Abstract
The pH-Low Insertion Peptide (pHLIP) offers the potential to deliver drugs selectively to the cytoplasm of cancer cells based on tumor acidosis. The WT pHLIP inserts into membrane with a pH50 of 6.1 while most solid tumors have extracellular pH (pHe) of 6.5-7.0. To close this gap, a SAR study was carried out to search for pHLIP variants with improved pH-response. We learned that (a) replacing Asp25 with α-aminoadipic acid (Aad) adjusts the pH50 to 6.74, matching average tumor acidity, and (b) replacing Asp14 with γ-carboxyglutamic acid (Gla) increases the sharpness of pH-response (i.e. transition over 0.5 instead of 1 pH unit). These effects are additive — the Asp14Gla/Asp25Aad double variant shows a pH50 of 6.79, with sharper transition than Asp25Aad. Further, the advantage of the double variant over WT pHLIP in terms of cargo delivery was demonstrated in turn-on fluorescence assays and anti-proliferation studies (using paclitaxel as cargo) in A549 lung cancer cells at pH 6.6.
Keywords: Biosensor, Cancer, Drug Delivery, Peptides, pHLIP
One approach to targeted cancer therapy relies on the use of a drug carrier that can distinguish between normal and cancer cells. The recent success of antibody-drug conjugates validates this approach.[1] However, targeting specific tumor-associated proteins can be hampered by heterogeneity among cells within a tumor [2] and rapid development of resistance.[3] An alternative approach is to target cancer cells based on universal features of the tumor microenvironment (e.g. hypoxia),[4] which may have broad applications in many different types of cancers. Tumor acidosis is such a property that may be exploited.[5] Many solid tumors have interstitial acidity (pHe of 6.5-7.0), compared to normal tissue pHe of 7.2-7.5, whereas the intracellular pH of cancer and healthy cells are similar (both ~ pH 7.0-7.5).[5-6]
Tumor acidosis results from (a) the altered metabolism of cancer cells, i.e. ATP production via glycolysis with reduced rate of respiration — at first, in response to hypoxia (the Pasteur effect), but eventually even under conditions of plentiful oxygen (the Warburg effect), and (b) a fast rate of metabolism coupled with poor clearance of waste products.[7] Low tumor pHe may confer growth advantages on cancer cells as healthy tissue and their extracellular matrix remodel under acidic stress, clearing space for local tumor invasion and eventual metastasis.[8] Cancer cells living in acidic and hypoxic environments are especially resistant to radiation and chemotherapy, which in turn contribute to tumor recurrence.[4, 7a, 9] Thus using pHLIP to target acidic cancer cells for destruction can offer a complementary approach to established therapies.
At the physiological pH of 7.2-7.5, pHLIP is unstructured in solution (State I in Figure 1) or peripherally bound to membrane surfaces (State II); under slightly acidic conditions, pHLIP inserts across a lipid bilayer, forming a transmembrane (TM) helix (State III).[10] The insertion is unidirectional with C-terminus translocated across the membrane,[10c, 11] rapid (equilibrium reached in minutes),[12] and mediated mainly by the protonations of Asp14 and Asp25 carboxyl sidechains in the TM region.[10b, 13] Both D- and L-pHLIP peptides can target acidic tumors in vivo.[13-14] Further, pHLIP does not cause membrane leakage,[11a, 15] and no toxic effects have been oberved in cell culture (up to 10 μM) or mice (5 mg/kg).[11a, 13, 16] Due to its small size (36-38 amino acids, ~ 4 KD), pHLIP can penetrate to the core of tumor where hypoxia and acidosis are most prominent.[16] These properties favor pHLIP as a drug carrier worthy of further development.
Figure 1.
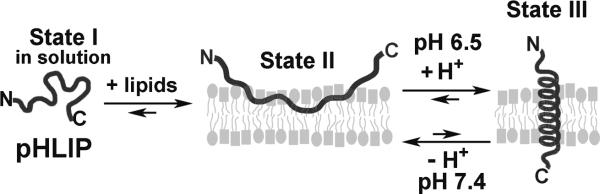
The pHLIP peptide inserts into membrane in response to acidity.
One challenge in targeting tumor acidosis is the small pH difference between healthy tissue and tumor, i.e. the drug carrier should be a pH sensor with sharp transition. Previous efforts in targeting tumor pHe have been focused on pH-responsive polymers. Taking advantage of the facts that the imidazole side-chain of histidine is protonated to the imidazonium cation with a pKa of 7.0 (neutral to positive at low pHe) whereas certain sulfonamides have pKa of 6.8 for the NH α-proton (negative to neutral at low pHe), Bae and co-workers engineered a wide variety of polymer-based molecular devices (e.g. micelles) and behaviors (e.g. unveiling of cell-penetrating peptides) to respond to the slightly acidic environment of tumors for drug delivery.[17] Another recently reported approach is based on the acid-catalyzed, β-carboxyl neighboring-group assisted hydrolysis of N-alkyl maleamic acids (negative to positive conversion in response to low pHe).[18] However, the stability of this charge reversal system at pH 7.4 is a concern.[18a, 18b, 18e] Compared to these polymer-based systems, pHLIP is simpler and more chemically defined, which can have significant practical advantages in drug carrier development.
When cargo, i.e. small dye molecule, cyclic peptide, or peptide nucleic acid (PNA), is attached to pHLIP's C-terminus, it can be carried across the membrane during pHLIP insertion.[11, 20] Thus, pHLIP not only can target cancer cells based on low tumor pHe but also deliver the cargo directly into the cytoplasm. Such insertion-mediated delivery of phalloidin (and other toxins) inhibited the proliferation of cancer cells in a pH-dependent fashion.[20a-c] Further, when many copies of pHLIP are attached to the surface of 13-nm gold [21] or 140-nm mesoporous silica nanoparticles,[22] they seem to be able to work in concert to bring such large cargo into cells. Recently, as the first example of in vivo efficacy, pHLIP-mediated delivery of PNA (anti-miR) silenced miR-155 onco-miR in a mouse lymphoma model.[23]
We believe pHLIP also presents an opportunity to improve cancer chemotherapy. Many drugs, such as paclitaxel (Taxol) or doxorubicin, have dose-limiting toxicity in off-target sites (e.g. bone marrow, heart). Our goal is to use pHLIP to deliver such drugs selectively to cancer cells. In the current study we aim to create pHLIP variants that insert more effectively in response to tumor pHe via incorporation of noncanonical amino acids. The best of these variants are further evaluated in cellular assays to demonstrate its advantage over WT pHLIP.
The pHLIP peptide is derived from the TM helix C of bacteriorhodopsin and has the following sequence: GGEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT.[10b] For the original ‘WT’ pHLIP, the apparent pH50 of insertion (i.e. the pH at which 50% of pHLIP are in the inserted State III) across 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) membrane is ~ 6.1 (Figure 3).[10b, 19, 24] When pHLIP peptides interact with cells, insertion may take place at plasmamembrane or endosomal membranes, and likely both.[25] Given its pH50 even the WT pHLIP is able to efficiently deliver cargo into the cytoplasm in response to endosomal acidity. Yet drug delivery via insertion in the plasma-membrane in response to tumor pHe can benefit from increased pH50 because most solid tumors exhibit average pHe of 6.8.[5-6]
Figure 3.
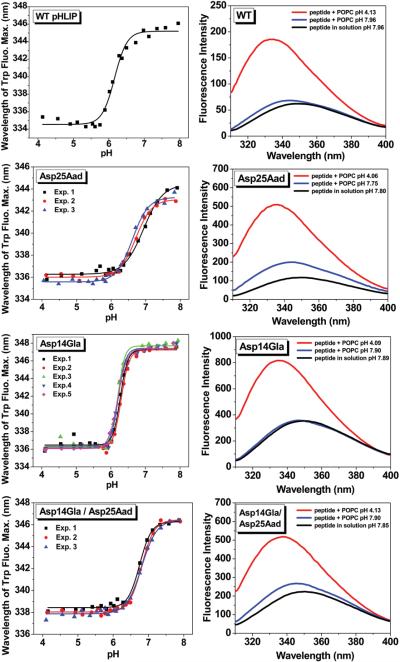
Trp fluorescence of Asp25Aad, Asp14Gla and Asp14Gla/Asp25Aad pHLIP variants show improved pH-response at tumor pHe range of 6.5-7.0. Left column (top to bottom): insertion is monitored via Trp fluorescence λmax blue-shift (different colors denote experimental repeats); compared to the WT, the Asp25Aad insertion shifted to a higher pH range closely matching tumor acidity but suffers from poor cooperativity; the Asp14Gla variant has strikingly sharp pH-response; the Asp14Gla/Asp25Aad double variant maintains a high pH50 of 6.8 with cooprativity restored to WT level. Right column: For the pHLIP variants studied, Trp fluorescence spectra of State I (black), II (blue) and III (red) show increase in fluorescence intensity and λmax blue-shift from State II to State III (i.e. pHLIP insertion into POPC liposome membrane).
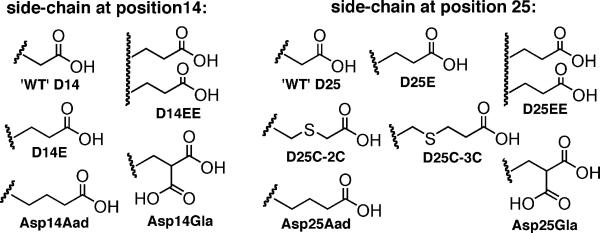
Our structure-activity relationship (SAR) study contains pHLIP variants whose structures are shown in Figure 2. Their insertion behavior into POPC membrane were characterized using established Trp fluorescence methods.[19-20, 24, 26] The basis of the assay is that pHLIP insertion (i.e. State II to III in Figure 1) leads to an increase in Trp fluorescence intensity and a blue-shift in emission λmax, reflecting the more hydrophobic environment the Trp side-chains experience after insertion (especially W15). Further, circular dichroism (CD) measurements were carried out to confirm the pH-dependent conformational change — random coil in States I and II but α-helical in State III.[10b, 19] The apparent pH50 values are calculated by fitting the transition curve of ‘pH vs. λmax’ (Figure 3 left column) to the Henderson-Hasselbalch equation (albeit with pH50 in place of pKa):
where n is the Hill coefficient (which reflects the sharpness or cooperativity of insertion into POPC membrane), and λmax-II and λmax-III are the wavelengths of maximum emission in the membrane-bound State II and the inserted State III, respectively. For each novel variant, the Trp fluorescence assay is repeated at least three times and the average pH50 and Hill coefficient values are reported along with standard deviations (s.d.) in Table 1.
Figure 2.
Side-chain structural variations at pHLIP position 14 and 25. WT, D14E, and D25E are previously known.[10b, 19]
Table 1.
The insertion pH50, Hill coefficient n and Trp fluorescence λmax-II/III of pHLIP variants studied.
| pHLIP variants | pH50 | Hill coefficient n | λmax-II (nm) | λmax-II (nm) |
|---|---|---|---|---|
| WT | 6.16 | 2.48 | 345.2 | 334.5 |
| D25E | 6.27 ± 0.03 | 2.14 ± 0.20 | 344.9 ± 0.3 | 334.9 ± 0.2 |
| D14E | 6.14 ± 0.05 | 2.47 ± 0.33 | 345.6 ± 0.3 | 335.6 ± 0.2 |
| D25C-2C | 6.05 ± 0.04 | 2.44 ± 0.10 | 344.6 ± 1.2 | 334.5 ± 0.5 |
| D25C-3C | 6.18 | 0.9 | 343.5 | 338.0 |
| Asp25Aad | 6.74 ± 0.14 | 1.73 ± 0.17 | 344.5 ± 1.9 | 336.1 ± 0.4 |
| Asp14Aad | 6.37 ± 0.03 | 2.05 ± 0.13 | 344.4 ± 1.5 | 336.0 ± 0.6 |
| Asp25Gla | 6.20 ± 0.02 | 1.90 ± 0.19 | 346.6 ± 0.6 | 335.1 ± 0.2 |
| Asp14Gla | 6.24 ± 0.05 | 3.98 ± 0.40 | 347.4 ± 0.2 | 336.2 ± 0.2 |
| D25EE | 6.60 ± 0.14 | 1.34 ± 0.26 | 345.0 ± 1.5 | 335.5 ± 1.6 |
| D14EE | 6.19 ± 0.04 | 1.41 ± 0.02 | 346.7 ± 1.0 | 335.6 ± 0.3 |
| Asp14Gla / Asp25Aad | 6.79 ± 0.04 | 2.38 ± 0.30 | 346.4 ± 0.1 | 338.1 ± 0.3 |
The D25E and D14E variants have been described to insert with pH50 of ~ 6.4-6.5.[19] To minimize aggregation, we carried out experiments with lower ionic strength (11 mM vs. 68 mM) and peptide concentration (2 μM vs. 7 μM) than reported procedures. Under such conditions, D25E and D14E showed pH50 of 6.27 ± 0.03 and 6.14 ± 0.05, respectively (Table 1, see supporting information for sequence details in Table S1, and Trp fluorescence and CD data in Figure S2). The D25E variant is an important precedent for our SAR study as it demonstrates that lengthening the D25 side-chain can increase pH50.
To find out to what extent can side-chain extension at position 25 be tolerated, the Cys side-chain of a D25C pHLIP was lengthened via reaction with bromoacetic acid or 3-bromopropionic acid to give variant D25C-2C or D25C-3C (Figure 2). The D25C-2C has a pH50 of 6.05 ± 0.04 and a Hill coefficient of 2.44 ± 0.10 (similar to WT), with pH-dependent helix formation characteristic of a pHLIP (as confirmed by CD, see Figure S2). However, the D25C-3C variant lost the coil-to-helix transition: its CD signal is already weakly helical at pH 8 and there is no increase in helicity at pH 4 (Figure S2). Compared to State I, its ‘State II’ Trp fluorescence shows significant increase in intensity and emission λmax blue-shift (Figure S2). Further λmax blue-shift from ‘State II’ to ‘III’ is unusually narrow (5.5 nm for D25C-3C vs. 8-12 nm for all other variants), and the pH vs. λmax transition is poorly defined (n = 0.9). Such data indicate that D25C-3C is not a typical pHLIP, possibly because the additional methylene group increases the tendency of aggregation. In D25C-2C, the electron withdrawing ability of the sulfur atom increases side-chain acidity, shifting the pH50 unfavorably down. However, D25C-2C gave hope that its carbon isostere Asp25Aad would still behave as a pHLIP.
Indeed, Asp25Aad pHLIP has clear coil-to-helix transition from State II to III (Figure S1). More importantly, Asp25Aad has a pH50 of 6.74 ± 0.14 (Figure 3), closely matching average tumor pHe in vivo. Asp25Aad is also consistent with the trend observed from WT to D25E — as the number of methylene groups in the side-chain increased, the pH50 rose (WT 6.16 to D25E 6.27 to Asp25Aad 6.74). Since the peptide backbone is electron-withdrawing, as it became more distant, the innate acidity of the side-chain carboxylic acid decreased, shifting pH50 up. When D14 instead of D25 is replaced with Aad, the resulting Asp14Aad variant has proper pH-dependent insertion (Figure S3), yet the pH50 is only 6.37 ± 0.03. Therefore, factors other than the inherent side-chain pKa, such as the precise location and the polarity of the protonation environment in State II, also influence the pH50. The side-chain at position 25 seems to dominate the setting of pH50.
However, the Hill coefficient of Asp25Aad insertion into POPC membrane is 1.73 ± 0.17, lower than that of WT (2.48). Insertion at neutral pH may hamper the selectivity of pHLIP-mediated drug delivery. Thus, a sharp pH-response, i.e. insertion over a narrow pH range, is desirable. Engelman and co-workers postulated that more carboxylic acid residues in the TM region may increase the insertion cooperativity.[19] Although their attempts at introducing additional carboxyl group were not successful, we thought to revisit this hypothesis. First, the Asp14 or Asp25 residue was replaced with a pair of Glu residues, giving the variants D14EE or D25EE. CD confirmed that D14EE and D25EE have pH-dependent coil-to-helix transition characteristic of a pHLIP (Figure S3). But to our disappointments, D14EE has a pH50 of 6.19 ± 0.04 with Hill coefficient of 1.41 ± 0.02 while D25EE has a pH50 of 6.60 ± 0.14 with Hill coefficient of 1.34 ± 0.26. Both of these variants insert over a wider range than WT pHLIP (Figure S3).
Next, the D14 or D25 residue was substituted with the noncanonical amino acid Gla, which has two carboxyl groups appended to the γ-carbon of the side-chain (Figure 2), resulting in the variants Asp14Gla or Asp25Gla. Compared with D14EE and D25EE variants, this modification allows for more precise introduction of the additional carboxyl group at locations critical for pHLIP insertion. CD showed that Asp14Gla and Asp25Gla both have clearly defined pH-dependent coil-to-helix transition (Figure S1 and S3). Asp25Gla turned out to be unremarkable, with pH50 of 6.20 ± 0.02 and Hill coefficient of 1.90 ± 0.19 (Figure S3). On the other hand, Asp14Gla has a pH50 of 6.24 ± 0.05 but with a transition Hill coefficient of 3.98 ± 0.40 (Figure 3). Thus, the Asp14Gla variant has the sharpest pH-response so far recorded for a pHLIP, with insertion occurring over half pH unit vs. one pH unit for the WT.
To see whether higher insertion pH50 and sharper pH-response could be tuned separately in an additive fashion, the Asp14Gla/Asp25Aad double variant was tested. The CD data showed clear pH-dependent coil-to-helix transition characteristic of a pHLIP (Figure S1); and indeed, this double variant maintains a high insertion pH50 of 6.79 ± 0.04, similar to that of Asp25Aad, while the transition Hill coefficient is restored to 2.38 ± 0.30, close to the WT level of sharpness in pH-response (Figure 3).
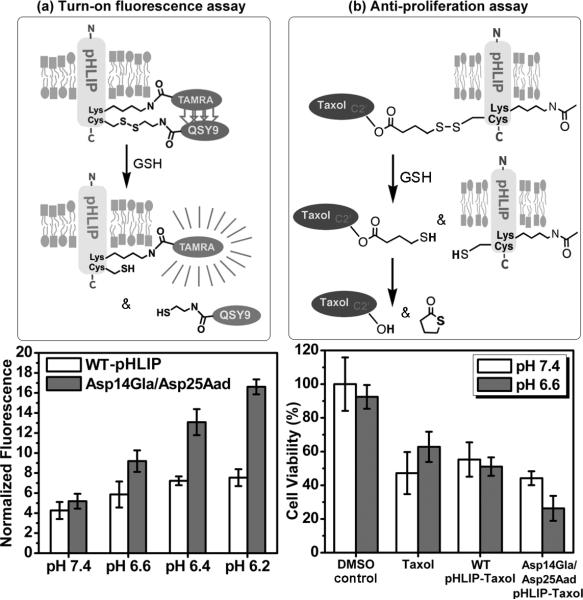
With this best variant in hand, we set out to demonstrate its advantage over WT pHLIP in cultures of A549 human lung cancer cells. WT or Asp14Gla/Asp25Aad pHLIP containing C-terminal Lys and Cys residues were used to synthesize turn-on fluorescence probes (with rhodamine TAMRA attached to the Lys side-chain via an amide bond and quencher QSY9 attached to the adjacent Cys side-chain via a disulfide bond) and pHLIP-paclitaxel conjugates (the drug paclitaxel is conjugated to the Cys side-chain via a disulfide bond traceless linker) (Figure 4). Details of their syntheses are described in Supporting Information (Schemes S1-S2).
Figure 4.
Evaluation of WT pHLIP vs. Asp14Gla/Asp25Aad variant in A549 cancer cells. (a) Turn-on fluorescence assays: At all acidic pHs, Asp14Gla/Asp25Aad pHLIP released more disulfide-linked QSY9 quencher than WT pHLIP. The TAMRA fluorescence levels plotted were measured 72 hr after treatment with 0.5 μM of self-quenched probe for 30 min (n = 8, error bar is s.d.). (b) Anti-proliferation assays: At pH 6.6, Asp14Gla/Asp25Aad pHLIP-paclitaxel inhibited the growth of A549 cancer cells to a greater extent than either WT pHLIP-paclitaxel or the free drug itself (30 min treatment with 0.5 μM of drug or pHLIP-drug, n = 8, error bar is s.d.).
In the self-quenched turn-on fluorescence probe, the quencher QSY9 serves as a model cargo of intracellular drug delivery. Since the disulfide linker is selectively cleaved inside of cells, presumably by glutathione (GSH), QSY9 release would lead to dequenching and increase in TAMRA fluorescence (Figure 4a), thus reporting the level of pHLIP insertion in cellular membranes (because the C-terminus is translocated across membrane during pHLIP insertion). In the WT vs. Asp14Gla/Asp25Aad comparison, data shown in Figure 4a establish that dequenching levels (and thus inferred pHLIP insertion levels) are considerably higher for the double variant at all acidic pHs but similar at neutral pH. In particular the pH 7.4 vs. 6.6 comparison reveals that the amount of pH-dependent cargo release, which represents the therapeutic window in pHLIP-mediated drug delivery in vivo, is much greater for the double variant (9.2- vs. 5.2-fold) than the WT (5.9- vs. 4.3-fold). Thus, the Asp14Gla/Asp25Aad variant should be able to deliver drug in a more pH-dependnt fashion than WT.
To test this hypothesis in antiproliferation assays, the drug paclitaxel is conjugated to pHLIP via an ester-disulfide linkage on its C2’ OH (Figure 4b and Scheme S2). Since attachment at this site would interfere with paclitaxel binding to tubulin,[27] a traceless linker is constructed following known methods[28] in which upon cleavage of the disulfide bond in cells, the resulting linker SH group would cyclize onto the paclitaxel C2’ ester carbonyl to release the cargo exactly as paclitaxel (Figure 4b). This trace-less linker strategy was tested in a chemical cleavage assay by incubating WT pHLIP-paclitaxel (4 μM) with glutathione (11 mM) in DPBS buffer (pH 7.6). Cargo release was monitored with HPLC, which showed that within 2 hr up to 90% of paclitaxel cargos were released as the free drug (Figure S4). In the growth inhibition assay, A549 cells were treated with WT or Asp14Gla/Asp25Aad pHLIP-paclitaxel (0.5 μM, 30 min) at the simulated tumor pH of 6.6-6.7 (pH measured before and after experiment) or the healthy tissue pH of 7.4. The results of these experiments are shown in Figure 4b. At pH 6.6, the double variant has decisive advantage over WT pHLIP in delivering paclitaxel into the cell, both in terms of absolute amount delivered (thus leading to more growth inhibition) and pH-dependence. Interestingly, even WT pHLIP-paclitaxel can inhibit cell growth as well as free drug but the pH-dependence is largely absent. On the other hand, Asp14Gla/Asp25Aad pHLIP-paclitaxel can inhibit cell growth to a greater extent than free drug (74% vs. 37% growth inhibition) at pH 6.6 and its anti-proliferative effects are very pH sensitive: 74% inhibition at pH 6.6 vs. 56% at pH 7.4.
In summary, to find pHLIP variants with improved insertion properties for drug delivery at tumor average pHe of 6.8, we carried out a SAR study in which the D14 and D25 residues critical for insertion were modified. Here we report the discovery of variants D25EE, Asp25Aad, and Asp14Gla/Asp25Aad, which insert with pH50 of 6.60, 6.74, and 6.79, respectively. These three variants insert into POPC membrane at higher pH than all previously known pHLIP peptides. Thus, they may insert more effectively at the tumor pHe range of 6.5-7.0, with the potential to deliver more drugs into the cytoplasm of cancer cells via plasmamembrane insertion. To reduce pHLIP-mediated drug delivery at neutral pH (which may lead to off-target effects in healthy tissues), insertion over a narrow pH range is desirable. To this end, we found that the Asp14Gla variant can insert into membrane with the sharpest pH-response observed so far for a pHLIP peptide — over half pH unit vs. one pH unit for WT. This Asp14Gla variant may improve pHLIP-based tumor imaging by reducing background signals. Thus, adding carboxyl group to the 14th position side-chain can sharpen the pH-response while lengthening the side-chain at position 25 can raise the pH50 of insertion. The Asp14Gla/Asp25Aad double variant shows that these effects are independent of each other and can be used additively to tune for desired pHLIP insertion properties. Further, the successful introduction of additional carboxyl groups in the TM region, as in D25EE and Asp14Gla, may also improve pHLIP solubility and reduce aggregation, which are parameters critical for future applications. In addition, we showed that the double variant Asp14Gla/Asp25Aad has considerble advantage over WT pHLIP in cargo delivery at true tumor pHe of 6.6, as demonstrated with turn-on fluorescence assays and anti-proliferation studies in A549 cells using paclitaxel as a model drug.
Experimental Section
Details of the sequences of pHLIP variants (and their characterization by mass spectrometry), synthetic derivatizations that led to D25C-2C and D25C-3C variants, capping of C-terminal Lys and Cys side-chains of D14E and D25E variants, syntheses of WT and Asp14Gla/Asp25Aad pHLIP turn-on fluorescence probes and pHLIP-paclitaxel conjugates, paclitaxel cargo release test, Trp fluorescence experiments, CD measurements, liposome preparation, cell culture (materials, instruments and cell line), turn-on fluorescence and anti-proliferation assays with A549 cells, can be found in the Supporting Information.
Supplementary Material
Footnotes
We thank Prof. Dr. Donald M. Engelman (Yale) for discussion and support; Prof. Dr. Eriks Rozners and Prof. Dr. Susan Bane (both of SUNY-Binghamton) for frequent use of their fluorimeter and plate reader, respectively. We also thank Raemer J. Lapid (for assistance in processing data), Rebecca A. Chandler, Meghan M. Bell, Vladyslav Nazarenko, Ilana G. Bandler (for assistance in cell assays) and Emma A. Gordon (for assistance in synthesis). This work was supported by SUNY-Binghamton University (BU) (Start-up funds to M.A. and L.Y., various financial supports to J.O., L.K. and M.C., HHMI-BU undergraduate summer fellowships to C-H. E. and R. L.), NIH (R01-GM073857 for training of M.A. and initial work), and NSF grant CHE-0922815 for the Regional NMR Facility (600 MHz instrument) at SUNY-Binghamton.
Contributor Information
Joab O. Onyango, Department of Chemistry, State University of New York (SUNY), Binghamton University P. O. Box 6000, Binghamton, NY 13902 (USA)
Michael S. Chung, Department of Chemistry, State University of New York (SUNY), Binghamton University P. O. Box 6000, Binghamton, NY 13902 (USA)
Chee-Huat Eng, Department of Chemistry, State University of New York (SUNY), Binghamton University P. O. Box 6000, Binghamton, NY 13902 (USA).
Lukas M. Klees, Department of Chemistry, State University of New York (SUNY), Binghamton University P. O. Box 6000, Binghamton, NY 13902 (USA)
Rachel Langenbacher, Department of Chemistry, State University of New York (SUNY), Binghamton University P. O. Box 6000, Binghamton, NY 13902 (USA).
Lan Yao, Department of Physics, Applied Physics and Astronomy State University of New York (SUNY), Binghamton University P. O. Box 6000, Binghamton, NY 13902 (USA).
Ming An, Department of Chemistry, State University of New York (SUNY), Binghamton University P. O. Box 6000, Binghamton, NY 13902 (USA).
References
- 1.a Senter PD, Sievers EL. Nat Biotechnol. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]; b Sievers EL, Senter PD. Annu Rev Med. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 2.Bae YH. J Control Release. 2009;133:2–3. doi: 10.1016/j.jconrel.2008.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 4.Wilson WR, Hay MP. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 5.a Gerweck LE, Seetharaman K. Cancer Res. 1996;56:1194–1198. [PubMed] [Google Scholar]; b Wike-Hooley JL, Haveman J, Reinhold HS. Radiother Oncol. 1984;2:343–366. doi: 10.1016/s0167-8140(84)80077-8. [DOI] [PubMed] [Google Scholar]
- 6.a Zhang X, Lin Y, Gillies RJ. J Nucl Med. 2010;51:1167–1170. doi: 10.2967/jnumed.109.068981. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bhujwalla ZM, Artemov D, Ballesteros P, Cerdan S, Gillies RJ, Solaiyappan M. NMR Biomed. 2002;15:114–119. doi: 10.1002/nbm.743. [DOI] [PubMed] [Google Scholar]
- 7.a Gatenby RA, Gillies RJ. Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]; b Tannock IF, Rotin D. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- 8.a Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF, Johnson J, Gatenby RA, Gillies RJ. Cancer Res. 2013;73:1524–1535. doi: 10.1158/0008-5472.CAN-12-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Robey IF, Baggett BK, Kirkpatrick ND, Roe DJ, Dosescu J, Sloane BF, Hashim AI, Morse DL, Raghunand N, Gatenby RA, Gillies RJ. Cancer Res. 2009;69:2260–2268. doi: 10.1158/0008-5472.CAN-07-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helmlinger G, Yuan F, Dellian M, Jain RK. Nat Med. 1997;3:177–182. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 10.a Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM. Proc Natl Acad Sci U S A. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hunt JF, Rath P, Rothschild KJ, Engelman DM. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]; c Reshetnyak YK, Segala M, Andreev OA, Engelman DM. Biophys J. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM. Proc Natl Acad Sci U S A. 2006;103:6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Thevenin D, An M, Engelman DM. Chem Biol. 2009;16:754–762. doi: 10.1016/j.chembiol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreev OA, Karabadzhak AG, Weerakkody D, Andreev GO, Engelman DM, Reshetnyak YK. Proc Natl Acad Sci U S A. 2010;107:4081–4086. doi: 10.1073/pnas.0914330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreev OA, Dupuy AD, Segala M, Sandugu S, Serra DA, Chichester CO, Engelman DM, Reshetnyak YK. Proc Natl Acad Sci U S A. 2007;104:7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adochite RC, Moshnikova A, Carlin SD, Guerrieri RA, Andreev OA, Lewis JS, Reshetnyak YK. Mol Pharm. 2014;11:2896–2905. doi: 10.1021/mp5002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoonens M, Reshetnyak YK, Engelman DM. Biophys J. 2008;95:225–235. doi: 10.1529/biophysj.107.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reshetnyak YK, Yao L, Zheng S, Kuznetsov S, Engelman DM, Andreev OA. Mol Imaging Biol. 2011;13:1146–1156. doi: 10.1007/s11307-010-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a Lee ES, Gao Z, Bae YH. J Control Release. 2008;132:164–170. doi: 10.1016/j.jconrel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hu J, Miura S, Na K, Bae YH. J Control Release. 2013;172:69–76. doi: 10.1016/j.jconrel.2013.08.007. [DOI] [PubMed] [Google Scholar]; c Lee ES, Gao Z, Kim D, Park K, Kwon IC, Bae YH. J Control Release. 2008;129:228–236. doi: 10.1016/j.jconrel.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a Du JZ, Du XJ, Mao CQ, Wang J. J Am Chem Soc. 2011;133:17560–17563. doi: 10.1021/ja207150n. [DOI] [PubMed] [Google Scholar]; b Du JZ, Sun TM, Song WJ, Wu J, Wang J. Angew Chem Int Ed Engl. 2010;49:3621–3626. doi: 10.1002/anie.200907210. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2010;122:3703–3708. [Google Scholar]; c Zhou ZX, Shen YQ, Tang JB, Fan MH, Van Kirk EA, Murdoch WJ, Radosz M. Advanced Functional Materials. 2009;19:3580–3589. [Google Scholar]; d Lee Y, Miyata K, Oba M, Ishii T, Fukushima S, Han M, Koyama H, Nishiyama N, Kataoka K. Angew Chem Int Ed Engl. 2008;47:5163–5166. doi: 10.1002/anie.200800963. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2008;120:5241–5244. [Google Scholar]; e Lee Y, Fukushima S, Bae Y, Hiki S, Ishii T, Kataoka K. J Am Chem Soc. 2007;129:5362–5363. doi: 10.1021/ja071090b. [DOI] [PubMed] [Google Scholar]
- 19.Musial-Siwek M, Karabadzhak A, Andreev OA, Reshetnyak YK, Engelman DM. Biochim Biophys Acta. 2010;1798:1041–1046. doi: 10.1016/j.bbamem.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a An M, Wijesinghe D, Andreev OA, Reshetnyak YK, Engelman DM. Proc Natl Acad Sci U S A. 2010;107:20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wijesinghe D, Engelman DM, Andreev OA, Reshetnyak YK. Biochemistry. 2011;50:10215–10222. doi: 10.1021/bi2009773. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Moshnikova A, Moshnikova V, Andreev OA, Reshetnyak YK. Biochemistry. 2013;52:1171–1178. doi: 10.1021/bi301647y. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Karabadzhak AG, An M, Yao L, Langenbacher R, Moshnikova A, Adochite RC, Andreev OA, Reshetnyak YK, Engelman DM. ACS Chem Biol. 2014;9:2545–2553. doi: 10.1021/cb500388m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies A, Lewis DJ, Watson SP, Thomas SG, Pikramenou Z. Proc Natl Acad Sci U S A. 2012;109:1862–1867. doi: 10.1073/pnas.1112132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z, Meng H, Wang N, Donovan MJ, Fu T, You M, Chen Z, Zhang X, Tan W. Angew Chem Int Ed Engl. 2013;52:7487–7491. doi: 10.1002/anie.201302557. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2013;125:7635–7639. [Google Scholar]
- 23.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, Saltzman WM, Slack FJ. Nature. 2014 doi: 10.1038/nature13905. doi: 10.1038/nature13905, published online 17 November 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fendos J, Barrera FN, Engelman DM. Biochemistry. 2013;52:4595–4604. doi: 10.1021/bi400252k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao L, Daniels J, Wijesinghe D, Andreev OA, Reshetnyak YK. J Control Release. 2013;167:228–237. doi: 10.1016/j.jconrel.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a Barrera FN, Fendos J, Engelman DM. Proc Natl Acad Sci U S A. 2012;109:14422–14427. doi: 10.1073/pnas.1212665109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Caputo GA, London E. Biochemistry. 2004;43:8794–8806. doi: 10.1021/bi049696p. [DOI] [PubMed] [Google Scholar]
- 27.Kingston DG. J Nat Prod. 2000;63:726–734. doi: 10.1021/np000064n. [DOI] [PubMed] [Google Scholar]
- 28.Dubikovskaya EA, Thorne SH, Pillow TH, Contag CH, Wender PA. Proc Natl Acad Sci U S A. 2008;105:12128–12133. doi: 10.1073/pnas.0805374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.