A review and historical perspective covering the many different aspects of antiparasitic drug discovery, in particular targeting protists, is presented. The key role of structural studies in the process is highlighted and specific high-profile examples are given.
Keywords: parasitic protozoa, antiparasitic drug discovery
Abstract
Parasitic protozoa cause a range of diseases which threaten billions of human beings. They are responsible for tremendous mortality and morbidity in the least-developed areas of the world. Presented here is an overview of the evolution over the last three to four decades of structure-guided design of inhibitors, leads and drug candidates aiming at targets from parasitic protozoa. Target selection is a crucial and multi-faceted aspect of structure-guided drug design. The major impact of advances in molecular biology, genome sequencing and high-throughput screening is touched upon. The most advanced crystallographic techniques, including XFEL, have already been applied to structure determinations of drug targets from parasitic protozoa. Even cryo-electron microscopy is contributing to our understanding of the mode of binding of inhibitors to parasite ribosomes. A number of projects have been selected to illustrate how structural information has assisted in arriving at promising compounds that are currently being evaluated by pharmacological, pharmacodynamic and safety tests to assess their suitability as pharmaceutical agents. Structure-guided approaches are also applied to incorporate properties into compounds such that they are less likely to become the victim of resistance mechanisms. A great increase in the number of novel antiparasitic compounds will be needed in the future. These should then be combined into various multi-compound therapeutics to circumvent the diverse resistance mechanisms that render single-compound, or even multi-compound, drugs ineffective. The future should also see (i) an increase in the number of projects with a tight integration of structural biology, medicinal chemistry, parasitology and pharmaceutical sciences; (ii) the education of more ‘medicinal structural biologists’ who are familiar with the properties that compounds need to have for a high probability of success in the later steps of the drug-development process; and (iii) the expansion of drug-development capabilities in middle- and low-income countries.
1. Parasitic protozoa and their devastating effects
In the course of the last ∼130 years, the major parasitic protozoa responsible for human diseases have gradually been discovered thanks to the often heroic efforts of numerous microbiologists (de Kruif, 1926 ▶; Cox, 2002 ▶). The tragic impact that many of these parasites have on human health has been realised for at least a century. Among these, Giardia duodenalis, also known as G. lamblia or G. intestinalis, was the first of the human parasitic protozoa to be observed, by Antoni van Leeuwenhoek in 1681 (Dobell, 1932 ▶). In contrast, the full impact of Cryptosporidium species on the suffering and death of humans, in particular young children in developing countries, has only gradually been revealed in recent decades (Shirley et al., 2012 ▶; Tzipori & Widmer, 2008 ▶).
The diseases caused by parasitic protozoa (or protists) mentioned in this overview are the following:
(i) Chagas disease caused by Trypanosoma cruzi;
(ii) African sleeping sickness by T. brucei;
(iii) various forms of leishmaniasis, ranging from skin, mucous membrane to liver diseases, by a spectrum of different Leishmania species;
(iv) toxoplasmosis by Toxoplasma gondii, which is of particular importance for pregnant women;
(v) malaria by a number of Plasmodium species, mainly P. falciparum and P. vivax; and
(vi) diarrheal diseases by Cryptosporidium, Entamoeba and Giardia species.
The impact of these parasites on human life has been and still is profound (Murray et al., 2012 ▶; Lozano et al., 2012 ▶; Table 1 ▶). Diseases caused by these protozoa occur most frequently in tropical areas, most intensively and tragically in the poorest populations. Underlying causes include poor water sanitation, intensive contacts with intermediate vectors, lack of public health infrastructure and other factors (Hotez et al., 2009 ▶).
Table 1. Parasites and diseases.
| Disease | Parasitic protozoa causing the disease | Year parasite was discovered† | Mortality (in thousands in 2010; Lozano et al., 2012 ▶) | DALY‡ (in thousands in 2010; Murray et al., 2012 ▶) |
|---|---|---|---|---|
| Chagas disease | Trypanosoma cruzi | 19071912 | 10 | 546 |
| Sleeping sickness | Trypanosoma brucei | 1894, 1902, 1910 | 9 | 560 |
| Leishmaniasis | Leishmania spp. | 18981911 | 52 | 3317 |
| Malaria | Plasmodium spp. | 1880 | 1170 | 82 |
| Toxoplasmosis | Toxoplasma gondii | 19081923 | Not listed separately | Not listed separately |
| Cryptosporidiosis | Cryptosporidium spp. | 19071970 | 100 | 8372 |
| Amebiasis | Entamoeba spp. | 1873 | 55 | 2237 |
| Giardiasis | Giardia spp. | 1954 | Not listed separately | Not listed separately |
A major concern is that the number of well tolerated therapeutics is very limited, or even absent, for essentially all of these parasitic diseases. The exception is malaria, where in particular artemisinin combination therapies have been a success since about 2000 (Miller & Su, 2011 ▶), although resistance is building up (see below). The available therapeutics were almost all discovered many decades ago and often have severe side effects, while several have to be administered intravenously. This is a serious handicap in the rural regions where the parasites occur most frequently. At the same time, the available drugs are continuously subject to various resistance mechanisms employed by these parasites. Hence, not only is there an urgent need to develop novel therapeutics, but constant vigilance is required to minimize the loss of available and future compounds to resistance.
A pioneering international effort to combat tropical maladies, including protozoan diseases, has been the creation of the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases, abbreviated as TDR. This program celebrated 40 years of existence in 2014 (http://www.who.int/tdr/news/2015/40-year-anniversary/en/). The magic of TDR was that, with limited resources, it made the world much more aware than ever before of a whole set of largely unrecognized tropical diseases. TDR was also able to leverage additional funds to promote research in this field.
Protozoan parasites have developed sophisticated mechanisms to circumvent human defense systems and, for those parasites which require intermediate vectors, also to avoid the countermeasures of these organisms. The most sophisticated immune-evasion method might be the use of variable surface glycoproteins (VSGs) by T. brucei. The capability of this parasite to express a successive series of VSGs allows them to survive the onslaught of antibodies in the bloodstream. VSG crystal structures (Freymann et al., 1984 ▶; Metcalf et al., 1987 ▶; Blum et al., 1993 ▶) have given remarkable insights into the architecture of the coat of the sleeping-sickness parasite. Variable surface proteins of other parasites include the proteins encoded by the var genes of Plasmodium species and variant-specific surface proteins (VSPs) in G. lamblia (Rivero et al., 2010 ▶). T. cruzi employs molecular mimicry tricks, hides inside various human cells and has a sophisticated mechanism to evade complement lysis (Bonney et al., 2011 ▶; Gironès et al., 2005 ▶; Joiner et al., 1986 ▶). Leishmania species live within macrophages, the very cells that are supposed to kill them. These features make the development of effective and affordable vaccines for the parasitic protozoa an enormous challenge.
Therefore, therapeutic compounds, and in particular combinations of compounds, are likely to remain a cornerstone of antiparasitic strategies for a long time. Structural information on drug targets can contribute to many stages of the long road leading to such compounds. Hence, it is encouraging that the number of three-dimensional structures of proteins from human parasites is approaching 2000 (Table 2 ▶). The challenge is to increase this body of knowledge and also to convert this three-dimensional information into compounds which prevent the diseases caused by these organisms. Three-dimensional structures can not only guide the design of compounds with great potency, but can also be of assistance in lead-optimization stages of drug discovery, when the selectivity, bioavailability, pharmacodynamic, pharmacokinetic, safety, formulation and other properties of the compound have to be improved (Wermuth, 2008 ▶; Nicolaou, 2014 ▶). As we shall see below, three-dimensional structures of target proteins can also be of assistance in designing compounds which are less likely to be the victim of resistance mechanisms.
Table 2. Three-dimensional structures per parasite.
The number of entries includes multiple structures of the same proteins, different domains from the same protein etc. The author is grateful to Gerard Kleywegt for providing the numbers in this table.
| Parasite | No. of PDB entries with proteins | No. of unique proteins with PDB entries |
|---|---|---|
| Trypanosoma cruzi | 578 | 222 |
| Trypanosoma brucei | 175 | 74 |
| Leishmania spp. | 251 | 98 |
| Plasmodium spp. | 627 | 290 |
| Toxoplasma gondii | 118 | 50 |
| Cryptosporidium spp. | 71 | 37 |
| Entamoeba histolytica | 111 | 52 |
| Giardia lamblia | 48 | 25 |
| Total | 1979 | 848 |
Here, we sketch an overview of three-dimensional structures of parasite proteins determined over the decades, and link these to the development of therapeutics by structure-guided drug design (SGDD). This is not a comprehensive review of the numerous projects where structure is guiding antiparasitic drug development. Instead, we will focus on a few selected examples.
2. What a difference 30 years make
The progress made over the last 30 years in the structural biology of pathogenic protozoa is remarkable. The first attempts to initiate structure determinations with the development of new therapeutics as a goal occurred in the early 1980s. A collaboration between groups in Europe focused on glycolytic proteins from the sleeping-sickness parasite T. brucei. Glycolysis in the bloodstream form of this parasite is essential and, remarkably, sequesters several glycolytic enzymes in a unique organelle, the glycosome (Opperdoes & Borst, 1977 ▶; Barros-Alvarez et al., 2014 ▶). The purification involved isolating parasites from Wistar rats, preparing a glycosome fraction, and employing several biochemical steps to obtain multiple glycosomal glycolytic proteins simultaneously (Misset & Opperdoes, 1984 ▶). From 500 µg purified protein, the first high-resolution crystal structure of a parasite protein could be elucidated, that of T. brucei triosephosphate isomerase (Wierenga et al., 1987 ▶). Studies on T. brucei triosephosphate isomerase also led to the first steps in fragment-cocktail crystallography (Verlinde et al., 2009 ▶).
Using the same purification procedure, a precious few milligrams of T. brucei glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were obtained. This allowed the growth of three crystals, which was not sufficient for a structure determination since the wet-capillary crystal-mounting procedure at room temperature that was then in vogue could not prevent rapid crystal deterioration in the X-ray beam. Fortunately, with the help of Janus Hajdu, it was possible to obtain an initial 33% complete data set for T. brucei GAPDH using the Laue method. Since there were six subunits (one and a half tetramers) per asymmetric unit, this was, per subunit, a larger number of observations than for a complete data set with one subunit per asymmetric unit (Vellieux et al., 1993 ▶, 1995 ▶). The resultant structure showed interesting differences between trypanosomatid GAPDH and its human counterpart near the ribose ring of the cofactor NAD which were exploited for selective inhibitor design (Aronov et al., 1999 ▶; Bressi et al., 2000 ▶; Suresh et al., 2001 ▶). Structure-guided studies focusing on trypanosomatid glycosomal enzymes and receptors are still actively being pursued (Barros-Alvarez et al., 2014 ▶; Guido et al., 2009 ▶; Lozano et al., 2014 ▶).
Interestingly, in recent years, proteins and large complexes from major parasites have been the subject of new methods to determine three-dimensional structures, with the extra interesting feature that the T. brucei cysteine protease cathepsin B crystals used were grown in insect cells. The X-ray free- electron laser (XFEL) methodology was able to solve the crystal structure using these in vivo-grown crystals (Redecke et al., 2013 ▶), as was serial crystallography using synchrotron radiation and parallel helical scans (Gati et al., 2014 ▶). In both cases the determination of the parasite enzyme structures represented important methodological progress. The remarkable resolution revolution in cryo-electron microscopy (cryoEM) of macromolecular assemblies is the result of a combination of high-speed direct detectors, particle-shift correction and maximum-likelihood treatment of errors. A fabulous result by cryoEM, and highly relevant for arriving at novel antimalarials, is the recent determination of the structure of the ribosome of the malaria parasite with the inhibitor emetine bound to it (Wong et al., 2014 ▶).
Protein crystallography in drug design poses special challenges, one of these being the need to take great care in the interpretation of electron-density features representing ligands. Surprises can occur when purchased or synthesized compounds are simply not what they were supposed to be. Challenges can also arise from ligands with the same chemical composition but different chirality or from overlapping binding modes of a single ligand. Fortunately, there are an increasing number of tools available for analyzing and reporting the quality of ligand densities (Pozharski et al., 2013 ▶; Sehnal et al., 2015 ▶).
In future, we can expect increased attention to membrane proteins, with the structure of Plasmodium aquaporin (PDB code 3c02; Newby et al., 2008 ▶) as an encouraging example. The current impressive progress in membrane-protein structure determination in general will gradually be expanded further to structure determinations of membrane proteins from parasites. Parasite membrane proteins are obviously attractive as drug targets in that compounds binding to them from the extracellular side have one membrane less to cross to exert a detrimental effect on the parasite.
3. Genome sequences
Just after the turn of the millennium, the genome sequences of several parasitic protozoa became available. By the end of 2014, the genome sequences of all of the parasites mentioned in this overview were completed or nearly completed and made available in EuPathDB (http://eupathdb.org/eupathdb/). The genome sizes range from about 9 to 35 megabases, encoding approximately 4000–11 000 proteins (Table 3 ▶). It is good to realise that the parasitic protozoa also have a mitochondrial genome, with the remarkable exception of Giardia and Entamoeba, where the mitosomes, which are relic mitochondria, have no detectable genome. Also, the Apicomplexa contain a unique organelle, the apicoplast, which contains a third genome.
Table 3. Parasites and genome sequences.
Numbers were obtained from http://eupathdb.org/eupathdb/.
| Parasitic protozoan | Name of organism sequenced | Genome size (Mbases) | No. of protein genes |
|---|---|---|---|
| Trypanosoma cruzi | T. cruzi CL Brener Non-Esmeraldo-like | 32.53 | 10834 |
| Trypanosoma brucei | T. brucei Lister strain 427 | 26.75 | 8833 |
| T. brucei gambiense DAL972 | 22.15 | 9895 | |
| T. brucei TREU927 | 35.83 | 11567 | |
| Leishmania spp. | L. braziliensis MHOM/BR/75/M2903 | 35.21 | 8567 |
| L. donovani BPK282A1 | 32.44 | 8195 | |
| L. major strain Friedlin | 32.86 | 8400 | |
| L. mexicana MHOM/GT/2001/U1103 | 32.11 | 8250 | |
| Plasmodium spp. | P. falciparum 3D7 | 23.33 | 5542 |
| P. vivax Sal-1 | 27.01 | 5586 | |
| Toxoplasma | T. gondii ME49 | 65.57 | 8920 |
| Cryptosporidium | C. hominis TU502 | 8.74 | 3886 |
| C. parvum Iowa II | 9.10 | 3805 | |
| Entamoeba | E. histolytica HM-1:IMSS | 20.08 | 8333 |
| Giardia | Giardia assemblage A isolate WB | 12.83 | 9667 |
Even though the functions of ∼50% or more of the ORFs remain to be determined (Peter Myler, personal communication), and the algorithms for identifying the start and end points of all the ORFs have scope for improvement, the effects of the genome sequences on structure-guided drug design are numerous. One of the major effects is that the nucleotide sequence of ORFs can be obtained from databases and be used for overexpressing proteins. Even though protein expression is highly idiosyncratic, compared with the dark days before the genome sequences were available, the situation has dramatically improved.
4. Target selection
Which protein to target is a key decision in every structure-guided drug-design project. Unraveling the importance of specific proteins in infection can start from an in silico analysis of genome sequences. An example is the discovery of the nonmevalonate pathway of isoprenoid synthesis in the apicoplast of the malaria parasite (Hunter, 2007 ▶, and references therein). Such discoveries from a genome analysis obviously have to be followed up by functional studies, which cover a wide diversity of techniques far beyond the scope of this overview. Aspects of target selection for structure-guided projects, several of which are specific for parasitic protozoa, are the following.
(i) Essentiality. Targets need to be essential so that blocking their function is detrimental to the parasite. For parasites transmitted via an intermediate host, it has to be established whether or not a particular protein is of relevance for life stages in the human host to avoid time and effort being wasted on targeting proteins that are irrelevant for the human patient. However, in certain cases it can be beneficial to target proteins important for the next step in the life cycle of the parasite, i.e. aiming for transmission-blocking therapeutics.
(ii) Redundancy. As with many infectious agents, redundant proteins and pathways occur in parasites. Hence, careful genome-sequence analysis and functional studies are needed to avoid targeting metabolic, transport or signaling pathways for which an alternative route is available to the parasite without a substantial loss of vitality.
(iii) Cell entry. For those parasites which are engaged in entering one or more types of human cells, the invasion machinery is an attractive source of potential targets, with a likelihood of unique proteins and of variants of well known proteins with new functions. Of particular interest for drug design are essential cell-entry proteins accessible to small molecules in the bloodstream, since this implies that such compounds may not have to cross one or more parasite membranes to exert an inhibitory action on cell entry. Given also the possible importance of such structures for vaccine design, it is no wonder that over 75 structures have been determined of proteins and domains involved in cell adhesion and invasion of Apicomplexans (Boucher & Bosch, 2015 ▶).
(iv) Selectivity. Ideally, a drug target in the parasite is absent in the human host. In many cases, however, parasite drug-target proteins have human homologs. The difference between the host and parasite proteins then needs careful attention to estimate the probability of success in obtaining selective inhibitors. Even when no structure of a parasite drug-target protein is available, in some cases structural information from homologues can be combined with the sequences from parasite and host, making it possible to estimate the likelihood of success in arriving at highly selective inhibitors. However, in exceptional cases little or no selectivity of a compound for purified targets from the pathogen and host exists, while the compound is still a useful drug. In the field of parasitic protozoa, the ‘resurrection drug’ eflornithine (or difluoromethylornithine; DFMO) for advanced stages of sleeping sickness is a well known example (Ebikeme, 2014 ▶). It is assumed that the higher turnover rate of the human ornithine decarboxylase enzyme compared with that of the parasite enzyme is key to human tolerance and allows the use of DFMO in patients despite displaying no selectivity at the enzyme level. Moreover, the fact that DFMO has more effect on T. brucei rhodesiense than on T. brucei gambiense is the result of a slower turnover rate of the former enzyme (Iten et al., 1997 ▶). Hence, testing compounds at the cellular level, and even in animal models, in the early stages of a drug-design process can have major advantages.
(v) Druggability. When three-dimensional structures of the drug target or of homologous proteins are available, the ‘druggability’ of the targeted site can be explored. Druggability can be defined as the probability that typical oral drugs, as first defined by Lipinski’s rule of five (Lipinski et al., 2001 ▶; Lipinski & Hopkins, 2004 ▶), will be able to make favorable interactions with the intended site of interaction (Volkamer et al., 2012 ▶; Schmidtke & Barril, 2010 ▶; Dorrestein & Carroll, 2011 ▶). This is an important consideration in target selection in many cases, which holds for compounds designed to block active sites, to inhibit or strengthen protein–protein interfaces, to prevent conformational changes or to bind to allosteric sites and induce conformational changes at critical points away from the actual binding site.
(vi) Protein flexibility as a challenge. Small-molecule binding can be accompanied by surprising and large conformational changes in protein structure (Teague, 2003 ▶). Flexibility is probably the major hurdle in the application of current computational methods for correctly predicting the binding poses of compounds bound to their targets and for calculating the affinities of compounds for their targets with reasonable accuracy. Recent published successes in structure-based selectivity prediction (e.g. Rodríguez et al., 2014 ▶) and in affinity rank ordering (e.g. Voet et al., 2014 ▶) have benefited from considerable human expertise even in those cases with limited conformational changes. Fortunately, protein crystallography is a powerful method to reveal conformational changes by solving structures with compounds bound to the target. Such structures of protein–inhibitor complexes form a new starting point to guide the drug-design process.
(vii) Protein flexibility as an opportunity, in particular when combined with high-throughput screening (HTS). The development of improved non-nucleoside reverse-transcriptase inhibitors (NNRTIs) for HIV is a nice example of exploiting the surprising flexibility of a target protein. The original NNRTIs were found by HTS of small-compound libraries (Pauwels et al., 1990 ▶). The binding site of NNRTIs is only apparent in structures with NNRTIs bound (Kohlstaedt et al., 1992 ▶; Smerdon et al., 1994 ▶). In structures of reverse transcriptase without compounds the binding site is absent. The subsequent development of new NNRTIs, which are major components of multi-compound anti-HIV drugs at present (Das et al., 2005 ▶, 2008 ▶; Janssen et al., 2005 ▶; De Clercq, 2012 ▶), points to an important opportunity for synergy between HTS and SGDD approaches in the design of therapeutics. HTS can not only lead to compounds with high to medium affinity for the target but, when followed up by structural studies, also to the discovery of unexpected target sites.
5. High-throughput screening and SGDD
HTS brings together chemistry, robotics, biophysics and biology. Carefully selected and maintained libraries of a few thousand to a few million compounds are tested using robots to mix compounds and purified proteins, or cell cultures, while biophysical or biochemical read-out methods measure the effect of each compound. With careful analysis of the results to weed out false positives (Baell & Walters, 2014 ▶; Dahlin & Walters, 2014 ▶), compounds of great value for further development can be discovered. The crystal structures of target proteins in complex with compounds discovered by the screens provide important information for follow-up studies. Combining HTS with SGDD is a powerful approach in arriving at potential drugs, as mentioned above in the case of the NNRTIs and as will be seen below.
HTS screens targeting purified proteins from parasitic protozoa have been carried out for several targets [for example, T. cruzi cruzain (Wiggers et al., 2013 ▶), T. cruzi and T. brucei phosphofructokinase (Brimacombe et al., 2014 ▶), T. brucei N-myristoyltransferase (Brand et al., 2012 ▶), T. brucei glycogen synthase kinase 3 short (Ojo et al., 2008 ▶), T. brucei ornithine decarboxylase (Smithson et al., 2010 ▶), T. brucei hexokinase (Sharlow et al., 2010 ▶) and P. falciparum dihydroorotate dehydrogenase (Baldwin et al., 2005 ▶)]. Mass spectrometry has also been used for screening fragment-sized natural products binding to P. falciparum 2′-deoxyuridine 5′-triphosphate nucleotidohydrolase (Vu et al., 2013 ▶).
HT screens investigating the effect of compounds in phenotypic screens on parasites in vivo are also important. The chemical structures of active compounds obtained from screens on the malaria parasite have generously been made available to the drug-development community (Guiguemde et al., 2010 ▶, 2012 ▶; Gamo et al., 2010 ▶). This has resulted in 13 533 compounds in the Tres Cantos Antimalarial Set (TCAMS). All of the public data sets are searchable (https://www.ebi.ac.uk/chemblntd). Another follow-up is the so-called ‘malaria box’, a collection of 400 diverse compounds selected from extensive screening campaigns of around four million compounds from three different screens and available for initiating antimalarial research (http://www.mmv.org/malariabox; Spangenberg et al., 2013 ▶). Several other phenotypic high-throughput and medium-throughput screens involving parasitic protozoa have been carried out, including studies targeting T. cruzi (Andriani et al., 2011 ▶), T. brucei (MacGregor et al., 2014 ▶, Mackey et al., 2006 ▶, Sharlow et al., 2009 ▶), L. donovani (Zhu et al., 2012 ▶), P. falciparum (Baniecki et al., 2007 ▶) and G. lamblia (Tejman-Yarden et al., 2013 ▶). Recently, a parallel HTS of a 1.8 million compound collection has been carried out against L. donovani, T. brucei and T. cruzi. The outcomes of this investigation are fully disclosed while three ‘kinetoplastid boxes’, with ∼250 carefully selected chemicals each, are available for collaborators upon request (Peña et al., 2015 ▶).
The targets of compounds obtained by phenotypic screening are often not known, which is a disadvantage, even though the hits can be optimized by chemical modifications without knowledge of the target or mode of action. However, given the increasingly robust ability to identify targets from screening hits by combining genetics and chemical proteomics, it is likely that structures of targets identified will be determined and SGDD can be effectively employed in follow-up studies.
The University of Dundee’s Drug Discovery Unit supported by the Wellcome Trust (Frearson et al., 2007 ▶) has developed a chemical library which has been used for screens against potential trypanosomatid targets. Another screening center with an emphasis on tropical parasites has been created at the Brazilian Biosciences National Laboratory (LNBio), Brazilian Center for Research in Energy and Materials, Campinas, São Paulo, Brazil. LNBio started screening the T. cruzi glucose-6-phosphate dehydrogenase against a commercial library of 30 000 compounds (Mercaldi et al., 2014 ▶) and efforts are under way to set up a collection of local natural products to feed the HTS campaigns. The Journal of Biomolecular Screening devoted its January issue of 2015 to ‘Novel Therapeutic Approaches for Neglected Infectious Diseases’, illustrating the growing activity in this field.
6. Structure-guided drug design
As mentioned above, almost 2000 structures of parasitic protozoan proteins have been deposited in the PDB (Table 2 ▶). Several hundred of these are the result of initiatives such as the Structural Genomics of Pathogenic Protozoa (SGPP) consortium, the MEPHITIS collaboration (http://www.mephitis.eu/), the Structural Genomics Consortium (SGC) and the Seattle Structural Genomics Center for Infectious Diseases (SSGCID).
Some proteins from parasitic protozoa have been used in fragment-cocktail crystallography approaches for initial steps in ligand discovery. In such studies, protein crystals are soaked in cocktails of high concentrations of 3–10 low-molecular-weight molecules. The basic idea is that it is easier for small molecules to find a pocket on the surface of the protein to bind to than for the typically larger molecules used in HTS. The more complex nature of the latter decreases the probability of finding a complementary binding site on a protein surface (Hann et al., 2001 ▶; Hall et al., 2014 ▶). A variety of biophysical techniques are employed to establish the binding site and affinity (Larsson et al., 2011 ▶). Once the binding site has been established and if it is close to an active site, as often appears to be the case, then the next steps can involve ‘growing’ the fragment; that is, adding substituents which fit into neighboring pockets with favorable interactions (infectious disease examples include Bauman et al., 2013 ▶; Tiefendbrunn & Stout, 2014 ▶). If multiple fragments bind reasonably close together these can be linked, followed by additional iterations of growing and structure determination, combined with testing many other properties of the molecules as required in all drug-design projects.
The number of cases published in which fragment-based approaches have been used for parasitic protozoa seems to still be quite small, with a study on T. brucei nucleoside 2-deoxyribosyltransferase having been described in detail (Bosch et al., 2006 ▶). In the Medical Structural Genomics of Parasitic Protozoa (MSGPP) project, an in-house library of 68 cocktails with 8–10 compounds each was used. Of 26 target proteins, seven proteins showed electron-density maps with evidence of a bound ligand (Verlinde et al., 2009 ▶). This emphasises the need for robust crystals which can survive the relatively high DMSO levels that are often required to obtain sufficiently high concentrations of the usually weak-binding fragments.
Several structures of proteins from parasitic protozoa in the PDB have initiated and/or are part of SGDD campaigns. A small selection from the many projects, with an emphasis on studies in which compounds were tested in animal models or beyond, identifies challenges and successes in the field.
6.1. Cruzipain, a cysteine protease from T. cruzi
An early SGDD project targeted cruzipain, an essential cysteine protease in T. cruzi (McGrath et al., 1995 ▶). Presently, there are 42 depositions of cruzipain with different compounds in the PDB. These studies include complexes with phenyl-containing vinyl-sulfone inhibitors (Brinen et al., 2000 ▶; Chen et al., 2010 ▶). One of these, compound K11777, has been shown to be safe and efficacious in animal models of acute and chronic Chagas disease, while a related inhibitor, WRR-483, also had trypanocidal activity in cell cultures and an animal model (Chen et al., 2010 ▶; McKerrow et al., 2009 ▶). Recently, HTS targeting cruzipain resulted in a new series of compounds with attractive features. A follow-up crystal structure of compound Neq176 in complex with cruzipain has revealed information which can be used for further development of this class of noncovalent inhibitors (Wiggers et al., 2013 ▶).
6.2. Protein farnesyltransferase (PFT)
Therapeutic compounds have to combine a large number of favorable characteristics, and this complex situation is illustrated by studies targeting P. falciparum protein farnesyltransferase (PFT; Buckner et al., 2005 ▶). This enzyme catalyzes an essential post-translational modification by adding a farnesyl moiety to the C-terminus of specific proteins and thereby anchoring these proteins to the membrane. Extremely promising tetrahydroquinoline derivatives were obtained by starting from known inhibitors of PFT from other species and docking the inhibitors into a homology model (Van Voorhis et al., 2007 ▶; Bendale et al., 2007 ▶). The most potent compound had a subnanomolar enzyme IC50, with an ED50 for P. falciparum growth in human cells of 15 nM. These compounds unfortunately turned out to be cleared too rapidly to be of practical use. A key weakness appeared to be an oxidative N-dealkylation, likely to be carried out by human cytochrome P450 enzymes, causing rapid compound clearance. This oxidation affected the zinc-chelating substituent of the tetrahydroquinoline inhibitors. Subsequent studies elucidated the metabolic pathways used by the human host (Bulbule et al., 2008 ▶), while the crystal structure of a mammalian PFT allowed modeling studies to arrive at likely binding modes in P. falciparum PFT. Combining this information resulted in 2-oxotetrahydroquinoline PFT inhibitors which had tenfold increased half-lives in the human host, but the uptake of these compounds in animal models when administered orally still needs further improvement.
6.3. Aminoacyl-tRNA synthetases (aaRS)
Protein synthesis is an attractive target for antiparasitic agents since protein synthesis is essential in critical steps of the life cycle in the human host. Many proteins, RNAs and the ribosome are involved in protein synthesis, with the aminoacyl-tRNA synthetases (aaRS) from parasitic protozoa receiving considerable attention recently. For instance, the current number of depositions in the PDB of tRNA synthetases with complete enzymes or domains from parasitic protozoa, often in complex with substrates and inhibitors, is 35 for trypanosomatid species, nine for Plasmodium spp., one for T. gondii, one for Cryptosporidium spp., nine for Entamoeba spp. and four for Giardia spp., giving a total of 56. The aaRS catalyze the critical step of attaching, with the help of ATP, an aminoacyl moiety to the 2′- or 3′-hydroxyl of the ribose from the nucleotide at the 3′-end of a cognate tRNA. Humans have two sets of canonical aaRS: 20 function in the cytoplasm and a second set of 20 in the mitochondria. In parasitic protozoa there are some surprising differences compared with the human host. For instance, each trypanosomatid species contains 23 annotated tRNA synthetase genes in its genome, i.e. these parasites contain only a single set of 20 aaRS, with redundancy of the aaRS for three amino acids (Charrière et al., 2009 ▶). This set has to function in the cytoplasm as well as in the mitochondrion. Moreover, certain aaRS from parasitic protozoa have unusual features (Gowri et al., 2012 ▶). Targeting parasitic aaRS is inspired by and benefits from the results of past and ongoing studies on inhibiting bacterial tRNA synthetases. The prime example is the natural product mupirocin (pseudomonic acid), an IleRS inhibitor, which is used for the topical treatment of bacterial skin infections (Nakama et al., 2001 ▶).
A recent review focuses on studies targeting aaRS in eukaryotic pathogens, mainly in protozoa (Pham et al., 2014 ▶). Interesting aaRS inhibitors interfering with the growth of parasitic protozoa and with the binding mode known from crystal structure determinations include the following.
(i) Cladosporin, a fungal secondary metabolite which inhibits cytoplasmic P. falciparum LysRS and P. falciparum proliferation in the blood and liver in the nanomolar range (Hoepfner et al., 2012 ▶). The crystal structure of cladosporin in complex with LysRS has recently been determined (Khan et al., 2014 ▶) and provides information which may assist in improving the poor bioavailability of this metabolite.
(ii) Aminoquinolone derivatives, synthetic compounds which inhibit MetRS from trypanosomatids with enzyme IC50 and in vivo cell-culture ED50 values in the low nanomolar range (Shibata et al., 2011 ▶, 2012 ▶). Several promising compounds clear parasites in animal models of Chagas disease without toxicity but combating recrudescence is still a challenge. Crystal structures with aminoquinolone derivatives (Koh et al., 2012 ▶) and urea-based inhibitors (Koh et al., 2014 ▶) revealed major conformational changes upon inhibitor binding. These structures are guiding the development of antitrypanosomatid compounds with improved pharmacokinetic and pharmacodynamic properties.
(iii) Halofuginone, a halogenated derivative of the natural product febrifugine obtained from the roots of the chang shan herb used as an antimalarial in traditional Chinese medicine (Hirai et al., 2003 ▶), is a ProRS inhibitor in P. falciparum (Keller et al., 2012 ▶). A crystal structure of unligated cytoplasmic P. falciparum ProRS has been determined (Jain et al., 2014 ▶) and also the structure of human ProRS in complex with halofuginone and with ATP (Zhou et al., 2013 ▶). This complex revealed that halofuginone is a remarkable dual-site inhibitor that simultaneously occupies the proline binding site and the pocket occupied by the 3′-end of the tRNA. The inhibitor acts synergistically with ATP. Structures of the P. falciparum enzyme in complex with halofuginone have recently been deposited in the PDB (PDB entry 4q15 by the SSGCID and PDB entry 4ydq by Jain et al., 2015 ▶). These studies form a platform for the structure-guided development of compounds with little affinity for the host ProRS enzymes while inhibiting Plasmodium ProRS effectively.
6.4. N-Myristoyltransferase (NMT)
N-Myristoyltransferase (NMT) transfers the myristate moiety from myristoyl-CoA to the N-terminal Gly residue of a large number of proteins. This affects the localization and/or the activity of the modified proteins. Initial studies on parasite NMTs focused on T. brucei, where RNAi studies showed that knockdown of NMT is lethal in cell cultures of this parasite and diminishes its infectivity in animal models (Price et al., 2003 ▶, 2010 ▶).
An HTS identified a series of N-pyrazole arylsulfonamides with low micromolar IC50 values for T. brucei NMT (Frearson et al., 2010 ▶; Brand et al., 2012 ▶). A pragmatic chemistry-driven approach then resulted in compounds with much improved enzyme IC50 values of 2 nM and similar EC50 values for inhibiting in vitro parasite cell growth (Brand et al., 2012 ▶). The lack of ability to penetrate the blood–brain barrier needed further attention. Crystal structures of homologous enzymes, including that of L. major NMT, assisted in further optimization of the initial compounds (Brand et al., 2014 ▶). In particular, capping the sulfonamide moiety, which was shown to be solvent-accessible in a crystal structure of a compound bound to NMT, was effective. Specifically, difluoromethylation increased CNS penetration while maintaining affinity for the enzyme and potency to kill parasites (Brand et al., 2014 ▶). The much improved, most promising compound described still needs further tweaking to be sufficiently effective in stage 2 animal models. This paper shows the broad spectrum of conditions that compounds have to fulfill in order to be suitable for the treatment of second-stage African sleeping sickness.
NMT is not only under investigation as a drug target for T. brucei but also for other protozoa, including P. falciparum, P. vivax, Leishmania spp. and T. cruzi (Wright et al., 2014 ▶; Olaleye et al., 2014 ▶; Tate et al., 2014 ▶; Brannigan et al., 2014 ▶; Hutton et al., 2014 ▶; Roberts et al., 2014 ▶). This recent explosion of investigations on parasite NMTs using a combination of HTS, medicinal chemistry, parasitology and structural studies may possibly lead to useful compounds.
6.5. Protein kinases, in particular calcium-dependent protein kinases (CDPKs)
Protein kinases affect numerous activities in a wide variety of cells. Owing to the presence of hundreds of different protein kinases in humans, targeting them for the design of therapeutics has long been considered to be an enormous challenge in view of the requirement of endowing compounds with sufficient specificity. Nevertheless, targeting specific protein kinases has resulted in important anticancer therapeutics. As a result of these studies, an enormous amount of knowledge has accumulated regarding the characteristics and the mode of action of effective protein kinase inhibitors (Capdeville et al., 2002 ▶; Cruzalegui, 2010 ▶; Zhang et al., 2009 ▶).
The potential of protein kinases as drug targets in parasitic protozoa was recognized early on from genome sequence-analysis studies (see, for example, Doerig et al., 2002 ▶; Parsons et al., 2005 ▶). The drug-design opportunities offered by this class of enzymes are reflected in the large number of PDB depositions of protein kinases from parasitic protozoa: approximately ten from trypanosomatids, Giardia spp. and Entamoeba spp. together, 55 from T. gondii, 32 from Plasmodium spp. and 14 from Cryptosporidium spp., giving a total of 111. Importantly, there appear to be a few classes of Ser/Thr protein kinases with features that make it likely that inhibitors can be obtained with higher affinity for parasite protein kinases than for their human homologs. These classes include the calcium-dependent kinases (CDPKs) in Apicomplexa. Some of these CDPKs play an essential role in the life cycle of parasites (Green et al., 2008 ▶; Billker et al., 2009 ▶; Wei et al., 2013 ▶; Lim et al., 2012 ▶). This combination of characteristics makes the CDPKs attractive drug targets.
A unique feature of CDPKs is a large calmodulin-like domain that can block access to substrate proteins, but which, after a major conformational change, can occupy a completely different position which then allows access of substrate proteins to the active site of the kinase domain. This change is promoted by the presence of calcium ions (Wernimont et al., 2010 ▶; Ojo et al., 2010 ▶). A second important feature of some apicomplexan CDPKs is a small ‘gatekeeper residue’: e.g. a serine in P. falciparum CDPK4 and a glycine in T. gondii CDPK1 and C. parvum CDPK1. The gatekeeper residue controls access to a hydrophobic pocket adjacent to the nucleotide-binding site. ATP analogs with an extra substituent (called the ‘bump’) that can fit into this hydrophobic pocket of protein kinases are potent inhibitors and fail to bind to kinases with a larger gatekeeper side chain. Such ‘bumped kinase inhibitors’ (BKIs) appeared to be potent and selective inhibitors of T. gondii CDPK1, C. parvum CDPK1 and P. falciparum CDPK4, with the binding modes of BKIs revealed by crystallographic studies (Ojo et al., 2010 ▶; Murphy et al., 2010 ▶; Vidadala et al., 2014 ▶). In the case of T. gondii CDPK1, a second generation of compounds with enhanced selectivity was obtained by probing the ribose-binding pocket with additional substituents. For the best compounds, single-digit or better nanomolar IC50 values for T. gondii CDPK1 were accompanied by IC50 values greater than 10 µM for human SRC and ABL kinases (Larson et al., 2012 ▶). A large selectivity index at the enzyme level was obtained, as the initial structural analysis had anticipated.
It will be of great interest to see whether a combination of (i) the unique characteristics of the CDPKs in protozoa, (ii) the immense accumulated chemical knowledge about protein kinase inhibitors in general and (iii) the increasing collection of parasite protein kinase crystal structures will lead to novel, effective and affordable oral drugs for parasitic diseases. Although CDPK inhibitors of apicomplexan parasites have not yet progressed to clinical trials in humans, a recent report is encouraging in that CDPK inhibitors have proven to be effective in treating cryptosporidiosis in animal trials (Lendner et al., 2015 ▶).
6.6. Plasmodium dihydroorotate dehydrogenase (DHODH)
An example where structural knowledge had a significant impact on the design of new compounds is the development of DSM265 and related inhibitors of P. falciparum DHODH. Briefly, the rationale for targeting P. falciparum DHODH is as follows.
(i) Functional studies revealed that pyrimidine synthesis is essential for P. falciparum since the parasite cannot scavenge pyrimidines from the host.
(ii) In the pyrimidine-synthesis pathway, the step catalyzed by DHODH is critical.
(iii) The human host can synthesize pyrimidines but can also salvage preformed bases and nucleosides.
(iv) The P. falciparum enzyme is related to the mitochondrial human DHODH. This human enzyme had been the target in the development of immunosuppressants and shown to be ‘druggable’. Hence, by analogy, the same would most likely hold for the P. falciparum enzyme.
(v) Inspection of the crystal structures of DHODH from various species and the P. falciparum DHODH amino-acid sequence revealed differences in the active site, which appeared promising for achieving good selectivity (Hurt et al., 2006 ▶). Additional structural and biochemical comparisons of inhibitor-binding properties of the DHODH enzymes from the parasite and host have confirmed this difference (Deng et al., 2014 ▶; Bedingfield et al., 2012 ▶).
This background formed the basis of a high-throughput screen against P. falciparum DHODH, which yielded a series of triazolopyrimidine-based compounds with IC50 values for the parasite enzyme of less than 500 nM (see also Phillips & Rathod, 2010 ▶). One of these compounds, DSM1, had an IC50 of ∼50 nM for the parasite enzyme and little activity against the human enzyme (Phillips et al., 2008 ▶). Subsequent testing of a series of related compounds revealed an excellent correlation between the IC50 for the parasite enzyme and the ED50 for parasite growth inhibition, providing chemical validation of the target. A related compound, DSM74, suppressed parasite growth in a P. berghei mouse model (Gujjar et al., 2009 ▶), yet additional rounds of lead optimization were needed to improve its effectiveness.
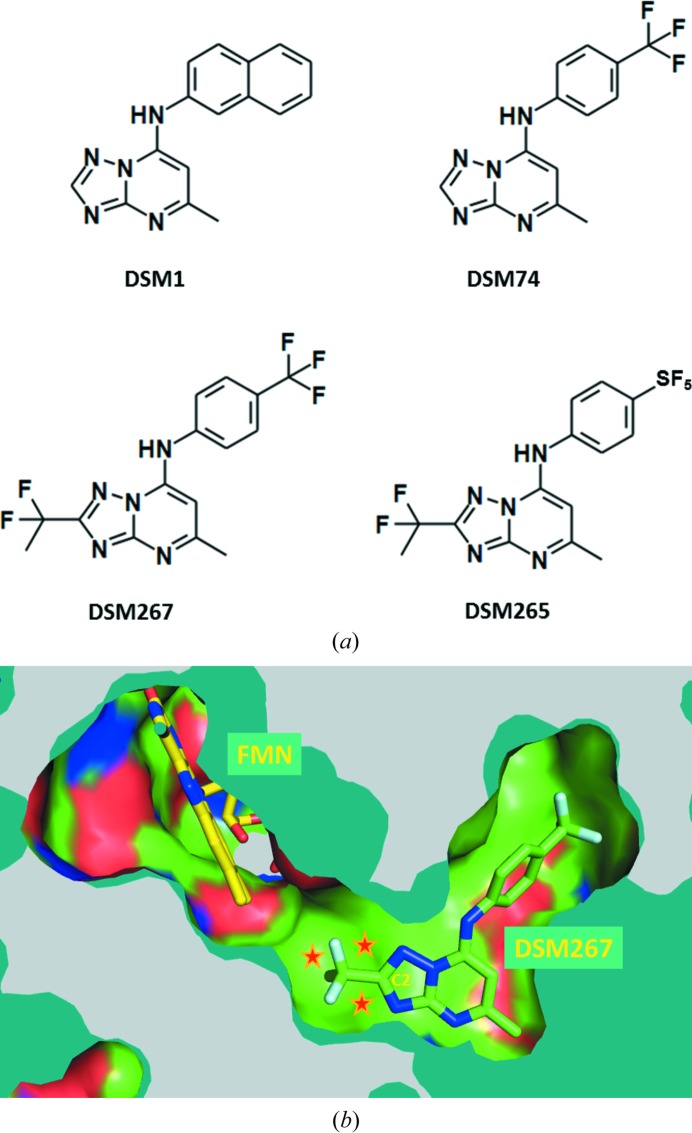
At about this stage, crystal structures of P. falciparum DHODH in complex with DSM1 and related compounds became available. They showed unexpected protein flexibility, resulting in a distinct pocket occupied by DSM1. Of critical importance was the fact that a narrow channel leads from the C2 atom of the triazolopyrimidine ring towards the binding site of the FMN cofactor (Deng et al., 2009 ▶; Fig. 1 ▶). Taking the hydrophobic nature of this channel into account, several substituents were explored. Two compounds obtained, both with a –CF2CH3 moiety at the C2 position, were DSM265 and DSM267, which differ only by an –SF5 or a –CF3 substituent, respectively, on the phenyl ring (Coteron et al., 2011 ▶; Fig. 1 ▶). DSM265 is presently in phase I human clinical trials for the treatment of malaria (http://www.mmv.org).
Figure 1.
Structure-guided development of a potent and selective inhibitor of P. falciparum DHODH. (a) Compounds which led to the P. falciparum DHODH inhibitor DSM265. Compound DSM1 was the result of an HTS screen (Phillips et al., 2008 ▶). DSM74 displayed improved properties (Gujjar et al., 2009 ▶). Exploiting the hydrophobic ‘tunnel’ to the FMN binding site [shown in (b)] enabled the structure-guided design of novel inhibitors. Of these, DSM265 and DSM267 had many favorable pharmacologic and pharmacodynamic properties (Coteron et al., 2011 ▶). The inhibitor DSM265 is in clinical trials. (b) Crystal structure of P. falciparum DHODH in complex with DSM267 and FMN. The hydrophobic channel (stars) leading towards the FMN binding site was initially observed in structures of P. falciparum DHODH in complex with DSM1 and DSM74 (Deng et al., 2009 ▶). This channel was explored by varying the substituent at the C2 position, leading to DSM265 (in clinical trials) and DSM267 (shown; PDB entry 3sfk; Coteron et al., 2011 ▶).
These examples show the successes and challenges in arriving at compounds with the properties needed to be useful. However, in the long term, there are other obstacles ahead, even after drugs become widely used. The major one is the wide spectrum of mechanisms by which parasites can avoid, or at least diminish, the effects of these compounds. This brings us to the threat of drug resistance in general, which is a very important issue.
7. Minimizing the occurrence of resistance
Drug resistance is a prominent and often disastrous phenomenon diminishing the effectiveness of therapeutics for the treatment and prevention of infectious diseases and cancers. In the field of parasitic protozoa, once-effective malaria therapeutics such as chloroquine and pyrimethamine have become useless in some regions (Fidock et al., 2000 ▶). Resistance of P. falciparum against artemisinin, the current front-line drug for treating malaria, is becoming more widespread (Ariey et al., 2014 ▶; Mok et al., 2015 ▶; Straimer et al., 2015 ▶; Miotto et al., 2015 ▶). This development threatens the gains made over the last few decades against the most devastating parasitic protozoan on our planet. Resistance is also undermining the efficacy of other compounds used for the treatment of diseases caused by parasitic protozoa (Croft et al., 2006 ▶; Upcroft et al., 1990 ▶; Mead, 2002 ▶; Barrett et al., 2011 ▶).
The resistance mechanisms used by parasites include point mutations in drug targets, increased gene expression, avoiding drug import, promoting drug efflux and changing prodrug-conversion enzymes. The occurrence of drug resistance can be diminished to a certain degree by a combination of measures. The first is by legal action, such as outlawing the use of specific drugs in agriculture. The second is by public health measures; for instance, by emphasizing to medical staff that drugs should only be used when definitely needed and by educating patients to take drugs for the full duration of the treatment. The latter may even require ‘direct observation therapy’, where healthcare workers bring drugs to the homes of patients and ensure that they take them. The third is by scientific strategies. These include the following.
(i) Aiming for compounds that hit multiple targets. Such compounds might be discovered by phenotypic screening and, in principle, also by SGDD, but this is challenging.
(ii) Characterizing in vitro resistance mechanisms and designing compounds which block frequently occurring early variants in the resistance pathway (Ross et al., 2014 ▶). This is an interesting approach, but in patients other resistance pathways may be employed by the pathogen.
(iii) Specific structure-guided strategies.
(iv) The multi-compound approach.
The latter two strategies will be discussed below.
7.1. Structure-guided strategies to decrease the probability of the occurrence of drug resistance
Viruses evolve at a rapid pace, hence strategies to combat drug resistance in viruses are worth looking into (see also, for example, Goldberg et al., 2012 ▶). The degree at which mutations occur in viruses to avoid the effect of inhibitors is astonishing. In HIV protease about half of the amino acids are involved in resistance mutations. In HIV reverse transcriptase, the binding site of NNRTIs is under constant mutational pressure at multiple positions. From these and many other examples, a few strategies are emerging which can in some cases decrease the likelihood that drug resistance will occur. One has been called ‘stay within the substrate envelope’ (Nalam et al., 2013 ▶) and another ‘incorporate strategic flexibility’ (Das et al., 2008 ▶).
‘Staying within the substrate envelope’ is based on the idea that inhibitors which only contact residues involved in substrate binding will be less prone to point mutations since such mutations are likely to decrease the efficiency of catalysis by the target enzyme. The same idea holds for drugs preventing protein–protein interactions: if the affinity of the drug for the interaction site is only owing to contacts with residues which are also essential for the protein–protein contact, alterations to avoid drug binding are likely to decrease the strength of the protein–protein interaction as well. If, instead, drugs depend for their affinity to a large extent on residues which are not involved in substrate or partner protein binding, then changes in the drug binding site can be made without substantial penalties for the functioning of the drug target. Such changes can occur without affecting the fitness of the protein targeted and are more likely to occur rapidly.
Here, a problem of specificity may occur when the parasite drug target is an enzyme with a homolog in the human host. Usually, the core of the active site is highly similar in homologous enzymes. Therefore, the requirements of staying within the substrate envelope and of obtaining sufficient selectivity need a careful balancing act. The larger the substrate, the more likely that this strategy will be of use.
‘Incorporate strategic flexibility’ is an idea to provide inhibitors with a certain degree of flexibility such that the effect of point mutations of residues lining the binding site can be met by subtle changes in inhibitor conformation. A completely rigid drug would be more sensitive to small changes in the binding pocket than a more flexible compound. This approach needs to be taken with care since highly flexible compounds lose conformational entropy upon target binding, and in drug design a strategy to increase affinity is ‘conformational restriction’, i.e. to decrease the number of rotatable bonds of a compound. Clearly, the right balance has to be found regarding the number of rotatable bonds in a compound.
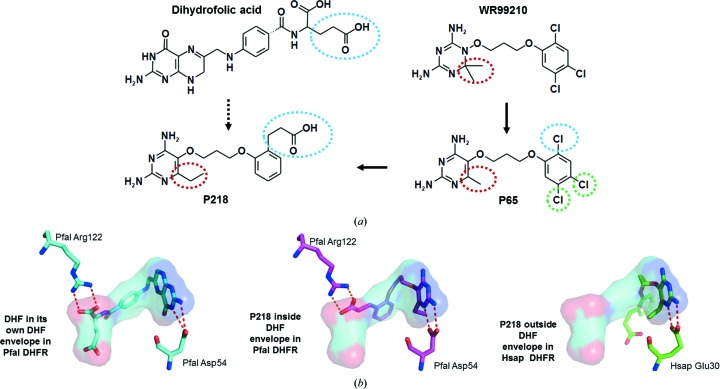
In drug design for parasitic protozoa, the case of P218, a novel inhibitor of P. falciparum dihydrofolate reductase (DHFR), may serve as an example in which both strategies mentioned above have been taken into consideration (Yuthavong et al., 2012 ▶). DHFR is an essential enzyme in folate synthesis and a target of drugs used in the treatment of certain cancers, bacterial infections and malaria. In Plasmodium species the drug pyrimethamine inhibits the DHFR activity of the fusion DHFR-thymidylate synthase (DHFR-TS). The structure determination of P. falciparum DHFR-TS was a tour de force, with multiple structures revealing the binding modes of several inhibitors and substrates in the DHFR active site (Yuvaniyama et al., 2003 ▶).
In the study leading to P218, the key points were as follows (Fig. 2 ▶).
(i) The compound WR99210 is an excellent inhibitor but has poor bioavailability since its triazine ring has a pK a of 10−11 and is fully charged at neutral pH values.
(ii) The pyrimidine group of P65 instead has a pK a of 6–7. The oral bioavailability of P65 in rats was found to be 83%, compared with less than 1% for WR99210.
(iii) Although P65 is 200-fold less potent than WR99210 in an in vitro cell-based infectivity assay with a pyrimethamine-resistant P. falciparum strain harboring a quadruple mutant DHFR, it has a far greater in vivo activity than WR99210 by the oral route because of its superior oral bioavailability.
(iv) A crystal structure of P65 in complex with P. falciparum DHFR indicated that the 2-Cl atom was pointing in the same direction as the glutamate moiety of the substrate dihydrofolate. This moiety interacts with a conserved arginine.
(v) The glutamate moiety of the substrate is quite flexible.
(vi) An ethylcarboxylate moiety at C2 of P65, resulting in the new inhibitor P218, would (1) point in the direction of and interact with the conserved arginine, (2) stay within the substrate envelope, (3) accommodate multiple point mutations of P. falciparum DHFR and (4) be a rather flexible part of the new inhibitor, i.e. have the capability to avoid point mutations by changing dihedral angles.
(vii) The other chloro groups of P65 are simply replaced by hydrogens (green dashed circles in Fig. 2 ▶ a).
Figure 2.
P. falciparum DHFR: design of the selective inhibitor P218. (a) The evolution of compounds in the development of the P. falciparum DHFR inhibitor P218. The high-affinity inhibitor WR99210 had poor bioavailability. The lower pK a of P65 resulted in much better bioavailability than WR99210 at the expense of affinity (the red dashed circle indicates the crucial difference between these two compounds). By incorporating features of the substrate dihydrofolate (blue dashed lines) and removing all Cl atoms, compound P218 was obtained which had many favorable properties. P218 was also designed to be sufficiently flexible and small so that it might be less prone to escape mutations (Yuthavong et al., 2012 ▶). (b) Binding modes of P218 and the substrate dihydrofolate. Left, the substrate dihydrofolate (DHF) bound to wild-type P. falciparum DHFR (PDB entry 4dpd; Yuthavong et al., 2012 ▶). The substrate envelope of DHF is also depicted. Note the interactions of the tetrahydropteridine ring of DHF with the essential Asp54 of the Plasmodium enzyme, anchoring this part of the inhibitor to the center of the active site, a feature common to many DHFR inhibitors. A carboxylate group of DHF makes a salt bridge with Arg122. Middle, the inhibitor P218 [see (a)] bound to P. falciparum DHFR with four resistant mutations (PDB entry 4dp3; Yuthavong et al., 2012 ▶). The compound P218 essentially resides inside the DHF substrate envelope. It makes very similar interactions with Asp54 as DHF, while a carboxylate moiety of the compound interacts with the guanidinium group of Arg122. Right, the inhibitor P218 [see (a)] bound to human DHFR (PDB entry 4ddr; Yuthavong et al., 2012 ▶). The interactions with the essential catalytic carboxylate Glu30 of the human enzyme (equivalent to Asp54 of P. falciparum DHFR) are essentially the same as those in the complex of P218 with the P. falciparum enzyme. However, the rest of P218 binds in a dramatically different way to the P. falciparum enzyme, explaining the substantial difference in affinity for the parasite and human enzymes. The substrate envelope of dihydrofolate as bound to P. falciparum DHFR is shown for reference.
Subsequent structure determinations of P218 bound to the enzymes from the parasite and the host showed that the binding mode of P218 to the parasite enzyme was as expected. In contrast, the mode of binding to human DHFR was substantially different (Fig. 2 ▶). Additional studies showed a high in vivo efficacy in a mouse model of P. falciparum malaria, good oral bioavailability, favorable enzyme selectivity and good safety characteristics. At present, P218 is in preclinical studies.
It might also be mentioned here that DHFR is also the target of studies aiming at novel antifolates in other parasitic protozoa such as Cryptosporidium (Anderson, 2005 ▶; Bolstad et al., 2008 ▶).
7.2. Multi-compound therapeutics
Designing single compounds, even if these have all of the properties needed to be a safe and efficacious drug, is not sufficient. There are at least two reasons why the future should only see the application of multi-compound antiparasitics for treating patients.
Firstly, the probability that resistance will appear in multi-compound drugs is much smaller than in single-compound drugs (Gassis & Rathod, 1996 ▶). This holds at least for mechanisms such as drug-target point mutations, decrease in drug influx facilitated by membrane proteins and changes in drug-conversion enzymes. The major threat here is the possibility that efflux pumps, and combinations of efflux pumps with different but overlapping specificities, are able to overcome even the inhibition by cocktails of compounds. Therefore, studies on efflux pumps in infectious organisms and how these could be inhibited should have a high priority.
Secondly, the biology of the parasites. Several parasitic protozoa reside in different organs during their life cycle in the human host. Plasmodium sporozoites first enter liver cells, where an explosive multiplication occurs, after which the emerging merozoites enter red blood cells. In infections by P. vivax, a reservoir of parasites may remain in the liver cells as hypnozoites and may cause a relapse, which can happen years later. The causative agent of Chagas disease, T. cruzi, hides in various cells, being largely undetectable for decades. T. brucei survives in the bloodstream and gradually enters the CNS by crossing the blood–brain barrier. From studies on Mycobacterium tuberculosis, a bacterium which is an expert in avoiding the effects of drugs by multiple mechanisms, it appears that drug cocktails are needed to hit the bacteria in various stages and in various places, where they may be in different metabolic states (Dartois, 2014 ▶). Given the sophistication of parasitic protozoa, it is likely that many of these parasites also need to be ferreted out by different drugs in different hiding places.
In order to prevent losing new drugs to the always looming resistance mechanisms, it is absolutely essential that multi-compound drugs receive top priority for all antiparasitics of the future. The need for multi-compound drugs has been advocated for antileishmaniasis drugs (Olliaro, 2010 ▶). Since 2000, the World Health Organization has recommended artemisinin combination therapies for malaria. Remarkably, over the last 15 years a gradual resistance against drugs containing artemisinin plus a second antimalarial has evolved near the Cambodia–Thailand border in spite of this multi-compound strategy (Ariey et al., 2014 ▶; Mok et al., 2015 ▶; Miotto et al., 2015 ▶). Future years could see the spread of artemisinin resistance into sub-Saharan Africa, which would have tragic consequences.
In view of this looming threat to the current front-line antimalarial drug combination as well as to several other anti-parasitic drugs, there is a desperate need to step up the development of new antiparasitic compounds. A substantial collection of at least ten compounds per parasite should be available, with each compound by itself having powerful antiparasitic properties and enabled with resistance-evading characteristics. Importantly, such compounds should never be administered as single-compound drugs. Also, they should not be combined with drugs that have already been used in the field where resistance has either already been observed or very probably exists. Combining two or three of the new compounds into future drugs should give a considerable likelihood of delaying resistance. Moreover, there are instances where the combination of two compounds is synergistic [e.g. NECT for the treatment of sleeping sickness (Ebikeme, 2014 ▶) and malarone for malaria (Srivastava & Vaidya, 1999 ▶)] such that lower doses can be given than for single-compound drugs, resulting in fewer side effects. The goal should be to have multiple compound mixtures available, such that backup combinations can come to the rescue when front-line compound combinations start to fail.
8. The future
In spite of all this progress, worldwide suffering owing to parasitic diseases is still continuing more than a century after the identification of most of the causative protozoan agents. This is a situation which needs to be addressed with the greatest urgency. Key steps in moving more rapidly from basic scientific knowledge, including three-dimensional structural knowledge, to compounds that help patients and prevent disease requires action at multiple levels, several of which fall outside the scope of this overview. Let us consider just three here.
8.1. Medicinal structural biology
A critical factor in the design of new therapeutics is to enhance the efficiency of the translation of structural insights into useful compounds. In drug development the efficacy of compounds in actual patients is what matters, and it is in the later stages where the greatest challenges and most frequent disappointments occur. Parasitic diseases pose extra challenges since (i) many of these parasites hide inside human cells, (ii) the costs of therapy have to be very low and (iii) oral administration of compounds is essential since healthcare centers in rural areas are generally unable to provide complex treatments.
Hence, incorporating biological tests for efficiency of compounds in in vitro and in vivo models in early stages of development makes sense in most cases. Such tests quickly reveal metabolic weaknesses or membrane-crossing difficulties of certain categories of compounds. It is crucially important that structural biologists in SGDD for parasitic protozoa are as familiar with terms such as IC50, ED50, ‘ligand efficiency’ (Abad-Zapatero, 2007 ▶), ‘lipophilic efficiency’ (Leeson & Springthorpe, 2007 ▶), polar surface area (PSA), C max, AUC, t 1/2, clogP, ‘% plasma protein binding’ and ‘Lipinski’s rule’ (Lipinski et al., 2001 ▶) as with their beloved crystallographic parameters. In other words, new generations of medicinal structural biologists have to be trained.
8.2. Involve more medicinal chemists
For foreign chemicals, the human body is usually not a friendly place. Compounds are easily degraded, modified and removed. Collaborations with medicinal chemists play a central role. Their knowledge of chemical transformations in the human host and the probability of compounds passing membranes, combined with their synthetic skills, are of the essence. The November 2014 special issue of Chemical Reviews, devoted to ‘Drug Discovery and Development for Neglected Diseases’ , is a sign of the increasing interest of the international medicinal chemistry community in parasitic protozoa. The role of chemists and the importance of collaborations between medicinal chemists and scientists from numerous other disciplines, including structural biologists, to promote future drug design in general has also been stressed in a recent essay (Nicolaou, 2014 ▶).
8.3. Capacity building in low- and middle-income countries: parasitic protozoa offer an opportunity
The countries in which these parasites are indigenous are increasingly creating research institutions and fund substantial projects focusing on these protozoa (Auparakkitanon, 2014 ▶). However, in many instances there are immense obstacles that challenge even the most talented scientists in these countries. Often these obstacles are owing to complex bureaucracies. It might be helpful in pointing out to the political and administrative leaders of these bureaucracies that the parasitic diseases in their countries are not only a burden, but are actually also an opportunity; specifically, an opportunity to develop in the long term in their own country a competitive industry in the area of design of therapeutics in general. The competition from rich countries in the area of parasitic protozoa is presently not (yet?) as formidable as for many other diseases. Therefore, low- and medium-income countries have a window of opportunity to develop expertise in the very complex area of the design of therapeutic compounds for the treatment and prevention of diseases caused by parasitic protozoa. Developing this expertise can be of tremendous value a few decades hence in the design of therapeutics for treating diseases occurring in rich and poor countries alike, including diabetes, cancers and Alzheimer’s disease, for example. Increasing the research and development capacities for antiparasitic drug design in countries where parasitic protozoa have a terrible impact is a major avenue towards making the world a fairer place.
9. Conclusions
There are substantial worldwide efforts taking place in the design of antiparasitic drugs. In light of the very large numbers of patients involved, these efforts are far from sufficient, but looking back a few decades there has been tremendous progress involving many disciplines, including structural biology for SGDD, as we can see from the number of structures solved from parasitic protozoa and from contributions from structures to drug-design campaigns. Yet, the number of new compounds which are far advanced in drug-design pipelines for parasitic protozoa remains limited, and the development of drug resistance is relentless. The future requires increasing scientific and financial contributions from governmental, nonprofit and for-profit institutions. While there is reason for considerable optimism for success in the coming years, formidable obstacles remain to be overcome before we are anywhere near the situation that was once the case in the field of antibiotics, where a broad arsenal of drugs was available. Even that arsenal is now hardly sufficient any more. This is a major lesson and emphasizes the need for the design of multiple new therapeutics for patients infected by parasitic protozoa.
Acknowledgments
I am grateful to Bill Hunter for the invitation to write this overview. I would like to thank Paul Michels, Christophe Verlinde, Ethan Merritt, Fred Buckner and Cho Yeow Koh for numerous comments and corrections, Rafael Guido, Penchit Chitnumsub, Wes Van Voorhis, David Matthews and Xiayang Qiu for helpful suggestions and Stewart Turley for assistance with preparing the manuscript. I am deeply indebted to many group members of the last 35 years for their dedication to structure-guided drug design efforts focusing on parasitic protozoa. Financial support from institutions in The Netherlands and the USA is also gratefully acknowledged.
References
- Abad-Zapatero, C. (2007). Exp. Opin. Drug. Discov. 2, 469–488. [DOI] [PubMed]
- Ajioka, J. W. & Morrissette, N. S. (2009). Int. J. Parasitol. 39, 859–860. [DOI] [PMC free article] [PubMed]
- Anderson, A. C. (2005). Acta Cryst. F61, 258–262. [DOI] [PMC free article] [PubMed]
- Andriani, G., Chessler, A. D., Courtemanche, G., Burleigh, B. A. & Rodriguez, A. (2011). PLoS Negl. Trop. Dis. 5, e1298. [DOI] [PMC free article] [PubMed]
- Ariey, F. et al. (2014). Nature (London), 505, 50–55.
- Aronov, A. M., Suresh, S., Buckner, F. S., Van Voorhis, W. C., Verlinde, C. L. M. J., Opperdoes, F. R., Hol, W. G. J. & Gelb, M. H. (1999). Proc. Natl Acad. Sci. USA, 96, 4273–4278. [DOI] [PMC free article] [PubMed]
- Auparakkitanon, S. (2014). Southeast Asian J. Trop. Med. Public Health, 45, 761–782. [PubMed]
- Baell, J. & Walters, M. A. (2014). Nature (London), 513, 481–483. [DOI] [PubMed]
- Baldwin, J., Michnoff, C. H., Malmquist, N. A., White, J., Roth, M. G., Rathod, P. K. & Phillips, M. A. (2005). J. Biol. Chem. 280, 21847–21853. [DOI] [PubMed]
- Baniecki, M. L., Wirth, D. F. & Clardy, J. (2007). Antimicrob. Agents Chemother. 51, 716–723. [DOI] [PMC free article] [PubMed]
- Barrett, M. P., Vincent, I. M., Burchmore, R. J., Kazibwe, A. J. & Matovu, E. (2011). Future Microbiol. 6, 1037–1047. [DOI] [PubMed]
- Barros-Alvarez, X., Gualdrón-López, M., Acosta, H., Cáceres, A. J., Graminha, M. A., Michels, P. A., Concepción, J. L. & Quiñones, W. (2014). Curr. Med. Chem. 21, 1679–1706. [DOI] [PubMed]
- Bauman, J. D., Patel, D., Baker, S. F., Vijayan, R. S., Xiang, A., Parhi, A. K., Martínez-Sobrido, L., LaVoie, E. J., Das, K. & Arnold, E. (2013). Chem. Biol. 8, 2501–2508. [DOI] [PMC free article] [PubMed]
- Bedingfield, P. T., Cowen, D., Acklam, P., Cunningham, F., Parsons, M. R., McConkey, G. A., Fishwick, C. W. & Johnson, A. P. (2012). J. Med. Chem. 55, 5841–5850. [DOI] [PubMed]
- Bendale, P. et al. (2007). J. Med. Chem. 50, 4585–4605. [DOI] [PMC free article] [PubMed]
- Billker, O., Lourido, S. & Sibley, L. D. (2009). Cell Host Microbe, 5, 612–622. [DOI] [PMC free article] [PubMed]
- Blum, M. L., Down, J. A., Gurnett, A. M., Carrington, M., Turner, M. J. & Wiley, D. C. (1993). Nature (London), 362, 603–609. [DOI] [PubMed]
- Bolstad, D. B., Bolstad, E. S., Frey, K. M., Wright, D. L. & Anderson, A. C. (2008). J. Med. Chem. 51, 6839–6852. [DOI] [PMC free article] [PubMed]
- Bonney, K. M., Taylor, J. M., Daniels, M. D., Epting, C. L. & Engman, D. M. (2011). PLoS One, 6, e14571. [DOI] [PMC free article] [PubMed]
- Bosch, J. et al. (2006). J. Med. Chem. 49, 5939–5946. [DOI] [PubMed]
- Boucher, L. E. & Bosch, J. (2015). J. Struct. Biol. /10.1016/j.jsb.2015.02.008.
- Brand, S. et al. (2012). J. Med. Chem. 55, 140–152.
- Brand, S. et al. (2014). J. Med. Chem. 57, 9855–9869. [DOI] [PMC free article] [PubMed]
- Brannigan, J. A., Roberts, S. M., Bell, A. S., Hutton, J. A., Hodgkinson, M. R., Tate, E. W., Leatherbarrow, R. J., Smith, D. F. & Wilkinson, A. J. (2014). IUCrJ, 1, 250–260. [DOI] [PMC free article] [PubMed]
- Bressi, J. C., Choe, J., Hough, M. T., Buckner, F. S., Van Voorhis, W. C., Verlinde, C. L. M. J., Hol, W. G. J. & Gelb, M. H. (2000). J. Med. Chem. 43, 4135–4150. [DOI] [PubMed]
- Brimacombe, K. R., Walsh, M. J., Liu, L., Vásquez-Valdivieso, M. G., Morgan, H. P., McNae, I., Fothergill-Gilmore, L. A., Michels, P. A., Auld, D. S., Simeonov, A., Walkinshaw, M. D., Shen, M. & Boxer, M. B. (2014). ACS Med. Chem. Lett. 5, 12–17. [DOI] [PMC free article] [PubMed]
- Brinen, L. S., Hansell, E., Cheng, J., Roush, W. R., McKerrow, J. H. & Fletterick, R. J. (2000). Structure, 8, 831–840. [DOI] [PubMed]
- Buckner, F. S., Eastman, R. T., Yokoyama, K., Gelb, M. H. & Van Voorhis, W. C. (2005). Curr. Opin. Investig. Drugs, 6, 791–797. [PubMed]
- Bulbule, V. J., Rivas, K., Verlinde, C. L. M. J., Van Voorhis, W. C. & Gelb, M. H. (2008). J. Med. Chem. 51, 384–387. [DOI] [PubMed]
- Capdeville, R., Buchdunger, E., Zimmermann, J. & Matter, A. (2002). Nature Rev. Drug. Discov. 1, 493–502. [DOI] [PubMed]
- Charrière, F., O’Donoghue, P., Helgadóttir, S., Maréchal-Drouard, L., Cristodero, M., Horn, E. K., Söll, D. & Schneider, A. (2009). J. Biol. Chem. 284, 16210–16217. [DOI] [PMC free article] [PubMed]
- Chen, Y. T., Brinen, L. S., Kerr, I. D., Hansell, E., Doyle, P. S., McKerrow, J. H. & Roush, W. R. (2010). PLoS Negl. Trop. Dis. 4, e825. [DOI] [PMC free article] [PubMed]
- Coteron, J. M. et al. (2011). J. Med. Chem. 54, 5540–5561. [DOI] [PMC free article] [PubMed]
- Cox, F. E. (2002). Clin. Microbiol. Rev. 15, 595–612. [DOI] [PMC free article] [PubMed]
- Croft, S. L., Sundar, S. & Fairlamb, A. H. (2006). Clin. Microbiol. Rev. 19, 111–126. [DOI] [PMC free article] [PubMed]
- Cruzalegui, F. (2010). Ann. Pharm. Fr. 68, 254–259. [DOI] [PubMed]
- Dahlin, J. L. & Walters, M. A. (2014). Future Med. Chem. 6, 1265–1290. [DOI] [PMC free article] [PubMed]
- Dartois, V. (2014). Nature Rev. Microbiol. 12, 159–167. [DOI] [PMC free article] [PubMed]
- Das, K., Bauman, J. D., Clark, A. D. Jr, Frenkel, Y. V., Lewi, P. J., Shatkin, A. J., Hughes, S. H. & Arnold, E. (2008). Proc. Natl Acad. Sci. USA, 105, 1466–1471. [DOI] [PMC free article] [PubMed]
- Das, K., Lewi, P. J., Hughes, S. H. & Arnold, E. (2005). Prog. Biophys. Mol. Biol. 88, 209–231. [DOI] [PubMed]
- De Clercq, E. (2012). Biochem. Pharmacol. 84, 241–248. [DOI] [PubMed]
- Deng, X., Gujjar, R., El Mazouni, F., Kaminsky, W., Malmquist, N. A., Goldsmith, E. J., Rathod, P. K. & Phillips, M. A. (2009). J. Biol. Chem. 284, 26999–27009. [DOI] [PMC free article] [PubMed]
- Deng, X., Kokkonda, S., El Mazouni, F., White, J., Burrows, J. N., Kaminsky, W., Charman, S. A., Matthews, D., Rathod, P. K. & Phillips, M. A. (2014). J. Med. Chem. 57, 5381–5394. [DOI] [PMC free article] [PubMed]
- Dobell, C. (1932). Antony van Leeuwenhoek and his Little Animals. London: John Bales & Sons.
- Doerig, C., Meijer, L. & Mottram, J. C. (2002). Trends Parasitol. 18, 366–371. [DOI] [PubMed]
- Dorrestein, P. C. & Carroll, K. S. (2011). Curr. Opin. Chem. Biol. 15, 3–4. [DOI] [PubMed]
- Ebikeme, C. (2014). PLoS Negl. Trop. Dis. 8, e2910. [DOI] [PMC free article] [PubMed]
- Fidock, D. A., Nomura, T., Talley, A. K., Cooper, R. A., Dzekunov, S. M., Ferdig, M. T., Ursos, L. M. B., bir Singh Sidhu, A., Naudé, B., Deitsch, K. W., Su, X.-Z., Wootton, J. C., Roepe, P. D. & Wellems, T. E. (2000). Mol. Cell, 6, 861–871. [DOI] [PMC free article] [PubMed]
- Frearson, J. A. et al. (2010). Nature (London), 464, 728–732. [DOI] [PMC free article] [PubMed]
- Frearson, J. A., Wyatt, P. G., Gilbert, I. H. & Fairlamb, A. H. (2007). Trends Parasitol. 23, 589–595. [DOI] [PMC free article] [PubMed]
- Freymann, D. M., Metcalf, P., Turner, M. & Wiley, D. C. (1984). Nature (London), 311, 167–169. [DOI] [PubMed]
- Gamo, F. J., Sanz, L. M., Vidal, J., de Cozar, C., Alvarez, E., Lavandera, J. L., Vanderwall, D. E., Green, D. V., Kumar, V., Hasan, S., Brown, J. R., Peishoff, C. E., Cardon, L. R. & Garcia-Bustos, J. F. (2010). Nature (London), 465, 305–310. [DOI] [PubMed]
- Gassis, S. & Rathod, P. K. (1996). Antimicrob. Agents Chemother. 40, 914–919. [DOI] [PMC free article] [PubMed]
- Gati, C., Bourenkov, G., Klinge, M., Rehders, D., Stellato, F., Oberthür, D., Yefanov, O., Sommer, B. P., Mogk, S., Duszenko, M., Betzel, C., Schneider, T. R., Chapman, H. N. & Redecke, L. (2014). IUCrJ, 1, 87–94. [DOI] [PMC free article] [PubMed]
- Gironès, N., Cuervo, H. & Fresno, M. (2005). Curr. Top. Microbiol. Immunol. 296, 89–123. [DOI] [PubMed]
- Goldberg, D. E., Siliciano, R. F. & Jacobs, W. R. Jr (2012). Cell, 148, 1271–1283. [DOI] [PMC free article] [PubMed]
- Gowri, V. S., Ghosh, I., Sharma, A. & Madhubala, R. (2012). BMC Genomics, 13, 621. [DOI] [PMC free article] [PubMed]
- Green, J. L., Rees-Channer, R. R., Howell, S. A., Martin, S. R., Knuepfer, E., Taylor, H. M., Grainger, M. & Holder, A. A. (2008). J. Biol. Chem. 283, 30980–30989. [DOI] [PMC free article] [PubMed]
- Guido, R. V. C., Balliano, T. L., Andricopulo, A. D. & Oliva, G. (2009). Lett. Drug. Des. Discov. 6, 210–214.
- Guiguemde, W. A. et al. (2010). Nature (London), 465, 311–315.
- Guiguemde, W. A., Shelat, A. A., Garcia-Bustos, J. F., Diagana, T. T., Gamo, F. J. & Guy, R. K. (2012). Chem. Biol. 19, 116–129. [DOI] [PMC free article] [PubMed]
- Gujjar, R., Marwaha, A., El Mazouni, F., White, J., White, K. L., Creason, S., Shackleford, D. M., Baldwin, J., Charman, W. N., Buckner, F. S., Charman, S., Rathod, P. K. & Phillips, M. A. (2009). J. Med. Chem. 52, 1864–1872. [DOI] [PMC free article] [PubMed]
- Hall, R. J., Mortenson, P. N. & Murray, C. W. (2014). Prog. Biophys. Mol. Biol. 116, 82–91. [DOI] [PubMed]
- Hann, M. M., Leach, A. R. & Harper, G. (2001). J. Chem. Inf. Comput. Sci. 41, 856–864. [DOI] [PubMed]
- Hirai, S., Kikuchi, H., Kim, H.-S., Begum, K., Wataya, Y., Tasaka, H., Miyazawa, Y., Yamamoto, K. & Oshima, Y. (2003). J. Med. Chem. 46, 4351–4359. [DOI] [PubMed]
- Hoepfner, D. et al. (2012). Cell Host Microbe, 11, 654–663. [DOI] [PMC free article] [PubMed]
- Hotez, P. J., Fenwick, A., Savioli, L. & Molyneux, D. H. (2009). Lancet, 373, 1570–1575. [DOI] [PubMed]
- Hunter, W. N. (2007). J. Biol. Chem. 282, 21573–21577. [DOI] [PubMed]
- Hurt, D. E., Widom, J. & Clardy, J. (2006). Acta Cryst. D62, 312–323. [DOI] [PubMed]
- Hutton, J. A., Goncalves, V., Brannigan, J. A., Paape, D., Wright, M. H., Waugh, T. M., Roberts, S. M., Bell, A. S., Wilkinson, A. J., Smith, D. F., Leatherbarrow, R. J. & Tate, E. W. (2014). J. Med. Chem. 57, 8664–8670. [DOI] [PMC free article] [PubMed]
- Iten, M., Mett, H., Evans, A., Enyaru, J. C., Brun, R. & Kaminsky, R. (1997). Antimicrob. Agents Chemother. 41, 1922–1925. [DOI] [PMC free article] [PubMed]
- Jain, V., Kikuchi, H., Oshima, Y., Sharma, A. & Yogavel, M. (2014). J. Struct. Funct. Genomics, 15, 181–190. [DOI] [PubMed]
- Jain, V., Yogavel, M., Oshima, Y., Kikuchi, H., Touquet, B., Hakimi, M.-A. & Sharma, A. (2015). Structure, 10.1016/j.str.2015.02.011. [DOI] [PubMed]
- Janssen, P. A. et al. (2005). J. Med. Chem. 48, 1901–1909. [DOI] [PubMed]
- Joiner, K., Sher, A., Gaither, T. & Hammer, C. (1986). Proc. Natl Acad. Sci. USA, 83, 6593–6597. [DOI] [PMC free article] [PubMed]
- Keller, T. L., Zocco, D., Sundrud, M. S., Hendrick, M., Edenius, M., Yum, J., Kim, Y.-J., Lee, H.-K., Cortese, J. F., Wirth, D. F., Dignam, J. D., Rao, A., Yeo, C.-Y., Mazitschek, R. & Whitman, M. (2012). Nature Chem. Biol. 8, 311–317. [DOI] [PMC free article] [PubMed]
- Khan, S., Sharma, A., Belrhali, H., Yogavel, M. & Sharma, A. (2014). J. Struct. Funct. Genomics, 15, 63–71. [DOI] [PubMed]
- Koh, C. Y., Kim, J. E., Shibata, S., Ranade, R. M., Yu, M., Liu, J., Gillespie, J. R., Buckner, F. S., Verlinde, C. L. M. J., Fan, E. & Hol, W. G. J. (2012). Structure, 20, 1681–1691. [DOI] [PMC free article] [PubMed]
- Koh, C. Y., Kim, J. E., Wetzel, A. B., de van der Schueren, W. J., Shibata, S., Ranade, R. M., Liu, J., Zhang, Z., Gillespie, J. R., Buckner, F. S., Verlinde, C. L. M. J., Fan, E. & Hol, W. G. J. (2014). PLoS Negl. Trop. Dis. 8, e2775. [DOI] [PMC free article] [PubMed]
- Kohlstaedt, L. A., Wang, J., Friedman, J. M., Rice, P. A. & Steitz, T. A. (1992). Science, 256, 1783–1790. [DOI] [PubMed]
- Kruif, P. de (1926). Microbe Hunters. New York: Blue Ribbon Books.
- Larson, E. T. et al. (2012). J. Med. Chem. 55, 2803–2810. [DOI] [PMC free article] [PubMed]
- Larsson, A., Jansson, A., Åberg, A. & Nordlund, P. (2011). Curr. Opin. Chem. Biol. 15, 482–488. [DOI] [PubMed]
- Leeson, P. D. & Springthorpe, B. (2007). Nature Rev. Drug Discov. 6, 881–890. [DOI] [PubMed]
- Lendner, M., Böttcher, D., Delling, C., Ojo, K. K., Van Voorhis, W. C. & Daugschies, A. (2015). Parasitol. Res. 114, 335–336. [DOI] [PMC free article] [PubMed]
- Lim, D. C., Cooke, B. M., Doerig, C. & Saeij, J. P. J. (2012). Int. J. Parasitol. 42, 21–32. [DOI] [PMC free article] [PubMed]
- Lipinski, C. & Hopkins, A. (2004). Nature (London), 432, 855–861. [DOI] [PubMed]
- Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. (2001). Adv. Drug Deliv. Rev. 46, 3–26. [DOI] [PubMed]
- Lozano, N. B., Oliveira, R. F., Weber, K. C., Honorio, K. M., Guido, R. V., Andricopulo, A. D., de Sousa, A. G. & da Silva, A. B. (2014). Int. J. Mol. Sci. 15, 3186–3203. [DOI] [PMC free article] [PubMed]
- Lozano, R. et al. (2012). Lancet, 380, 2095–2128. [DOI] [PMC free article] [PubMed]
- MacGregor, P., Ivens, A., Shave, S., Collie, I., Gray, D., Auer, M. & Matthews, K. R. (2014). Eukaryot. Cell, 13, 412–426. [DOI] [PMC free article] [PubMed]
- Mackey, Z. B., Baca, A. M., Mallari, J. P., Apsel, B., Shelat, A., Hansell, E. J., Chiang, P. K., Wolff, B., Guy, K. R., Williams, J. & McKerrow, J. H. (2006). Chem. Biol. Drug Des. 67, 355–363. [DOI] [PubMed]
- McGrath, M. E., Eakin, A. E., Engel, J. C., McKerrow, J. H., Craik, C. S. & Fletterick, R. J. (1995). J. Mol. Biol. 247, 251–259. [DOI] [PubMed]
- McKerrow, J. H., Doyle, P. S., Engel, J. C., Podust, L. M., Robertson, S. A., Ferreira, R., Saxton, T., Arkin, M., Kerr, I. D., Brinen, L. S. & Craik, C. S. (2009). Mem. Inst. Oswaldo Cruz, 104, Suppl. 1, 263–269. [DOI] [PMC free article] [PubMed]
- Mead, J. R. (2002). Drug Resist. Updates, 5, 47–57. [DOI] [PubMed]
- Mercaldi, G. F., Ranzani, A. T. & Cordeiro, A. T. (2014). J. Biomol. Screen. 19, 1362–1371. [DOI] [PubMed]
- Metcalf, P., Blum, M., Freymann, D., Turner, M. & Wiley, D. C. (1987). Nature (London), 325, 84–86. [DOI] [PubMed]
- Miller, L. H. & Su, X. (2011). Cell, 146, 855–858. [DOI] [PMC free article] [PubMed]
- Miotto, O. et al. (2015). Nature Genet. 47, 226–233.
- Misset, O. & Opperdoes, F. R. (1984). Eur. J. Biochem. 144, 475–483. [DOI] [PubMed]
- Mok, S. et al. (2015). Science, 347, 431–435. [DOI] [PMC free article] [PubMed]
- Murphy, R. C., Ojo, K. K., Larson, E. T., Castellanos-Gonzalez, A., Perera, B. G., Keyloun, K. R., Kim, J. E., Bhandari, J. G., Muller, N. R., Verlinde, C. L., White, A. C. Jr, Merritt, E. A., Van Voorhis, W. C. & Maly, D. J. (2010). ACS Med. Chem. Lett. 1, 331–335. [DOI] [PMC free article] [PubMed]
- Murray, C. J. et al. (2012). Lancet, 380, 2197–2223. [DOI] [PubMed]
- Nakama, T., Nureki, O. & Yokoyama, S. (2001). J. Biol. Chem. 276, 47387–47393. [DOI] [PubMed]
- Nalam, M. N., Ali, A., Reddy, G. S., Cao, H., Anjum, S. G., Altman, M. D., Yilmaz, N. K., Tidor, B., Rana, T. M. & Schiffer, C. A. (2013). Chem. Biol. 20, 1116–1124. [DOI] [PMC free article] [PubMed]
- Newby, Z. E., O’Connell, J. III, Robles-Colmenares, Y., Khademi, S., Miercke, L. J. & Stroud, R. M. (2008). Nature Struct. Mol. Biol. 15, 619–625. [DOI] [PMC free article] [PubMed]
- Nicolaou, K. C. (2014). Angew. Chem. Int. Ed. 53, 9128–9140. [DOI] [PubMed]
- Ojo, K. K., Gillespie, J. R., Riechers, A. J., Napuli, A. J., Verlinde, C. L. M. J., Buckner, F. S., Gelb, M. H., Domostoj, M. M., Wells, S. J., Scheer, A., Wells, T. N. & Van Voorhis, W. C. (2008). Antimicrob. Agents Chemother. 52, 3710–3717. [DOI] [PMC free article] [PubMed]
- Ojo, K. K. et al. (2010). Nature Struct. Mol. Biol. 17, 602–607. [DOI] [PMC free article] [PubMed]
- Olaleye, T. O., Brannigan, J. A., Roberts, S. M., Leatherbarrow, R. J., Wilkinson, A. J. & Tate, E. W. (2014). Org. Biomol. Chem. 12, 8132–8137. [DOI] [PMC free article] [PubMed]
- Olliaro, P. L. (2010). Curr. Opin. Infect. Dis. 23, 595–602. [DOI] [PubMed]
- Opperdoes, F. R. & Borst, P. (1977). FEBS Lett. 80, 360–364. [DOI] [PubMed]
- Parsons, M., Worthey, E. A., Ward, P. N. & Mottram, J. C. (2005). BMC Genomics, 6, 127. [DOI] [PMC free article] [PubMed]
- Pauwels, R., Andries, K., Desmyter, J., Schols, D., Kukla, M. J., Breslin, H. J., Raeymaeckers, A., Gelder, J. V., Woestenborghs, R., Heykants, J., Schellekens, K., Janssen, M. A. C., Clercq, E. D. & Janssen, P. A. J. (1990). Nature (London), 343, 470–474. [DOI] [PubMed]
- Peña, I. et al. (2015). Sci. Rep. 5, 8771. [DOI] [PMC free article] [PubMed]
- Pham, J. S., Dawson, K. L., Jackson, K. E., Lim, E. E., Pasaje1, C. F. A., Turner, K. E. C. & Ralph, S. A. (2014). Int. J. Parasitol. Drugs Drug Resist. 4, 1–13. [DOI] [PMC free article] [PubMed]
- Phillips, M. A., Gujjar, R., Malmquist, N. A., White, J., El Mazouni, F., Baldwin, J. & Rathod, P. K. (2008). J. Med. Chem. 51, 3649–3653. [DOI] [PMC free article] [PubMed]
- Phillips, M. A. & Rathod, P. K. (2010). Infect. Disord. Drug Targets, 10, 226–239. [DOI] [PMC free article] [PubMed]
- Pozharski, E., Weichenberger, C. X. & Rupp, B. (2013). Acta Cryst. D69, 150–167. [DOI] [PubMed]
- Price, H. P., Güther, M. L., Ferguson, M. A. & Smith, D. F. (2010). Mol. Biochem. Parasitol. 169, 55–58. [DOI] [PMC free article] [PubMed]
- Price, H. P., Menon, M. R., Panethymitaki, C., Goulding, D., McKean, P. G. & Smith, D. F. (2003). J. Biol. Chem. 278, 7206–7214. [DOI] [PubMed]
- Redecke, L. et al. (2013). Science, 339, 227–230.
- Rivero, F. D., Saura, A., Prucca, C. G., Carranza, P. G., Torri, A. & Lujan, H. D. (2010). Nature Med. 16, 551–557. [DOI] [PubMed]
- Roberts, A. J., Torrie, L. S., Wyllie, S. & Fairlamb, A. H. (2014). Biochem. J. 459, 323–332. [DOI] [PMC free article] [PubMed]
- Rodríguez, D., Brea, J., Loza, M. I. & Carlsson, J. (2014). Structure, 22, 1140–1151. [DOI] [PubMed]
- Ross, L. S., Gamo, F. J., Lafuente-Monasterio, M. J., Singh, O. M., Rowland, P., Wiegand, R. C. & Wirth, D. F. (2014). J. Biol. Chem. 289, 17980–17995. [DOI] [PMC free article] [PubMed]
- Schmidtke, P. & Barril, X. (2010). J. Med. Chem. 53, 5858–5867. [DOI] [PubMed]
- Sehnal, D., Svobodová Vařeková, R., Pravda, L., Ionescu, C. M., Geidl, S., Horský, V., Jaiswal, D., Wimmerová, M. & Koča, J. (2015). Nucleic Acids Res. 43, D369–D375. [DOI] [PMC free article] [PubMed]
- Sharlow, E. R., Close, D., Shun, T., Leimgruber, S., Reed, R., Mustata, G., Wipf, P., Johnson, J., O’Neil, M., Grogl, M., Magill, A. J. & Lazo, J. S. (2009). PLoS Negl. Trop. Dis. 3, e540. [DOI] [PMC free article] [PubMed]
- Sharlow, E. R., Lyda, T. A., Dodson, H. C., Mustata, G., Morris, M. T., Leimgruber, S. S., Lee, K.-H., Kashiwada, Y., Close, D., Lazo, J. S. & Morris, J. C. (2010). PLoS Negl. Trop. Dis. 4, e659. [DOI] [PMC free article] [PubMed]
- Shibata, S., Gillespie, J. R., Kelley, A. M., Napuli, A. J., Zhang, Z., Kovzun, K. V., Pefley, R. M., Lam, J., Zucker, F. H., Van Voorhis, W. C., Merritt, E. A., Hol, W. G., Verlinde, C. L., Fan, E. & Buckner, F. S. (2011). Antimicrob. Agents Chemother. 55, 1982–1989. [DOI] [PMC free article] [PubMed]
- Shibata, S., Gillespie, J. R., Ranade, R. M., Koh, C. Y., Kim, J. E., Laydbak, J. U., Zucker, F. H., Hol, W. G. J., Verlinde, C. L. M. J., Buckner, F. S. & Fan, E. (2012). J. Med. Chem. 55, 6342–6351. [DOI] [PMC free article] [PubMed]
- Shirley, D. A., Moonah, S. N. & Kotloff, K. L. (2012). Curr. Opin. Infect. Dis. 25, 555–563. [DOI] [PMC free article] [PubMed]
- Smerdon, S. J., Jäger, J., Wang, J., Kohlstaedt, L. A., Chirino, A. J., Friedman, J. M., Rice, P. A. & Steitz, T. A. (1994). Proc. Natl Acad. Sci. USA, 91, 3911–3915. [DOI] [PMC free article] [PubMed]
- Smithson, D. C., Lee, J., Shelat, A. A., Phillips, M. A. & Guy, R. K. (2010). J. Biol. Chem. 285, 16771–16781. [DOI] [PMC free article] [PubMed]
- Spangenberg, T., Burrows, J. N., Kowalczyk, P., McDonald, S., Wells, T. N. & Willis, P. (2013). PLoS One, 8, e62906. [DOI] [PMC free article] [PubMed]
- Srivastava, I. K. & Vaidya, A. B. (1999). Antimicrob. Agents Chemother. 43, 1334–1339. [DOI] [PMC free article] [PubMed]
- Straimer, J. et al. (2015). Science, 347, 428–431. [DOI] [PMC free article] [PubMed]
- Suresh, S., Bressi, J. C., Kennedy, K. J., Verlinde, C. L. M. J., Gelb, M. H. & Hol, W. G. J. (2001). J. Mol. Biol. 309, 423–435. [DOI] [PubMed]
- Tate, E. W., Bell, A. S., Rackham, M. D. & Wright, M. H. (2014). Parasitology, 141, 37–49. [DOI] [PubMed]
- Teague, S. J. (2003). Nature Rev. Drug Discov. 2, 527–541. [DOI] [PubMed]
- Tejman-Yarden, N., Miyamoto, Y., Leitsch, D., Santini, J., Debnath, A., Gut, J., McKerrow, J. H., Reed, S. L. & Eckmann, L. (2013). Antimicrob. Agents Chemother. 57, 2029–2035. [DOI] [PMC free article] [PubMed]
- Tiefendbrunn, T. & Stout, C. D. (2014). Prog. Biophys. Mol. Biol. 116, 124–140. [DOI] [PubMed]
- Tzipori, S. & Widmer, G. (2008). Trends Parasitol. 24, 184–189. [DOI] [PMC free article] [PubMed]
- Upcroft, J. A., Upcroft, P. & Boreham, P. F. (1990). Int. J. Parasitol. 20, 489–496. [DOI] [PubMed]
- Van Voorhis, W. C. et al. (2007). Antimicrob. Agents Chemother. 51, 3659–3671. [DOI] [PMC free article] [PubMed]
- Vellieux, F. M. D., Hajdu, J. & Hol, W. G. J. (1995). Acta Cryst. D51, 575–589. [DOI] [PubMed]
- Vellieux, F. M., Hajdu, J., Verlinde, C. L. M. J., Groendijk, H., Read, R. J., Greenhough, T. J., Campbell, J. W., Kalk, K. H., Littlechild, J. A., Watson, H. C. & Hol, W. G. J. (1993). Proc. Natl Acad. Sci. USA, 90, 2355–2359. [DOI] [PMC free article] [PubMed]
- Verlinde, C. L. M. J. et al. (2009). Curr. Top. Med. Chem. 9, 1678–1687. [DOI] [PMC free article] [PubMed]
- Vidadala, R. S. et al. (2014). Eur. J. Med. Chem. 74, 562–573. [DOI] [PMC free article] [PubMed]
- Voet, A. R. D., Kumar, A., Berenger, F. & Zhang, K. Y. J. (2014). J. Comput. Aided Mol. Des. 28, 363–373. [DOI] [PubMed]
- Volkamer, A., Kuhn, D., Grombacher, T., Rippmann, F. & Rarey, M. (2012). J. Chem. Inf. Model. 52, 360–372. [DOI] [PubMed]
- Vu, H., Roullier, C., Campitelli, M., Trenholme, K. R., Gardiner, D. L., Andrews, K. T., Skinner-Adams, T., Crowther, G. J., Van Voorhis, W. C. & Quinn, R. J. (2013). Chem. Biol. 8, 2654–2659. [DOI] [PubMed]
- Wei, F., Wang, W. & Liu, Q. (2013). Parasitol. Res. 112, 2121–2129. [DOI] [PubMed]
- Wermuth, C. (2008). The Practice of Medicinal Chemistry, 3rd ed. New York: Academic Press.
- Wernimont, A. K., Artz, J. D., Finerty, P. Jr, Lin, Y.-H., Amani, M., Allali-Hassani, A., Senisterra, G., Vedadi, M., Tempel, W., Mackenzie, F., Chau, I., Lourido, S., Sibley, L. D. & Hui, R. (2010). Nature Struct. Mol. Biol. 17, 596–601. [DOI] [PMC free article] [PubMed]
- Wierenga, R. K., Kalk, K. H. & Hol, W. G. J. (1987). J. Mol. Biol. 198, 109–121. [DOI] [PubMed]
- Wiggers, H. J., Rocha, J. R., Fernandes, W. B., Sesti-Costa, R., Carneiro, Z. A., Cheleski, J., da Silva, A. B., Juliano, L., Cezari, M. H., Silva, J. S., McKerrow, J. H. & Montanari, C. A. (2013). PLoS Negl. Trop. Dis. 7, e2370. [DOI] [PMC free article] [PubMed]
- Wong, W., Bai, X.-C., Brown, A., Fernandez, I. S., Hanssen, E., Condron, M., Tan, Y. H., Baum, J. & Scheres, S. H. W. (2014). eLife, 3, e03080. [DOI] [PMC free article] [PubMed]
- Wright, M. H. et al. (2014). Nature Chem. 6, 112–121. [DOI] [PMC free article] [PubMed]
- Yuthavong, Y. et al. (2012). Proc. Natl Acad. Sci. USA, 109, 16823–16828.
- Yuvaniyama, J., Chitnumsub, P., Kamchonwongpaisan, S., Vanichtanankul, J., Sirawaraporn, W., Taylor, P., Walkinshaw, M. D. & Yuthavong, Y. (2003). Nature Struct. Biol. 10, 357–365. [DOI] [PubMed]
- Zhang, J., Yang, P. L. & Gray, N. S. (2009). Nature Rev. Cancer, 9, 28–39. [DOI] [PMC free article] [PubMed]
- Zhou, H., Sun, L., Yang, X.-L. & Schimmel, P. (2013). Nature (London), 494, 121–124. [DOI] [PMC free article] [PubMed]
- Zhu, X., Pandharkar, T. & Werbovetz, K. (2012). Antimicrob. Agents Chemother. 56, 1182–1189. [DOI] [PMC free article] [PubMed]