Abstract
The SEMA3B gene is located in the 3p21.3 LUCA region, which is frequently affected in different types of cancer. The objective of our study was to expand our knowledge of the SEMA3B gene as a tumor suppressor and the mechanisms of its inactivation. In this study, several experimental approaches were used: tumor growth analyses and apoptosis assays in vitro and in SCID mice, expression and methylation assays and other. With the use of the small cell lung cancer cell line U2020 we confirmed the function of SEMA3B as a tumor suppressor, and showed that the suppression can be realized through the induction of apoptosis and, possibly, associated with the inhibition of angiogenesis. In addition, for the first time, high methylation frequencies have been observed in both intronic (32-39%) and promoter (44-52%) CpG-islands in 38 non-small cell lung carcinomas, including 16 squamous cell carcinomas (SCC) and 22 adenocarcinomas (ADC), and in 83 clear cell renal cell carcinomas (ccRCC). Correlations between the methylation frequencies of the promoter and the intronic CpG-islands of SEMA3B with tumor stage and grade have been revealed for SCC, ADC and ccRCC. The association between the decrease of the SEMA3B mRNA level and hypermethylation of the promoter and the intronic CpG-islands has been estimated in renal primary tumors (P < 0.01). Using qPCR, we observed on the average 10- and 14-fold decrease of the SEMA3B mRNA level in SCC and ADC, respectively, and a 4-fold decrease in ccRCC. The frequency of this effect was high in both lung (92-95%) and renal (84%) tumor samples. Moreover, we showed a clear difference (P < 0.05) of the SEMA3B relative mRNA levels in ADC with and without lymph node metastases. We conclude that aberrant expression and methylation of SEMA3B could be suggested as markers of lung and renal cancer progression.
Introduction
Semaphorins are negative mediators of axonal guidance in the central nervous system [1]. Semaphorins comprise a large family of glycoproteins (8 classes, including 5 vertebrate classes, of more than 30 members), but only class 3 (SEMA3) represents secreted soluble molecules. Members of the SEMA family are differentially expressed in cancer, and either promote or suppress cell proliferation, migration and angiogenesis, and the induction of drug resistance. Thus, the roles of separate members of semaphorin family can be quite different [2–9].
Class 3 semaphorins (SEMA3s, also known as collapsins) comprise one of five vertebrate families of semaphorins and play an important role in tumor biology, including regulating cellular processes, such as endothelial cell proliferation, apoptosis, migration and angiogenesis [10]. Recently, the involvement of this protein class in carcinogenesis has been intensively studied. SEMA3s are secreted by cells of multiple lineages, including epithelial cells, neurons, and specific tumor cells [10]. Neuropilins (NRP) represent the primary receptors of SEMA3s. The binding of SEMA3s to NRP1/2 initiates their downstream signaling but prevents the interaction between NRP1/2 and vascular endothelial growth factor (VEGF) and the subsequent induction of a pro-angiogenic transcriptional program. However, it is not clear whether SEMA3s inhibit tumor growth by competing with VEGF for neuropilins ligand-binding sites, by acting independently of VEGF, or by a combination of these effects [10–13].
Previous studies, including ours, of human chromosome 3 in renal, lung, breast and cervical carcinomas revealed frequent allelic losses (up to 40%) in the LUCA region (3p21.3), which harbors two semaphorins—SEMA3B and SEMA3F. This region (hg38/chr3: 50.0–50.5Mb) comprised of 445 Kb contains about 20 tumor suppressors (TSG) and TSG-candidates: RASSF1, NPRL2, TUSC2, CACNA2D2 and others. Surprisingly, these genes playing roles in cellular processes and exerting tumor suppression by several different ways (cell cycle block, inhibition of angiogenesis, induction of apoptosis etc.) are located in the compact region [14–18].
Important evidence of tumor suppressor activity includes the identification of cell regulatory pathways and other mechanisms that are affected by SEMA3B. Using MDA-MB435 (breast carcinoma) and A549 (lung adenocarcinoma) cells it was previously shown that SEMA3B suppressed tumor growth but triggered a pro-metastatic program by releasing interleukin 8 [19, 20]. Furthermore, it was found that the induction of apoptosis by SEMA3B in tumor cells was mediated by inactivation of the Akt signaling pathway [21]. Therefore, it was important to further elucidate particular aspects of SEMA3B tumor suppression.
Methylation is an important mechanism of SEMA3B gene inactivation [17, 22]. However, the majority of previous research focused on methylation studies of the intronic CpG-island, that was incorrectly considered as located in the promoter region.
The objective of our study was to elucidate the distinct roles of SEMA3B in tumor suppression, particularly in apoptosis and angiogenesis. Furthermore we aimed to evaluate frequencies of promoter (hg38/chr3: 50,267,308–50,267,797) and intronic (hg38/chr3: 50,268,972–50,269,271) CpG-island hypermethylation correlations with SEMA3B expression, and tumor progression in lung and renal cancers.
Materials and Methods
Cell lines
Genomic DNA was isolated from 14 cancer cell lines: 3 squamous cell lung cancers (SCLC: ACC-LC5, NCI-N417, U2020), 2 non-small cell lung cancers (NSCLC: NCI-H157, NCI-H647) and 9 renal cell cancers (RCC: A498, ACHN, Caki-1, Caki-2, HN-51, KH-39, KRC/Y, TK-10, TK-164). The cell line U2020 was described earlier [23]. The ACC-LC5 cell line that carries a deletion in 3p21.3 [24] was kindly provided by Dr. Yusuke Nakamura (University of Tokyo, Tokyo, Japan). Renal A498, Caki1, and Caki2 and lung NCI-N417, NCI-H157, and NCI-H647 cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). Cell lines KRC/Y, ACHN, TK-164, HN-51, TK-10, and KH-39 were obtained from the Karolinska Institute (Stockholm, Sweden) cell line collection [25]. All human cell lines were grown as monolayer cultures in IMDM/RPMI or DMEM (with 4.5 g/l glucose) supplemented with 10% fetal calf serum (FCS).
Tissue samples
In total, 70 paired tumor/normal samples of NSCLC (29 ADC and 41 SCC) and 133 clear cell RCC (ccRCC) were obtained from the N.N. Blokhin Cancer Research Center, Russian Academy of Medical Sciences (Moscow, Russia). The set of 38 NSCLC (16 ADC and 22 SCC) and 83 ccRCC was used in the methylation studies and the expression or copy number studies by semi-quantitative RT-PCR. The additional set of 32 NSCLC (13 ADC and 19 SCC) and 50 ccRCC was used for validation by qPCR expression studies. The sample information is presented in Table 1 and S1 Table. The samples were collected in accordance with the guidelines issued by the Ethics Committee of N.N. Blokhin Cancer Research Center, Russian Academy of Medical Sciences (Moscow, Russia). All patients gave written informed consent (available upon request). The Ethics Committee of N.N. Blokhin Cancer Research Center, Russian Academy of Medical Sciences, specifically approved this study. The study was performed in accordance with the principles outlined in the Declaration of Helsinki. Tumor tissues and paired morphologically normal tissues were obtained from patients after surgical resection prior to radiation or chemotherapy and were stored in liquid nitrogen. The diagnosis was verified by histopathology, and only samples with 70–80% or more tumor cells were used in the study. “Normal” controls were obtained at a minimum of 2 cm from the tumor and were confirmed histologically as normal epithelial cells. Tumor specimens were characterized according to the International System of Classification of Tumors, based on the tumor-node-metastasis (TNM) and staging classification of the Union for International Cancer Control (UICC, version 2002) [26] and World Health Organization (WHO) criteria classification [27, 28]. Blood samples from 15 healthy donors were also used in the study.
Table 1. Pathological and histological characteristics of the tumors.
| Stage | Number of samples | ||
|---|---|---|---|
| ADC | SCC | ccRCC | |
| I | 6/7 | 7/6 | 20/22 |
| II | 2/1 | 8/6 | 23/12 |
| III | 7/5 | 7/7 | 30/14 |
| IV | 1/0 | 0/0 | 10/2 |
| All | 16/13 | 22/19 | 83/50 |
Note: The slash separates the number of samples used in the methylation studies and the expression or copy number studies by semi-quantitative RT-PCR and the number of samples used in the qPCR expression studies.
SCID mice
Twelve SCID mice were used in the experiments. The mice were obtained from Scanbur (Sollentuna, Sweden). Animal euthanasia was performed by CO2 asphyxiation followed by cervical dislocation. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (NRC 2011), the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes, Council of Europe (ETS 123), and the guidelines of the North Stockholm Ethical Committee for Care and Use of Laboratory Animals. The experiments with the SCID mice were approved by the North Stockholm Ethical Committee.
Transfection and selection of stably transfected SEMA3B-U2020 cell clones
The cDNA encoding the SEMA3B gene was cloned into an episomal tetracycline—regulated vector, pETE. The resulting plasmid was sequenced. To obtain stable cell clones expressing SEMA3B, U2020 cells (SCLC) were transfected with empty pETE or pETE/SEMA3B plasmid DNA (0.5 mg DNA per well) in 12-well plates using Lipofectamine and Plus Reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. Transiently transfected pETE/SEMA3B-U2020 and pETE-U2020 cells were cultured for 2–3 weeks in IMDM medium containing Bsd (5 μg/ml) to select stable clones. The expression of SEMA3B was regulated by doxycycline.
Colony formation assay
Transiently transfected U2020 cells (with pETE and pETE/SEMA3B) were stripped 24–48 h after transfection and plated on 100 mm2 cell culture dishes at a density of 500–1000 cells per plate. After selection with Bsd (5 μg/ml), Giemsa-stained colonies were photographed and counted using Quantity One software (version 4.4.0; Bio-Rad, Hercules, CA, USA). Cell viabilities were estimated by FACS analysis with propidium iodide (PI-FACS), following the manufacturer’s guidelines (FACSCalibur, BD Bioscience).
Tumor growth in SCID mice
The tumorigenicity of pETE/SEMA3B-transfected U2020 and empty pETE-transfected U2020 cells (control) was assessed by subcutaneous injecting the cells into 6–8-week-old SCID mice as previously described [29]. The cells were collected by centrifugation at 800 rpm for 2 min and resuspended in serum-free IMDM medium at a concentration of 2–3×106 cells per 100 μl injection. The cells were embedded into a Matrigel matrix (BD Biosciences, Erembodegem, Belgium) according to the manufacturer’s protocol. The mice were observed for tumor formation twice per week and the tumor size was measured using calipers. Then we used the multi-gene inactivation test (MGIT) for three genes (ZMYND10/BLU, TUSC2/FUS1 and SEMA3B) in SCID mice. MGIT is based on the monitoring of tumor suppressor candidate gene inactivation in cells and tumors. It was accomplished according to a published method [29, 30]. Briefly, U2020 cells were transfected either with pETE/SEMA3B, pETE/ZMYND10, pETE/TUSC2 or empty pETE plasmids. Mixes of cell clones were subcutaneously injected in SCID mice. Subsequently, if a tumor is formed, the expression of SEMA3B, ZMYND10 and TUSC2 is evaluated. The knock-down of the genes suggests their importance for tumor suppression.
Immunohistochemical analysis of SCLC tumor cell line U2020 and tumor angiogenesis in SCID mice
The plasmid pETE/SEMA3B was introduced into SCLC cell line U2020, which constitutively produced a tetracycline transactivator (tTA) [29]. The resulting sub-line was inoculated subcutaneously into SCID mice. The mice were given drinking water with doxycycline (+dox, gene is OFF) or without (−dox, gene is ON). The mice were euthanized after 1 month. The tumors or places of cell injections were excised and embedded with paraffin. The sections were 5 μm in thickness. Paraffin was dissolved in xylene (Sigma-Aldrich, St. Louis, MO, USA), and the tissue sections were treated sequentially with 99, 95, 75 and 30% ethanol. Epitopes were recovered by heating in a microwave oven for 5 min in citrate buffer. Anti-CD31 mouse antibody, together with rabbit-anti-mouse FITC-conjugated secondary antibody (Dako, Karlstrup, Denmark), was used to stain microvessels. The TUNEL assay (In Situ Cell Death Detection Kit, Boehringer Mannheim, Germany) for the detection of apoptosis was performed according to the manufacturer’s protocol.
DNA and total RNA isolation, reverse transcription-PCR
Nitrogen-frozen tissues were disrupted using a Mikro-Dismembrator (Sartorius, Germany). The DNA from human tissues and cell cultures was isolated by phenol extraction according to the standard protocols. Total RNA was isolated using the RNeasy mini kit as recommended by Qiagen (Netherlands). Purified RNA was quantified using NanoDrop-1000 (NanoDrop Technologies Inc., DE, USA). RNA quality was assessed using 28S and 18S rRNA bands after electrophoresis in a 1% denaturing agarose gel and analyzed using a Bioanalyzer 2100 (Agilent Technologies, CA, USA). The lack of DNA contamination was checked by semi-quantitative PCR with primers for the main histocompatibility complex I gene (MHCI, designed using Vector NTI, see S2 Table). All RNA samples were treated with RNase-free DNase I (Fermentas, Lithuania). RNA samples containing over 0.1% DNA were discarded. The cDNA was synthesized from 1 μg of total RNA using M-MuLV reverse transcriptase and random hexamers, according to the standard manufacturer’s protocol (Fermentas, Lithuania).
Bisulfite sequencing of the promoter CpG-island of SEMA3B gene
Bisulfite DNA conversion was conducted as described in [31, 32] with the use of 1–2 μg DNA (lung and renal cell lines and tumor/normal tissues). The modified DNA was purified using a JETquick PCR Purification Spin Kit (Genomed, Sweden). Modified DNA was maintained at -20°C and used as a template for PCR with the designed primers (listed in S2 Table), whose product was sequenced. Amplification of the SEMA3B promoter CpG-island fragment was performed in a 50 μl reaction mixture containing PCR buffer (67 mM Tris-HCl pH 8.8, 16.6 mM ammonium sulfate, 0.01% Tween 20); 2.0 mM MgCl2; 0.25 mM of each dNTP; 25 pM of each primer; 1 unit Hot Start Taq DNA polymerase (SibEnzyme, Russia); and 5–20 ng of modified DNA in a DNA Engine Dyad Cycler (Bio-Rad, United States) using the following program: 95°C, 5 min; 35 cycles of 95°C, 15 s; 62°C, 30 s; 72°C, 30 s and 72°C, 7 min. The PCR amplified product was purified using 1.5% agarose gel electrophoresis and the JETquick Gel Extraction Spin Kit (Genomed, Sweden). For sequencing, 5–10 ng of the purified DNA fragment and 25 pM of one of the primers were used. Sequencing was conducted using an automatic sequencing machine (Beckman-Coulter).
Methylation specific PCR (MSP)
The bisulfite-treated DNA, dissolved in twice-distilled water, was also used as a template for MSP. The PCR conditions and primers for the methylated and unmethylated allele of intronic [17] and promoter (designed by DNASTAR Lasergene 2000 program) CpG-islands are given in S2 Table. In case of the promoter CpG-island, 6 CpG-dinucleotides were analyzed (2 by the forward primer and 4 by the reverse), and in case of the intronic—3 (1 by the forward primer and 2 by the reverse). PCR was performed on a DNA Engine Dyad Cycler amplifier (Bio-Rad, United States) using the following program: 95°C, 5 min; 35 cycles of 95°C, 10 s; Tann (see S2 Table), 20 s; 72°C, 30 s and 72°C, 3 min. The absence of PCR product on unconverted DNA was checked for each pair of primers. DNA of the human fibroblast cell line L-68 served as an unmethylated allele control; L-68 SssI DNA from L-68 fibroblasts treated with SssI methyltransferase (SibEnzyme, Russia) served as a positive control for 100% methylation.
Semi-quantitative RT-PCR
To control the reverse transcription, primers for the transcript of the beta2-microglobulin (B2M) gene were used [33]. Semi-quantitative RT-PCR was performed with equal quantities of cDNA using the primers and conditions listed in S2 Table. Multiplex PCR with primers to the SEMA3B [17] and B2M genes was performed under conditions optimized for the SEMA3B primers, and the concentration of B2M primers was 1.5 times lower. The products of the RT-PCR were separated on 2% agarose gels, and the band pattern was analyzed using GelImager software (DNA Technology, Russia). A semi-quantitative copy number assay for the markers of LUCA region was used as described elsewhere [14].
qPCR
Quantitative PCR was performed with the primers and TaqMan probes listed in S2 Table using a 7500 Real-Time PCR System (Applied Biosystems, CA, USA). Each reaction was repeated three times. The nucleotide sequences of the amplicons were verified by sequencing in a 3730 DNA Analyzer automated sequencer (Applied Biosystems, CA, USA). QPCR data were analyzed using the reference genes GAPDH, GUSB and RPN1 [34] and the relative quantification or ΔΔCt-method as described earlier [35]. At least 2-fold mRNA level changes were considered as significant because of mRNA level variability of the reference genes.
Statistical analysis
The nonparametric Wilcoxon test was used to compare mRNA expression differences in the target and reference genes in the ccRCC and NSCLC samples. Kruskal-Wallis and Mann-Whitney rank-sum tests, Fisher’s exact test and χ2 criteria were used for analysis of mRNA level and methylation status changes in ccRCC and NSCLC groups with different pathological and histological characteristics. Student's t-test was used to compare the data obtained for groups of replicates. P-values ≤ 0.05 were considered statistically significant. Spearman’s rank correlation coefficient (r s) was used for revealing correlations.
Results
In vitro growth suppression of SCLC cells U2020 by SEMA3B
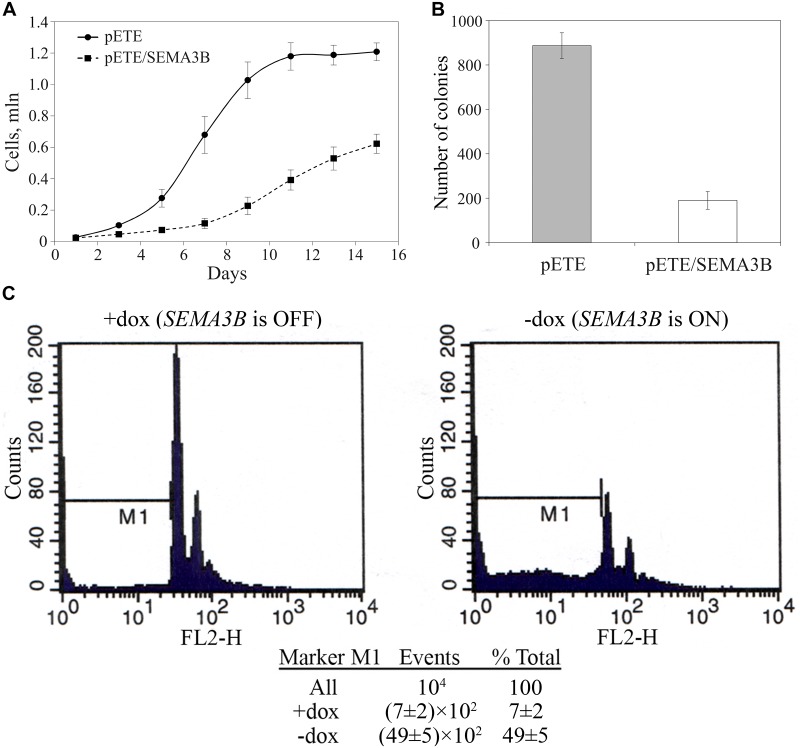
The small-cell lung carcinoma cell line U2020 was transfected with pETE/SEMA3B or pETE as the control. The transfected cells were cultured for 15 days. The growth rate of U2020 cells expressing SEMA3B was lower than the control (P < 0.01 since the day 5, see Fig 1A). The colony formation assay showed that the number of colonies of U2020 cells containing pETE/SEMA3B was lower after re-expression of the doxycycline-suppressed SEMA3B gene in comparison to the control cells (890 ± 60 vs 190 ± 40, P < 0.01, Fig 1B). Based on PI-FACS analysis, the abundance of apoptotic and necrotic cells expressing SEMA3B (without doxycycline) was increased significantly in comparison with SEMA3B-off cells (with doxycycline): from (7±2)×102 to (49±5)×102, P < 0.01, see Fig 1C. Taken together, these data suggest that SEMA3B is the inhibitor of human SCLC cells growth via induction of apoptosis in vitro.
Fig 1. In vitro growth of U2020 cells (U7111 clone) depends on the expression of SEMA3B.
A—The growth rate of U2020 cells: dashed line with squares—U7111 clone with pETE/SEMA3B, solid line with circles—U2020 cells with pETE (control); B—colony formation assay; C—PI-FACS analysis of cells with and without expression of SEMA3B. Mean values ± standard deviations for 4 replicates are represented in each case.
Multi-gene inactivation test in SCID mice
We have previously reported that the GIT technique allows for the efficient and controlled induction of various genes in cells [29, 30]. For the MGIT experiment, we used the SCLC cell line U2020 for the conditional expression of three TSG-candidates located in 3p21.3: ZMYND10 (BLU), TUSC2 (FUS1) and SEMA3B. Mixes of cell clones carrying different genes were inoculated subcutaneously into six-week-old SCID mice using a Matrigel (basement membrane matrix) implantation technique in the absence of doxycycline (the genes were ON).
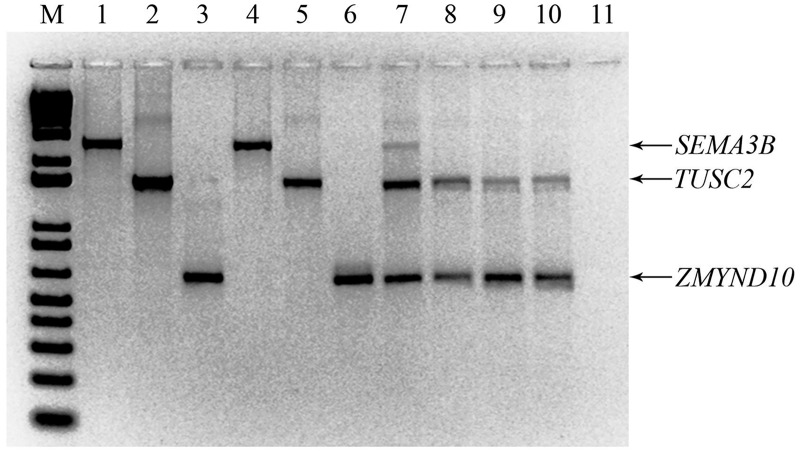
A PCR-based comparison of SCID mice tumors and primary cell clones showed unequivocally that SEMA3B was selectively knocked-down in all three tumors grown in vivo (see samples 8–10 in Fig 2), whereas the expression of ZMYND10 and TUSC2 genes did not change under these conditions. These two genes most likely did not antagonize the tumor growth of U2020 cells in SCID mice (we leave this question open because the search for mutation through retained genes was not included in the MGIT). These data suggest that SEMA3B is a growth inhibitor of human SCLC cells in vivo.
Fig 2. Absence of SEMA3B expression in tumors grown in vivo.
Electropherogram of multiplex PCR from plasmids, clones and SCID mice tumors of three genes. M—marker, 1—PCR from plasmid pETE/SEMA3B, 2—PCR from plasmid pETE/TUSC2, 3—PCR from plasmid pETE/ZMYND10, 4—PCR from U7111/SEMA3B cell clone 1, 5—PCR from U7111/TUSC2 cell clone 3, 6—PCR from U7111/ZMYND10 cell clone 4, 7—mixed cell clones, 8—PCR from tumor 1, 9—PCR from tumor 2, 10—PCR from tumor 3, 11—negative control.
Effect of SEMA3B transgenes on tumor growth in SCID mice and angiogenesis
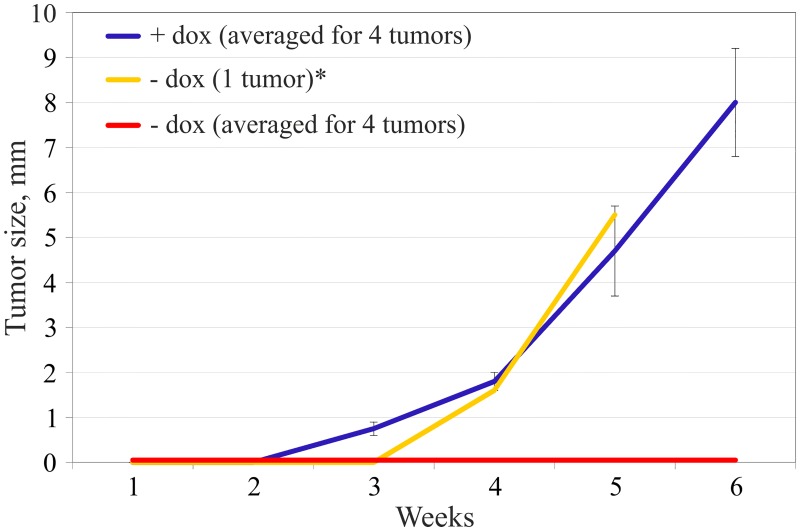
The U2020 sub-line with conditional SEMA3B expression under doxycycline control was inoculated in SCID mice subcutaneously. Four mice received doxycycline in drinking water (+dox mice, control) and five were not administered doxycycline (-dox mice). The onset of solid tumors actively expressing SEMA3B was not observed in 4 out of 5 cases (Fig 3, red line). In the fifth—dox mouse active tumor growth (Fig 3, yellow line) began one week later compared to the control (Fig 3, blue line), in spite of the presence of SEMA3B in the construct. However, the expression of the SEMA3B gene was not detected in this tumor according to the Northern blot analysis (data not shown) suggesting loss of SEMA3B in these cells. The tumors (observed in SEMA3B-OFF mice) had areas of active cell proliferation, in contrast to the tissues taken from sites of cell injections (in SEMA3B-ON mice), in which abundant fibrous and poorly differentiated cellular stroma and necrotic areas were observed. These data demonstrate the possibility of SEMA3B tumor suppression activity in SCID mice.
Fig 3. Inhibition of tumor growth by SEMA3B re-expression.
The growth rate of U2020 cells (U7111 clone) in SCID mice: blue line—U2020 cells without SEMA3B expression (+ doxycycline, 4 mice), red and yellow line—U2020 cells with SEMA3B expression (- doxycycline, 4 mice and 1 mouse respectively). *—no expression of SEMA3B gene according to the Northern blot (data not shown). One +dox and one—dox mice were withdrawn from the study after one month.
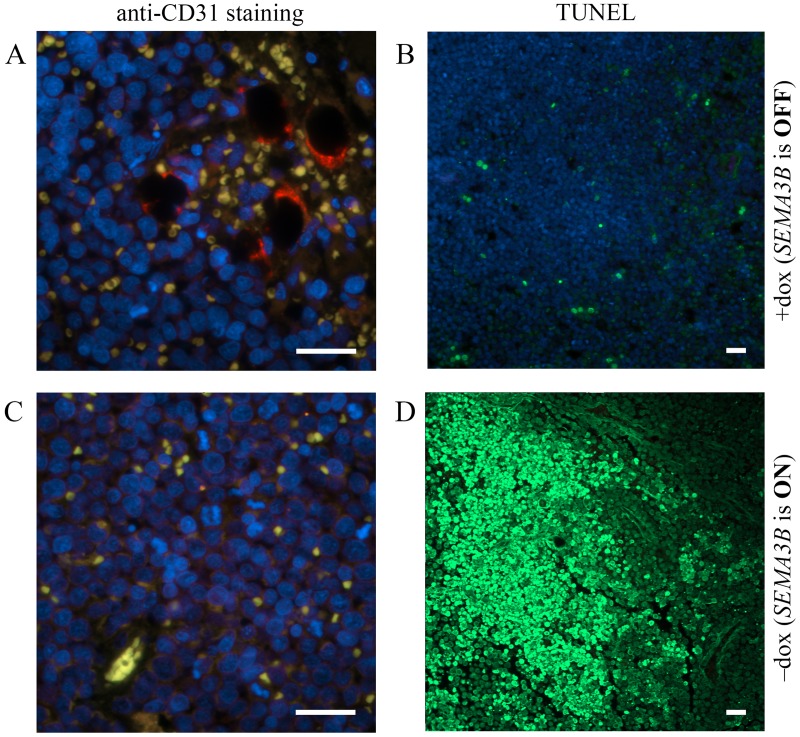
Tissues from two sites of U2020 cells inoculation were analyzed using CD31 staining and TUNEL assay: one SEMA3B-negative (Fig 4A and 4B) and one SEMA3B-positive (Fig 4C and 4D) mouse. Anti-CD31 mouse antibodies were used to stain blood microvessels. A very few number of microvessels was detected in SEMA3B-positive tissues. A fragment containing one of the microvessel-like objects is shown at Fig 4C. Microvessel signal (green channel) was co-localized with signal from erythrocytes (red channel) resulting to yellow colored areas in Fig 4A. However, SEMA3B-negative solid tumors demonstrated abundance of elongated, compressed blood ducts with fluorescence typical to epithelia. These tumor ducts were also co-localized with erythrocytes (Fig 4A). This could support a role of SEMA3B as inhibitor of angiogenesis. However, further experiments on the extended sampling are needed to prove this suggestion.
Fig 4. Tumor sections from SCID mice.
(A, B)—SEMA3B is OFF (mouse received doxycycline in drinking water). (C, D)—SEMA3B is ON (mouse were not administered doxycycline). (A, C)—staining with anti-CD31 to monitor blood vessels (green signal). When SEMA3B is OFF, areas filled with erythrocytes (red signal) are seen. Yellow signal indicates co-localization of green and red signals. Blue signal corresponds to DNA. (B, D)—TUNEL assay. Notice the area with apoptotic cells (green signal) where the SEMA3B gene was expressed.
We hypothesized that the SEMA3B-positive cells in the areas without blood vessels and erythrocytes were undergoing cell death. To test this hypothesis, sections of the same tissue fragments were analyzed by a TUNEL assay, a technique enabling detection of apoptotic cells. (Fig 4B, 4D). A huge area of apoptotic cells was observed in SEMA3B-positive tissue sample whereas no apoptotic area was seen in SEMA3B-negative tumors. In this case, we assumed that the expression of SEMA3B suppressed tumor growth in vivo, likely by the induction of apoptosis. Inhibition of angiogenesis could be suggested also.
Methylation of the promoter and intronic SEMA3B CpG-islands in lung and renal cell lines and primary tumors by bisulfite sequencing and methylation-specific PCR
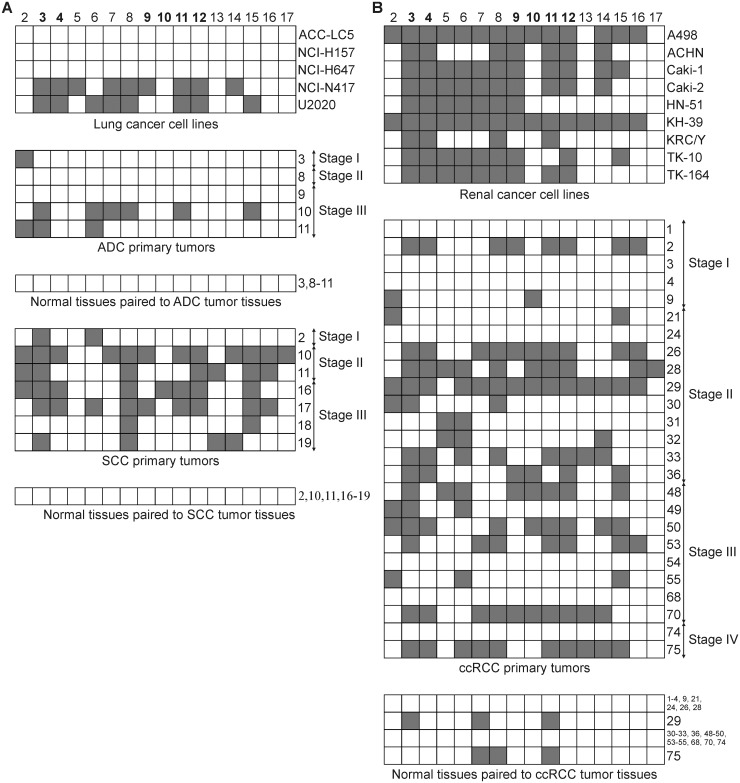
The SEMA3B gene is comprised of 18 exons and contains two CpG-islands: one is located in the promoter region (1-st CpG-island, hg38/chr3: 50,267,308–50,267,797, 22 CpG-dinucleotides) and the other one in the first intron (2-nd CpG-island, hg38/chr3: 50,268,972–50,269,271, 12 CpG-dinucleotides). We analyzed the methylation profile of 16 CpG-dinucleotides of the promoter CpG-island in 5 lung cancer cell lines (3 SCLC and 2 NSCLC) and 12 NSCLC primary tumors using bisulfite sequencing (5 ADC and 7 SCC, see Fig 5A). Dense methylation (> 40% of the analyzed CpGs were methylated) of the promoter CpG-island of the SEMA3B gene was observed in 2 of 3 SCLC cell lines, but in none of the NSCLC cell lines (NCI-H157 and NCI-H647). However, in all 7 SCC primary tumors and in 2 of 5 ADC primary tumors, methylation was detected in 2–12 of the CpGs. In addition, we examined the methylation profile in 9 renal cancer cell lines and 25 ccRCC primary tumors (Fig 5B). Dense methylation of the promoter CpG-island was observed in 8 of 9 cell lines and 11 of 25 ccRCC primary tumors. Among matched histological normal tissues, methylated CpGs were detected only in 2 of 25 ccRCC cases and in none of 12 NSCLC cases (Fig 5A and 5B).
Fig 5. Methylation profile of the promoter CpG-island of the SEMA3B gene in lung (A) and renal (B) cancer cell lines and primary tumors.
Bisulfite sequencing data, 16 CpG-dinucleotides (2–17) of the CpG-island are given. Grey squares show methylated CpG-dinucleotides, white squares—unmethylated. Numbers of primary tumors correspond to those in S1 Table. The bold numbers of CpG-dinucleotides (3–4 and 9–12) indicate the location of the primers that were used for MSP method.
Next, we evaluated the methylation frequencies of both CpG-islands of the SEMA3B gene by the MSP method using a representative set of primary tumors. The methylation frequency of the promoter CpG-island was 44% (7/16) in ADC, 45% (10/22) in SCC and 52% (43/83) in ccRCC. The intronic CpG-island was methylated slightly less than the promoter island in both histological types of NSCLC (ADC −38%, 6/16; SCC −32%, 7/22) and in ccRCC (39%, 32/83; Table 2). The methylation frequencies of both islands were significantly higher in tumor tissues than in paired histologically normal tissues (P ≤ 0.04, see Table 2). Thus, the methylation of both CpG-islands of the SEMA3B gene is a hallmark of lung and renal cancer. As expected, the methylation of neither the 1-st nor the 2-nd SEMA3B CpG-island was detected in any DNA samples isolated from blood lymphocytes of healthy donors (n = 15). The MSP data were in agreement with the bisulfite sequencing results for each type of cancer investigated.
Table 2. Methylation frequencies of two CpG-islands of the SEMA3B gene in NSCLC (ADC and SCC) and ccRCC in groups of samples with different pathological and histological characteristics.
| Stage | ADC | SCC | ccRCC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-st CpG | 2-nd CpG | 1-st CpG | 2-nd CpG | 1-st CpG | 2-nd CpG | |||||||
| T | N | T | N | T | N | T | N | T | N | T | N | |
| I | 33% | 0% | 33% | 0% | 14% | 14% | 0% | 0% | 40% | 0% | 45% | 0% |
| (2/6) | (0/6) | (2/6) | (0/6) | (1/7) | (1/7) | (0/7) | (0/7) | (8/20) | (0/20) | (9/20) | (0/20) | |
| II | 50% | 0% | 50% | 0% | 75% | 13% | 38% | 0% | 44% | 4% | 30% | 9% |
| (1/2) | (0/2) | (1/2) | (0/2) | (6/8) | (1/8) | (3/8) | (0/8) | (10/23) | (1/23) | (7/23) | (2/23) | |
| III | 43% | 14% | 29% | 0% | 43% | 0% | 57% | 0% | 60% | 3% | 33% | 17% |
| (3/7) | (1/7) | (2/7) | (0/7) | (3/7) | (0/7) | (4/7) | (0/7) | (18/30) | (1/30) | (10/30) | (5/30) | |
| IV | 100% | 0% | 100% | 0% | - | - | - | - | 70% | 10% | 60% | 0% |
| (1/1) | (0/1) | (1/1) | (0/1) | (7/10) | (1/10) | (6/10) | (0/10) | |||||
| All | 44% | 6% | 38% | 0% | 45% | 9% | 32% | 0% | 52% | 4% | 39% | 8% |
| (7/16) | (1/16) | (6/16) | (0/16) | (10/22) | (2/22) | (7/22) | (0/22) | (43/83) | (3/83) | (32/83) | (7/83) | |
| P = 0.04 | P = 0.02 | P = 0.02 | P = 0.01 | P = 6×10 –13 | P = 6×10 –6 | |||||||
Note: MSP data. 1-st CpG—promoter CpG-island; 2-nd CpG—intronic CpG-island; T—primary tumors; N—morphologically normal (conventional “normal”) tissues paired to tumor tissues. The P-values show the significance of the methylation frequencies distinction between tumor and normal tissues (Fisher’s exact test and χ2 criteria).
The use of a representative set of primary tumors allowed us to reveal possible correlations between the methylation frequency of the promoter and intronic CpG-islands with the pathological and histological parameters of the tumors. Advanced ccRCC and NSCLC tumors had a higher frequency of CpG-island methylation compared to the early stages (Tables 2 and 3). The strongest correlation was shown for SCC (Spearman’s rank correlation coefficients were equal 0.37, P = 0.09, and 0.60, P < 0.01, for the 1-st and for the 2-nd CpG-island, respectively). A positive correlation was observed between tumor grade and the frequency of CpG-island methylation for NSCLC and ccRCC (Table 3). This correlation was stronger than the correlation between tumor stage and the frequency of CpG-island methylation for both histological types of NSCLC and almost equal to those for ccRCC (Table 3). The strongest correlation was shown for SCC (0.49, P = 0.02, and 0.68, P < 0.01, for the 1-st and for the 2-nd CpG-island, respectively). These results suggest that methylation of both SEMA3B CpG-islands is a frequent event in renal and lung cell lines and primary tumors and contributes to the progression of tumors of these locations, especially in the lung.
Table 3. Correlations between the frequency of CpG-island methylation and tumor stage or grade.
| Feature | ADC | SCC | ccRCC | ||||
|---|---|---|---|---|---|---|---|
| 1-st CpG | 2-nd CpG | 1-st CpG | 2-nd CpG | 1-st CpG | 2-nd CpG | ||
| Stage | r s | 0.36 | 0.30 | 0.37 | 0.60 | 0.34 | 0.21 |
| P | 0.17 | 0.26 | 0.09 | < 0.01 | < 0.01 | 0.05 | |
| Grade | r s | 0.46 | 0.37 | 0.49 | 0.68 | 0.33 | 0.33 |
| P | 0.07 | 0.16 | 0.02 | < 0.01 | < 0.01 | < 0.01 | |
Note: Based on MSP data. 1-st CpG—promoter CpG-island; 2-nd CpG—intronic CpG-island; r s—Spearman’s rank correlation coefficient; P—p-value.
Correlation between methylation status of two CpG-islands and SEMA3B gene expression level in ccRCC tumors
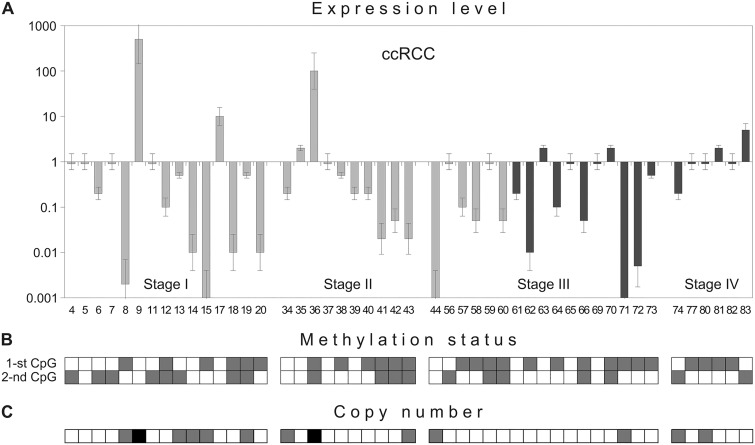
To investigate the consequences of CpG-island methylation, the mRNA level in 48 ccRCC samples was evaluated by semi-quantitative PCR. In ccRCC samples, the expression of SEMA3B was 5–1000 times lower than the matched normal samples in 24 of 48 cases (Fig 6A). In 4 cases, a 5-300-fold up-regulation of expression was observed. No correlation between mRNA level changes and stage, grade or presence of metastases was observed.
Fig 6. SEMA3B gene expression level (A), copy number (C) and methylation status of its two CpG-islands (B) in the same ccRCC samples.
Semi-quantitative PCR (A, C) and MSP (B) data. Numbers of primary tumors correspond to those in S1 Table and Fig 5B. (A) Light grey columns—samples without metastases, dark grey columns—samples with lymph node or distant metastases. (B) 1-st CpG—promoter CpG-island, 2-nd CpG—intronic CpG-island. Grey squares show methylated CpG-islands, white squares—unmethylated. (C) Grey squares show hemi- or homozygous deletions of the 5’Sema5 marker, black—amplification, white squares—retention. Assessed mean values ± error bars are represented in the “A” part.
Next, we compared the changes in mRNA levels of the SEMA3B gene in ccRCC tumors (semi-quantitative PCR data) with the methylation status of two CpG-islands (methylation-specific PCR data). Methylation of the promoter CpG-island was observed in 27 of 48 (56%) cases and methylation of the intronic CpG-island—in 19 of 48 (40%) (Fig 6B). As shown in Fig 6A and 6B, both islands were methylated primarily in the ccRCC samples with decreased SEMA3B expression. The Spearman’s correlation coefficient between expression level down-regulations and methylation status was equal to 0.50 (P < 0.01) for the 1-st CpG-island and 0.25 (P < 0.01) for the 2-nd CpG-island. These data suggest that inactivation of SEMA3B in carcinomas could be associated primarily with methylation of the promoter CpG-island.
Amplifications and deletions of the SEMA3B gene can contribute to mRNA level alterations in ccRCC
A correlation between changes in SEMA3B mRNA expression and the methylation status of its promoter CpG-island was shown in ccRCC, but for several samples, significant aberrations in expression were not associated with methylation of either the 1-st or the 2-nd CpG-island. In one sample, a 100-fold up-regulation was observed when both CpG-islands were methylated. Consequently, we investigated an alternative mechanism of expression regulation, copy number alterations, by semi-quantitative PCR. The increase in SEMA3B mRNA level in ccRCC samples #9 and #36 (300- and 100-fold up-regulation, respectively) was associated with amplification of the 5’Sema5 marker [14] (Fig 6A and 6C). A five-fold increase in mRNA level in ccRCC sample #83 was associated with amplification of the D3S1573 marker (data not shown). Decreased SEMA3B expression (100–1000 times) in ccRCC samples #8, #14, #15, #44 and #71 was associated with hemi- or homozygous deletions of the 5’Sema5 marker (Fig 6A and 6C). A hemizygous deletion was observed in sample #34 with 5-fold down-regulation and no methylation. The Spearman’s correlation coefficient between expression level alterations and copy number changes was equal to 0.61 (P < 0.01) Thus, in addition to the epigenetic modifications, genetic events (e.g., deletion or amplification of the SEMA3B gene locus) can also contribute to the alteration of SEMA3B gene expression in tumors. However, in samples #39 and #61, with 4-5-fold down-regulation, and sample #17, with a 10-fold up-regulation, no copy number changes were observed. We propose that other mechanisms may be responsible for these mRNA level alterations; for example, regulation via miRNA.
Quantitative evaluation of SEMA3B expression in lung and renal tumors
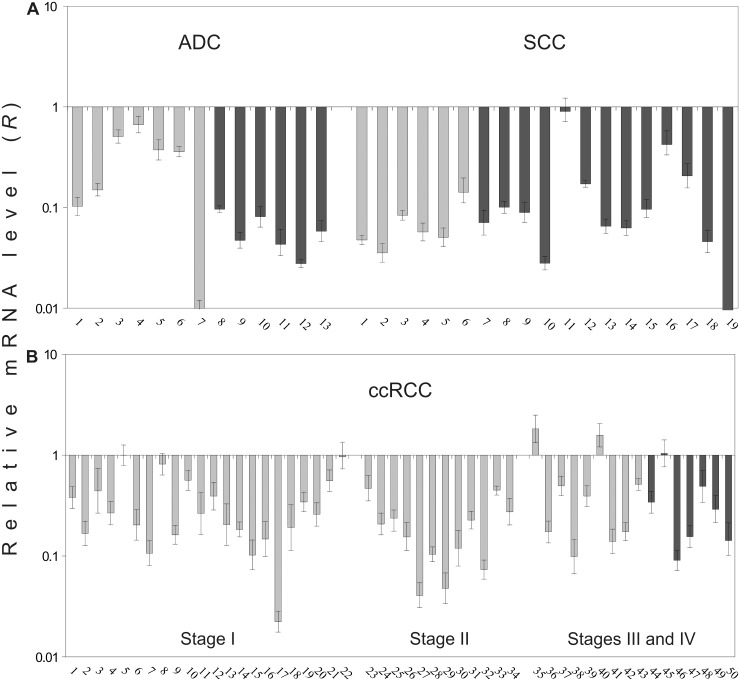
To confirm the mRNA level differences, we performed qPCR analysis on an additional set of lung and renal tumors (Table 1). We found a noticeable (up to 300 times) and frequent (94%, 30/32) down-regulation of SEMA3B gene expression in NSCLC primary tumors using qPCR (Fig 7A). The overall frequency and extent of the decrease in SEMA3B mRNA level was similar in the tumors of the two main histological types of NSCLC; ADC and SCC. An average 10-fold decrease and 92% (12/13) was observed in ADC, and a 14-fold and 95% (18/19) was observed in SCC (Table 4). Other differences were also observed. In ADC with lymph node metastases compared to ADC without metastases, the average mRNA level was considerably decreased (19-fold decrease vs. 3-fold, P < 0.05), but in SCC, significant expression down-regulation was observed in the early stages of tumor development.
Fig 7. Relative mRNA level of the SEMA3B gene in NSCLC (A) and ccRCC (B).
QPCR data, additional samplings. Light grey columns—samples without metastases, dark grey columns—samples with lymph node or distant metastases. The numbers of primary tumors correspond to those in S1 Table. Mean values ± standard deviations for 3 replicates are represented.
Table 4. Frequency and mRNA level changes of the SEMA3B gene in NSCLC (ADC and SCC) and ccRCC in groups of samples with different pathological and histological characteristics.
| Stage | Median of R, n-fold decrease* | Frequency of R decrease, % | ||||
|---|---|---|---|---|---|---|
| ADC | SCC | ccRCC | ADC | SCC | ccRCC | |
| I | 3 | 19 | 4 | 86 | 100 | 77 |
| (1–102) | (7–28) | (1–45) | (6/7) | (6/6) | (17/22) | |
| II | 10 | 11 | 6 | 100 | 83 | 100 |
| (1–36) | (2–25) | (1/1) | (5/6) | (12/12) | ||
| III+IV | 21 | 15 | 3 | 100 | 100 | 81 |
| (12–36) | (2–3×102) | (1–11) | (5/5) | (7/7) | (13/16) | |
| All | 10 | 14 | 4 | 92 | 95 | 84 |
| (1–102) | (1–3×102) | (1–45) | (12/13) | (18/19) | (42/50) | |
Note: QPCR data. R—relative mRNA level.
*—in parentheses a range of mRNA level changes is shown.
In ccRCC samples we also observed frequent (84%, 42/50) SEMA3B gene expression down-regulation (4-fold on average, Fig 7B). However, these values were not as high as for NSCLC. No correlation was observed between mRNA level changes and stage (Table 4) or the presence of metastases (Fig 7B).
Based on these results, we conclude that significant and frequent decreases in SEMA3B mRNA level are in agreement with the semi-quantitative PCR and methylation data and confirm that SEMA3B down-regulation is a frequent event both in NSCLC and ccRCC. However, the frequency of down-regulation was higher than the frequency of CpG-island methylation (Tables 2 and 4). Thus, other mechanisms of inactivation are also likely contributors: for example, deletions in renal, lung and breast cancers, according to our results and the data of others [14, 15, 17].
Discussion
The SEMA3B gene is located in the LUCA region (3p21.3) that harbors 19 genes, the majority of which are involved in carcinogenesis [16, 36–38]. Some of these genes, e.g., RASSF1, NPRL2, and SEMA3B, suppress the growth of tumor cells in vitro and in vivo [36–38]. However, SEMA3B may have both tumor-suppressive and pro-invasive properties. It has been reported that SEMA3B is dramatically down-regulated in the cell line H460-M, derived from the large cell lung tumor cell line NIH-H460, which induces spontaneous metastasis in nude mice [39]. On the other hand, it is highly expressed in many invasive and metastatic human cancers; for example, colorectal carcinoma, neuroblastoma, melanoma and acute myeloid leukemia [20, 40]. SEMA3B induces apoptosis and the production of interleukin 8 by tumor cells by initiating the p38-mitogen-activated protein kinase pathway. In turn, the release of interleukin 8 induces the recruitment of tumor-associated macrophages, which may lead to metastatic dissemination of the tumor [20, 41]. In this study, we confirmed the tumor-suppressive activity of SEMA3B both in vitro (U2020 SCLC cells) and in vivo (SCID mice models, by both tumor growth assay and multi-gene inactivation tests). Our studies in vitro are in agreement with previous research in NSCLC, breast and ovarian ADC cell lines [19, 22, 42]. However, in this study, the suppressive effect of SEMA3B in vivo has been shown for the first time.
Semaphorins 3 encode proteins with axonal guidance properties that generally affect the motility and migration of tumor and endothelial cells by inducing a collapse of the actin cytoskeleton via binding to neuropilins/plexins complexes [43, 44]. Earlier, the class-3 semaphorins were implicated in tumor progression and metastasis [39, 45, 46]. However, tumor suppression properties of distinct semaphorins differ across various cancer types. For example SEMA3D and SEMA3E displayed strong inhibitory effect on the glioblastoma cells whereas SEMA3A and SEMA3B expression just caused persistent cell shape contraction [47]. On the other hand, SEMA3B was previously demonstrated to inhibit proliferation of breast and lung cancer cell lines in vitro [20, 22]. Some studies suggest that the mechanism of the tumor- and angio-suppressive properties of SEMA3 proteins is competition between SEMA3 and VEGF for binding to neuropilin receptors [19, 48]. Other studies reveal SEMA3 growth-inhibitory properties independent of competition with VEGF [10]. Several studies indicate that SEMA3A and SEMA3F are potent inhibitors of metastasis and angiogenesis [10, 49, 50]. Semaphorins 3 were suggested as potential angiogenesis inhibitory agents for triple negative breast cancer treatment [51]. However, Varshavsky et al. have determined that point mutations at the cleavage site of SEMA3B can inhibit angiogenesis in vitro and in vivo [52]. Joseph et al. demonstrated that forced expression of SEMA3B, but not SEMA3F, inhibited the viability of ovarian cancer cells lines in vitro, and presence of SEMA3F but not SEMA3B significantly inhibited the production of endothelial tubes formed by normal human umbilical vein endothelial cells (HUVEC) in vitro [53]. In this study, using PI-FACS we have shown that tumor growth suppression by SEMA3B is associated with induction of apoptosis in vitro. In addition, we have suggested that SEMA3B could be able to induce apoptosis and, possibly, inhibit angiogenesis in vivo, which was previously demonstrated only in vitro [20, 22].
One of the hallmarks of tumor-suppressor genes is the down-regulation of their expression in various tumors at the mRNA or protein level. Inactivation of suppressor genes in tumors can be caused by methylation of their promoter regions. Hypermethylation often reduces the mRNA levels not only of SEMA3B but also of some other genes of the LUCA region in tumors; e.g., RASSF1A, BLU, and CACNA2D2 [36–38]. SEMA3B methylation has been observed in various types of cancer, including lung, liver, gallbladder, gastric, breast and oral carcinomas, and neuroblastoma [17, 54–61]. However, these studies included only the CpG-island, which, according to the NCBI database (http://www.ncbi.nlm.nih.gov/), belongs to the first intron (+1350..+1700 bp from SEMA3B 5’-end) of SEMA3B. However, there is a true promoter region, which includes CpG-dinucleotides with higher density (-300..+50 bp from SEMA3B 5’-end) and is capable of binding transcription factors, such as Lyf−1, DeltaEF−1, Tcf−11, c−Myb, SP1, C/EBP, and AhR/Arnt (according to the recent ENCODE data, http://genome.ucsc.edu/ENCODE/).
Here, for the first time, methylation of both intronic and promoter CpG islands has been analyzed in 14 lung and renal cancer cell lines and representative sets of primary tumors (38 NSCLC and 83 ccRCC cases). We observed frequent methylation of intronic and promoter CpG-islands in both lung (SCC and ADC) and renal primary tumors. In addition, the methylation frequency detected in the promoter CpG-island was higher than in the intronic island in SCC, ADC, and ccRCC (44–52% vs. 32–39%). Thus, methylation of both promoter and intronic SEMA3B CpG-islands is a feature of lung and renal tumors.
In addition, an association was observed between the decrease of the SEMA3B mRNA level and the methylation of the promoter and intronic CpG-islands in ccRCC primary tumors (the Spearman’s correlation coefficient was equal to 0.50 (P < 0.01) for the promoter CpG-island and 0.25 (P < 0.01) for the intronic island). These data imply a primary contribution of promoter CpG-island methylation to down-regulation of the SEMA3B suppressor gene.
The frequency of SEMA3B promoter hypermethylation (44–52%) is approximately two times lower than the frequency of expression decreases (84–95% cases, according to semi-quantitative PCR data) in NSCLC and ccRCC. Thus, other mechanisms could lead to SEMA3B down-regulation. For example, we have found hemi- or homozygous deletions of the 5’Sema5 marker in some ccRCC samples. There are several reports, including ours, of chromosome 3 allelic losses resulting in decreased SEMA3B mRNA levels in lung and renal tumors [14, 55]. The impact of non-coding RNA, especially miRNA, should not be ignored; e.g., miR-137 or miR-193a, as predicted by miRWalk (http://www.ma.uni-heidelberg.de/apps/zmf/mirwalk/).
The use of a representative set of primary tumors allowed us to reveal the correlations between the methylation frequency of both the promoter and the intronic CpG-islands of SEMA3B with tumor stage and grade for histological types of NSCLC (SCC and ADC) and for ccRCC. The highest Spearman’s rank correlation coefficients between mentioned features were observed for SCC (r s was localized over the range 0.37–0.68).
SEMA3B expression was evaluated by quantitative PCR using an additional set of NSCLC (SCC and ADC) and ccRCC samples. We observed frequent and significant SEMA3B down-regulation in SCC and ADC, and somewhat less in ccRCC. SEMA3B down-regulation has been shown in lung, liver, breast, ovarian, renal and colon primary tumors using different semi-quantitative methods [2, 62]. Down-regulation of SEMA3B gene was negatively correlated with tumor size and gastric cancer staging [61]. Here, a large decrease of SEMA3B mRNA level in primary NSCLC and ccRCC was shown for the first time using qPCR. We also observed an association of the frequency and extent of the decrease in SEMA3B mRNA level with the development of ADC metastases (P < 0.05). In SCC, we revealed 10-fold and above down-regulation of the expression of this gene already at early stages of tumor development.
The methylation analysis revealed a significant association of SEMA3B hypermethylation with tumor progression in terms of tumor stage and grade for both subtypes of NSCLC (SCC and ADC) and ccRCC. Moreover, our qPCR expression studies have established a strong reverse correlation between the SEMA3B mRNA level and the presence of metastases in lung ADC. Therefore, hypermethylation of SEMA3B CpG-islands could be suggested as a progression marker for ccRCC and NSCLC (especially SCC), and for the SEMA3B mRNA level as a marker of metastasis development in lung ADC.
Other members of class 3 semaphorins also reveal distinct features, such as expression level variations or tumor suppressive ability, depending on the tumor type or the model system. Mouse melanoma (B16F10) cells overexpressing Sema3A show a significant inhibition of cell motility, invasiveness and proliferation, as well as suppression of tumor growth in vivo, and angiogenesis and metastasis in mice models [50]. Sema3A/B/C/E are also involved in the lymph node metastasis of prostate cancer, but they are likely to modulate the behavior of prostate cancer with a pro-tumor or anti-tumor effect, depending on the subtype [63].
In conclusion, our in vitro and in vivo results reveal the tumor suppressor role of SEMA3B, which could acts by inducing apoptosis or, possibly, inhibiting angiogenesis. Our data also show that methylation of both the promoter and intronic CpG-islands of SEMA3B is a frequent event in ccRCC and two major histological types of NSCLC, but correlation of SEMA3B down-regulation and hypermethylation is stronger for the promoter CpG-island. Significant decreases in the level of SEMA3B mRNA in the majority of the lung and renal tumor samples were revealed by qPCR. Association of SEMA3B hypermethylation and down-regulation with tumor progression could serve as prognostic markers. An understanding of the role of SEMA3s, including SEMA3B, in carcinogenesis will help to use their tumor suppressive and anti-angiogenic properties for the development of new agents for cancer therapy in future.
Supporting Information
(DOC)
(DOC)
Acknowledgments
Part of this work was performed using the equipment of EIMB RAS "Genome" center (http://www.eimb.ru/RUSSIAN_NEW/INSTITUTE/ccu_genome_c.php). This article is dedicated to the memory of our friend and our teacher—Eugene R. Zabarovsky
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants 13-04-00828a, 13-04-02072a, and 14-04-32084 mol_a from the Russian Foundation for Basic Research; grant 28.12.07-6888 from Istituto Toscano Tumori; a grant from CNR-RAS Joint Projects 2008-2010; the grant from RAS Presidium Program "Molecular and Cellular Biology"; contracts 14.604.21.0117 (project's unique identifier RFMEFI60414X0117) and 14.621.21.0001 (project's unique identifier RFMEFI62114X0001) from the Ministry of Education and Science of the Russian Federation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Luo Y, Raible D, Raper JA. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75(2): 217–27. [DOI] [PubMed] [Google Scholar]
- 2. Rehman M, Tamagnone L. Semaphorins in cancer: biological mechanisms and therapeutic approaches. Seminars in cell & developmental biology. 2013;24(3): 179–89. [DOI] [PubMed] [Google Scholar]
- 3. Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19(10): 2590–7. [DOI] [PubMed] [Google Scholar]
- 4. Kigel B, Rabinowicz N, Varshavsky A, Kessler O, Neufeld G. Plexin-A4 promotes tumor progression and tumor angiogenesis by enhancement of VEGF and bFGF signaling. Blood. 2011;118(15): 4285–96. 10.1182/blood-2011-03-341388 [DOI] [PubMed] [Google Scholar]
- 5. Miyato H, Tsuno NH, Kitayama J. Semaphorin 3C is involved in the progression of gastric cancer. Cancer science. 2012;103(11): 1961–6. 10.1111/cas.12003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basile JR, Castilho RM, Williams VP, Gutkind JS. Semaphorin 4D provides a link between axon guidance processes and tumor-induced angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(24): 9017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dorfman DM, Shahsafaei A, Nadler LM, Freeman GJ. The leukocyte semaphorin CD100 is expressed in most T-cell, but few B-cell, non-Hodgkin's lymphomas. The American journal of pathology. 1998;153(1): 255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma Y, Visser L, Roelofsen H, de Vries M, Diepstra A, van Imhoff G, et al. Proteomics analysis of Hodgkin lymphoma: identification of new players involved in the cross-talk between HRS cells and infiltrating lymphocytes. Blood. 2008;111(4): 2339–46. [DOI] [PubMed] [Google Scholar]
- 9. Bielenberg DR, Hida Y, Shimizu A, Kaipainen A, Kreuter M, Kim CC, et al. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. The Journal of clinical investigation. 2004;114(9): 1260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaur P, Bielenberg DR, Samuel S, Bose D, Zhou Y, Gray MJ, et al. Role of class 3 semaphorins and their receptors in tumor growth and angiogenesis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(22): 6763–70. 10.1158/1078-0432.CCR-09-1810 [DOI] [PubMed] [Google Scholar]
- 11. Gu C, Limberg BJ, Whitaker GB, Perman B, Leahy DJ, Rosenbaum JS, et al. Characterization of neuropilin-1 structural features that confer binding to semaphorin 3A and vascular endothelial growth factor 165. The Journal of biological chemistry. 2002;277(20): 18069–76. [DOI] [PubMed] [Google Scholar]
- 12. Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M. Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. The Journal of cell biology. 1999;146(1): 233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pan Q, Chanthery Y, Liang WC, Stawicki S, Mak J, Rathore N, et al. Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth. Cancer cell. 2007;11(1): 53–67. [DOI] [PubMed] [Google Scholar]
- 14. Senchenko VN, Liu J, Loginov W, Bazov I, Angeloni D, Seryogin Y, et al. Discovery of frequent homozygous deletions in chromosome 3p21.3 LUCA and AP20 regions in renal, lung and breast carcinomas. Oncogene. 2004;23(34): 5719–28. [DOI] [PubMed] [Google Scholar]
- 15. Senchenko V, Liu J, Braga E, Mazurenko N, Loginov W, Seryogin Y, et al. Deletion mapping using quantitative real-time PCR identifies two distinct 3p21.3 regions affected in most cervical carcinomas. Oncogene. 2003;22(19): 2984–92. [DOI] [PubMed] [Google Scholar]
- 16. Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer research. 2000;60(21): 6116–33. [PubMed] [Google Scholar]
- 17. Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, Williams NN, et al. Allelic loss on chromosome 3p21.3 and promoter hypermethylation of semaphorin 3B in non-small cell lung cancer. Cancer research. 2003;63(12): 3352–5. [PubMed] [Google Scholar]
- 18. Ji L, Minna JD, Roth JA. 3p21.3 tumor suppressor cluster: prospects for translational applications. Future oncology. 2005;1(1): 79–92. [DOI] [PubMed] [Google Scholar]
- 19. Castro-Rivera E, Ran S, Thorpe P, Minna JD. Semaphorin 3B (SEMA3B) induces apoptosis in lung and breast cancer, whereas VEGF165 antagonizes this effect. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(31): 11432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rolny C, Capparuccia L, Casazza A, Mazzone M, Vallario A, Cignetti A, et al. The tumor suppressor semaphorin 3B triggers a prometastatic program mediated by interleukin 8 and the tumor microenvironment. The Journal of experimental medicine. 2008;205(5): 1155–71. 10.1084/jem.20072509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castro-Rivera E, Ran S, Brekken RA, Minna JD. Semaphorin 3B inhibits the phosphatidylinositol 3-kinase/Akt pathway through neuropilin-1 in lung and breast cancer cells. Cancer research. 2008;68(20): 8295–303. 10.1158/0008-5472.CAN-07-6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tomizawa Y, Sekido Y, Kondo M, Gao B, Yokota J, Roche J, et al. Inhibition of lung cancer cell growth and induction of apoptosis after reexpression of 3p21.3 candidate tumor suppressor gene SEMA3B. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24): 13954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heppell-Parton AC, Nacheva E, Carter NP, Rabbitts PH. A combined approach of conventional and molecular cytogenetics for detailed karyotypic analysis of the small cell lung carcinoma cell line U2020. Cancer genetics and cytogenetics. 1999;108(2): 110–9. [DOI] [PubMed] [Google Scholar]
- 24. Murata Y, Tamari M, Takahashi T, Horio Y, Hibi K, Yokoyama S, et al. Characterization of an 800 kb region at 3p22-p21.3 that was homozygously deleted in a lung cancer cell line. Human molecular genetics. 1994;3(8): 1341–4. [DOI] [PubMed] [Google Scholar]
- 25. Alimov A, Kost-Alimova M, Liu J, Li C, Bergerheim U, Imreh S, et al. Combined LOH/CGH analysis proves the existence of interstitial 3p deletions in renal cell carcinoma. Oncogene. 2000;19(11): 1392–9. [DOI] [PubMed] [Google Scholar]
- 26. Sobin LH, Wittekind C. TNM Classification of Malignant Tumours, 6th edition New York, USA: Wiley; 2002. [Google Scholar]
- 27. Eble JN, Sauter GS, Epsten JI, Sisterhenn IA. World Health Organization Classification of Tumours Pathology and Genetics of Tumors of the Urinary System and Male Genital Organs. Lyon, France: IARC Press; 2004. [Google Scholar]
- 28. Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. World Health Organization Classification of Tumours Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2004. [Google Scholar]
- 29. Protopopov AI, Li J, Winberg G, Gizatullin RZ, Kashuba VI, Klein G, et al. Human cell lines engineered for tetracycline-regulated expression of tumor suppressor candidate genes from a frequently affected chromosomal region, 3p21. The journal of gene medicine. 2002;4(4): 397–406. [DOI] [PubMed] [Google Scholar]
- 30. Li J, Protopopov AI, Gizatullin RZ, Kiss C, Kashuba VI, Winberg G, et al. Identification of new tumor suppressor genes based on in vivo functional inactivation of a candidate gene. FEBS letters. 1999;451(3): 289–94. [DOI] [PubMed] [Google Scholar]
- 31. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18): 9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, et al. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13): 7504–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angeloni D, ter Elst A, Wei MH, van der Veen AY, Braga EA, Klimov EA, et al. Analysis of a new homozygous deletion in the tumor suppressor region at 3p12.3 reveals two novel intronic noncoding RNA genes. Genes, chromosomes & cancer. 2006;45(7): 676–91. [DOI] [PubMed] [Google Scholar]
- 34. Krasnov GS, Oparina NY, Dmitriev AA, Kudryavtseva AV, Anedchenko EA, Kondrat'eva TT, et al. RPN1, a new reference gene for quantitative data normalization in lung and kidney cancer. Mol Biol+. 2011;45(2): 211–20. [PubMed] [Google Scholar]
- 35. Senchenko VN, Krasnov GS, Dmitriev AA, Kudryavtseva AV, Anedchenko EA, Braga EA, et al. Differential expression of CHL1 gene during development of major human cancers. PloS one. 2011;6(3): e15612 10.1371/journal.pone.0015612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21(45): 6915–35. [DOI] [PubMed] [Google Scholar]
- 37. Angeloni D. Molecular analysis of deletions in human chromosome 3p21 and the role of resident cancer genes in disease. Briefings in functional genomics & proteomics. 2007;6(1): 19–39. [DOI] [PubMed] [Google Scholar]
- 38. Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene. 2007;26(52): 7283–301. [DOI] [PubMed] [Google Scholar]
- 39. de Lange R, Dimoudis N, Weidle UH. Identification of genes associated with enhanced metastasis of a large cell lung carcinoma cell line. Anticancer research. 2003;23(1A): 187–94. [PubMed] [Google Scholar]
- 40. Klimov EA, Selivanova NL, Razumnova GI, Rudko OI, Golovatenko-Abramov PK. RHOA, SEMA3B, and CKAP2 expression in leukaemia of different types: the results of a pilot experiment. Folia biologica. 2013;59(5): 204–6. [PubMed] [Google Scholar]
- 41. Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer research. 2007;67(6): 2649–56. [DOI] [PubMed] [Google Scholar]
- 42. Tse C, Xiang RH, Bracht T, Naylor SL. Human Semaphorin 3B (SEMA3B) located at chromosome 3p21.3 suppresses tumor formation in an adenocarcinoma cell line. Cancer research. 2002;62(2): 542–6. [PubMed] [Google Scholar]
- 43. Janssen BJ, Malinauskas T, Weir GA, Cader MZ, Siebold C, Jones EY. Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nature structural & molecular biology. 2012;19(12): 1293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shimizu A, Mammoto A, Italiano JE Jr, Pravda E, Dudley AC, Ingber DE, et al. ABL2/ARG tyrosine kinase mediates SEMA3F-induced RhoA inactivation and cytoskeleton collapse in human glioma cells. The Journal of biological chemistry. 2008;283(40): 27230–8. 10.1074/jbc.M804520200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Neufeld G, Lange T, Varshavsky A, Kessler O. Semaphorin signaling in vascular and tumor biology. Advances in experimental medicine and biology. 2007;600: 118–31. [DOI] [PubMed] [Google Scholar]
- 46. Neufeld G, Kessler O. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nature reviews Cancer. 2008;8(8): 632–45. 10.1038/nrc2404 [DOI] [PubMed] [Google Scholar]
- 47. Sabag AD, Bode J, Fink D, Kigel B, Kugler W, Neufeld G. Semaphorin-3D and semaphorin-3E inhibit the development of tumors from glioblastoma cells implanted in the cortex of the brain. PloS one. 2012;7(8): e42912 10.1371/journal.pone.0042912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Osada R, Horiuchi A, Kikuchi N, Ohira S, Ota M, Katsuyama Y, et al. Expression of semaphorins, vascular endothelial growth factor, and their common receptor neuropilins and alleic loss of semaphorin locus in epithelial ovarian neoplasms: increased ratio of vascular endothelial growth factor to semaphorin is a poor prognostic factor in ovarian carcinomas. Human pathology. 2006;37(11): 1414–25. [DOI] [PubMed] [Google Scholar]
- 49. Bielenberg DR, Klagsbrun M. Targeting endothelial and tumor cells with semaphorins. Cancer metastasis reviews. 2007;26(3–4): 421–31. [DOI] [PubMed] [Google Scholar]
- 50. Chakraborty G, Kumar S, Mishra R, Patil TV, Kundu GC. Semaphorin 3A suppresses tumor growth and metastasis in mice melanoma model. PloS one. 2012;7(3): e33633 10.1371/journal.pone.0033633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bender RJ, Mac Gabhann F. Expression of VEGF and semaphorin genes define subgroups of triple negative breast cancer. PloS one. 2013;8(5): e61788 10.1371/journal.pone.0061788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Varshavsky A, Kessler O, Abramovitch S, Kigel B, Zaffryar S, Akiri G, et al. Semaphorin-3B is an angiogenesis inhibitor that is inactivated by furin-like pro-protein convertases. Cancer research. 2008;68(17): 6922–31. 10.1158/0008-5472.CAN-07-5408 [DOI] [PubMed] [Google Scholar]
- 53. Joseph D, Ho SM, Syed V. Hormonal regulation and distinct functions of semaphorin-3B and semaphorin-3F in ovarian cancer. Molecular cancer therapeutics. 2010;9(2): 499–509. 10.1158/1535-7163.MCT-09-0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tischoff I, Markwarth A, Witzigmann H, Uhlmann D, Hauss J, Mirmohammadsadegh A, et al. Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. International journal of cancer Journal international du cancer. 2005;115(5): 684–9. [DOI] [PubMed] [Google Scholar]
- 55. Ito M, Ito G, Kondo M, Uchiyama M, Fukui T, Mori S, et al. Frequent inactivation of RASSF1A, BLU, and SEMA3B on 3p21.3 by promoter hypermethylation and allele loss in non-small cell lung cancer. Cancer letters. 2005;225(1): 131–9. [DOI] [PubMed] [Google Scholar]
- 56. Nair PN, McArdle L, Cornell J, Cohn SL, Stallings RL. High-resolution analysis of 3p deletion in neuroblastoma and differential methylation of the SEMA3B tumor suppressor gene. Cancer genetics and cytogenetics. 2007;174(2): 100–10. [DOI] [PubMed] [Google Scholar]
- 57. Riquelme E, Tang M, Baez S, Diaz A, Pruyas M, Wistuba II, et al. Frequent epigenetic inactivation of chromosome 3p candidate tumor suppressor genes in gallbladder carcinoma. Cancer letters. 2007;250(1): 100–6. [DOI] [PubMed] [Google Scholar]
- 58. Bernal C, Aguayo F, Villarroel C, Vargas M, Diaz I, Ossandon FJ, et al. Reprimo as a potential biomarker for early detection in gastric cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14(19): 6264–9. 10.1158/1078-0432.CCR-07-4522 [DOI] [PubMed] [Google Scholar]
- 59. da Costa Prando E, Cavalli LR, Rainho CA. Evidence of epigenetic regulation of the tumor suppressor gene cluster flanking RASSF1 in breast cancer cell lines. Epigenetics: official journal of the DNA Methylation Society. 2011;6(12): 1413–24. 10.4161/epi.6.12.18271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang K, Ling T, Wu H, Zhang J. Screening of candidate tumor-suppressor genes in 3p21.3 and investigation of the methylation of gene promoters in oral squamous cell carcinoma. Oncology reports. 2013;29(3): 1175–82. 10.3892/or.2012.2213 [DOI] [PubMed] [Google Scholar]
- 61. Chen R, Zhuge X, Huang Z, Lu D, Ye X, Chen C, et al. Analysis of SEMA3B methylation and expression patterns in gastric cancer tissue and cell lines. Oncology reports. 2014;31(3): 1211–8. 10.3892/or.2014.2972 [DOI] [PubMed] [Google Scholar]
- 62. Pronina IV, Loginov VI, Prasolov VS, Klimov EA, Khodyrev DS, Kazubskaya TP, et al. Altered expression of the SEMA3B gene in epithelial tumors. Mol Biol+. 2009;43(3): 403–9. [PubMed] [Google Scholar]
- 63. Li K, Chen MK, Li LY, Lu MH, Shao CK, Su ZL, et al. The predictive value of semaphorins 3 expression in biopsies for biochemical recurrence of patients with low- and intermediate-risk prostate cancer. Neoplasma. 2013;60(6): 683–9. 10.4149/neo_2013_087 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.