wider coverage, and RNase One is reported to cleave single stranded RNA.18a It is surprising then that the reactivity profile of protein-free tTER using RNase One varies so much from SHAPE reactivity, particularly in the stem III domain of the protein free structure (residues A44 – U102, see Table S1). This difference requires an explanation. One possibility for decreased RNase One reactivity at non-base-paired nucleotides is steric restriction on RNase One binding. Additionally, since RNase One binding could shift the structural equilibrium from double-stranded to single-stranded RNA. This would result in increases reactivity at base-paired residues. Since SHAPE is governed almost exclusively by flexibility, these secondary effectors of RNase One reactivity may explain the differences between SHAPE and RNase One and would challenge accurate structural interpretation of RNase One experiments when compared to SHAPE chemistry. The model proposed here for the protein-free tTER structure also rationalizes FRET data of tTER at labeled pairs U63 and U92 and pairs U73 and U99, which are lower than expected for a folded pseudoknot.31 The FRET data are, however, consistent with an extended stem III structure.
The SHAPE inferred-secondary structure of tTER bound to tTERT, in contrast to the protein-free structure, is consistent with the accepted secondary structure. However, the specific base-pairing pattern of the pseudoknot domain remains challenging to define. In an attempt to better address this, we compared the SHAPE profile of the pseudoknot residues with several proposed models (Figure S8). We found that not one model was entirely consistent with the SHAPE profile; instead each model is partially consistent with the data. The G-C rich regions are predicted by each model to be base-paired, which is consistent with the SHAPE profile. However, the A-U rich regions do not appear to form a consistent base-pairing pattern. It seems that the pseudoknot domain forms a triple helix with the reactive A-U-rich nucleotides bound to the minor or major groove of the stems IIIa and IIIb, similar to the model forwarded by the Tzfati lab.35 To account for the high SHAPE reactivity of the A-U rich strands of the pseudoknot domain, we propose that either several base-pairing configurations of the pseudoknot are present or the pseudoknot is flexible. One possibility is that during catalysis or in the presence of the telomerase holoenzyme component p65, the pseudoknot forms a more stable structure.
The decrease in SHAPE reactivity of the apical loops of stem-loops II and IV upon binding tTERT is evidence for decreased flexibility resulting from either increased stability of the secondary structure elements or direct tTERT interaction. Specifically, the loop of stem II, which displayed decreased SHAPE reactivity, is unlikely to bind directly to tTERT since mutating or extending the length of stem II is well tolerated.18b, 39 Accordingly, we conclude that association of tTER with tTERT stabilizes stem II resulting in decreased nucleotide flexibility. The reduced reactivity of 15-CAUU-18 and 39-UC-40 are consistent with predicted direct and stable interactions of these nucleotides with tTERT. Binding to tTERT could reduce nucleotide flexibility or sterically block reaction with NMIA.
Like stem-loop II, stem-loop IV residues displayed decreased SHAPE reactivity within the loop region. Several previous reports suggest a direct interaction between loop IV and tTERT. Therefore direct interactions with tTERT as well as increased structural order are likely to contribute to the observed decrease in SHAPE reactivity. Interestingly, RNase One was reported to display the opposite distribution of reactivities in loop IV with A136, U137, and U138 exhibiting resistance to RNase One cleavage but high SHAPE reactivity (Table S2). One model that is consistent with the data is that residues C132, A133, and C134 form a rigid platform to constrain the range of motion of flexible nucleotides 135-UAUU-138. Evidence from several mutagenesis studies suggest that stem IV binds tTERT and that this interaction is stabilized by p65. Interestingly, when the C132-U138 base pair is mutated to an A-U base-pair, SHAPE reactivity decreases for U138 commensurate with dramatically reduced catalytic activity.28a Flexibility in these nucleotides therefore seems in part related to their biochemical role in telomerase assembly. Interestingly mutation of A136, U137, and U138 causes a significant decrease in assembly of active telomerase even in the presence of p65, but do not appear to negatively affect activity of properly assembled complexes. We propose that the UAUU nucleotides are flexible to allow an induced fit with tTERT. In addition, we predict that tTERT binds tTER in the major groove of stem IV. Binding the major groove would likely protect A136, U137, and U138 from RNase One cleavage but not block reaction of these nucleotides with NMIA, assuming RNase ONE cleavage is governed by sterics and NMIA by nucleotide flexibility.
We were surprised that four template residues, 46-CCCC-49 remained resistant to SHAPE reactivity after assembly. These nucleotides are also resistant to RNase One cleavage when tTER is bound to the N-terminus of tTERT (amino acids 1-516). Since a primer must bind these residues, we expected that they were single stranded and would exhibit high SHAPE reactivity. The low reactivity suggests that these nucleotides are directly bound to tTERT in a rigid conformation in the active site, perhaps providing a platform for primer binding.
A three dimensional model of tTER
The three dimensional model of tTER generated by DMD simulations predicts all base pairs within stems I, II, and IV and displays relatively low RMSD alignments to NMR generated models of stems II and IV. Though the biochemical data do not allow assignment of a specific base-pairing pattern for the pseudoknot, DMD simulation suggests a compact structure with several triple base-triples. The DMD simulations also allow insight into tTER dynamism. Overall, simulations reveal that the template region is remarkably flexile (compare 6B, 6C to 6D). The simulations suggest that one important aspect of this flexibility is rotation of the single-stranded joining region between stems I and IV. One possibility is that the lack of FRET constraints for any nucleotides in the distal loop of the stem II domain may account for this dynamic positioning of the template. An alternative and more interesting interpretation is that the observed motion captures necessary movement of the template during successive rounds of nucleotide addition and repeat addition processivity. Based on this model, stem IV remains docked to tTERT in an allosteric activating site while the template can cycle through its required positions, a motion allowed by rotation about the linker between stem IV and stem I (Figure 6) perhaps coupled with scrunching of the template recognition element.40 Alternatively, the motion may allow proper docking of stem IV during assemblage.
A biological model for the tTER structural rearrangement
The significant conformational change we detected in tTER that attends telomerase assemblage can be interpreted in many ways. One possibility is that the alternative structure is an artifact of in vitro transcription, and tTER does not fold into a biologically relevant structure owing to the lack of tTER binding partners, for example p65, which may be present during its transcription in vivo. Alternatively, it can be proposed that tTER folds as we show for the protein free tTER in vivo prior to p65 binding, which can induce a conformational changes in tTER,18 followed by association with tTERT. If this is the case, does the misfolded tTER structure serve a purpose? We propose a model that protein free-tTER folds with a large stem III to sequester the template cytosine residues in a double stranded helix until assembly in order to protect the integrity of the telomere sequence and may serve other purposes as well. Since tTER codes for the DNA sequence at chromosome termini, damage to the templating residues could have significant negative consequences. For example, mutation of the human TER templating residues results in cell death.41 Because the deamination rate of cytosine in single stranded oligonucleotides is faster than that of cytosine in double stranded duplexes,42 the misfolded tTER would protect the coding cytosine residues.
CONCLUSIONS
In summary, high-resolution footprinting of protein-free and tTERT-bound tTER revealed a significant conformational change in tTER. In the absence of tTERT, tTER does not form a pseudoknot but instead forms a large stem that encompasses the pseudoknot and template nucleotides. Importantly, the data provide critical evidence that the previous solution structure models of stem II and IV derived from NMR constraints are consistent with the structure of tTERT-bound tTER, offer robust evidence for the pseudoknot structure in tTERT-bound tTER, and provide new hypotheses for telomerase RNA function during assemblage and catalysis.
METHODS AND MATERIALS
Preparation of tTER and pFLAG-tTERT
RNAs were transcribed in vitro using Ampliscribe T7 Transcription Kit was used (Epicentre Technologies). Templates were generated by PCR using the plasmid pTET-telo, a pUC19-bsaed plasmid containing the tTER gene, a T7 RNA polymerase promoter, and a self-cleaving hammerhead ribozyme that processes the 5′-end of the RNA. Primers are listed in Table S4. PCR products were gel purified using Wizard PCR Prep Kits and RNAs gel purified and stored in TE (pH 7.5) at −80 °C.
A sequence coding the FLAG eiptope was ligated into a pET-28a plasmid containing tTERT cloned into the BamH1 and Xho1. Oligonucleotides were gel purified and annealed before ligation into the Nco1 and BamH1 sites in pET-28a-tTERT. This removed the Nco1 site and an Nde1 site, allowing for easy screening of positive clones and removed the N-terminal His- and T7-tags.
Reconstitution and Affinity Purification of Tetrahymena telomerase
Tetrahymena telomerase was reconstituted in rabbit reticulocyte lysates following standard protocols (Promega) and affinity purified using Anti-FLAG M2 Agarose beads (Sigma). Beads were prewashed with WB1 (20 mM Tris-acetate pH 7.5, 100 mM potassium glutamate, 5 mM MgCl2, 1 mM EDTA, 0.1 mM DTT, and 10% glycerol) and blocked with blocking buffer (WB1 with 0.5 mg/mL lysozyme, 0.5 mg/mL BSA, 0.05 mg/mL glycogen, and 0.1mg/mL yeast RNA). 400 μL of crude telomerase complex in rabbit reticulocyte lysates were mixed with 400 μL of blocking buffer and the mixture was centrifuged at 15,000g for 10 min at 4 °C to remove any precipitates. The supernatant was then added to the 100 μL of pre-blocked Anti-FLAG beads and the resultant slurry was mixed on an orbital shaker for 2 h at 4 oC. The beads were washed 4 times with 1400 μL of WB1 containing 300 mM potassium glutamate, 2 times with 1400 μL of TMG (10 mM Tris-Acetate pH 8.0, 1mM MgCl2, 0.1 mM DTT, and 10% glycerol) and resuspended in 100 μL of TMG to afford a 1:1 slurry. The telomerase complexes were eluted in 1.5 mL Protein LoBind Tube (Eppendorf). The bead slurry containing telomerase complexes were washed 2 times with 1200 μL of WB2 (20 mM Tris-acetate pH 7.5, 5 mM MgCl2, 1 mM EDTA, 0.1 mM DTT, and 10% glycerol. 12 μL of 10 mg/ml BSA was added directly to the beads, followed by 200 μL of 3xFLAG peptide solution (WB2 with 0.75 mg/mL of 3xFLAG peptide (Sigma)). This slurry was incubated on an orbital shaker for 1 hr at 4 °C. The slurry was centrifuged at 1,500g for 2 min at 4 °C, and the supernatant containing soluble telomerase was gently removed and transferred to a fresh LoBind tube. Samples were flash frozen in a dry ice/ethanol bath and stored at −80 °C.
SHAPE analysis of tTER-3′-Ext
A 7 μl solution of tTER-3′-Ext (1 pmol) in deionized water was snap annealed by heating at 95 °C for 2 min then cooling on ice for 5 min before 2 μl of 5× folding buffer (250 mM Hepes pH 8.0, 10 mM MgCl2) was added. The solution was then incubated at 30 °C for 5 min. The RNA was then treated with 1 μl of NMIA (100 mM in anhydrous DMSO) or 1 μl of anhydrous DMSO as a control, incubated at 30 °C for 90 min, precipitated with ethanol in the presence of 0.2 M NaCl and 200 μg/ml glycogen, washed once with 70% ethanol, speed vacuumed till dry, and reconstituted in 5 μl of pH 8.0 TE buffer. Sites of modification were mapped reverse transcription using two separate 5′-[32P]-labeled primers: con-RT, which binds the primer binding site in the SHAPE cassette, and C103, which binds to tTER to begin reverse transcription at C103. cDNA extension products were separated by electrophoresis and compared to dideoxythymidine sequencing ladders, visualized by phosphorimaging using ImageQuant 5.1, and quantified using SAFA. For greater detailed descritption, see Supporting Information.
SHAPE analysis of tTER in Complex with tTERT
Affinity purified telomerase (25 μL, ~125 fmol) was incubated in folding buffer (50 mM Hepes pH 8.0, 2 mM MgCl2) (50 μL total reaction volume) was incubated at 30 °C for 2 min. NMIA or DMSO was added to separate sample at a final concentration of 10 mM NMIA or 10% DMSO and incubated for 17.5 min (1 half life). The reaction was immediately quenched by the addition of dithiothreitol (5 mM). The solution was proteolyzed for 10 min at 37 °C with 160 μg/mL of proteinase K in 1X TES (40 mM Tris pH 8.0, 4 mM EDTA and 0.15% SDS), phenol/chloroform extracted, precipitated with ethanol and reconstituted in 5 μl of RNase Free TE pH 8.0 (Ambion). Sites of modification were mapped reverse transcription as described above.
Structural models of tTER
Secondary structures were modeled with SHAPE constraints using RNAstructure. Because RNAstructure could not predict the tTER pseudoknot, we compared the SHAPE reactivities to tTER pseudoknots predicted using conserved sequence analysis and heuristic folding prediction methods.
Model of tTERT
The tTERT model was generated with the crystal structure of the tTERT residues (the RNA binding domain, PDB-2R4G ) and homology modeling of the remaining tTERT RT domain using the T. castaneum TERT crystal structure with model of the primer-template duplex bound to the active site (PDB-3KYL). The N-terminal domain of tTERT was not included. A large domain, D624-D688, in tTERT is absent in the T. castaneum sequence. This insertion was modeled using ab initio folding methods and included in the tTERT model.43 PDB-3KYL contains a RNA-DNA chimeric hairpin the mimics the template-primer duplex. Only the nucleotides representing the DNA primer were maintained in the tTERT model.
Discrete Molecular Dynamics Modeling of tTER
Sequence information and base pairs established by SHAPE were subjected to one round of refinement by DMD23 at (T) = 0.3 for 105 time units (tu), where T is the reduced temperature in units of kcal/(mol • kB).13 After base pair formation was visually confirmed, files were prepared for incorporation of potential energy functions describing distances between FRET fluorophores. We also model the base pairing between tTER and a nine nucleotide sequence complementary to the template, and a penalty for base pairing of nucleotides 10-18.
We estimated distances between four pairs of TER nucleotides using the following equation where R0 is the
Förster radius and r is the distance between FRET fluorophores. FRET values were obtained from published single molecule FRET efficiencies between four fluorophore-labeled TER nucleotide pairs.18b Å Forster radius of 50 Å was used to estimate the distance between fluorophores in active telomerase observed at maximum FRET efficiency. Similarly, a Forster radius of 60 Å was used to estimate the distance between fluorophores in active telomerase observed at half maximal FRET efficiency. It is important to note that the four labeled uridines were in full length TER when the RNA was assembled in the telomerase complex. It is also important to note that the labeled RNAs were used by telomerase successfully as templates despite being labeled with bulky Cy3 and Cy5 adducts. We then used a potential function to restrict the distances between the four pairs of labeled TER uridines to within distances calculated from the FRET efficiencies (Figure S10).
We introduced a nine-nucleotide RNA sequence that was complimentary to tTER template nucleotides 43-51 to mimic association of TER with its primer and to provide a steric block of the template from other tTER domains. The fifth nucleotide in the primer mimic was constrained to be less than 10 Å from C47. We also used a potential function to maintain the distance between nucleotides 10-18 and 38-46 to a minimum of 10 Å because nucleotides 15-18 are predicted to function as a protein-binding site (see Figures S10 and S11 for the potential function and algorithm used). Once constraints were incorporated, the RNA was allowed to cool at T = 0.25 for 3×104 tu before confirming the primer mimic approached the template nucleotides. The RNA was cooled in two additional steps at T = 0.15 for 104 tu; and T = 0.15 for 105 tu. One complete three-dimensional refinement of the 159 nucleotide TER required <2 h on a Linux computational node (3.2 GHz Intel Xeon IBM BladeCenter node, Red Hat Linux v5, 64-bit OS).
Distance-based hierarchical clustering was performed without user intervention on 4,500 predominant RNA conformations using OC software (available at http://www.compbio.dundee.ac.uk/downloads/oc).44 Final conformations were divided into 10 clusters, subject to the requirement that structures within a cluster agree to better than 6 Å RMSD. From the 10 clusters, we focused on the most highly populated ensemble, which contained ~ 65% of the total representative models. We focused our analysis on the most central structure in each of these final clusters because the Boltzmann distribution dictates that these clusters represent the lowest free energy state. DMD model verification by RMSD alignments were computed on the basis of superposition of backbone phosphate atoms at base paired positions when compared to stem II (PDB ID 2FRL) and stem IV (PDB ID 2FEY) NMR models.
To model tTER bound to tTERT, we inserted the most populated tTER model from DMD simulations with the tTERT homology model described above. tTER was aligned with the tTERT active site by setting nucleotides 51-AAG-49 to as base paired to the DNA primer. Then, the molecular system was relaxed with all-atom DMD simulations,45 where the protein and template are kept fixed, the secondary structure, and FRET-based tertiary structure are maintained. The all-atom relaxation simulations were performed at room temperature (300K). The lowest energy structure from the 100 ns simulations was used as the model structure of tTER bound to tTERT.
Supplementary Material
Acknowledgments
The authors would like to thank Professor Kevin Weeks for helpful discussions.
Funding Sources
This work was supported by NSF grant MCB-0751372 (M.B.J.), Predoctoral fellowship 1-F31-GM086084-01 (D.I.C), the UNC Research Council (F.D.), and NIH grant R01GM080742 (ND).
ABBREVIATIONS
- CSA model
comparative sequence analysis model for tTER secondary structure
- DMD
discrete molecular dynamics
- SHAPE
selective 2′-hydroxyl acylation analyzed by primer extension
- TER
telomerase RNA
- TERT
telomerase reverse transcriptase
Footnotes
Author Contributions
All authors have given approval to the final version of the manuscript.
Supporting Information. Supplemental figures referred to in the text and detailed experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–44. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 2.Woodson SA. Compact intermediates in RNA folding. Annu Rev Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482(7385):322–30. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekaran VG, Soares J, Jarstfer MB. Structures of telomerase subunits provide functional insights. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Fouche N, Moon IK, Keppler BR, Griffith JD, Jarstfer MB. Electron Microscopic Visualization of Telomerase from Euplotes aediculatus Bound to a Model Telomere DNA. Biochemistry. 2006;45(31):9624–31. doi: 10.1021/bi060313s. [DOI] [PubMed] [Google Scholar]
- 6.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116(2):273–9. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 7.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413(6854):432–5. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 8.Donate LE, Blasco MA. Telomeres in cancer and ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366(1561):76–84. doi: 10.1098/rstb.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8(3):167–79. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 10.Mason M, Schuller A, Skordalakes E. Telomerase structure function. Curr Opin Struct Biol. 2011;21(1):92–100. doi: 10.1016/j.sbi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 11.(a) Robart AR, O’Connor CM, Collins K. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. Rna. 2010;16(3):563–71. doi: 10.1261/rna.1936410. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- 12.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 13.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Legassie JD, Jarstfer MB. The unmasking of telomerase. Structure. 2006;14(11):1603–9. doi: 10.1016/j.str.2006.09.004. [DOI] [PubMed] [Google Scholar]; (b) Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]; (c) Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7(7):484–94. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402(6761):551–5. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]; (b) Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol. 2010;30(11):2775–86. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18(10):1107–18. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Berman AJ, Gooding AR, Cech TR. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol. 2010;30(20):4965–76. doi: 10.1128/MCB.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stone MD, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446(7134):458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell. 2012;47(1):16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461(7261):230–5. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Legassie JD, Jarstfer MB. Telomerase as a DNA-dependent DNA polymerase. Biochemistry. 2005;44(43):14191–201. doi: 10.1021/bi050628s. [DOI] [PubMed] [Google Scholar]
- 20.Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1(3):1610–6. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 23.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. Rna. 2008;14(6):1164–73. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17(4):513–8. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 25.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15(11):1403–12. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 26.McGinnis JL, Dunkle JA, Cate JH, Weeks KM. The Mechanisms of RNA SHAPE Chemistry. J Am Chem Soc. 2012;134(15):6617–24. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106(1):97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25(13):3156–66. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Richards RJ, Theimer CA, Finger LD, Feigon J. Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res. 2006;34(3):816–25. doi: 10.1093/nar/gkj481. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Richards RJ, Wu H, Trantirek L, O’Connor CM, Collins K, Feigon J. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. Rna. 2006;12(8):1475–1485. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130(37):12244–5. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharyya A, Blackburn EH. Architecture of telomerase RNA. EMBO J. 1994;13(23):5721–31. doi: 10.1002/j.1460-2075.1994.tb06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mihalusova M, Wu JY, Zhuang X. Functional importance of telomerase pseudoknot revealed by single-molecule analysis. Proc Natl Acad Sci USA. 2011;108(51):20339–44. doi: 10.1073/pnas.1017686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67(2):343–53. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 33.Lu ZJ, Gloor JW, Mathews DH. Improved RNA secondary structure prediction by maximizing expected pair accuracy. Rna. 2009;15(10):1805–13. doi: 10.1261/rna.1643609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellaousov S, Mathews DH. ProbKnot: fast prediction of RNA secondary structure including pseudoknots. Rna. 2010;16(10):1870–80. doi: 10.1261/rna.2125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulyanov NB, Shefer K, James TL, Tzfati Y. Pseudoknot structures with conserved base triples in telomerase RNAs of ciliates. Nucleic Acids Res. 2007;35(18):6150–60. doi: 10.1093/nar/gkm660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.(a) Cao S, Giedroc DP, Chen SJ. Predicting loop-helix tertiary structural contacts in RNA pseudoknots. Rna. 2010;16(3):538–52. doi: 10.1261/rna.1800210. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sato K, Kato Y, Hamada M, Akutsu T, Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27(13):i85–93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ren J, Rastegari B, Condon A, Hoos HH. HotKnots: heuristic prediction of RNA secondary structures including pseudoknots. Rna. 2005;11(10):1494–504. doi: 10.1261/rna.7284905. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bindewald E, Kluth T, Shapiro BA. CyloFold: secondary structure prediction including pseudoknots. Nucleic Acids Res. 2010;38:W368–72. doi: 10.1093/nar/gkq432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.(a) Ding F, Lavender CA, Weeks KM, Dokholyan NV. Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat Methods. 2012;9(6):603–8. doi: 10.1038/nmeth.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. Journal of the American Chemical Society. 2009;131(7):2541–6. doi: 10.1021/ja805460e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cruz JA, Blanchet MF, Boniecki M, Bujnicki JM, Chen SJ, Cao S, Das R, Ding F, Dokholyan NV, Flores SC, Huang L, Lavender CA, Lisi V, Major F, Mikolajczak K, Patel DJ, Philips A, Puton T, Santalucia J, Sijenyi F, Hermann T, Rother K, Rother M, Serganov A, Skorupski M, Soltysinski T, Sripakdeevong P, Tuszynska I, Weeks KM, Waldsich C, Wildauer M, Leontis NB, Westhof E. RNA-Puzzles: a CASP-like evaluation of RNA three-dimensional structure prediction. Rna. 2012;18(4):610–25. doi: 10.1261/rna.031054.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.(a) Cunningham DD, Collins K. Biological and biochemical functions of RNA in the tetrahymena telomerase holoenzyme. Mol Cell Biol. 2005;25(11):4442–54. doi: 10.1128/MCB.25.11.4442-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.(a) Autexier C, Greider CW. Mutational analysis of the Tetrahymena telomerase RNA: identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 1998;26:787–795. doi: 10.1093/nar/26.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18(12):1371–5. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc Natl Acad Sci USA. 2001;98(14):7982–7. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29(10):2532–7. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 43.Sharma S, Ding F, Nie H, Watson D, Unnithan A, Lopp J, Pozefsky D, Dokholyan NV. iFold: a platform for interactive folding simulations of proteins. Bioinformatics. 2006;22(21):2693–4. doi: 10.1093/bioinformatics/btl460. [DOI] [PubMed] [Google Scholar]
- 44.Barton GJO-Acap. 2002 www.compbio.dundee.ac.uk/downloads/oc.
- 45.Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16(7):1010–8. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.