Abstract
The discovery of microbial expansins emerged from studies of the mechanism of plant cell growth and the molecular basis of plant cell wall extensibility. Expansins are wall-loosening proteins that are universal in the plant kingdom and are also found in a small set of phylogenetically diverse bacteria, fungi, and other organisms, most of which colonize plant surfaces. They loosen plant cell walls without detectable lytic activity. Bacterial expansins have attracted considerable attention recently for their potential use in cellulosic biomass conversion for biofuel production, as a means to disaggregate cellulosic structures by non-lytic means (‘amorphogenesis’). Evolutionary analysis indicates that microbial expansins originated by multiple horizontal gene transfers from plants. Crystallographic analysis of BsEXLX1, the expansin from Bacillus subtilis, shows that microbial expansins consist of two tightly-packed domains: the N-terminal domain D1 has a double-ψ β-barrel fold similar to glycosyl hydrolase family-45 enzymes, but lacks catalytic residues usually required for hydrolysis; the C-terminal domain D2 has a unique β-sandwich fold with three co-linear aromatic residues that bind β-1,4-glucans by hydrophobic interactions. Genetic deletion of expansin in Bacillus and Clavibacter cripples their ability to colonize plant tissues. We assess reports that expansin addition enhances cellulose breakdown by cellulase and compare expansins with distantly related proteins named swollenin, cerato-platanin and loosenin. We end in a speculative vein about the biological roles of microbial expansins and their potential applications. Advances in this field will be aided by a deeper understanding of how these proteins modify cellulosic structures.
Keywords: Amorphogenesis, Biofuels, Cellulase synergism, Expansin, Plant-microbe interactions, Swollenin
Introduction: Discovery of plant and bacterial expansins
Expansins were originally identified as plant cell wall loosening proteins isolated from growing cucumber and oat tissues (Cosgrove 2000; Li et al. 1993; McQueen-Mason et al. 1992). These proteins act as mediators of ‘acid growth’, which refers to the pH-dependent enlargement of plant cells (Rayle and Cleland 1992). Expansins bind to plant cell walls and induce stress relaxation and creep of primary cell walls, but without detectable hydrolytic or other enzymatic modification of the cell walls (McQueen-Mason and Cosgrove 1994; McQueen-Mason and Cosgrove 1995). Cloning of cDNAs for the cucumber proteins eventually led to the discovery that expansins are encoded by two major gene families in plant genomes (Sampedro and Cosgrove 2005; Shcherban et al. 1995). The first group is now called α-expansin, designated EXPA, whereas the second group is called β-expansin, designed EXPB.
Plant expansins function in cell growth and in other developmental processes that involve wall loosening, such as fruit softening, abscission, seed germination and pollen tube invasion of the stigma (Cosgrove 2000). Plant genomes also contain one or more expansin-like genes named EXLA and EXLB with uncertain function. A recent study showed that ectopic expression of EXLA2 in Arabidopsis increased root and hypocotyl lengths by 10–20% (Boron et al. 2014), so these proteins may have a wall loosening function as well, although the evidence is indirect at this point. Plant expansins accumulate to very low protein levels in cell walls of growing tissues (approximately 1:5,000; protein:wall on a dry mass basis), with the sole known exception being grass pollen where they accumulate to exceptionally high levels (Cosgrove et al. 1997; Li et al. 2003; Sampedro et al. 2015). These pollen proteins are a subset of divergent β-expansins and are also known as group-1 grass pollen allergens in the immunology field. They are potent elicitors of hay fever and seasonal asthma in a large sector of the human population (Chabre et al. 2010). The loosening action of the pollen β-expansins is selective for grass cell walls, with a selectivity towards the polymeric glue (the middle lamella) that bonds plant cell walls together (Sampedro et al. 2015; Tabuchi et al. 2011). In addition to causing cell wall creep, these proteins solubilize the middle lamella and facilitate cell separation, thereby aiding invasion of the pollen tube through the maternal tissues of grass tissues. Yet, remarkably, they have little or no loosening effect on dicot cell walls. The activity of other β-expansins is likely to differ from the pollen proteins, but this has not yet been assessed experimentally (Sampedro et al. 2015).
A weak resemblance of plant expansins to certain microbial endoglucanases and to cellulose binding domains was noted early on, based on distant sequence similarity (Cosgrove 1996; Jahr et al. 2000; Laine et al. 2000; Li et al. 2002; Saloheimo et al. 2002; Sampedro and Cosgrove 2005; Shcherban et al. 1995). This similarity was firmly established by the crystallographic structure of ZmEXPB1, a maize pollen β-expansin, which revealed the two-domain structure characteristic of all expansins (Yennawar et al. 2006). Domain D1 resembles family-45 glycosyl hydrolases (GH45; www.cazy.org), which also have a double-ψ β-barrel fold and are found in a wide range of microbes. The expansin D1 domain characteristically preserves some features of the GH45 catalytic cite, but lacks a residue corresponding to the catalytic base that is normally required for GH45 hydrolytic activity (we return to the topic of enzymatic activity later in this review). Domain D2 has a β-sandwich fold, resembling some carbohydrate binding modules (CBMs), but with a distinctive β-strand folding pattern. CBMs are often linked to catalytic domains of polysaccharide hydrolases and lyases by a flexible linker, potentiating their activity by targeting and proximity effects (Boraston et al. 2004; Gilbert 2010; Herve et al. 2010).
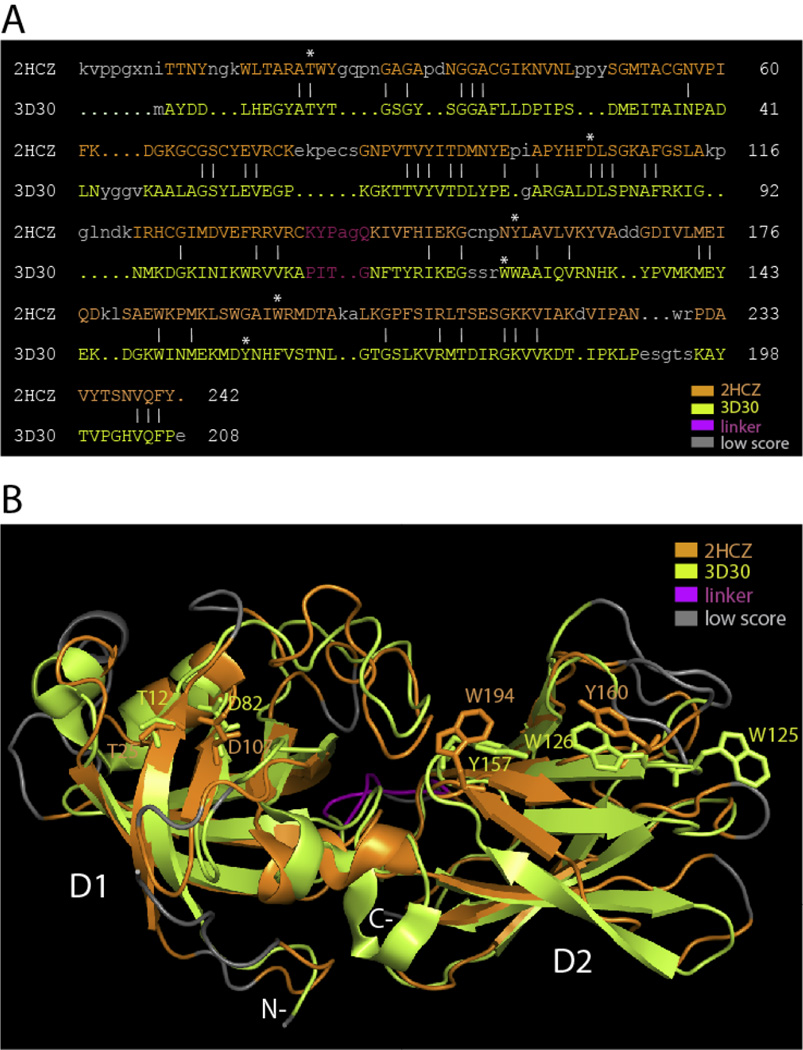
The existence of expansin homologs (i.e. proteins with both domains, not only the GH45-like domain) in bacteria became unequivocal with the crystallographic structure of the YOAJ protein (renamed BsEXLX1) from Bacillus subtilis (Kerff et al. 2008). B. subtilis is a soil organism that in nature colonizes plant roots (Beauregard et al. 2013) and is also used in agriculture as a biocontrol agent to reduce fungal diseases (Ashwini and Srividya 2013). BsEXLX1 has the same two-domain structure as plant expansins with identical folds for each domain (Figure 1). Moreover, BsEXLX1 can induce creep in plant cell walls, thus possessing both the characteristic activity of plant expansins and a homologous structure.
Figure 1.
Structure-based sequence alignment and structure superposition of BsEXLX1 (PDB ID: 3D30; light green color) with maize β-expansin (PDB ID: 2HCZ; orange color). A: The alignment was made with DaliLite (http://www.ebi.ac.uk/Tools/structure/dalilite/); upper case letters indicate close alignment of the protein backbones (structurally equivalent residues); identical residues are marked by vertical bars); lower case letters indicate poor alignment of protein backbones (low score alignments of structurally non-equivalent residues). B: Corresponding superposition of protein structures shows they have the same two-domain structure (D1 and D2), with each domain folded in the same way. The linker is shown in magenta and low score alignments are in grey. Key amino acid residues required for wall loosening activity are mapped on the structures in stick representation.
The discovery that BsEXLX1 has expansin activity has proved to be a major watershed for studies of how expansins work. This is so because bacterial expansins are readily expressed in E. coli whereas plant expansins have proved remarkably recalcitrant to heterologous expression in active form. As a result, several experimental approaches that require heterologous protein expression became feasible for analyzing expansin function. The ease of BsEXLX1 expression in E.coli has facilitated the detailed study of its activity and binding characteristics. Consequently, BsEXLX1 has served as a model for understanding structure-function relationships of microbial expansins and, by extension, plant expansins. In the following sections we summarize the structure, activities and evolution of bacterial expansins before assessing the potential of bacterial expansins for biotechnological applications, particularly in the biofuel field. We defer discussion of some expansin-related proteins named swollenin and cerato-platanin until the end.
Classification and nomenclature of expansins
The definition of expansins and the convention for naming them was settled a decade ago (Kende et al. 2004). The terms ‘expansin’ and ‘expansin-like’ were reserved for proteins containing the two-domain structure characteristic of plant expansins. They typically are made of ~225 amino acids (~26 kD) plus an N-terminal signal peptide that directs the protein to the secretory pathway. Four families of expansins and expansin-like genes and their protein products were recognized in plants at that time (designated EXPA, EXPB, EXLA, and EXLB) based on sequence-based phylogenetic analysis, and this grouping has not changed, although the number of sequenced genes falling into these families has increased enormously. The designation EXLX was reserved for genes that fulfilled three criteria: (i) they occur in non-plant organisms; (ii) their protein products are homologous to both expansin domains; and (iii) they do not fall within the established expansin gene families by phylogenetic analysis. In cases where multiple expansin genes are found in a genome, they would be numbered consecutively EXLX1, EXLX2, and so on, in order of discovery. An abbreviation for the species name may be prepended when needed for clarity, e.g. BsEXLX1 for the expansin from Bacillus subtilis. In most microbes examined to date, only a single expansin gene is found, but there are exceptions: the slime mold Dictyostelium discoideum has multiple expansin genes (Darley et al. 2003) and the actinomycete Clavibacter michiganensis has an expansin gene found on the main chromosome and an expansin as part of a plasmid-borne modular endoglucanase (Gartemann et al. 2003; Nikolaidis et al. 2014). Because the plasmid-borne expansin was studied first (Gartemann et al. 2003; Georgelis et al. 2014; Jahr et al. 2000; Laine et al. 2000), it is designated CmEXLX1, while the nuclear gene should be designated CmEXLX2. Adherence to this naming convention will help avoid massive naming confusion, as found in the cellulase field prior to its nomenclature standardization (Henrissat et al. 1998) and establishment of the CAZY database (WWW.CAZY.ORG). The EXLX name is a catch-all polyphyletic grouping; at some point in the future the evolutionary relationships of the non-plant sequences may be well-enough worked out and accepted to begin subdividing it into distinctive evolutionary families, but large sequence divergence and the nature of the transmission of expansins in the microbial world present difficulties for this scheme, as described below.
Phylogenetic distribution and evolution of bacterial and other microbial expansins
A recent report surveyed all the available prokaryotic and eukaryotic genomes and identified numerous sequences from a diverse set of bacteria, fungi, and amoebozoa that align to both expansin domains (Nikolaidis et al. 2014). This study expands on an earlier phylogenetic analysis based on a more limited database and simpler analytical tools (Li et al. 2002). Sequence alignments, phylogenetic tools, as well as protein domain and fold recognition programs confirmed that all identified proteins contain both expansin domains and showed that the predicted protein structures resemble both B. subtilis EXLX1 and plant expansins. Modeling of bacterial expansins and surface charge properties by Pastor et al. (2014) likewise support structural homology between plant and bacterial expansins. This study identified three clades among the bacterial expansins. Clades normally imply an evolutionary descent from a common ancestor but such an interpretation in this case is complicated by large divergence of the protein sequences, low bootstrap support of many branches, and evidence of horizontal gene transfer (Nikolaidis et al. 2014).
The argument that bacterial and plant expansins are homologous, that is, arose from common descent rather than by independent sequence convergence, is based on their shared structural features and activity. These include the spatial arrangement of two tightly packed domains, each of which has the same fold in the plant and bacterial proteins, the same missing catalytic residue in domain D1 of the plant and bacterial proteins, and the existence of a conserved, open surface that spans the two domains and is populated by residues suitable for binding a long polysaccharide chain (up to 12 residues). Support for homology also comes from the similar and unusual ability to induce plant cell wall creep without lytic activity. Moreover, although many classes of CBM have a β-sandwich fold, the specific β-strand arrangement of D2 appears to be unique and is not shared with other CBMs (Carvalho et al. 2014). Shoseyov et al. (2006) reported that expansin D2 resembles the fold of CBM family 3, but our structural comparison as well as that of Carvalho et al. (2014) indicate that D2 has a distinct β-strand arrangement, which makes homology with CBM3 unlikely. Expansin domain D2 is now classified as CBM family 63 in the CAZY.ORG database (Georgelis et al. 2012). The case for convergent evolution of plant and bacterial expansins would require the independent loss of the same catalytic residues in a GH45 enzyme, the independent creation of a uniquely folded β-sandwich, the fusion of the two proteins in the same geometry to form a similar binding surface, and the independent development of cell wall loosening activity that is unique to expansins. The probability of such multiple independent events to create expansin proteins by convergence is astronomically small.
While the foregoing observations indicate that microbial and plant expansins are homologous, transmission among the diverse phylogenetic groups may have occurred by vertical or horizontal transmission or a combination of the two mechanisms. Nikolaidis et al. (2014) concluded that expansins most likely originated in the ancestor of plants (all of which contain expansins genes) and subsequently were transferred to microbes that colonize or digest plant tissues. Five lines of evidence support this concept. First, expansin proteins are sparsely and diversely distributed in bacteria (3% of surveyed genomes contained a putative expansin), fungi (5% of fungal genomes in GenBank include a putative expansin), and amoebozoa. Second, microbes with expansin genes are mostly plant pathogens, soil inhabitants, or cellulose producers, but their close relatives with other life styles typically lack expansins. Third, the phylogenetic tree derived from microbial expansins does not follow the established bacterial or fungal species trees. Fourth, microbial expansins are found in some cases to be part of modular enzymes containing GH5 cellulases and carbohydrate-binding domains, apparently arising from multiple independent fusion events. Fifth, the microbial and plant expansins share many functional and structural similarities, but cell wall creep activity is comparatively weak in the microbial homologs tested to date (Georgelis et al. 2014). The relatively low wall loosening activity of bacterial expansins may be important as a means to avoid tripping plant cell wall integrity sensors and triggering plant defense (Wolf et al. 2012).
Although, horizontal gene transfer (HGT) from plants is the likeliest origin of microbial expansins, the number and the timing of HGT events as well as the donor and recipient species remain obscure. The HGT events were probably quite ancient, based on the lack of differences in GC-content, codon usage, and other genomic features in the BsEXLX1 homologs compared with other genes in the respective genomes (Nikolaidis et al. 2014). This is the case even in the emerging plant pathogen, the actinomycete Streptomyces acidiscabies, which has an expansin sequence (Genbank accession WP_010353361) that rather closely resembles plant expansins, suggesting a relatively recent HGT. Another expansin sequence from the genome of the actinomycete Kutzneria sp. 744 (Genbank accession EWM10128) appears to be a second candidate for a relatively recent HGT event. Nikolaidis et al. (2014) proposed that microbial expansins originated from multiple HGTs from plants and that these initial events were followed by further vertical and horizontal transfers to selective bacterial, fungal and amoebozoan species. HGTs of expansins appear to be ongoing.
In addition to HGTs of expansins from plants to microbes, the phylogenetic survey revealed several instances where an expansin was fused with sequences coding for GH5 endoglucanases and various CBMs. Modular cellulases with multiple domains are common in microbes, particularly fungi. The GH5 fusions with expansin appear to have occurred multiple times independently, apparent cases of convergent evolution (Georgelis et al. 2014; Nikolaidis et al. 2014). It is curious that such fusions only involve GH5 enzymes, and not other families of cellulases. We note that some GH5 enzymes have the ability to loosen key sites of biomechanical importance in plant cell walls via their hydrolytic activity (Cosgrove 2014; Park and Cosgrove 2012). The fusion with expansin may confer additional selectivity or activity to the modular GH5 enzymes; however, in an analysis of the modular GH5 enzyme from Clavibacter the deletion of the expansin did not reduce the hydrolysis of cellulose, whereas deletion of both the CBM and expansin substantially reduced hydrolytic activity (Georgelis et al. 2014). Thus, the function of expansin in this modular protein is unclear. Fusion of expansin modules with other modules has not been seen in plants.
Microbial expansins are found in the genomes of several phylogenetically-diverse plant pathogenic bacteria, including species of Xanthomonas, Clavibacter, Streptomyces, and Xylella, but are not found in Pseudomonas, Agrobacterium, or Rhizobium. The first group, known as vascular wilts, are primarily xylem dwellers within vascular systems of plants, whereas the second group have different modes of interaction with the plant hosts. It is possible that expansins confer a particular advantage to xylem-dwelling bacteria. Fungal genomes with expansin sequences include plant pathogenic species of Magnaporthe, Gibberella, Fusarium, Schlerotina, Puccinia, and other genera (Georgelis et al. 2014; Nikolaidis et al. 2014). Expansin-like sequences have been reported from the white-rot fungus Phanerochaete carnosa (Suzuki et al. 2014), but our inspection of the sequences indicate they so divergent from expansins that it is unclear how expansin-like they really are. They are more closely related to a protein named ‘loosenin’.
The presence of microbial expansins in organisms that colonize or digest plant tissues or residues in the soil suggests that expansins may confer an advantage for certain modes of interaction with plants or plant residues. This concept is supported by the results of expansin gene deletions in B. subtilis (Kerff et al. 2008), Clavibacter michiganensis (Jahr et al. 2000) and the saprophytic fungus Trichoderma reesei (Brotman et al. 2008). In all three cases, mutants lacking expansin genes were less able to colonize or infect plants than the wild-type strain. These findings suggest that microbial expansins bind to and possibly modify plant cell walls in the course of plant infection. These inferences gain support from studies showing that bacterial expansins bind to cellulose in vitro (Georgelis et al. 2014; Georgelis et al. 2011; Georgelis et al. 2012; Kim et al. 2013a; Kim et al. 2013b; Lee et al. 2010; Olarte-Lozano et al. 2014). In species not known to interact with plants, these proteins may have been adopted for other cellular roles, as in Aspergillus nidulans (Bouzarelou et al. 2008) and Dictyostelium (Ogasawara et al. 2009). The absence of expansins from Pseudomonas, Agrobacterium and Rhizobium suggests that expansins confer an advantage only with selective modes of plant infection and colonization.
In addition to microbial homologs of plant expansins, there are some examples of proteins with expansin-like activity that likely evolved independently, although the state of our knowledge on this point is too sketchy to be certain. One example would be the fungal proteins known as cerato-platanins which may have evolved from an ancestral protein with a GH45 fold (see below). Moreover, a nematode protein that has been called an expansin (Kikuchi et al. 2009; Lee et al. 2012; Qin et al. 2004) is likely an example of independent evolution; it has a family-2 CBM at its N-terminus and a GH45-like domain at its C-terminus, a structure indicative of separate evolutionary origin, not consistent with canonical expansins (Nikolaidis et al. 2014).
Structure-function analysis of bacterial expansins and their interactions with β1,4-D-glucans
Crystallographic structures
Alignments between BsEXLX1 and plant expansin protein sequences show statistically significant alignment, but with multiple short gaps and overall low identity throughout the length of the protein (~12% or less, depending on the proteins selected) (Fig. 1). Despite the low sequence identity, the crystallographic structures can be superimposed with very low divergence (low root mean square distance between the backbone carbons), indicating congruent protein structure (Kerff et al. 2008). Both plant and bacterial expansins consist of two tightly-packed domains with an open surface suitable for docking a polysaccharide chain that could span the two domains.
Crystallographic structures have also been obtained for BsEXLX1 in complex with various cellulose-like β1,4-D-glucan oligosaccharides (Georgelis et al. 2012). Co-crystallization with cellohexaose (a six-residue oligosaccharide of cellulose) showed an uncommon sandwich-like arrangement, with one cellohexaose packed between the D2 domains of two proteins (Figure 2A). This sandwich structure was obtained with several cellulose-like ligands and was also obtained with another expansin from Clavibacter michiganensis (PDB numbers 4JS7, 4L48, 4JCW, and unpublished data of N. Yennawar, H. Yennawar, N. Georgelis and D.J. Cosgrove), indicating that this sandwich configuration is a favorable packing arrangement for expansin-oligosaccharide complexes. It is made possible by the open binding surface of D2; most other proteins that bind polysaccharides make us of a groove or cleft that facilitates protein-polysaccharide interaction, but sterically prevents the sandwich-like dimerization seen with expansins. Dynamic light scattering showed that in the presence of cellohexaose BsEXLX1 is a monomer in solution, so the dimer structure is most likely the result of packing during the crystallization process and may not be a stable structure found under biological conditions. The lack of ligand-mediated dimerization in solution is also consistent with the weak affinity (>1 mM) of BsEXLX1 for cellohexaose, as evaluated by isothermal titration calorimetry (Georgelis et al. 2012). BsEXLX1 binds more strongly to cellulose, with a Kd ~1 µM. Despite the weak cellohexaose-protein interaction, the structure has provided valuable insights into the nature of BsEXLX1 binding to cellulose.
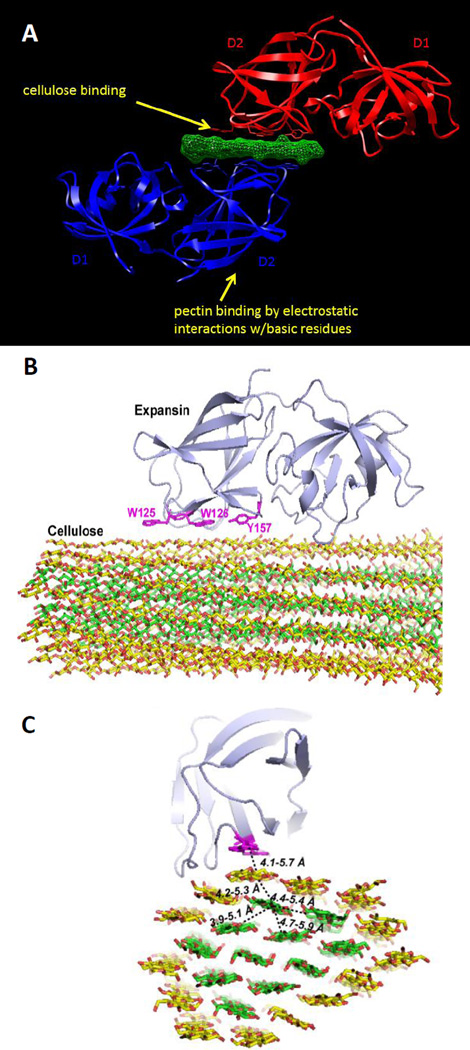
Figure 2.
Structure of BsEXLX1 bound to cellohexaose (A) and to a cellulose microfibril (B,C). A: In the crystallographic structure based on protein:ligand crystals, a single cellohexaose (green) is sandwiched between the D2 domains of two BsEXLX1 proteins (red, blue). The regions that control binding to cellulose and to pectins are located on opposite surface of the D2 domain. B and C show BsEXLX1 docked onto the surface of a cellulose microfibril (two views). This is a computational result that is based on molecular dynamic simulation and is validated by NMR evidence of interactions between the protein and internal glucan chains. Image credits: B and C from Wang et al. 2013, copyright PNAS, used with permission.
As seen in solved crystallographic structures of protein-ligand complexes, cellohexaose binds to BsEXLX1 primarily through hydrophobic CH-π interactions with three linearly arranged aromatic residues (W125, W126, Y157) in domain D2. The two proteins in the sandwich complex bind to opposite faces of alternating glucose residues in the glucan, consistently with the more hydrophobic of the two faces of the glucose ring, i.e. the face with three, rather than two, axial CH groups extending from the surface. It may be significant that there is a slight right-handed twist in the surface formed by W125, W126 and Y157 that is mirrored in the twist of cellohexaose (Georgelis et al. 2012). The slight twist of the protein surface suggests that EXLX1 may have a preference to bind to cellulose surfaces with a similar twist. Computational studies indicate that cellulose microfibrils develop a right-handed twist (Zhao et al. 2013), but it is not as steep as that seen in the expansin-ligand crystals. Hence BsEXLX1 may selectively bind to a microfibril site with a more accentuated twist than is generally the case. This would also be consistent with the relatively low density of binding sites estimated from binding isotherms (see below). Einding may also impart some torque on the surface glucan chain in an otherwise relatively straight microfibril. The only direct H-bonds are formed with amino acid residue K119. Similar interactions were seen with other β1,4-D-glucans such as cellotetraose, hemithiocellodextrin and oligosaccharides from mixed-linked β1,3;1,4-D-glucan. The dearth of direct H-bonds contrasts with so-called type-B CBMs where direct H bonds play a vital role in the protein:oligosaccharide interaction (Boraston et al. 2004). Consequently BsEXLX1:cellulose binding is largely an entropy-driven interaction, in contrast to the dominantly enthalpy-driven interaction of type-B CBMs with cellulose. In the crystal complexes, domain D1 did not make contact with cellohexaose or with other longer cellulose-like oligosaccharides, indicating negligible affinity of D1 with oligosaccharides. This behavior differs from bona fide GH45 enzymes where cello-oligosaccharides bind to the GH45 domain (Davies et al. 1995), although with less avidity than seen in enzymes with CBM domains (Eriksson et al. 2005).
In its natural context BsEXLX1 binds cellulose to loosen cell walls (see below). Crystallographic analysis of this interaction is not feasible because of the size and irregularity of cellulose microfibrils, but such interactions can be assessed by molecular dynamics simulations (Figure 2B,C). This approach shows direct binding of aromatic residues in the D2 domain to glucose rings exposed on the edge of the hydrophobic surface of cellulose microfibrils. Such binding brings the aromatic residues close enough to the internal residues of the microfibril to allow nuclear magnetic spin diffusion, which was verified by experimental means (Wang et al. 2013). The results also show that the D1 domain has little or no interaction with the cellulose surface, which is supported by in-vitro binding results.
Binding
Consistent with the crystallographic results described above, in-vitro binding experiments show that domain D2 is the dominant determinant of BsEXLX1 binding, but the binding to whole plant cell walls is rather complex and involves different surfaces for binding to cellulose and to acidic polysaccharides (Georgelis et al. 2011). Tests of individual D1 and D2 domains showed that D1 binding to cellulose and to plant cell walls was too low to be measured, whereas binding of D2 could account for the binding of the BsEXLX1 protein. Binding of BsEXLX1 to cellulose has an apparent Kd of ~1 µM (Georgelis et al. 2011; Kim et al. 2013a) and similar values were reported for other bacterial expansins (Bunterngsook et al. 2015; Olarte-Lozano et al. 2014). In contrast, native α-expansin from cucumber binds more tightly (McQueen-Mason and Cosgrove 1995), with a Kd of ~0.2 µM (this value is based on a recalculation of the published data). The structural basis for the higher affinity of α-expansin is not known because we do not have the structural insights and the ability to manipulate protein structure that has been possible with BsEXLX1 and other bacterial expansins. The higher affinity to cellulose may contribute to the stronger activity of α-expansins in wall extension assays. An attempt to express a functional α-expansin D2 domain in E. coli was reported by Nardi et al. (2013). However, the binding characteristics of the recombinant protein were anomalous: binding curves showed no evidence of saturation and the reported affinity for cellulose was 1000X lower than that of native α-expansin. It is possible that the recombinant protein was incorrectly folded and consequently dysfunctional (although this was not the conclusion of the authors). In our lab similar attempts at recombinant expression of plant expansins have likewise resulted in dysfunctional protein. Thus while insights gained from study of BsEXLX1 are a good basis for understanding structure-function relations of expansins, exploration of the distinctive properties of plant expansins awaits an effective means for altering the plant proteins and assessing changes in binding and activity.
Whole plant walls bound ~60X more BsEXLX1 than did cellulose (Avicel), when assessed at saturation on a dry mass basis (Georgelis et al. 2011). Some of this binding difference may be due to the difference in surface area in these two rather different binding materials, but much of the greater binding to whole plant walls was the result of electrostatic interactions between the positive charges on the protein (pI=9) and the negative charges in the cell wall from pectins and glucuronoarabinoxylan. Extractions to remove the matrix polysaccharides greatly reduced BsEXLX1 binding to the remaining cell wall material. Likewise CaCl2 strongly reduced BsEXLX1 binding to cell walls. These results indicate that the majority of BsEXLX1 binding is electrostatic binding to non-cellulosic components of the wall. However, productive binding (in terms of wall loosening activity) was correlated with binding to cellulose, not to the acidic polysaccharides in the cell wall matrix. This was demonstrated more clearly by site-directed mutagenesis to change the cellohexaose-binding residues W125, W126 and Y157 to alanine. The triple mutant, named ‘WWY’, showed greatly diminished binding to cellulose and lost its wall loosening activity, yet total binding to plant walls was hardly affected, evidently because of nonproductive binding to the acidic matrix polysaccharides. Conversely, mutation of basic residues on the ‘opposite’ face of BsEXLX1 domain D2 reduced binding to plant walls to very low levels, but did not affect binding to cellulose. For example the specific mutant named ‘RKK’ (for three stacked mutations R173Q, K180Q, K183Q) had two-fold greater wall loosening activity than wild type, yet its binding to whole walls was reduced to a level too low to quantify. This is additional evidence of the unproductive nature of binding to acidic components of the cell wall. These mutations altered the partitioning of BsEXLX1 within the cell wall, increasing or decreasing the interaction with a target containing cellulose.
The target site for wall loosening by BsEXLX1 within complex primary cell walls was further characterized by advanced solid-state nuclear magnetic resonance (ssNMR). Wang et al. (2013) developed a novel experimental design to measure the 13C shift of wall polymers within spin diffusion distance of expansin proteins. Their approach was to mix 15N, 13C-labeled protein with 13C-labeled plant cell walls and to apply a series of pulse sequences to drive proton spin transfer to 15N (in the protein), then through-bond spin transfer to adjacent 13C atoms (likewise in the protein), and finally through-space transfer to 13C atoms in the neighborhood of the protein. One technical challenge with this approach is that the concentration of expansin binding sites in the wall is so low that ssNMR signals would be too low to detect by standard methods. This limitation was overcome by use of a technique called dynamic nuclear polarization to enhance the sensitivity of ssNMR detection by ~25-fold (Maly et al. 2012; Takahashi et al. 2013). The results showed that BsEXLX1 bound to a rather specific site within the complex wall that contains cellulose, but with a slightly different structure from bulk cellulose. In addition, xyloglucan was in close proximity to the protein-binding site. This site resembles the biomechanical ‘hotspots’ that are loosened by bifunctional endoglucanases able to hydrolyze both cellulose and xyloglucan (Cosgrove 2014; Park and Cosgrove 2012). The ssNMR analysis also directly showed that protein interaction with cellulose within the cell wall was diminished in the WWY mutant but enhanced in the mutant that lacked pectin binding.
These studies established BsEXLX1 domain D2 as the founding member of family-63 CBM (WWW.CAZY.ORG). CBMs that bind to cellulose are divided into two classes: type-A bind to crystalline cellulose whereas type-B bind to the less organized areas of cellulose (Boraston et al. 2004). Competitive binding experiments showed that type-A CBMs compete with BsEXLX1 binding, supporting the conclusion from the crystallographic analysis that domain D2 of BsEXLX1 binds to the hydrophobic face of crystalline cellulose (Georgelis et al. 2012). This also supports the conclusion that type-A CBMs bind to the hydrophobic face of cellulose microfibrils, a conclusion based previously on inference from microscopic studies (Lehtio et al. 2003). BsEXLX1 binds to 5–10-fold fewer cellulose sites than do other well characterized type-A CBMs, indicating that BsEXLX1 does not cover the whole hydrophobic face of the cellulose microfibrils but it binds to selective areas of the microfibril (Georgelis et al. 2012). These sites might be junctions between crystalline and less organized cellulose, similar to the proposed binding sites of endoglucanase CtCel124, which has separate binding sites for crystalline and less organized cellulose (Bras et al. 2011). These cellulose target sites are likely to be relatively rare, judging from the low extent of binding when nonspecific electrostatic interactions are minimized. Figure 3 shows a plant cell wall surface decorated with a representative density of markers for expansin binding sites. It is obvious from this image that expansins bind only a small fraction of microfibril surface, even when most matrix polysaccharides have been removed (McQueen-Mason and Cosgrove 1995; Olarte-Lozano et al. 2014).
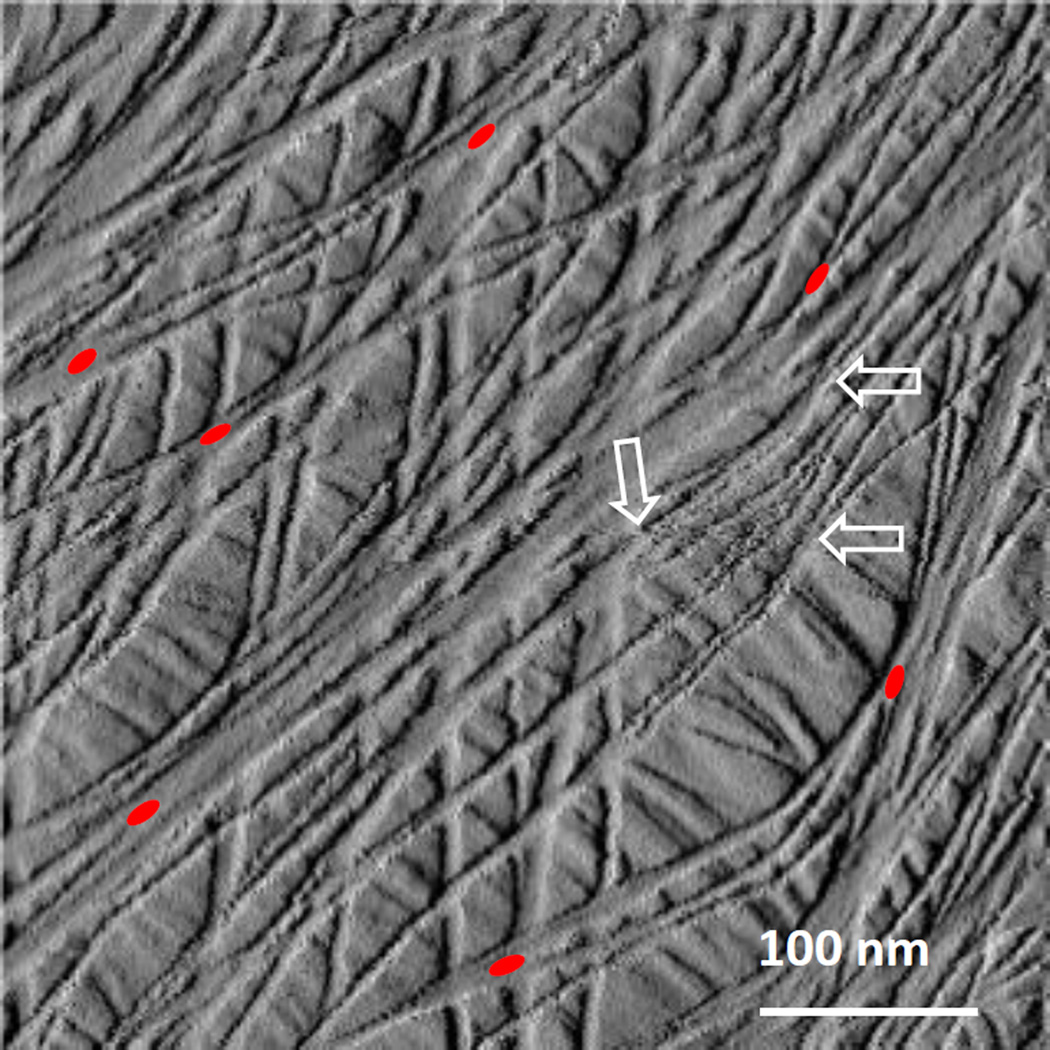
Figure 3.
Graphical representation of the density of expansin binding sites in the surface lamella of a plant cell wall. The image is an atomic force micrograph of a primary cell wall from onion scale, showing single cellulose microfibrils and microfibril bundles (arrows), decorated with red ellipses representing the density, but not the actual locations, of expansin binding sites in the surface lamella. This graphic was produced by transforming the binding isotherms of McQueen-Mason and Cosgrove (1995) into the maximum number of binding sites per µm of cellulose microfibril, assuming an 18-chain microfibril (Cosgrove, 2014). The density of microfibrils in the surface lamella was based on the AFM image. The image illustrates that expansins bind to limited sites on cellulose microfibrils. Micrograph adapted from Zhang et al. 2014, copyright Springer Verlag, used with permission.
The thermodynamics of BsEXLX1 binding to cellulose were also studied by isothermal titration calorimetry (ITC) (Georgelis et al. 2012). Mixing of cellulose with BsEXLX1 did not release or absorb detectable amounts of heat, indicating that BsEXLX1 binding to cellulose is mostly driven by changes in entropy rather than enthalpy. According to one interpretation, BsEXLX1 binding displaces structured water molecules on the protein surface and on the hydrophobic surface of cellulose, increasing the entropy of the system. ITC additionally showed that BsEXLX1 binds to soluble cello-oligosaccharides with low affinity, but the enthalpy change was too low to obtain accurate binding parameters (Georgelis et al. 2012). Tight binding to crystalline cellulose and weak binding to soluble cello-oligosaccharides is a characteristic feature of type-A CBMs and was also evident in the BsEXLX1 binding studies of Kim et al. (2013a) who additionally reported that binding to Avicel was increased >4-fold by 1 M NaCl, which is consistent with a dominantly hydrophobic mode of binding. Likewise consistent with hydrophobic binding, Lin et al. (2013) reported that BsEXLX1 binding to pretreated lignocellulosic materials might be greater at 30°C than 4°C. However, the exact temperature dependencies are obscured by the way the data were presented, so this point needs further study.
Enthalpy changes were not seen upon mixing of BsEXLX1 with soluble oligosaccharides and polysaccharides of the plant cell wall matrix, including xyloglucan hepta-saccharide, xylohexaose, mannohexaose, arabinan, galactan, arabinoxylan, and xyloglucan. These results suggest that these neutral matrix polysaccharides do not avidly bind to BsEXLX1, at least in their soluble form (Georgelis et al. 2012). However, binding to pectic oligo-uronides did release heat, presumably as a result of the dominantly polar interactions between the basic protein and the acid pectins (N. Georgelis and D.J. Cosgrove, unpublished).
BsEXLX1 is a basic protein (pI ~9), as are most of the plant expansins. Pastor et al. (2014) calculated the pI for a selection of 44 phylogenetically diverse bacterial expansins and found that half were acidic proteins (pIs of ~5), nearly half were basic proteins (pIs of ~9) and ~10% had neutral pIs. Protein pI generally correlated with the type of bacterial cell wall of the species, with some exceptions. Basic expansins were predominant in Gram-positive species (with a thick peptidoglycan wall external to the plasma membrane), whereas acidic expansins were mostly found in Gram-negative bacteria, which have a thin peptidoglycan layer between the plasma membrane and an outer cell membrane. The significance of this pattern is unclear. Perhaps it is related to delivery of expansins from bacterial surfaces to plant cell walls, but this process has not yet been studied. It is also possible that the surface charge facilitates expansin targeting to specific sites within the cell wall. The charged residues that contribute strongly to pI are not conserved residues, so electrostatic binding may be dominated by surface charge rather than specific binding interactions.
Olarte-Lozano et al. (2014) found that an acidic expansin from Pectobacterium carotovorum, named PcExl1, bound less to various plant cell walls compared with BsEXLX1. Part of the reason is likely to be the charge difference between these two proteins: at the normal pH of the cell wall (~5–6), PcExl1 has low net charge whereas BsEXLX1 carries a highly positive charge. As a consequence, celery walls, known to be rich in acidic pectins (Thimm et al. 2002), bound >10-fold more BsEXLX1 than PcExl1, as expected based on charge interactions. However, additional binding interactions may be involved because the binding to cucumber and bean cell walls, which are also pectin rich, was nearly the same for the two expansins, while wheat coleoptile cell wall, which contains only ~5% pectin and should therefore have a relatively low charge density, bound >3X more BsEXLX1 than PcExl1. Olarte-Lozano et al. (2014) also found that PcExl1 binding increased as matrix polysaccharides were extracted from wheat cell walls prior to expansin binding. This is consistent with PcExl1 selectively binding to cellulose rather than the matrix polysaccharides. In contrast BsEXL1 binding to coleoptile walls was strongly reduced as the matrix was removed, a consequence of the strong binding interaction based on charge (Georgelis et al. 2011). Kim et al. (2013b) also reported that BsEXLX1 bound to lignin-rich substrates, but the mechanism of binding to lignin (electrostatic or hydrophobic) was not elucidated. Mutants with reduced binding to cellulose or pectins would be useful tools to elucidate this question.
As mentioned above, the relationship between binding and wall-loosening activity was also explored by mutagenesis of BsEXLX1 residues that bind to cellulose and to acidic polysaccharides (Georgelis et al. 2011). Cellulose binding was essential for a productive interaction (wall loosening) whereas the electrostatic interactions were nonproductive. Pastor et al. (2014) speculated that electrostatic gradients on the expansin surface may facilitate its wall loosening action by promoting reorientation of H-bonds in cellulose. This idea remains to be carefully tested, but the altered activities of ‘RKK’ and other mutants seem difficult to explain by this theory.
Assays for activity
Because no enzymatic activity has been identified for expansins, their activity is typically measured by biomechanical assays. One such assay measures expansin’s effect on the strength of filter paper (a mat of pure cellulose fibers). A strip of paper is soaked in a solution before it is clamped and stretched to the breaking point (Georgelis et al. 2014; Georgelis et al. 2011; Kim et al. 2009; Olarte-Lozano et al. 2014). An early variant of this assay showed that plant α-expansins weakened filter paper (McQueen-Mason and Cosgrove 1994). The method has the advantage of substrate purity, which simplifies conclusions about expansin action, but it lacks sensitivity and dynamic range, and it is possible that contaminating cellulases or other proteins may interfere with the assay by weakening the paper. A second assay measures the creep activity with isolated plant cell walls, usually from sensitized wheat or maize coleoptiles (Georgelis et al. 2014; Georgelis et al. 2011), but other materials, including cellulosic composites from Gluconacetobacter pellicles (Georgelis et al. 2014; Whitney et al. 2000), have been used. The creep response is diagnostic for expansins and is more specific and less variable than the paper breakage assay. There have also been reports that BsEXLX1 causes changes in the morphology of cellulosic fibers (Kim et al. 2009; Lin et al. 2013), but such effects seem highly variable and difficult to quantify. An assay based on release of suspended particles from filter paper or cotton may be useful alternative assays worth exploring (Baccelli et al. 2014; Din et al. 1991).
The fact that microbial expansins can weaken filter paper demonstrates that they are able to act on cellulose, but it does not preclude the possibility that they may act on other polysaccharides in the plant cell wall as well. No soluble material is released from plant cell walls or cellulose after incubation with microbial expansins (Georgelis et al. 2014; Georgelis et al. 2011; Kerff et al. 2008; Kim et al. 2009; Olarte-Lozano et al. 2014), indicating a non-lytic mechanism of action, which may involve loosening of non-covalent bonds between cellulose microfibrils in the plant cell wall (Cosgrove 2014). In this respect, bacterial expansins are very similar to plant expansins, which likewise lack lytic activity.
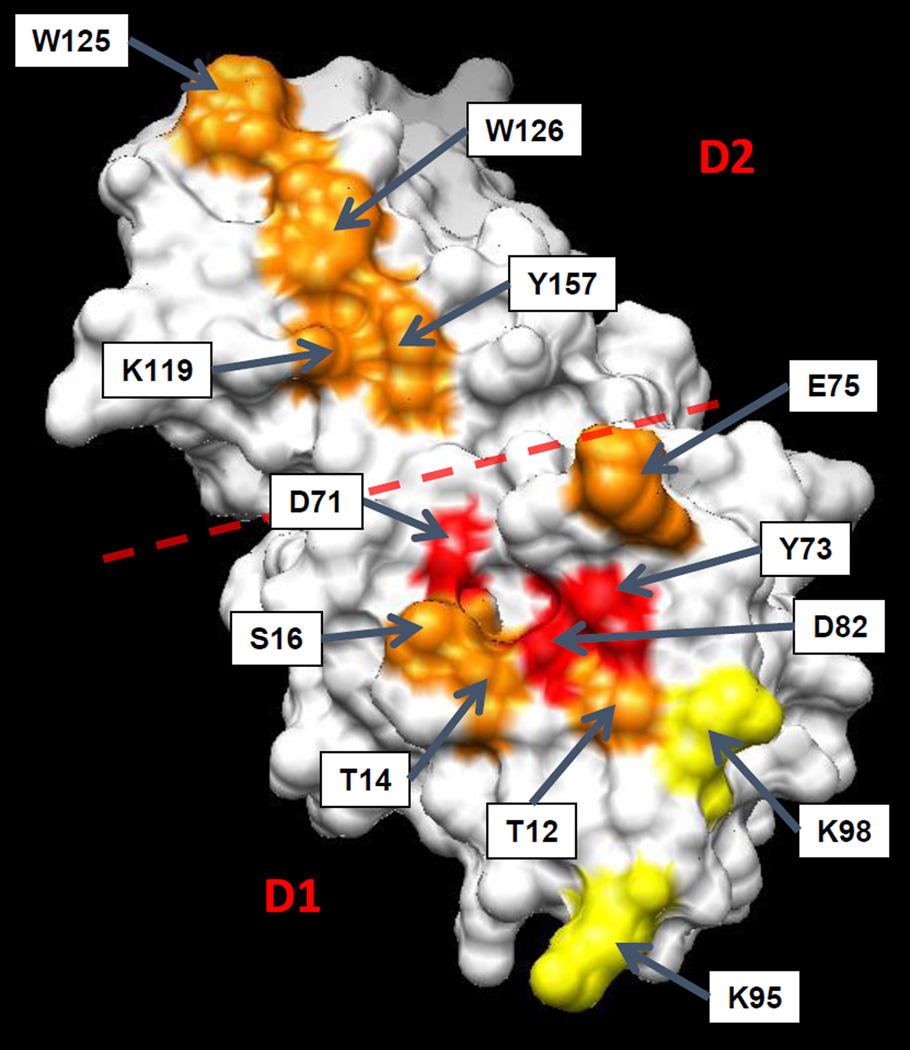
Site-directed mutagenesis showed that the planar aromatic surface of domain D2 is essential for binding to cellulose and for wall-loosening activity (Georgelis et al. 2011). Additional mutagenesis experiments showed that residues D71, Y73 and D82 in domain 1 are required for high creep activity and paper weakening (Figure 4). The complete loss of wall loosening activity in the D82A mutant is particularly intriguing because this residue corresponds to the catalytic proton donor of GH45 endoglucanases (Davies et al. 1995; Kerff et al. 2008) and because the D82A mutation does not affect the cellulose binding properties of the protein; indeed, domain 1 has very weak or no binding interactions with cellulose or cell walls. In contrast, cello-oligosaccharides stably bind to the catalytic cleft of GH45 enzymes (Davies et al. 1995). One might infer a cryptic enzymatic activity from the loss of activity of the D82A mutant, but extensive search for such activity has not yielded a convincing positive result. BsEXLX1 lacks a second acidic residue in a suitable position to serve as a catalytic base, offering a likely basis for the lack of endoglucanase activity. As discussed elsewhere (Georgelis et al. 2011), a catalytic base is sometimes missing in some inverting endoglucanases with clear hydrolytic activity, which is lacking in expansins. To serve as a proton donor, D82 would have to be protonated, but BsEXLX1 is active at pH values as high as 9.5 where D82 is unlikely to be protonated. Finally, it is worth noting that GH45 enzymes with clear hydrolytic activity against plant cell walls lack expansin’s ability to induce cell wall extension (Yennawar et al. 2006).
Figure 4.
Colored-coded activity map of key amino acid residues in BsEXLX1 that are required for wall loosening activity, based upon parallel site-directed mutagenesis and creep activity studies (Georgelis et al. 2011). Domains D1 and D2 are demarked with a red dotted line. Color code: red: loss of most or all activity; orange: partial loss of activity; yellow: little or no loss of activity. Image credit: from Georgelis et al. 2011, copyright American Society for Biochemistry and Molecular Biology, used with permission.
Another potential activity for expansin is suggested by the distant structural similarity to the catalytic site of the peptidoglycan lytic transglycosylase (LT) called MltA (Kerff et al. 2008). LTs cleave the bacterial cell wall peptidoglycan backbone, forming an intramolecular 1,6-anhydro loop where the normal reducing end would be formed by a hydrolase (Scheurwater et al. 2008). D82 corresponds to the acidic residue used by MltA to cleave β1,4-glycosidic bonds in peptidoglycan (van Straaten et al. 2005). This mechanism requires D82 to be protonated and the N-acetyl group of the muramoyl residue in peptidoglycan to assist in cleaving the β1,4-glycosidic bond in the case of MltA (Reid et al. 2007; Scheurwater et al. 2008). Such a group is not present in β1,4-D-glucans. Furthermore, a large portion of the MltA structure is needed to stabilize the glycan and enable lytic action, whereas there is no such stabilizing structure in expansins.
Thus, BsEXLX1 is likely to be acting differently than enzymes of the GH45 or LT type. Of course, negative results can never prove that expansins lack any form of enzymatic activity. The recent discovery of lytic polysaccharide mono-oxygenases (LPMOs), some of which were considered nonenzymatic cellulose binding modules until recently (Hemsworth et al. 2013; Hemsworth et al. 2014), is a case in point. Do expansins have LPMO activity? LPMOs contain copper and their activity is reduced by copper chelators, whereas expansins do not contain copper or other metals and their activity is not increased by addition of such metals; indeed, it is stabilized by metal chelators which reduce oxidative inhibition of plant expansins. Moreover, LPMOs produce lytic oligosaccharide fragments, something not observed with expansins. Thus, there is little reason to think expansins act like LPMOs.
Nevertheless, the requirement for D82 and some nearby residues for wall extension activity (Figure 4) is suggestive of a cryptic enzymatic activity, yet hardly conclusive. With a substrate as simple as β1,4-D-glucan (cellulose), there are limited enzymatic possibilities, and every direct test for enzymatic activity has so far proved negative. It might be informative to use atomic force microscopy to look for subtle changes in cellulose microfibril ultrastructure under expansin action; such an approach has proven informative for characterizing cellulytic enzymes (Brunecky et al. 2013; Eibinger et al. 2014). Until an enzymatic activity for expansin is demonstrated, alternative mechanisms of wall loosening, however repugnant to enzymologists, must be considered. The disruption of noncovalent interactions, which are important for the higher-order structural integrity of cellulose and complex cell wall, remains a viable working hypothesis.
Based on current evidence, our current model is as follows: BsEXLX1 binds to specific junctions of crystalline and non-crystalline cellulose where cellulose microfibrils make contact with one another, perhaps with the assistance of xyloglucan or similar flexible cellulose-binding polysaccharides in the case of complex cell walls (Georgelis et al. 2012; Zhao et al. 2014). These ‘biomechanical hotspots’ are important load-bearing junctions in plant cell walls and cellulosic materials such as paper (Cosgrove 2014). Domain D2 directs the protein to these sites and D1 is needed for local loosening of the cellulose-cellulose connections, perhaps in conjunction with cooperative action of D2, by a physicochemical process yet to be identified. Resolution of the molecular mechanism of expansin-mediated wall loosening is likely to reveal important aspects of primary cell wall structure.
Applications for biomass conversion
There is intense interest in processes to enhance biochemical conversion of cellulosic biomass to liquid transportation fuels as an alternative to petroleum-based fuels. Economic barriers include the high cost of enzymes and the need for expensive, energy-intensive pretreatments to overcome the recalcitrance of cellulose for enzymatic breakdown to sugars. Bacterial expansins have recently drawn considerable attention for their potential in processing of cellulosic biomass with reduced cellulase applications (Arantes and Saddler 2010; Kim et al. 2014). Experimental results have not been uniformly positive, however. Several studies report that expansin addition enhances the hydrolytic activity of cellulases by 20% to >100%, whereas other studies report only nonspecific effects (see below).
The earliest work in this vein made use of plant expansins in conjunction with cellulases to hydrolyze Avicel, a commercial cellulose product made from wood, as a means of investigating the mechanism of expansin-mediated cell wall loosening (Cosgrove 2001). This study found substantial enhancement of cellulase action at low levels of cellulase and % conversion. Another study showed that at high conversion the addition of extremely small amounts of β-expansin (0.1 mg per g cellulose) provided a small but apparently real enhancement of the rate of cellulose conversion to sugars (Baker et al. 2000). However, a major obstacle to industrial exploitation of this effect is the limited availability of plant expansins, which have proved difficult to express in active form in recombinant systems.
A recent study reported recombinant expression in E. coli of plant expansins from rice and examined their potential use as a cellulase synergist (Seki et al. 2014). As is common with hydrolysis studies carried out at low cellulase concentrations and low conversion (~1%), similar synergistic effects were also found with addition of bovine serum album (BSA) as a nonspecific protein blocker. Such nonspecific enhancements may stem from increases in cellulase stability or decreases in nonproductive enzyme binding to cellulose or to the side walls of the reaction chamber. While BSA showed significant enhancement of cellulose hydrolysis, somewhat greater effectiveness was observed with addition of expansins. Because expansin has separate hydrophobic and hydrophilic regions, it may act partly as a surfactant, thereby increasing cellulase activity (Eriksson et al. 2002). Using the same expression clones (a kind gift of Dr. Yasutaka Seki), we tested the recombinant protein for cell wall extension activity, but found it to be inactive in cell wall creep assays (D.J. Cosgrove, unpublished results), which is characteristic for plant expansins expressed in E. coli. Thus the cellulase enhancement is not likely to be the result of the specific wall loosening activity by expansin, but potentially by some other feature of the protein. In a similar study Liu et al. (2014) expressed a tomato expansin (named LeEXP2 or more correctly LeEXPA2, as it a member of the α-expansin family) in the methanotrophic yeast Pichia pastoris. They reported that expansin addition to cellulase/filter paper mixtures enhanced cellulose hydrolysis (at <1% conversion), but no blocking agents such as BSA were used in control experiments to distinguish specific from nonspecific effects. It would be of interest to know whether the recombinant plant expansins produced by Pichia have wall extension activity.
Unlike plant expansins, bacterial expansins are readily expressed in E. coli in active form, and consequently many synergism studies have been carried out with these forms of expansin. We reported that BsEXLX1 enhanced Avicel hydrolysis when small amounts of cellulase were applied, but we found slightly greater synergism with BSA, leading to the conclusion that the synergism at these low conversion rates was a nonspecific effect (Kerff et al. 2008). Kim et al. (2009) reported a similar synergistic effect of BsEXL1 for hydrolysis of filter paper by very low levels of cellulase, but did not find an enhancement by BSA. The cellulase used in one of their experiments was applied in such low amounts that it resulted in undetectable cellulose hydrolysis on its own, but addition of BsEXLX1 resulted in 0.6% net hydrolysis after 36 h of incubation. Additional experiments showed diminishing enhancement at higher cellulase loadings: approximately 2X enhancement when the cellulase by itself hydrolyzed ~2% of the filter paper; a 20% enhancement at 5% hydrolysis and no enhancement at 20% hydrolysis. The lack of BSA effect in these assays is puzzling because it is regularly observed in such studies, including a later study by the same group using the same materials for the assay (Lee et al. 2010). In this last report, BSA addition enhanced hydrolysis by a cellulase mixture (at 1% conversion over 48 h) by about 2.5-fold, while addition of an expansin from Hahella chejuensis resulted in an even greater enhancement. Another study reported very high synergism with several bacterial expansins when used with low concentration of cellulases (Bunterngsook et al. 2014). This study is unusual in that no BSA enhancement was seen in the data reported and moreover the cellulase-only treatments gave quite variable results from one experiment to another, ranging from 1% conversion to 0.02% conversion over 48 h. Georgelis et al. (2014) tested for synergism with three different cellulase cocktails and expansins from Xanthomonas campestris, Ralstonia solanacearum, Clavibacter michiganensis and Aspergillus niger: there was no enhancement by addition of either expansin or BSA (at 2% hydrolysis). Additionally, Olarte-Lozano et al. (2014) working with an expansin from Plectobacterium, concluded that expansin did not have a special synergistic effect beyond the nonspecific effect seen with BSA addition. Several other reports document a cellulase synergism by BsEXLX1 with various lignocellulosic materials, but at low hydrolysis levels and sometimes without BSA comparisons to assess the effect of nonspecific protein (Bunterngsook et al. 2015; Lin et al. 2013; Wang et al. 2014; Yan et al. 2012). One study reported that cellulase synergism by BsEXLX1 survived denaturing treatments such as SDS and 90°C treatment for 24 h (Wang et al. 2014); in our hands the wall extension activity of BsEXLX1 is irreversibly destroyed by a few minutes at 90°C. It seems unlikely the cellulase synergism depends on expansin activity in this case.
From this survey of recent results it should be clear that addition of expansin often enhances cellulose hydrolysis when low enzyme levels are used (giving <1–5% cellulose hydrolysis). The case for a specific synergistic effect of expansin arising from its wall-loosening activity is more difficult to make, however, because addition of BSA mimics much of the effect and expansins seem to be particularly effective at blocking nonspecific protein binding by lignin (Kim et al. 2013b). Whether there is anything more to the expansin enhancement beyond its nonspecific action is unclear. In this regard it would be very informative to compare the wild-type BsEXLX1 with the D82A mutant for evidence that the cellulase synergism depends on the wall loosening activity of expansin. None of the studies published to date can make a strong case on this important point.
Furthermore, for industrial purposes of biofuel production, complete hydrolysis of cellulose is essential. The few studies that have used higher cellulase loadings to raise the % conversion have generally reported that the enhancement by expansin disappears at higher conversion (Kim et al. 2009; Kim et al. 2014). Perhaps the most promising study in this light is that by Suwannarangsee et al. (2012) who reported optimization studies for hydrolysis of rice straw by ternary mixtures of two complex enzyme cocktails and BsEXLX1. At moderately high conversion rates (up to 78%) they reported a substantial synergistic effect by addition of BsEXLX1. This result however seems to be an outlier and further work is needed to test whether such effects can be replicated more generally with other materials and to test whether inactive forms of BsEXLX1 such as the D82A mutant have similar effects.
Swollenins, cerato-platanins and other expansin-like proteins
Genome surveys indicate that a few fungi have canonical expansins (Nikolaidis et al. 2014), but their biological roles and activities have drawn little study up to this point (Georgelis et al. 2014). Fungi also have more distantly related proteins named swollenin, cerato-platanin, EXPN, and loosenin that have been called expansin-like because of their sequence relatedness and their ability to fragment cellulose aggregates without evidence of lytic activity.
Cerato-platanins (CPs) are small fungal proteins widely found in basidiomycetes and ascomycetes (Yu and Li 2014). Their name derives from the plant pathogenic fungus Ceratocystis platani (Pazzagli et al. 1999). They also go by other names such as EPL1 in other studies (Frischmann et al. 2013). CPs have been connected with pathogen virulence in plants (Frias et al. 2011), with fungal development (Boddi et al. 2004), with hydrophobin-like action (Frischmann et al. 2013) and with the ability to disrupt cellulose aggregates without lytic activity (Baccelli et al. 2014). The NMR-based structure of CP shows it to be a double-ψ β-barrel and thus structurally related to GH45 (de Oliveira et al. 2011) and to expansin domain D1 (Figure 5). It lacks both of the catalytic aspartic acid residues needed for GH45 hydrolytic activity, but contains two of the three conserved disulfide bridges found in plant β-expansins. In one study CP was reported to disrupt filter paper and other forms of cellulose but without hydrolytic or LPMO activity and without detectable binding to cellulose (Baccelli et al. 2014). When added to a cellulase-filter paper mixture, CP enhanced cellulase action but to the same extent as BSA, so no evidence of specific synergism was seen.
Figure 5.
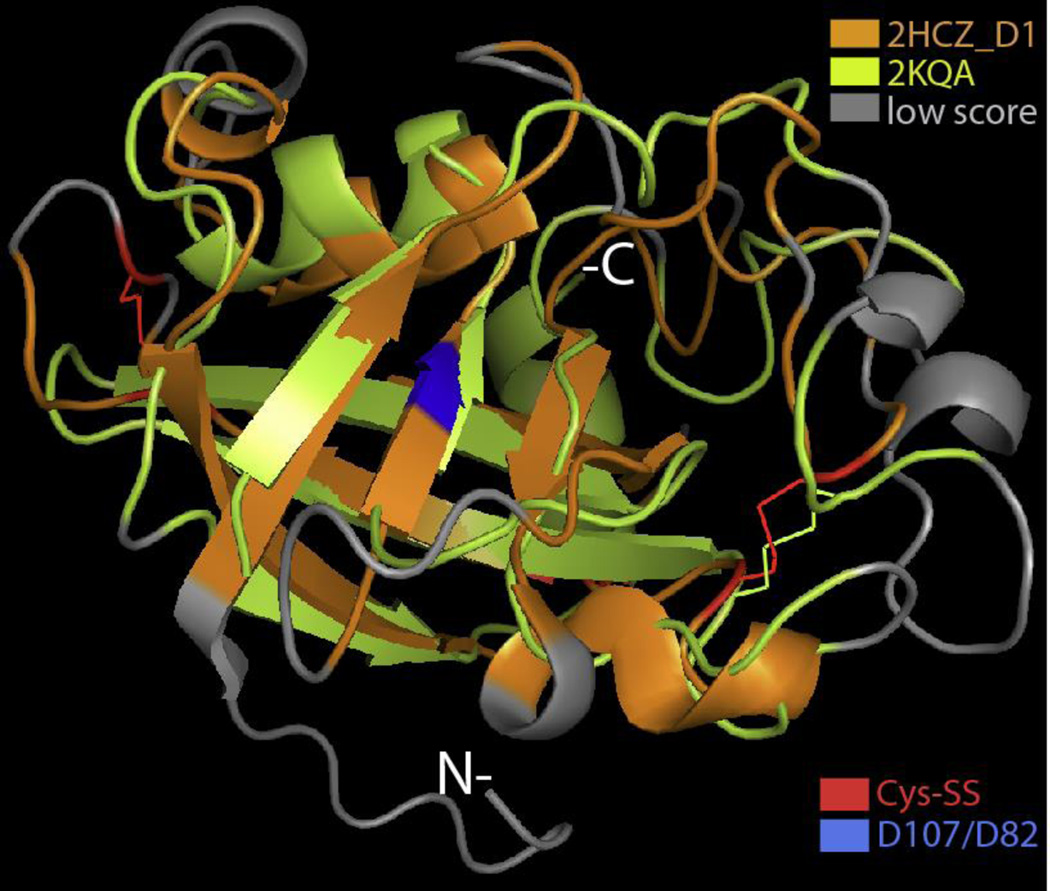
Structural superposition of domain D1 from the plant β-expansin ZmEXPB1 (P DB ID: 2HCZ; orange) with cerato-platanin (PDB ID: 2KQA; light green). The backbone superimposition was performed with DaliLite (http://www.ebi.ac.uk/Tools/structure/dalilite/). The grey segments indicate poor backbone alignment; the colored segments indicate at least a moderate score for alignment. The red residues indicate cysteines forming a disulfide bond; the blue residue is the conserved aspartic acid (D107) residue corresponding to D82 in BsEXLX1; in cerato-platanin it is replaced with asparagine. The N- and C-termini are indicate with white letters.
Fungal proteins dubbed ‘loosenin’ reportedly have cellulose disrupting activity similar to cerato-platanin (Quiroz-Castaneda et al. 2011). Cellulase synergism assays showed remarkably strong effects of loosenin addition, but comparison with BSA was not made, so it is unclear whether the synergism is specific or of the more mundane nonspecific type. The ‘expansin-related proteins’ studied by Suzuki et al. (2014) in Phanerochaete were actually proteins of the loosenin class and like CP they have distant sequence relatedness to the GH45-like domain of expansins. In ectomycorrhizal fungus Laccaria bicolor, a protein named EXPN is located in the fungal cell walls specifically in regions of close contact with the cells of plant roots (Veneault-Fourrey et al. 2014). EXPN, loosenin and CPs are single-domain fungal proteins related to the N-terminal domain of expansin, but lacking expansin’s C-terminal binding domain.
Swollenin (SWO) is a fungal protein that is about twice the size of expansin (493 versus 225 amino acids) and is likewise reported to disrupt cellulose aggregates (Saloheimo et al. 2002). SWO comprises a modular protein containing an expansin-like sequence at the C-terminus, an additional CBM1 domain at the N-terminus and a putative linker or fibronectin-III like domain (Fn-III) between the two modules. The function of the linker/Fn-III domain is uncertain but may aid in formation of multi-enzyme complexes for plant cell wall digestion (da Silva et al. 2012). It seems too long to be a simple linker region. The expansin-like region in SWO contains many conserved insertions, relative to bona fide expansins, and consequently bears very low similarity with expansin, yet it is predicted to have a similar two-domain structure (Nikolaidis et al. 2014; Saloheimo et al. 2002). An experimental structure of SWO would go a long way towards resolving remaining questions about the similarities of expansins, swollenins and GH45 enzymes, but none has been reported yet.
SWO reportedly hydrolyzes soluble β(1,4)-glucans, but lacks detectable hydrolytic activity against cellulose, yet it strongly disrupts cellulose aggregates such as Avicel and cotton fibers (Gourlay et al. 2014; Jager et al. 2011; Kang et al. 2013; Saloheimo et al. 2002; Wang et al. 2011). According to Yao et al. (2008), recombinant SWO had hydrolytic activity against an undefined xylan and yeast cell wall, but not against various forms of soluble and solid celluloses. In a related vein, Gourlay et al. (2013) reported that SWO by itself increased release of xylan oligomers from steam-pretreated corn stover. These activities were not characterized in detail and not claimed to be based on enzymatic activity. How oligosaccharide release might relate to the cellulose-disrupting activity reported for SWO is unclear, but Gourlay et al. (2013) suggested that it was a consequence of nonenzymatic surfactant action by SWO. Because the SWO was expressed in Trichoderma reesei, it is also possible that the recombinant SWO was contaminated with trace amounts of other lytic enzymes that co-purified with SWO. SWO exhibited particularly strong synergism with xylanases (Gourlay et al. 2013), which is reminiscent of the action of pollen β-expansins (Sampedro et al. 2015). Nakatani et al. (2013) reported a synergistic effect for degradation of phosphoric acid swollen cellulose by the yeast Saccharomyces cerevisiae genetically engineered to express a cellulase and SWO. These reports on SWO activity seem promising for commercial applications, but synergism at high rates of conversion have not yet been demonstrated.
A recent study examined the β(1,4)-glucanase activity of SWO, which showed very odd kinetics, with cutting action that appears to be partly like an endoglucanase and partly like a cellobiohydrolase (Andberg et al. 2015). Whether the hydrolytic action of SWO is necessary for its cellulose disruption activity has not been ascertained. Years ago we tested SWO (a kind gift of Dr. Colin Mitchinson, Genencor) for cell wall creep activity, but did not detect any creep of walls from cucumber hypocotyls and wheat coleoptiles (Yennawar et al. 2006). Similarly two GH45 enzymes did not induce creep of these walls. Thus, the wall-loosening actions of swollenin and GH45 enzymes differ from that of plant and bacterial expansins.
Prospectus
Although we have learned a great detail about molecular aspects of bacterial expansin structure and function, many questions about the biological roles of expansins in the microbial world remain wide open. In view of their relatively weak wall loosening activity, the mechanism by which these proteins promote plant colonization and pathogenesis is uncertain. Would a form of expansin with stronger wall loosening activity result in a more virulent pathogen or would it deter pathogenesis by activating plant defenses? Better understanding of this question could lead to methods to curtail the spread of bacterial wilt diseases and well as deeper molecular insights into the limiting steps in wall loosening by expansins.
Whether expansins and swollenin will find a place in commercial deconstruction of cellulosic biomass for biofuel production depends on many factors, both economic and scientific. While there are some indications that they may enhance cellulase action under some conditions, the approaches used to date have been empirical rather than based on a mechanistic understanding of what limits lignocellulose deconstruction and how these proteins modify cell walls to promote enzymatic attack. Many studies have been published showing effects at low levels of cellulose conversion, but these have little value unless they generate novel insights into the mechanism by which these proteins loosen cellulosic materials or how such effects can be made useful under conditions of full conversion of cellulose. This might be an area where in-vitro evolution of expansin properties might prove useful as well as informative.
Finally, expansins and similar proteins might also find application for their cellulose-binding characteristics, in the manner of more traditional CBMs (Shoseyov et al. 2006). The field of nanocellulosics is currently under rapid development for the production of novel materials based on renewable cellulose microfibrils (Habibi et al. 2010; Klemm et al. 2011). This is an area where the novel action of expansins and related proteins might find a role for constructing unique nanocellulosic structures, perhaps ones that are readily dissembled by application of expansins.
Acknowledgments
This work was supported by United States Department of Energy Grant DE-FG02-84ER13179 to D.J.C. from the Office of Basic Energy Sciences.
References
- Andberg M, Penttila M, Saloheimo M. Swollenin from Trichoderma reesei exhibits hydrolytic activity against cellulosic substrates with features of both endoglucanases and cellobiohydrolases. Bioresour Technol. 2015;181c:105–113. doi: 10.1016/j.biortech.2015.01.024. [DOI] [PubMed] [Google Scholar]
- Arantes V, Saddler JN. Access to cellulose limits the efficiency of enzymatic hydrolysis: the role of amorphogenesis. Biotechnol Biofuels. 2010;3:4. doi: 10.1186/1754-6834-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwini N, Srividya S. Potentiality of Bacillus subtilis as biocontrol agent for management of anthracnose disease of chilli caused by Colletotrichum gloeosporioides OGC1. 3 Biotech. 2013;4(2):127–136. doi: 10.1007/s13205-013-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccelli I, Luti S, Bernardi R, Scala A, Pazzagli L. Cerato-platanin shows expansin-like activity on cellulosic materials. Appl Microbiol Biotechnol. 2014;98(1):175–184. doi: 10.1007/s00253-013-4822-0. [DOI] [PubMed] [Google Scholar]
- Baker JO, King MR, Adney WS, Decker SR, Vinzant TB, Lantz SE, Nieves RE, Thomas SR, Li LC, Cosgrove DJ, Himmel ME. Investigation of the cell-wall loosening protein expansin as a possible additive in the enzymatic saccharification of lignocellulosic biomass. Appl Biochem Biotechnol. 2000;84-86:217–223. doi: 10.1385/abab:84-86:1-9:217. [DOI] [PubMed] [Google Scholar]
- Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A. 2013;110(17):E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddi S, Comparini C, Calamassi R, Pazzagli L, Cappugi G, Scala A. Cerato-platanin protein is located in the cell walls of ascospores, conidia and hyphae of Ceratocystis fimbriata f. sp. platani . FEMS Microbiol Lett. 2004;233(2):341–346. doi: 10.1016/j.femsle.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron AK, Van Loock B, Suslov D, Markakis MN, Verbelen J-P, Vissenberg K. Over-expression of AtEXLA2 alters etiolated arabidopsis hypocotyl growth. Ann Botany. 2014 doi: 10.1093/aob/mcu221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzarelou D, Billini M, Roumelioti K, Sophianopoulou V. EglD, a putative endoglucanase, with an expansin like domain is localized in the conidial cell wall of Aspergillus nidulans . Fungal Gen Bio. 2008;45(6):839–850. doi: 10.1016/j.fgb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Bras JL, Cartmell A, Carvalho AL, Verze G, Bayer EA, Vazana Y, Correia MA, Prates JA, Ratnaparkhe S, Boraston AB, Romao MJ, Fontes CM, Gilbert HJ. Structural insights into a unique cellulase fold and mechanism of cellulose hydrolysis. Proc Natl Acad Sci U S A. 2011;108(13):5237–5242. doi: 10.1073/pnas.1015006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman Y, Briff E, Viterbo A, Chet I. Role of swollenin, an expansin-like protein from trichoderma, in plant root colonization. Plant Physiol. 2008;147(2):779–789. doi: 10.1104/pp.108.116293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, Yang SJ, Resch MG, Adams MW, Lunin VV, Himmel ME, Bomble YJ. Revealing nature’s cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science. 2013;342(6165):1513–1516. doi: 10.1126/science.1244273. [DOI] [PubMed] [Google Scholar]
- Bunterngsook B, Eurwilaichitr L, Thamchaipenet A, Champreda V. Binding characteristics and synergistic effects of bacterial expansins on cellulosic and hemicellulosic substrates. Bioresour Technol. 2015;176:129–135. doi: 10.1016/j.biortech.2014.11.042. [DOI] [PubMed] [Google Scholar]
- Bunterngsook B, Mhuantong W, Champreda V, Thamchaiphenet A, Eurwilaichitr L. Identification of novel bacterial expansins and their synergistic actions on cellulose degradation. Bioresour Technol. 2014;159C:64–71. doi: 10.1016/j.biortech.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Carvalho CC, Phan NN, Chen Y, Reilly PJ. Carbohydrate binding module tribes. Biopolymers. 2014 doi: 10.1002/bip.22584. [DOI] [PubMed] [Google Scholar]
- Chabre H, Gouyon B, Huet A, Baron-Bodo V, Nony E, Hrabina M, Fenaille F, Lautrette A, Bonvalet M, Maillere B, Bordas-Le Floch V, Van Overtvelt L, Jain K, Ezan E, Batard T, Moingeon P. Molecular variability of group 1 and 5 grass pollen allergens between Pooideae species: implications for immunotherapy. Clin Exp Allergy. 2010;40(3):505–519. doi: 10.1111/j.1365-2222.2009.03380.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Plant cell enlargement and the action of expansins. BioEssays. 1996;18:533–540. doi: 10.1002/bies.950180704. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407(6802):321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Enhancement of accessibilty of cellulose by expansins. US Patent. 2001;6:326–470. [Google Scholar]
- Cosgrove DJ. Re-constructing our models of cellulose and primary cell wall assembly. Curr Opin Plant Biol. 2014;22:122–131. doi: 10.1016/j.pbi.2014.11.001. doi: http://dx.doi.org/10.1016/j.pbi.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94(12):6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva AJ, Gomez-Mendoza DP, Junqueira M, Domont GB, Ximenes Ferreira Filho E, de Sousa MV, Ricart CA. Blue native-PAGE analysis of Trichoderma harzianum secretome reveals cellulases and hemicellulases working as multienzymatic complexes. Proteomics. 2012;12(17):2729–2738. doi: 10.1002/pmic.201200048. [DOI] [PubMed] [Google Scholar]
- Darley CP, Li Y, Schaap P, McQueen-Mason SJ. Expression of a family of expansin-like proteins during the development of Dictyostelium discoideum . FEBS Letters. 2003;546(2-3):416–418. doi: 10.1016/s0014-5793(03)00598-2. [DOI] [PubMed] [Google Scholar]
- Davies GJ, Tolley SP, Henrissat B, Hjort C, Schulein M. Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolens at 1.9 A resolution. Biochemistry. 1995;34(49):16210–16220. doi: 10.1021/bi00049a037. [DOI] [PubMed] [Google Scholar]
- de Oliveira AL, Gallo M, Pazzagli L, Benedetti CE, Cappugi G, Scala A, Pantera B, Spisni A, Pertinhez TA, Cicero DO. The structure of the elicitor Cerato-platanin (CP), the first member of the CP fungal protein family, reveals a double-psi beta-barrel fold and carbohydrate binding. J Biol Chem. 2011;286(20):17560–17568. doi: 10.1074/jbc.M111.223644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N, Gilkes NR, Tekant B, Miller RC, Warren AJ, Kilburn DG. Non-hydrolytic disruption of cellulose fibers by the binding domain of a bacterial cellulase. Bio-Technology. 1991;9(11):1096–1099. [Google Scholar]
- Eibinger M, Ganner T, Bubner P, Rosker S, Kracher D, Haltrich D, Ludwig R, Plank H, Nidetzky B. Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J Biol Chem. 2014;289(52):35929–35938. doi: 10.1074/jbc.M114.602227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Malmsten M, Tiberg F, Callisen TH, Damhus T, Johansen KS. Model cellulose films exposed to H. insolens glucoside hydrolase family 45 endo-cellulase--the effect of the carbohydrate-binding module. J Colloid Interface Sci. 2005;285(1):94–99. doi: 10.1016/j.jcis.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Eriksson T, Borjesson J, Tjerneld F. Mechanism of surfactant effect in enzymatic hydrolysis of lignocellulose. Enzyme Microbial Technol. 2002;31(3):353–364. [Google Scholar]
- Frias M, Gonzalez C, Brito N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011;192(2):483–495. doi: 10.1111/j.1469-8137.2011.03802.x. [DOI] [PubMed] [Google Scholar]
- Frischmann A, Neudl S, Gaderer R, Bonazza K, Zach S, Gruber S, Spadiut O, Friedbacher G, Grothe H, Seidl-Seiboth V. Self-assembly at air/water interfaces and carbohydrate binding properties of the small secreted protein EPL1 from the fungus Trichoderma atroviride . J Biol Chem. 2013;288(6):4278–4287. doi: 10.1074/jbc.M112.427633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartemann KH, Kirchner O, Engemann J, Grafen I, Eichenlaub R, Burger A. Clavibacter michiganensis subsp michiganensis: first steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J Biotech. 2003;106(2–3):179–191. doi: 10.1016/j.jbiotec.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Georgelis N, Nikolaidis N, Cosgrove DJ. Biochemical analysis of expansin-like proteins from microbes. Carbohydrate Polymers. 2014;100:17–23. doi: 10.1016/j.carbpol.2013.04.094. [DOI] [PubMed] [Google Scholar]
- Georgelis N, Tabuchi A, Nikolaidis N, Cosgrove DJ. Structure-function analysis of the bacterial expansin EXLX1. J Biol Chem. 2011;286(19):16814–16823. doi: 10.1074/jbc.M111.225037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgelis N, Yennawar NH, Cosgrove DJ. Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc Natl Acad Sci U S A. 2012;109(37):14830–14835. doi: 10.1073/pnas.1213200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert HJ. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010;153(2):444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay K, Hu J, Arantes V, Andberg M, Saloheimo M, Penttila M, Saddler J. Swollenin aids in the amorphogenesis step during the enzymatic hydrolysis of pretreated biomass. Bioresour Technol. 2013;142:498–503. doi: 10.1016/j.biortech.2013.05.053. [DOI] [PubMed] [Google Scholar]
- Gourlay K, Hu J, Arantes V, Penttila M, Saddler JN. The use of Carbohydrate Binding Modules (CBMs) to monitor changes in fragmentation and cellulose fibre surface morphology during Cellulase and Swollenin induced deconstruction of lignocellulosic substrates. J Biol Chem. 2014 doi: 10.1074/jbc.M114.627604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi Y, Lucia LA, Rojas OJ. Cellulose nanocrystals: chemistry, self-assembly, and applications. Chem Rev. 2010;110(6):3479–3500. doi: 10.1021/cr900339w. [DOI] [PubMed] [Google Scholar]
- Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nature Chem Biol. 2014;10(2):122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Teeri TT, Warren RAJ. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Letters. 1998;425(2):352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- Herve C, Rogowski A, Blake AW, Marcus SE, Gilbert HJ, Knox JP. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A. 2010;107(34):15293–15298. doi: 10.1073/pnas.1005732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager G, Girfoglio M, Dollo F, Rinaldi R, Bongard H, Commandeur U, Fischer R, Spiess AC, Buchs J. How recombinant swollenin from Kluyveromyces lactis affects cellulosic substrates and accelerates their hydrolysis. Biotechnol Biofuels. 2011;4(1):33. doi: 10.1186/1754-6834-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr H, Dreier J, Meletzus D, Bahro R, Eichenlaub R. The endo-beta-1,4-glucanase CelA of Clavibacter michiganensis subsp michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol Plant-Microbe Inter. 2000;13(7):703–714. doi: 10.1094/MPMI.2000.13.7.703. [DOI] [PubMed] [Google Scholar]
- Kang K, Wang S, Lai G, Liu G, Xing M. Characterization of a novel swollenin from Penicillium oxalicum in facilitating enzymatic saccharification of cellulose. BMC Biotechnol. 2013;13:42. doi: 10.1186/1472-6750-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, Bradford K, Brummell D, Cho HT, Cosgrove D, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose J, Voesenek LA. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol. 2004;55(3):311–314. doi: 10.1007/s11103-004-0158-6. [DOI] [PubMed] [Google Scholar]
- Kerff F, Amoroso A, Herman R, Sauvage E, Petrella S, Filee P, Charlier P, Joris B, Tabuchi A, Nikolaidis N, Cosgrove DJ. Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization. Proc Natl Acad Sci U S A. 2008;105(44):16876–16881. doi: 10.1073/pnas.0809382105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T, Li HM, Karim N, Kennedy MW, Moens M, Jones JT. Identification of putative expansin-like genes from the pine wood nematode, Bursaphelenchus xylophilus, and evolution of the expansin gene family within the Nematoda. Nematol. 2009;11:355–364. [Google Scholar]
- Kim ES, Lee HJ, Bang WG, Choi IG, Kim KH. Functional characterization of a bacterial expansin from Bacillus subtilis for enhanced enzymatic hydrolysis of cellulose. Biotechnol Bioeng. 2009;102(5):1342–1353. doi: 10.1002/bit.22193. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Ko HJ, Kim TW, Choi IG, Kim KH. Characteristics of the binding of a bacterial expansin (BsEXLX1) to microcrystalline cellulose. Biotechnol Bioeng. 2013a;110(2):401–407. doi: 10.1002/bit.24719. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Ko HJ, Kim TW, Nam KH, Choi IG, Kim KH. Binding characteristics of a bacterial expansin (BsEXLX1) for various types of pretreated lignocellulose. Appl Microbiol Biotechnol. 2013b;97(12):5381–5388. doi: 10.1007/s00253-012-4412-6. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Lee HJ, Choi IG, Kim KH. Synergistic proteins for the enhanced enzymatic hydrolysis of cellulose by cellulase. Appl Microbiol Biotechnol. 2014 doi: 10.1007/s00253-014-6001-3. [DOI] [PubMed] [Google Scholar]
- Klemm D, Kramer F, Moritz S, Lindstrom T, Ankerfors M, Gray D, Dorris A. Nanocelluloses: a new family of nature-based materials. Angewandte Chem Internl Ed. 2011;50(24):5438–5466. doi: 10.1002/anie.201001273. [DOI] [PubMed] [Google Scholar]
- Laine MJ, Haapalainen M, Wahlroos T, Kankare K, Nissinen R, Kassuwi S, Metzler MC. The cellulase encoded by the native plasmid of Clavibacter michiganensis ssp sepedonicus plays a role in virulence and contains an expansin-like domain. Physiol Molr Plant Path. 2000;57(5):221–233. [Google Scholar]
- Lee DW, Seo JB, Kang JS, Koh SH, Lee SH, Koh YH. Identification and characterization of expansins from Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) Plant Path J. 2012;28(4):409–417. [Google Scholar]
- Lee HJ, Lee S, Ko HJ, Kim KH, Choi IG. An expansin-like protein from Hahella chejuensis binds cellulose and enhances cellulase activity. Mol Cells. 2010;29(4):379–385. doi: 10.1007/s10059-010-0033-z. [DOI] [PubMed] [Google Scholar]
- Lehtio J, Sugiyama J, Gustavsson M, Fransson L, Linder M, Teeri TT. The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc Natl Acad Sci U S A. 2003;100(2):484–489. doi: 10.1073/pnas.212651999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Bedinger PA, Volk C, Jones AD, Cosgrove DJ. Purification and characterization of four beta-expansins (Zea m 1 isoforms) from maize pollen. Plant Physiol. 2003;132(4):2073–2085. doi: 10.1104/pp.103.020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002;128(3):854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZC, Durachko DM, Cosgrove DJ. An oat coleoptile wall protein that induces wall extension in vitro and that is antigenically related to a similar protein from cucumber hypocotyls. Planta. 1993;191:349–356. [Google Scholar]
- Lin H, Shen Q, Zhan JM, Wang Q, Zhao YH. Evaluation of bacterial expansin EXLX1 as a cellulase synergist for the saccharification of lignocellulosic agro-Industrial wastes. Plos One. 2013;8(9):e75022. doi: 10.1371/journal.pone.0075022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Liu C, Ma Y, Hong J, Zhang M. Heterologous expression and functional characterization of a novel cellulose-disruptive protein LeEXP2 from Lycopersicum esculentum . J Biotechnol. 2014;186:148–155. doi: 10.1016/j.jbiotec.2014.07.013. [DOI] [PubMed] [Google Scholar]
- Maly T, Cui D, Griffin RG, Miller AF. 1H dynamic nuclear polarization based on an endogenous radical. J Phys Chem B. 2012;116(24):7055–7065. doi: 10.1021/jp300539j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci U S A. 1994;91(14):6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 1995;107(1):87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Yamada R, Ogino C, Kondo A. Synergetic effect of yeast cell-surface expression of cellulase and expansin-like protein on direct ethanol production from cellulose. Microbial Cell Factories. 2013;12(1):66. doi: 10.1186/1475-2859-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi C, Escudero C, Villarreal N, Martinez G, Civello PM. The carbohydrate-binding module of Fragaria x ananassa expansin 2 (CBM-FaExp2) binds to cell wall polysaccharides and decreases cell wall enzyme activities “in vitro”. J Plant Res. 2013;126(1):151–159. doi: 10.1007/s10265-012-0504-8. [DOI] [PubMed] [Google Scholar]
- Nikolaidis N, Doran N, Cosgrove DJ. Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol Biol Evol. 2014;31(2):376–386. doi: 10.1093/molbev/mst206. [DOI] [PubMed] [Google Scholar]
- Ogasawara S, Shimada N, Kawata T. Role of an expansin-like molecule in Dictyostelium morphogenesis and regulation of its gene expression by the signal transducer and activator of transcription protein Dd-STATa. Devel Growth & Diff. 2009;51(2):109–122. doi: 10.1111/j.1440-169X.2009.01086.x. [DOI] [PubMed] [Google Scholar]
- Olarte-Lozano M, Mendoza-Nunez MA, Pastor N, Segovia L, Folch-Mallol J, Martinez-Anaya C. PcExl1 a novel acid expansin-like protein from the plant pathogen Pectobacterium carotovorum, binds cell walls differently to BsEXLX1. PLoS One. 2014;9(4):e95638. doi: 10.1371/journal.pone.0095638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012;158(4):1933–1943. doi: 10.1104/pp.111.192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor N, Davila S, Perez-Rueda E, Segovia L, Martinez-Anaya C. Electrostatic analysis of bacterial expansins. Proteins. 2014 doi: 10.1002/prot.24718. [DOI] [PubMed] [Google Scholar]
- Pazzagli L, Cappugi G, Manao G, Camici G, Santini A, Scala A. Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani . J Biol Chem. 1999;274(35):24959–24964. doi: 10.1074/jbc.274.35.24959. [DOI] [PubMed] [Google Scholar]
- Qin L, Kudla U, Roze EH, Goverse A, Popeijus H, Nieuwland J, Overmars H, Jones JT, Schots A, Smant G, Bakker J, Helder J. Plant degradation: a nematode expansin acting on plants. Nature. 2004;427(6969):30. doi: 10.1038/427030a. [DOI] [PubMed] [Google Scholar]
- Quiroz-Castaneda RE, Martinez-Anaya C, Cuervo-Soto LI, Segovia L, Folch-Mallol JL. Loosenin, a novel protein with cellulose-disrupting activity from Bjerkandera adusta . Microbial Cell Factories. 2011;10:8. doi: 10.1186/1475-2859-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99(4):1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CW, Legaree BA, Clarke AJ. Role of Ser216 in the mechanism of action of membrane-bound lytic transglycosylase B: further evidence for substrate-assisted catalysis. FEBS Lett. 2007;581(25):4988–4992. doi: 10.1016/j.febslet.2007.09.037. [DOI] [PubMed] [Google Scholar]
- Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem. 2002;269(17):4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome biology. 2005;6(12):242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Guttman M, Li LC, Cosgrove DJ. Evolutionary divergence of beta-expansin structure and function in grasses parallels emergence of distinctive primary cell wall traits. Plant J. 2015;81(1):108–120. doi: 10.1111/tpj.12715. [DOI] [PubMed] [Google Scholar]
- Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: bacterial space-making autolysins. Internl J Biochem & Cell Biol. 2008;40(4):586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Seki Y, Kikuchi Y, Yoshimoto R, Aburai K, Kanai Y, Ruike T, Iwabata K, Goitsuka R, Sugawara F, Abe M, Sakaguchi K. Promotion of crystalline cellulose degradation by expansins from Oryza sativa . Planta. 2014 doi: 10.1007/s00425-014-2163-6. [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, Mcqueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins - a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci U S A. 1995;92(20):9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov O, Shani Z, Levy I. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev. 2006;70(2):283–295. doi: 10.1128/MMBR.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwannarangsee S, Bunterngsook B, Arnthong J, Paemanee A, Thamchaipenet A, Eurwilaichitr L, Laosiripojana N, Champreda V. Optimisation of synergistic biomass-degrading enzyme systems for efficient rice straw hydrolysis using an experimental mixture design. Bioresource Technol. 2012;119:252–261. doi: 10.1016/j.biortech.2012.05.098. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Vuong TV, Gong Y, Chan K, Ho CY, Master ER, Kondo A. Sequence diversity and gene expression analyses of expansin-related proteins in the white-rot basidiomycete, Phanerochaete carnosa . Fungal Gen Biol. 2014;72:115–123. doi: 10.1016/j.fgb.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Li LC, Cosgrove DJ. Matrix solubilization and cell wall weakening by beta-expansin (group-1 allergen) from maize pollen. Plant J. 2011;68(3):546–559. doi: 10.1111/j.1365-313X.2011.04705.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Ayala I, Bardet M, De Paepe G, Simorre JP, Hediger S. Solid-state NMR on bacterial cells: selective cell wall signal enhancement and resolution improvement using dynamic nuclear polarization. J Am Chem Soc. 2013;135(13):5105–5110. doi: 10.1021/ja312501d. [DOI] [PubMed] [Google Scholar]
- Thimm JC, Burritt DJ, Sims IM, Newman RH, Ducker WA, Melton LD. Celery (Apium graveolens) parenchyma cell walls: cell walls with minimal xyloglucan. Physiol Plant. 2002;116(2):164–171. doi: 10.1034/j.1399-3054.2002.1160205.x. [DOI] [PubMed] [Google Scholar]
- van Straaten KE, Dijkstra BW, Vollmer W, Thunnissen AM. Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold. J Mol Biol. 2005;352(5):1068–1080. doi: 10.1016/j.jmb.2005.07.067. [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Commun C, Kohler A, Morin E, Balestrini R, Plett J, Danchin E, Coutinho P, Wiebenga A, de Vries RP, Henrissat B, Martin F. Genomic and transcriptomic analysis of Laccaria bicolor CAZome reveals insights into polysaccharides remodelling during symbiosis establishment. Fungal Gen Biol. 2014;72:168–181. doi: 10.1016/j.fgb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Wang T, Park YB, Caporini MA, Rosay M, Zhong L, Cosgrove DJ, Hong M. Sensitivity-enhanced solid-state NMR detection of expansin’s target in plant cell walls. Proc Natl Acad Sci U S A. 2013;110(41):16444–16449. doi: 10.1073/pnas.1316290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WC, Liu C, Ma YY, Liu XW, Zhang K, Zhang MH. Improved production of two expansin-like proteins in Pichia pastoris and investigation of their functional properties. Biochem Eng J. 2014;84:16–27. [Google Scholar]
- Wang Y, Tang R, Tao J, Gao G, Wang X, Mu Y, Feng Y. Quantitative investigation of non-hydrolytic disruptive activity on crystalline cellulose and application to recombinant swollenin. Appl Microbiol Biotechnol. 2011;91(5):1353–1363. doi: 10.1007/s00253-011-3421-1. [DOI] [PubMed] [Google Scholar]
- Whitney SE, Gidley MJ, McQueen-Mason SJ. Probing expansin action using cellulose/hemicellulose composites. Plant J. 2000;22(4):327–334. doi: 10.1046/j.1365-313x.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- Wolf S, Hematy K, Hofte H. Growth control and cell wall signaling in plants. Annu Rev Plant Biol. 2012;63:381–407. doi: 10.1146/annurev-arplant-042811-105449. [DOI] [PubMed] [Google Scholar]
- Yan Z, He M-X, Bo W, Hu Q-C, Li Q, Zhao J. Recombinant EXLX1 from Bacillus subtilis for enhancing enzymatic hydrolysis of corn stover with low cellulase loadings. African J Biotech. 2012;11:11126–11131. [Google Scholar]
- Yao Q, Sun TT, Liu WF, Chen GJ. Gene cloning and heterologous expression of a novel endoglucanase, swollenin, from Trichoderma pseudokoningii S38. Biosci Biotechnol Biochem. 2008;72(11):2799–2805. doi: 10.1271/bbb.80124. [DOI] [PubMed] [Google Scholar]
- Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, Cosgrove DJ. Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc Natl Acad Sci U S A. 2006;103(40):14664–14671. doi: 10.1073/pnas.0605979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Li L. Phylogeny and molecular dating of the cerato-platanin-encoding genes. Gen Mol Biol. 2014;37(2):423–427. doi: 10.1590/s1415-47572014005000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Crespi VH, Kubicki JD, Cosgrove DJ, Zhong L. Molecular dynamics simulation study of xyloglucan adsorption on cellulose surfaces: effects of surface hydrophobicity and side-chain variation. Cellulose. 2014;21:1025–1039. [Google Scholar]
- Zhao Z, Shklyaev OE, Nili A, Mohamed MNA, Kubicki JD, Crespi VH, Zhong LH. Cellulose microfibril twist, mechanics, and implication for cellulose biosynthesis. J Phys Chem A. 2013;117(12):2580–2589. doi: 10.1021/jp3089929. [DOI] [PubMed] [Google Scholar]