Abstract
In prokaryotes, RNA derived from type I and type III CRISPR loci direct large ribonucleoprotein complexes to destroy invading bacteriophage and plasmids. In Escherichia coli, this 405-kDa complex is called Cascade. Here we report the 3.03Å crystal structure of Cascade bound to a single-stranded DNA target. The structure reveals that the CRISPR RNA and target strands do not form a double helix but instead adopt an underwound ribbon-like structure. This non-canonical structure is facilitated by rotation of every sixth nucleotide out of the RNA-DNA hybrid and is stabilized by the highly interlocked organization of protein subunits. These studies provide insight into both the assembly and the activity of this complex and suggest a mechanism to enforce fidelity of target binding.
Main Text
Prokaryotes employ an RNA-based adaptive immune system called the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) system to prevent invasion by bacteriophage and plasmids (1, 2). Found in the host genome, CRISPR loci comprise an array of identical repeats interspersed by variable foreign DNA. CRISPR loci are transcribed and then cleaved within the repeat regions to generate small CRISPR RNAs (crRNAs) (3-5). In type I and type III CRISPR-Cas systems (6), crRNA and Cas proteins assemble into large multisubunit complexes (3, 7-12). Despite a common seahorse architecture (8, 13-17), these complexes employ different mechanisms to destroy their targets: type I complexes are surveillance complexes that bind DNA complementary to the crRNA guide and then recruit a trans-acting helicase-nuclease, Cas3, to unwind and degrade the invading DNA (18-20); type III-A complexes are effector complexes that destroy targets directly via their intrinsic nuclease activity (7, 10).
The type I-E surveillance complex in Escherichia coli is known as Cascade (CRISPR-associated complex for antiviral defense), a 405-kDa complex consisting of eleven subunits of five Cas proteins (Cse11, Cse22, Cas76, Cas51 and Cas6e1) and a 61-nucleotide (nt) crRNA. The crRNA consists of a 32-nt guide sequence flanked by 5′ and 3′ handles, the sequence of which are derived from the repeats (Fig. 1A). Cascade recognizes DNA as foreign if it contains a region complementary to the crRNA guide (protospacer) that is adjacent to a small 3 base pair (bp) sequence element called a protospacer adjacent motif (PAM). CRISPR arrays lack a PAM sequence, enabling discrimination between self and non-self DNA (21). Cascade binding to foreign DNA initiates at the PAM and then proceeds through base pairing between the crRNA and target strand, displacing the non-target strand to produce an R-loop (14). R-loop formation is unidirectional, proceeding 5′ to 3′ across the protospacer (22). Cryo electron microscopy (cryoEM) reconstructions of target-bound Cascade suggest base pairing between the crRNA guide and the target strand occurs step-wise through a series of discontinuous helical segments to form the guide-target hybrid (15). The structure of these discontinuous segments and how Cascade stabilizes this structure is currently unknown.
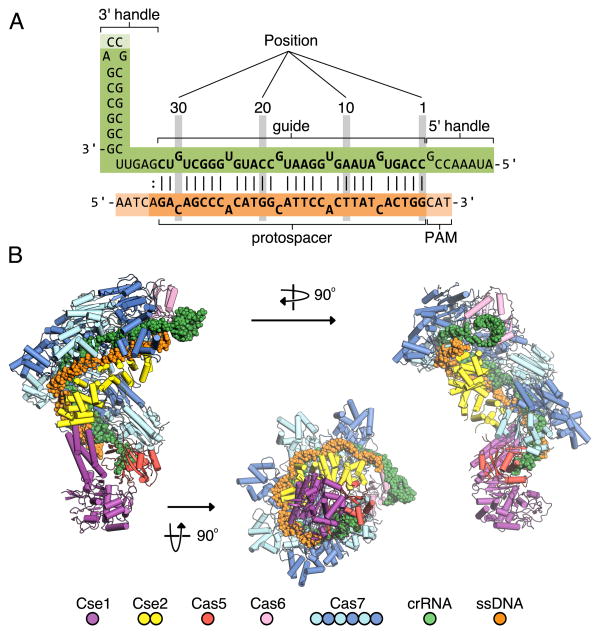
Fig. 1. Overall structure of ssDNA-bound Cascade.
(A) Cartoon schematic of the crRNA and ssDNA target. Watson-Crick base pairing between the two strands is indicated by lines, two dots indicates an observed shear base pair. The position of each base pair within the target-guide hybrid is also indicated. (B) Three orthogonal views of ssDNA-bound Cascade showing the seahorse architecture of the complex. Protein subunits are shown as a ribbon representation in the colors indicated. The crRNA (green) and ssDNA target (orange) are displayed in a spheres representation.
Crystal structure of the Cascade-ssDNA complex
To gain insights into the structural organization of Cascade and into target recognition, we have determined the crystal structure of a ∼425 kDa complex of E. coli Cascade bound to a complementary single-stranded DNA (ssDNA) target. We screened a variety of ssDNA and double-stranded (ds) DNA targets for crystallization with Cascade. The best crystals contained a complex of Cascade and a 40-nt ssDNA target consisting of a 32-nt protospacer sequence, a 5′ PAM sequence (5′-CAT-3′) and a 3′ extension (5-nt) (Fig. 1A). The structure, containing the ssDNA target strand, crRNA and all of the eleven Cascade subunits, was refined to an Rwork/Rfree of 0.225/0.269 at 3.03Å resolution (Table S1), has good stereochemistry (Table S1) and accounts for the vast majority of the expected residues (Table S2). Although the ssDNA target contained a PAM and a 5-nt 3′-extension, only the protospacer and the adjacent 3′ nucleotide of the extension were visible in the electron density map.
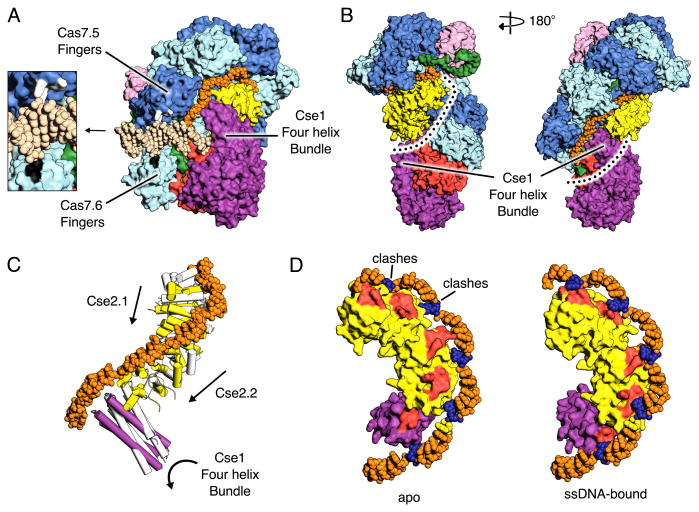
The crystal structure of ssDNA-bound Cascade has the same seahorse architecture observed by cryoEM (14, 15) (Fig. 1B). The body is formed by a helical filament of six Cas7 subunits (Cas7.1-7.6) wrapped around the crRNA guide, with a head-to-tail dimer of Cse2 (Cse2.1 and Cse2.2) at the belly. Cas6e and the 3′ handle of crRNA cap the Cas7 filament at the head, while Cas5 and the 5′ handle cap the tail. The N-terminal base of Cse1 is positioned at the tail of the filament while the C-terminal four-helix bundle contacts Cse2.2. The ssDNA target is juxtaposed to the guide region of the crRNA in a groove formed by the Cas7 filament, the four-helix bundle of Cse1, and the Cse2 dimer (Fig. 1B). The structures of the E. coli subunits are structurally similar to the stand-alone structures of their respective homologs (Fig. S1) (11, 12, 21, 23-30).
Two cryoEM reconstructions of target-bound Cascade are available, one bound to a 32-nt ssRNA target (15) and one bound to a 72-bp dsDNA target with a functional PAM sequence (31). The structural organization of Cascade in the two reconstructions is similar except for the position of the Cse1 base, which has been implicated in PAM binding (21). The crystal structure of Cascade fits best in the EM density of dsDNA-bound Cascade, requiring only small changes in the position of Cas6e and the 3′ handle for an optimal fit (Fig. S2). Lattice contacts appear to hold the base of Cse1 in the dsDNA-bound conformation. Thus, the structure of ssDNA-bound Cascade observed in the crystal is comparable to that bound to dsDNA, the in vivo target of Cascade (3, 31).
The guide-target hybrid adopts a ribbon conformation
The two strands of the guide-target hybrid do not twist around one another in a helix, but instead, adopt an underwound ribbon-like structure, reminiscent of a ladder (Fig. 2A). The 5′ and 3′ ends of the curved target strand are ∼102Å apart, roughly the length of straight B-form dsDNA with an identical sequence (∼107 Å). Under-winding is facilitated by kinks that occur every sixth base pair in the backbone of both strands of the hybrid (Fig. 2B). At each kink, complementary nucleotides are rotated ∼90°, in opposing directions, from the axis of the duplex (Fig. 2C). Thus, consistent with cryoEM studies, the guide-target ribbon contains five 5-bp segments separated by 1-bp gaps (15). The remaining 2 nucleotides of the guide region also form Watson-Crick base pairs with the target strand and are followed by a shear base pair between the nucleotide 5′ of the protospacer (A5) and the first nucleotide (G41) in the 5′ handle of the crRNA (Fig. S3A). The significance of this shear base pair is currently unclear as there is no reported conservation at this position of target sequence.
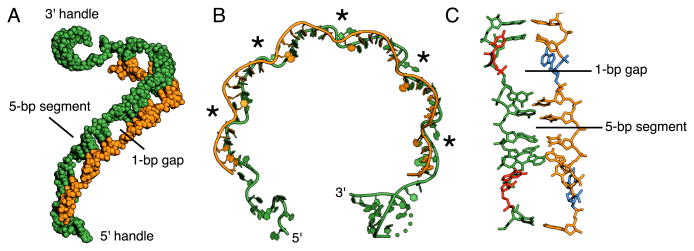
Fig. 2. Structure of the crRNA and ssDNA target.
(A) Overall structure of the two nucleic acids, highlighting the ribbon-like structure of the guide-target hybrid. The crRNA (green) and ssDNA target (orange) are displayed in a spheres representation. (B) Ribbon representation of the crRNA and ssDNA revealing the periodic kinking (marked by asterisks) in the backbone of both strands. (C) Structure of a 5-bp segment and disrupted base pairs. Disrupted RNA and DNA nucleotides are colored red and blue respectively. In all panels, the crRNA and ssDNA target are colored green and orange, respectively.
The structure of each repeating unit, a 5-bp segment plus the two displaced nucleotides, is essentially identical, superposing to an average root mean square deviation (rmsd) of ∼1.20Å over 209 atoms (Fig. S3B). The local conformation of the 5-bp segments is highly distorted from canonical A-form, owing to the non-uniform roll and twist angles between base pairs. However, this distortion is necessary for the hybrid to remain continuous throughout the guide region, as the geometry of an A-form duplex is incompatible with the spacing between kink sites. (Fig. S3C).
The observed structure of the guide-target hybrid predicts that mutation of the disrupted nucleotides would not affect binding by Cascade. Consistent with this, mutations at position 6 of the protospacer (Fig. 1A) have no effect on target binding, while mutations at positions 1-5 or 7-8 (Fig. 1A) greatly reduce affinity (32). In addition, recent high throughput genetic experiments revealed mutations at the sites of the disrupted bases are readily tolerated in vivo (33).
The Cas7 filament and its interaction with the head of Cascade
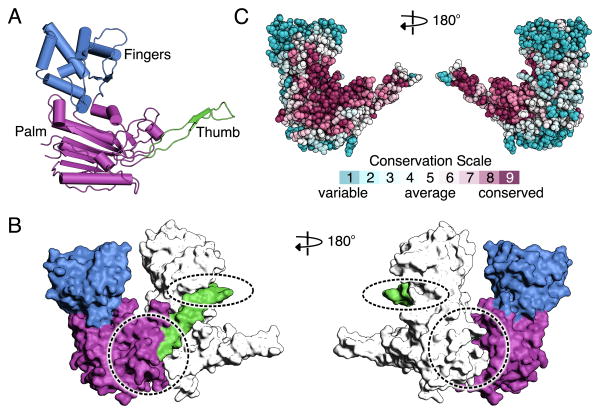
The structure of Cas7 resembles a right hand, consisting of fingers (residues 59-180), palm (residues 1-58, 181-189, and 224-263), and thumb (residues 190-223) domains (Fig. 3A). The palm contains the modified RNA-recognition motif (RRM) characteristic of many Cas protein families (34). The domain architecture and key surface features appear conserved among Cas7 family proteins (Fig. S4), suggesting that the structure of the Cas7 filament and consequently the ribbon-like structure of the guide-target hybrid (see below) are likely conserved in other type I and the type III CRISPR-Cas systems. Within Cascade, the six Cas7 subunits form a right-handed filament, with a pitch of ∼135 Å, around the guide-target hybrid (Fig. 1B). The filament is arranged such that the thumb of one Cas7 subunit, composed of an extended β-hairpin, extends towards the fingers of the adjacent subunit (Fig. 3B). The conformations of Cas7.2 to Cas7.6 are essentially identical, except that the fingers of Cas7.6 are disordered in the structure (Fig. S5A). Three points of contact stabilize the Cas7 filament, one between Cas7 and the guide region of the crRNA (see below), and two involving conserved protein-protein contacts (Fig. 3B and C) between Cas7 subunits. The larger of these contacts buries ∼1,500 Å2 of solvent-accessible surface area per subunit and is formed by packing the base of the thumb and the back of the palm of one subunit against the front of the palm of the neighboring subunit (Fig. 3B). The second, smaller interface buries ∼400 Å2 of solvent-accessible surface area per subunit and is formed between the tip of the thumb of one subunit and the fingers of the neighboring subunit (Fig. 3B).
Fig. 3. Structure of the Cas7 subunit.
(A) Ribbon representation of Cas7 colored by domain: thumb (green), fingers (blue) and palm (purple). (B) Surface representation of adjacent Cas7 subunits. The locations of the large and small interfaces are highlighted by dashed circles. (C) Surface representation of Cas7 colored according to evolutionary conservation.
At the head of Cascade, the Cas7 filament and the 3′-end of the crRNA are capped by Cas6e (Fig. S5B). This region appears to be flexible relative to the rest of Cascade as the corresponding electron density is quite weak. One face of Cas6e engages the 3′ hairpin of the crRNA, as observed with Thermus thermophilus Cas6e (28, 30), while the opposite face binds the thumb of Cas7.1 (Fig. S5C). Although a portion of the Cas7.1 thumb is disordered in the structure, it is clear that it has a conformation distinct to the other Cas7 subunits. Specifically, the Cas7 thumb rotates ∼90° towards Cas6e, adopting an open loop structure (Fig. S5D) that binds primarily with a conserved face of the C-terminal RRM of Cas6e (Fig. S5C).
Subunit interactions with the guide-target hybrid
A highly interdigitated network of protein-nucleic acid interactions stabilizes the ribbon conformation of the guide-target hybrid (Fig. 4A). Extensive contacts between the guide region of the crRNA and the Cas7 filament bury a large portion of the crRNA backbone, leaving the bases solvent exposed (Fig. 4B). The absence of direct contacts between protein side-chains and bases of the crRNA explains the lack of sequence specificity by Cascade for the guide sequence. The target strand is bound primarily through Watson-Crick hydrogen bonding with the guide but also interacts with the Cas7, Cse1 and Cse2 subunits (Fig. 1B). Few interactions are seen with the DNA backbone, rendering the majority of the backbone solvent exposed (Fig. 1B).
Fig. 4. Stabilization of the guide-target hybrid by Cas7, Cse1 and Cse2.
(A) Surface representation showing the interaction of Cas7 subunits with the guide-target hybrid. The 5-bp segment bound between the fingers of Cas7.4 and the palm of Cas7.4 is colored red. (B) Close-up view of the bound crRNA. The DNA target has been removed for clarity. Intercalation by Met166 from Cas7 is highlighted. (C) Surface representation showing that the thumb of Cas7 pushes through the guide-target hybrid at the 1-bp gaps. Boxed is the interaction of thumb residues with the minor groove face of the hybrid. (D) Binding site for the disrupted RNA bases. (E) Left: overview of the interactions between Cse2.1, Cse2.2 and Cse1 and the ssDNA target. The proteins are represented as ribbons, the DNA as a surface. The positions of the disrupted DNA nucleotides (dark blue) are indicated. Right: Close-up views of the DNA nucleotide binding pockets. In all panels, the proteins and nucleic acids are colored as in Fig. 1.
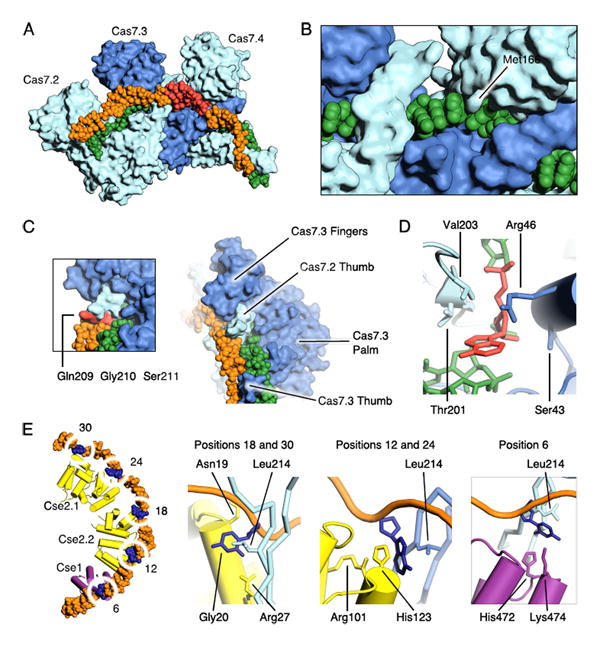
Each 5-bp segment of the hybrid is situated between the palm of one Cas7 subunit and the fingers of the adjacent subunit (Fig. 4A). Several highly conserved polar and positively charged residues (Arg20, Lys27, Ser40, Gln42, Lys45 and Lys49) from the palm of one Cas7 contact the RNA backbone, while the fingers from the adjacent Cas7 subunit (residues 109-111 and 163-169) contact both strands of the hybrid across the minor groove (Fig. 4C). In particular, the side chain of a highly conserved methionine (Met166) partially intercalates with the bases at the third and fourth position of each RNA segment (Fig. 4B). As a result, the bases at these positions are separated, which helps distort each segment away from A-form geometry, facilitating the under-wound structure of the crRNA guide.
The thumb of Cas7 both prohibits base pairing at the kink sites of the hybrid and stabilizes the conformation of the 5-bp segments. As the thumb of one Cas7 subunit extends towards the fingers of the adjacent subunit it passes directly between the strands of the guide-target hybrid, displacing the nucleotides and disrupting their base pairing (Fig. 4C). As a result, adjacent Cas7 subunits completely encircle the guide region of the crRNA (Fig. 4A-C). Residues from the middle of the Cas7 thumb (Trp199, Phe200, Thr201, Ala202, Leu214 and His213) (Fig. S6) also stabilize the position of the nucleotides that flank the ends of the 5-bp segments through van der Waals contacts. Residues at the tip of the thumb (Gln209, Gly210 and Ser211) contact the bases of the target strand on the minor groove face of the duplex (Fig. 4C). Cas7.6 is located at the 3′-end of the crRNA.
Distinct binding pockets accommodate the displaced RNA and DNA nucleotides, respectively. Each binding pocket is solvent exposed and provides minimal contacts with the Watson-Crick edges of the bases, consistent with the lack of sequence specificity at these positions. Each displaced RNA nucleotide adopts the syn conformation, is similarly positioned above the backbone of the downstream RNA, and is contacted by residues from both the Cas7 palm (Ser43 and Arg46) and thumb (Thr201 and Val203) (Fig. 4D). Each displaced DNA nucleotide adopts the anti conformation, is sandwiched on one side by Leu214 from the nearest Cas7 thumb, and, depending on its position along the hybrid, by residues from either Cse1, Cse2.1 or Cse2.2 (Fig. 4E). Cse1 contacts the first displaced DNA nucleotide (position 6) (Fig. 1A) with residues Lys474 and His472 from the four-helix bundle (Fig. 4E). This interaction is consistent with cryoEM studies showing ordering of this region of Cse1 (residues 406-414) on target binding (15). The second and fourth displaced DNA nucleotides (positions 12 and 24) stack with His123 and contact Arg101 from either Cse2.2 or Cse2.1, respectively (Fig. 4E). The third and fifth displaced nucleotides (positions 18 and 30) make contact with Asn19, Gly20 and Arg27 from either Cse2.2 or Cse2.1, respectively (Fig. 4E). We also observe a salt bridge between Arg26 from Cse2.1 and Cse2.2 and backbone phosphates at positions 17 and 29. Consistent with these observations, the highly conserved Arg26, Arg27 and Arg101 from Cse2 have been implicated as important for DNA binding by electrophoretic mobility shift assays (25).
Interactions capping the tail of Cascade
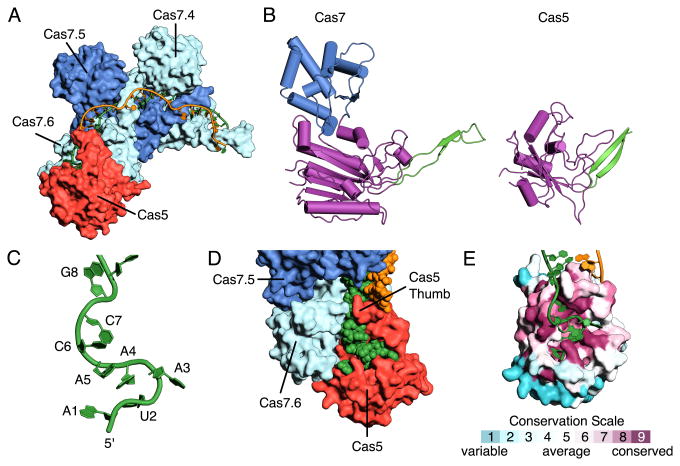
Cas5 caps the tail of the Cas7 filament at the 5′-end of the crRNA (Fig. 5A). The structure of Cascade reveals that Cas5 is structurally related to Cas7, as it consists of a palm (residues 1-78 and 115-224) and a thumb (residues 79-114) domain, but lacks a fingers domain (Fig. 5B). The Cas5 palm contains an RRM that superposes with the palm of Cas7 (rmsd of ∼3.3 Å between 123 Cα atoms). The thumb domains of Cas5 and Cas7 adopt a similar orientation with respect to their palm domains and appear to have similar functions within Cascade. The thumb of Cas5 displaces nucleotide G8 in the 5′ handle of the crRNA in a manner reminiscent of how the Cas7 thumb displaces RNA nucleotides in the guide. The thumb of Cas5 also provides similar residues (Leu89 and Thr87) to the binding pocket of the G8 nucleotide as the Cas7 thumb does for the displaced RNA nucleotides (Val203 and Thr201). Additionally, two residues from the thumb of Cas5 (Tyr85 and Gln105) stabilize the first duplex segment (positions 1-5) through van der Waals contacts with the exposed face of the base at position 1 of the hybrid (Fig. S6).
Fig. 5. Structure of Cas5 and the 5′ handle of the crRNA.
(A) Surface representation of Cas5 capping the end of the Cas7 filament. (B) Ribbon representation of Cas7 and Cas5 colored by domain, thumb (green) fingers (blue) and palm (purple). (C) Structure of the 5′ hook of the crRNA. (D) Close-up view of the interaction between Cas5, Cas7.6 and the 5′ hook. (E) Surface representation of Cas5 colored according to evolutionary conservation, highlighting the conservation of the residues that bind the 5′ hook.
The first seven nucleotides of the 5′-handle form a hook-like structure (Fig. 5C) that sits between Cas5 and Cas7.6 (Fig. 5D), forming extensive contacts between the RNA backbone and highly conserved charged and polar residues from both Cas5 (Arg25, Ser34, Arg48, Arg49, Arg148 and Arg149) and Cas7.6 (Lys45, Arg49 and Arg46) (Fig 5E). The first three nucleotides (A1, U2 and A3) are splayed and their bases sandwiched in three distinct pockets formed by conserved residues from Cas5 (Fig. S7). Sequence specific contacts are observed between nucleotide U2 and the backbone of Tyr145 (Fig. S7). The bases of nucleotides A4, A5 and C6 form a triplet stack that sits orthogonal to nucleotide A3 (Fig. S7). The backbone of this stack packs against the palm domains of both Cas5 and Cas7.6, leaving the bases solvent exposed. The side chain of Arg206 from the palm of Cas5 also stacks with the base of nucleotide A4 (Fig. S7). Finally, the position of C7 is stabilized through sequence specific hydrogen bonds with Arg108 from the Cas5 thumb (Fig. S7). Thus, only nucleotides U2 and C7 form sequence specific interactions with Cas5 in ssDNA-bound Cascade, consistent with sequence alignments showing these positions are the invariant in the 5′ handle (35).
Modeling interactions between Cascade, dsDNA target and Cas3
CryoEM studies (31) revealed that Cascade binds the PAM-proximal end of a dsDNA target between Cas7.5, Cas7.6 and the L1 loop of Cse1, which has been linked to PAM binding (21). The fingers of Cas7.6 and the L1 loop of Cse1 are disordered in the crystal structure of ssDNA-bound Cascade, suggesting that these regions are mobile in the absence of dsDNA. Using the cryoEM density as a guide, we modeled the position of this dsDNA and the fingers of Cas7.6 onto the crystal structure. The resulting model suggests interactions between the DNA backbone and a cluster of basic residues from the fingers of both Cas7.5 (Lys137, Lys138 and Lys141) and Cas7.6 (His67 and Lys105) (Fig 6A). The orientation of the modeled duplex and the path of target-strand agree with atomic force microscopy studies suggesting Cascade bends dsDNA upon binding (18).
Fig. 6. Modeling interactions with dsDNA and associated conformation changes.
(A) Surface representation of Cascade and the modeled dsDNA and Cas7.6 fingers domain. Boxed is a close up view of the residues predicted to bind the dsDNA: Lys137, Lys138 and Lys141 from the fingers of Cas7.5 (white) and His67 and Lys105 from the fingers of Cas7.6 (black). (B) Surface representation of Cascade, with the proposed path of the non-target strand indicated by a dotted line. (C) Ribbon representation of Cse2.1, Cse2.2 and the four-u8helix bundle of Cse1 highlighting the structural rearrangements that accompany ssDNA binding. The target strand is shown in orange. The subunits in the apo conformation are colored white; those in the ssDNA-bound conformation are colored as in Fig. 1. (D) Surface representations of Cse2.1, Cse2.2 and the four helix bundle of Cse1 in ssDNA-bound (right) and apo (left) conformations. The binding pockets for displaced DNA nucleotides (dark blue) are colored red. Clashes between Cse2.1 and the distal end of the ssDNA-target are indicated.
R-loop formation by Cascade displaces the non-target strand of the protospacer. Footprinting experiments have shown that the 5′-end but not the 3′-end of the displaced strand is protected by Cascade binding (14, 21). The structure of ssDNA-bound Cascade reveals a prominent basic groove that could serve as a binding site for the 5′-end of the displaced strand (Fig. 6B). Spanning from Cas6e to the four-helix bundle of Cse1, the groove is lined with conserved basic residues from Cas7 (Lys34, Lys299 and Lys301) and Cse2 (Arg53, Arg142, Arg143 and Arg110) (Fig. S8A). As the PAM is bound at the L1 loop of Cse1 (31), the displaced strand must first loop around the four-helix bundle of Cse1 to gain access to the groove, in agreement with the footprinting data that shows the 3′-end of the displaced strand is exposed. Following R-loop formation, Cascade recruits the Cas3 helicase-nuclease (18-20). Negative stain reconstruction of a complex between dsDNA-bound Cascade and Cas3 revealed Cas3 binds Cascade between the four-helix bundle and the base of Cse1 (31). Mapping this interaction onto the crystal structure reveals the residues that likely mediate Cas3 binding (Fig. S8B), these include Glu192, Glu280, Asn376, Thr383 and two conserved loops located at the base of Cse1 (residues 288-294 and residues 318-323).
CryoEM studies have shown that target binding triggers structural rearrangements of the Cse1 and Cse2 subunits (15). To further understand these changes, we fit each subunit of the crystal structure of ssDNA-bound Cascade into the ∼9Å cryoEM reconstruction of apo Cascade as a rigid body, except for Cse1, which required individual placement of its two domains (Fig. S9). As previously established, superposition of the apo Cascade model with the structure of ssDNA-bound Cascade reveals a concerted conformational rearrangement. The two Cse2 subunits each move ∼16Å towards the four-helix bundle of Cse1, that subsequently, undergoes a ∼30° rotation away from Cse2.2 (Fig. 6C). We also observe a ∼15° rotation of the base of Cse1 (31). Together, these rearrangements facilitate the formation of binding pockets for the displaced DNA nucleotides (Fig. 6D). Additionally, in the apo conformation, Cse2.1 blocks access to the distal end of the guide (positions 24-30) (Fig. 6D). Therefore, repositioning Cse2.1 facilitates base pairing in this region, consistent with previous binding data showing that short oligonucleotides complementary to the 5′ region of the target strand have lower affinity than short oligonucleotides that bind to the 3′ region (15).
Implications for target binding and recruitment of Cas3
Analysis of the crystal structure, in the context of the reported literature, provides a model for how Cascade recognizes foreign DNA and recruits Cas3. During target recognition, Cascade searches dsDNA for potential PAM sites, possibly facilitated by sequence-independent interactions between the DNA backbone and basic residues from the fingers of Cas7.5 and Cas7.6 (Fig. 6A) (31). Once a PAM is found, the adjacent duplex is destabilized and base pairing between the crRNA guide and the target strand proceeds along the guide in 5-bp increments separated by 1-bp gaps (15, 22, 32).
Cascade incrementally binding 5-nt segments of the target strand likely increases the fidelity of target recognition, by a mechanism similar to that employed by RecA (36-38). RecA catalyzes strand exchange in homologous recombination. Thus, despite differing cellular roles, Cascade and RecA both function by correctly base pairing a single strand of RNA or DNA with a complementary strand in a DNA duplex. Comparison of the structures of the guide-target hybrid in ssDNA-bound Cascade and the heteroduplex bound to the RecA filament (36) reveals globally similar distortions (Fig. S10), as previously predicted (2). RecA is thought to bind its target in 3-nt increments to enforce high fidelity during strand exchange, suggesting Cascade may employ a similar conformational proofreading mechanism (36-38). Specifically, conformational proofreading by Cascade involves optimizing the cost-benefit ratio of binding targets by introducing a structural difference between the crRNA guide and the foreign DNA. The energetics of target binding are the net sum of the energy gained by base pairing and the energy lost in conformational distortion, i.e. formation of the guide-target hybrid. Non-specific targets lacking favorable base pair interactions will not overcome the energetic costs associated with conformational distortion, while specific targets can. Thereby, enhanced specificity is achieved. Genetic and biochemical assays identified a seed region (positions 1-5, 7-8) in crRNA required for high affinity binding to target (32). Consistent with this proposed conformational fidelity mechanism, mismatches in the seed region would inhibit formation of the initial 5-nt segments, terminating binding.
Cascade undergoes structural rearrangement of its Cse1 and Cse2 subunits upon target binding. Propagation of base pairing between the crRNA guide and target strand across the target facilitates these conformation changes, which form the binding pockets for the disrupted DNA nucleotides (Fig. 6D). Moreover, movement of Cse2.1 relieves a steric block at the distal end of the guide (positions 24-30) (Fig. 6D), enabling base pairing in this region. Recent single molecule studies monitoring DNA supercoiling revealed Cascade binding to target DNA is unstable until base pairs form at the distal end of the guide (22). Our structural analysis suggests that that base pairing in this region would prevent Cse2.1 from reassuming its apo position, effectively locking Cascade on the DNA. This locking mechanism could also act as an additional proofreading step, as targets that cannot base pair across the entire length of the guide will not be stably bound (22).
Following target recognition, Cascade recruits the Cas3 helicase-nuclease, likely through interactions with two conserved loops at the base of Cse1 (Fig. S8B) (31). Once recruited, Cas3 nicks the displaced non-target strand ∼7-11 nucleotides from the 3′-end of the PAM (19, 20), consistent with the predicted path of the non-target strand (Fig. 6B and Fig S8A). Cas3 then loads onto the newly formed ssDNA end (31) and continues to progressively degrade the foreign DNA.
Supplementary Material
Acknowledgments
We thank R. McMacken, B. Learn, J. Berger, D. Leahy and J Kavran for helpful discussion. The authors are grateful to Jürgen Bosch for providing the Tungsten clusters used in the soaking experiments and to I. Mathews for help with data collection. We also thank J. Kavran for critical reading of the manuscript. This work was funded by the National Institute of Health (GM097330 to S.B). Data for this study were measured at beamline X25 of the National Synchrotron Light Source (NSLS) and at beamlines 7-1, 11-1 and 12-2 of the Stanford Synchrotron Radiation Lightsource (SSRL). Funding for X25 comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy, and from the National Center for Research Resources (P41RR012408) and the National Institutes of Health (P41GM103473). Use of the SSRL is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the Nation Institutes of Health (P41GM103393). The atomic coordinates and structure factors have been deposited into the Protein Data Bank with the accession code 4QYZ.
Footnotes
Supplementary Materials: Materials and Methods
Materials and Methods
References and Notes
- 1.Barrangou R, et al. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Sorek R, Lawrence CM, Wiedenheft B. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 3.Brouns SJJ, et al. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carte J, Wang R, Li H, Terns RM, Terns MP. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E, et al. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS, et al. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale CR, et al. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouillon C, et al. Mol Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, et al. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam KH, et al. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lintner NG, et al. J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidrich N, Vogel J. Mol Cell. 2013;52:4–7. doi: 10.1016/j.molcel.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Jore MM, et al. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 15.Wiedenheft B, et al. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staals RHJ, et al. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spilman M, et al. Mol Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westra ER, et al. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinkunas T, et al. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulepati S, Bailey S. J Biol Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sashital DG, Wiedenheft B, Doudna JA. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczelkun MD, et al. Proc Natl Acad Sci U S A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulepati S, Orr A, Bailey S. J Biol Chem. 2012;287:22445–22449. doi: 10.1074/jbc.C112.379503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agari Y, Yokoyama S, Kuramitsu S, Shinkai A. Proteins. 2008;73:1063–1067. doi: 10.1002/prot.22224. [DOI] [PubMed] [Google Scholar]
- 25.Nam KH, Huang Q, Ke A. FEBS Lett. 2012;586:3956–3961. doi: 10.1016/j.febslet.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo Y, Ka D, Kim EJ, Suh N, Bae E. J Mol Biol. 2013;425:3799–3810. doi: 10.1016/j.jmb.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 27.Hrle A, et al. RNA Biol. 2013;10:1670–1678. doi: 10.4161/rna.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sashital DG, Jinek M, Doudna JA. Nat Struct Mol Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 29.Ebihara A, et al. Protein Science. 2006;15:1494–1499. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gesner EM, Schellenberg MJ, Garside EL, George MM, MacMillan AM. Nat Struct Mol Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 31.Hochstrasser ML, et al. Proc Natl Acad Sci U S A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semenova E, et al. Proc Natl Acad Sci U S A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fineran PC, et al. Proc Natl Acad Sci U S A. 2014;111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunin V, Sorek R, Hugenholtz P. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, Yang H, Pavletich NP. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 37.Kowalczykowski SC. Nature. 2008;453:463–466. doi: 10.1038/453463a. [DOI] [PubMed] [Google Scholar]
- 38.Savir Y, Tlusty T. Mol Cell. 2010;40:388–396. doi: 10.1016/j.molcel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider TR, Sheldrick GM. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 41.Terwilliger T. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 42.Read RJ. Acta Crystallogr Sect A Found Crystallogr. 1986;42:140–149. [Google Scholar]
- 43.Emsley P, Cowtan K. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, et al. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen EF, et al. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Ashkenazy H, Erez E, Martz E, Pupko T. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The PyMOL Molecular Graphics System, Version 1.5.04. Schrödinger; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.