Abstract
Background
Chagas disease, caused by the parasite Trypanosoma cruzi, is a neglected tropical disease that causes severe human health problems. To develop a new chemotherapeutic agent for the treatment of Chagas disease, we predicted a pharmacophore model for T. cruzi dihydroorotate dehydrogenase (TcDHODH) by fragment molecular orbital (FMO) calculation for orotate, oxonate, and 43 orotate derivatives.
Methodology/Principal Findings
Intermolecular interactions in the complexes of TcDHODH with orotate, oxonate, and 43 orotate derivatives were analyzed by FMO calculation at the MP2/6-31G level. The results indicated that the orotate moiety, which is the base fragment of these compounds, interacts with the Lys43, Asn67, and Asn194 residues of TcDHODH and the cofactor flavin mononucleotide (FMN), whereas functional groups introduced at the orotate 5-position strongly interact with the Lys214 residue.
Conclusions/Significance
FMO-based interaction energy analyses revealed a pharmacophore model for TcDHODH inhibitor. Hydrogen bond acceptor pharmacophores correspond to Lys43 and Lys214, hydrogen bond donor and acceptor pharmacophores correspond to Asn67 and Asn194, and the aromatic ring pharmacophore corresponds to FMN, which shows important characteristics of compounds that inhibit TcDHODH. In addition, the Lys214 residue is not conserved between TcDHODH and human DHODH. Our analysis suggests that these orotate derivatives should preferentially bind to TcDHODH, increasing their selectivity. Our results obtained by pharmacophore modeling provides insight into the structural requirements for the design of TcDHODH inhibitors and their development as new anti-Chagas drugs.
Introduction
Chagas disease is an infectious disease caused by the parasitic protozoan Trypanosoma cruzi (T. cruzi) and is the third most common parasitic disease in the world [1]. It affects people from approximately 20 countries, particularly those living in the southern United States and Latin America [2–4], with 15 million people estimated to be infected [5]. T. cruzi is primarily transmitted by blood-sucking insects belonging to the subfamily Triatominae (family Reduviidae) or by infected blood transfusion [6]. Once infected, the host may experience influenza-like symptoms during the acute phase and gastrointestinal and cardiac disorders during the chronic phase [7–8]. Two drugs, nifurtimox and benznidazole, are currently available for the treatment of Chagas disease, but there are severe problems associated with their use, including adverse effects and limited effectiveness during the chronic phase of the disease [9–10]. Thus, developing new therapeutic agents against T. cruzi infection is desirable [11–12].
To develop a novel anti-Chagas drug, we focused on dihydroorotate dehydrogenase (DHODH) as the target protein. DHODH is an enzyme that takes part in the fourth step in the de novo biosynthesis of pyrimidines, which are heterocyclic compounds essential for RNA and DNA synthesis. This enzyme is an oxidoreductase that catalyzes the oxidation of dihydroorotate to orotate using flavin mononucleotide (FMN) as a cofactor. FMN can take either an oxidized form (FMN) or a reduced form (FMNH2), and the oxidized form serves as the oxidizing agent during orotate production. FMNH2 is re-oxidized by an electron acceptor that differs according to the cellular localization of DHODH [13]. In humans, DHODH is a mitochondrial inner-membrane protein that uses respiratory ubiquinone as the electron acceptor [14]. In contrast, T. cruzi DHODH (TcDHODH), a cytosolic protein, uses fumarate as the electron acceptor [15]. A previous study showed that a DHODH-knockout T. cruzi was not viable [16]. The differences in biochemical properties between human and T. cruzi DHODHs as well as its essentiality for the parasite make TcDHODH a promising target for developing novel therapeutic agents against Chagas disease.
DHODH is a validated drug target for humans [17–18], as an immunosuppressant and also against Plasmodium falciparum [17, 19, 20] and Helicobacter pylori [21]. Species-specific DHODH inhibitors have been developed and shown to be effective in vitro [22] and in vivo [23]. However, all inhibitors developed to date target the ubiquinone binding site and do not inhibit the cytosolic DHODHs, for which potent and selective inhibitors have never been reported.
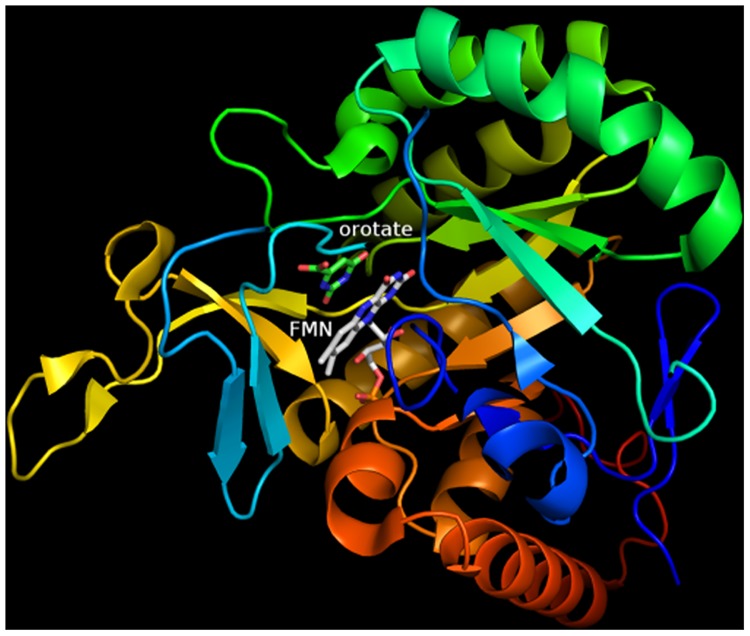
The atomic resolution crystal structures of TcDHODH in complexes with its substrates and products have been determined [15]. Based on analysis of overall structure and the active site region of the TcDHODH-orotate complex (Fig 1), it is thought that a strong π-π interaction between orotate and the isoalloxazine ring of FMN occurs.
Fig 1. TcDHODH overall structure.
Crystal structure of TcDHODH (PDB ID: 2E6A, A-chain).
A pharmacophore is defined as “an ensemble of steric and electronic features that ensures optimal supramolecular interactions with a specific biological target and the trigger (or inhibit) of its biological function” [24]. Based on this definition, we define pharmacophore modeling as a process for predicting pharmacophores with common or specific characteristics among compounds. This definition is applied not only to molecular design but to protein—ligand docking simulation and quantitative structure-activity relationships (QSAR) as well [25]. However, pharmacophore modeling without ligand structural alignment information is difficult. Thus, knowledge of protein—ligand structure is useful for predicting pharmacophores.
The fragment molecular orbital (FMO) method [26] employs ab initio quantum mechanical calculations for large biomolecules such as protein—ligand complexes. Intermolecular interaction energies typically can be determined on the basis of molecular mechanics. However, this method is not universally applicable to all compounds, because there is a limit to the determination of molecular potentials based on atom type, especially of quantum chemical elements such as π electrons. For this reason, in this study, we used the FMO method to analyze the interaction energies between the target proteins and ligands with the aim of identifying important amino acid residues for ligand binding. Amino acids and ligands in the system of interest are divided into fragments, and molecular orbital calculations are performed for individual fragments. Because the effects of interfragment potentials are taken into account in these molecular orbital calculations, the FMO method can estimate the interaction energy between each pair of fragments. The method can clearly describe the detailed interactions between the ligand and each amino acid residue, and is frequently used in the design of new drugs [27–34]. Moreover, the method can extract specific interaction from a wide variety of derivatives. FMO calculation is thus suitable for obtaining pharmacophore models and is useful for guiding molecular design.
In the present study, we identified pharmacophores using the FMO method with the aim of designing anti-Chagas drugs, via analysis of interaction energy between TcDHODH and orotate, oxonate [35] as a competitive inhibitor of DHODH, and 43 orotate derivatives.
Materials and Methods
Co-crystallization of TcDHODH with orotate derivatives
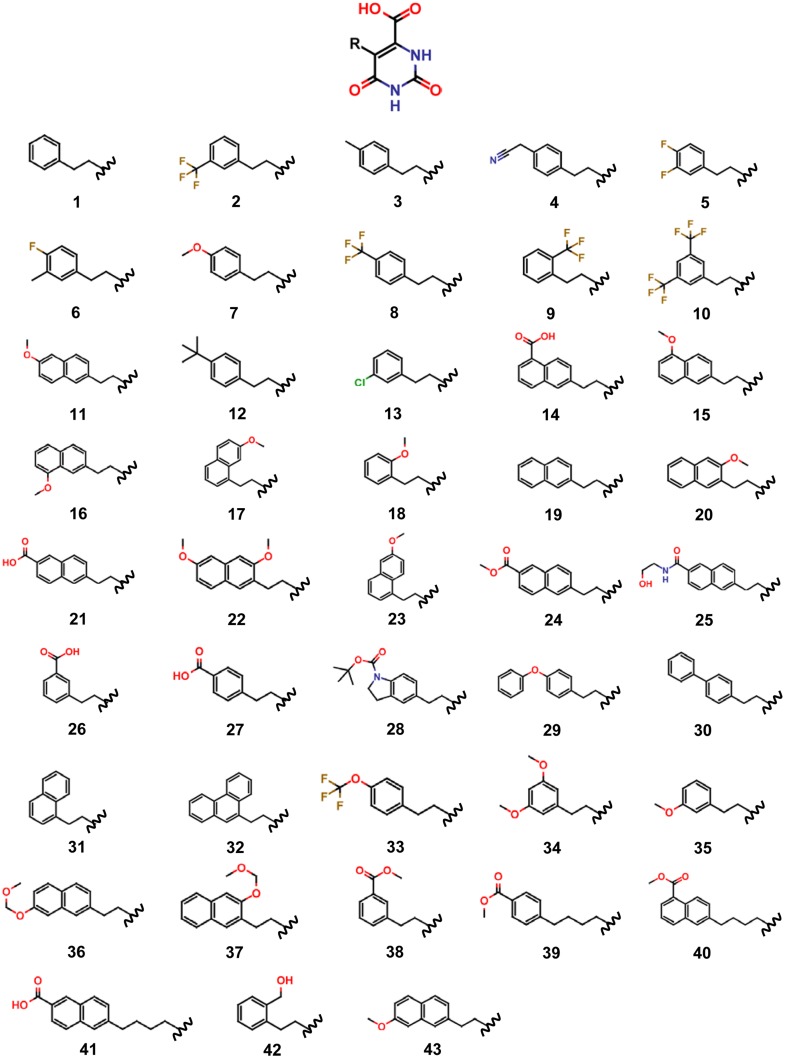
Recombinant TcDHODH expression, purification, and crystallization in complex with 43 orotate derivatives were performed essentially as previously reported [15]. Briefly, TcDHODH was purified to homogeneity from BL21(DE3)pET3a/TcDHODH using DEAE Sepharose Fast Flow (GE Healthcare) followed by Phenyl Sepharose H.P. (GE Healthcare) and TSK G3000SW (Tosoh). By this method, a total of 10–15 mg of TcDHODH with specific activity ranging from 12 to 18 μmol/min/mg could be purified from 10 L of culture. TcDHODH crystals formed in the presence of oxonate [15] were used to soak overnight into crystallization buffer containing 1 mM of orotate derivatives that are listed in Fig 2. X-ray diffraction data were collected at SPring-8 (beam lines BL32XU, BL41XU and BL44XU) or Photon Factory (beam lines AR-NW12A, AR-NE3A, BL-5A and BL-17A). All the data were processed and scaled with HKL2000 [36]. The co-crystal structures of TcDHODH in complex with orotate derivatives were initially solved by molecular replacement using coordinates from TcDHODH-oxonate complexed structure (3W1Q) and later by one of the open form structures (e.g., 3W1R). Manual building and crystallographic refinement were performed with the programs COOT [37] and REFMAC5 [38], respectively. The PDB IDs, resolution, R-value, R-Free, average of the occupancy of the ligands and average of the B-factors are listed in Table 1. The detailed synthetic process of the 43 orotate derivatives and the X-ray analysis performed in this study will be described elsewhere (Inaoka et al., manuscript in preparation).
Fig 2. Structures of orotate and R groups of synthesized derivatives.
Functional groups were introduced at the pyrimidine 5-position through an ethylene or butylene linker.
Table 1. Summary of co-crystal structures (UniProtKB: Q4D3W2).
| PDB ID | Derivative | Resolution(Å) | R-value | R-Free | Ligand Occupancy | B-factor (ligand) | B-factor (All) |
|---|---|---|---|---|---|---|---|
| 3W1R | 1 | 1.58 | 0.157 | 0.181 | 1.00 | 12.92 | 11.45 |
| 3W1T | 2 | 1.68 | 0.144 | 0.179 | 1.00 | 15.02 | 12.69 |
| 3W1U | 3 | 1.85 | 0.153 | 0.185 | 1.00 | 18.78 | 15.90 |
| 3W1X | 4 | 1.45 | 0.140 | 0.161 | 1.00 | 8.31 | 11.01 |
| 3W2J | 5 | 1.42 | 0.144 | 0.168 | 1.00 | 13.82 | 11.56 |
| 3W2K | 6 | 1.54 | 0.138 | 0.169 | 1.00 | 19.46 | 13.26 |
| 3W2L | 7 | 1.64 | 0.136 | 0.170 | 1.00 | 16.30 | 13.27 |
| 3W2M | 8 | 1.58 | 0.139 | 0.169 | 1.00 | 15.00 | 12.37 |
| 3W2N | 9 | 1.96 | 0.149 | 0.194 | 1.00 | 10.98 | 14.11 |
| 3W2U | 10 | 2.25 | 0.208 | 0.270 | 1.00 | 23.15 | 22.87 |
| 3W3O | 11 | 1.96 | 0.151 | 0.192 | 1.00 | 19.84 | 20.06 |
| 3W22 | 12 | 1.98 | 0.166 | 0.211 | 1.00 | 26.97 | 23.73 |
| 3W23 | 13 | 1.48 | 0.146 | 0.177 | 1.00 | 16.13 | 13.79 |
| 3W6Y | 14 | 2.68 | 0.199 | 0.282 | 1.00 | 40.63 | 23.14 |
| 3W7C | 15 | 1.75 | 0.144 | 0.195 | 1.00 | 14.68 | 13.99 |
| 3W7D | 16 | 1.52 | 0.146 | 0.174 | 1.00 | 14.87 | 13.93 |
| 3W7E | 17 | 1.56 | 0.143 | 0.173 | 1.00 | 13.05 | 12.60 |
| 3W7G | 18 | 1.55 | 0.138 | 0.165 | 0.67 | 11.01 | 12.85 |
| 3W7H | 19 | 1.67 | 0.145 | 0.176 | 0.67 | 18.94 | 15.19 |
| 3W7I | 20 | 1.69 | 0.145 | 0.179 | 1.00 | 19.22 | 13.84 |
| 3W7J | 21 | 1.58 | 0.149 | 0.182 | 0.67 | 10.33 | 10.79 |
| 3W7K | 22 | 1.61 | 0.150 | 0.180 | 1.00 | 15.93 | 15.69 |
| 3W7L | 23 | 1.88 | 0.145 | 0.189 | 1.00 | 14.73 | 14.66 |
| 3W7M | 24 | 2.40 | 0.175 | 0.246 | 1.00 | 33.44 | 29.54 |
| 3W7N | 25 | 2.39 | 0.188 | 0.255 | 1.00 | 53.08 | 37.34 |
| 3W7O | 26 | 1.68 | 0.142 | 0.178 | 1.00 | 14.63 | 13.76 |
| 3W7P | 27 | 1.70 | 0.142 | 0.175 | 1.00 | 18.19 | 13.29 |
| 3W7Q | 28 | 1.83 | 0.144 | 0.179 | 1.00 | 19.10 | 18.48 |
| 3W70 | 29 | 2.60 | 0.202 | 0.275 | 1.00 | 42.47 | 25.34 |
| 3W71 | 30 | 1.68 | 0.146 | 0.175 | 1.00 | 21.25 | 15.98 |
| 3W72 | 31 | 1.55 | 0.142 | 0.172 | 0.67 | 11.48 | 14.44 |
| 3W73 | 32 | 1.78 | 0.146 | 0.181 | 1.00 | 19.84 | 14.40 |
| 3W74 | 33 | 1.90 | 0.175 | 0.228 | 1.00 | 29.90 | 22.87 |
| 3W75 | 34 | 1.47 | 0.139 | 0.166 | 0.67 | 12.02 | 12.50 |
| 3W76 | 35 | 1.58 | 0.140 | 0.170 | 1.00 | 16.11 | 14.47 |
| 3W83 | 36 | 2.80 | 0.199 | 0.297 | 1.00 | 40.63 | 23.70 |
| 3W84 | 37 | 1.93 | 0.202 | 0.254 | 1.00 | 22.26 | 16.84 |
| 3W85 | 38 | 2.00 | 0.157 | 0.202 | 1.00 | 24.14 | 19.12 |
| 3W86 | 39 | 1.50 | 0.145 | 0.184 | 1.00 | 19.13 | 15.13 |
| 3W87 | 40 | 1.43 | 0.155 | 0.187 | 0.67 | 15.53 | 10.62 |
| 3W88 | 41 | 1.40 | 0.140 | 0.166 | 0.67 | 21.44 | 11.03 |
| 4JD4 | 42 | 1.51 | 0.138 | 0.162 | 1.00 | 15.53 | 14.13 |
| 4JDB | 43 | 1.82 | 0.169 | 0.211 | 1.00 | 26.24 | 20.87 |
B-factor: The average of B-factors.
Interaction energy analysis
Interaction energy analysis was performed using the analytical tool Facio [39] based on pair interaction energy decomposition analysis, as proposed by Fedorov et al. [40]. This analysis using the FMO method provides a quantitative evaluation of hydrogen bonding and hydrophobic interactions that are important for ligand binding to a protein, as well as π-π, π–cation, and CH–π interactions, which require quantum chemical calculations [41]. These analyses are sometimes applied in structure-based drug design [42].
Calculation procedure
All structures of the TcDHODH—compound complexes were visualized and computations were performed with hydrogenation. Optimization of the structures was performed only for the added hydrogen atoms, with all heavy atoms fixed at the positions given in the PDB using the CHARMM force field [43] by Discovery Studio (Accelrys, San Diego, CA). FMO calculation job files were generated using FMOutil version 2.1, and calculations were performed for each A-chain monomer using GAMESS [44] at the MP2/6-31G level. Alternate positions at except active site adopted a type-A conformation. Alternate positions in the active site adopted both. All calculations were performed with the TSUBAME2.5 supercomputer at Tokyo Tech (a HP Proliant SL390s G7 server with an Intel Xeon X5670 2.93-GHz processor). The approximate calculation time of each structure was 3 h at the MP2/6-31G level.
Definition of pharmacophore
In this paper, we define a common pharmacophore as that having characteristics common to more than 80% conserved interactions between TcDHODH and compounds, and a particular pharmacophore as that having characteristics that have interactions stronger than −10 kcal mol-1 between TcDHODH and compounds.
Results
Analysis of interaction energies between TcDHODH and orotate and oxonate
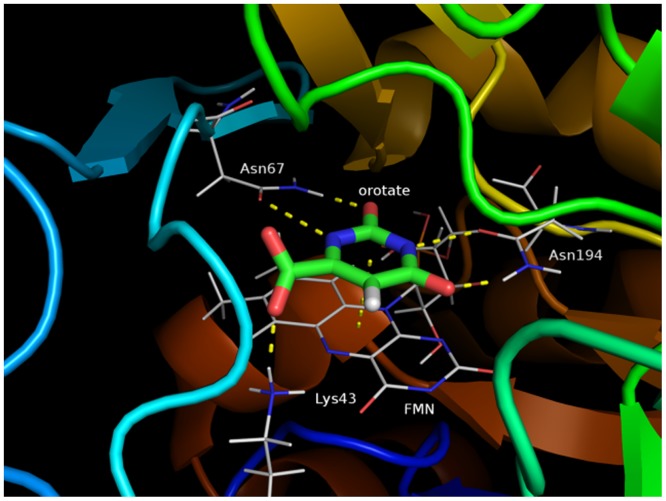
Orotate binds to the active center of TcDHODH, which is positioned parallel to an aromatic ring of FMN. Certain amino acid residues such as Lys43, Asn67, and Asn194 form hydrogen bonds with orotate and are important for substrate binding (Fig 3).
Fig 3. Orotate binding mode.
Crystal structure of the TcDHODH—orotate complex active site. Hydrogen bond distance with the Lys43 amine group: 2.93 Å, with the Asn67 amine group: 3.05 Å, with the Asn67 carbonyl group: 2.82, with the Asn194 amine group: 3.32 and with the Asn194 carbonyl group: 2.92.
The side chain of Asn67 interacts with the 1-position hydrogen atom and 2-position carbonyl group of orotate, while the side chain of Asn194 side chain interacts with orotate’s 3-position hydrogen atom and 4-position carbonyl group through two hydrogen bonds. Moreover, the Lys43 side chain amide interacts with orotate’s 6-position carboxylate group.
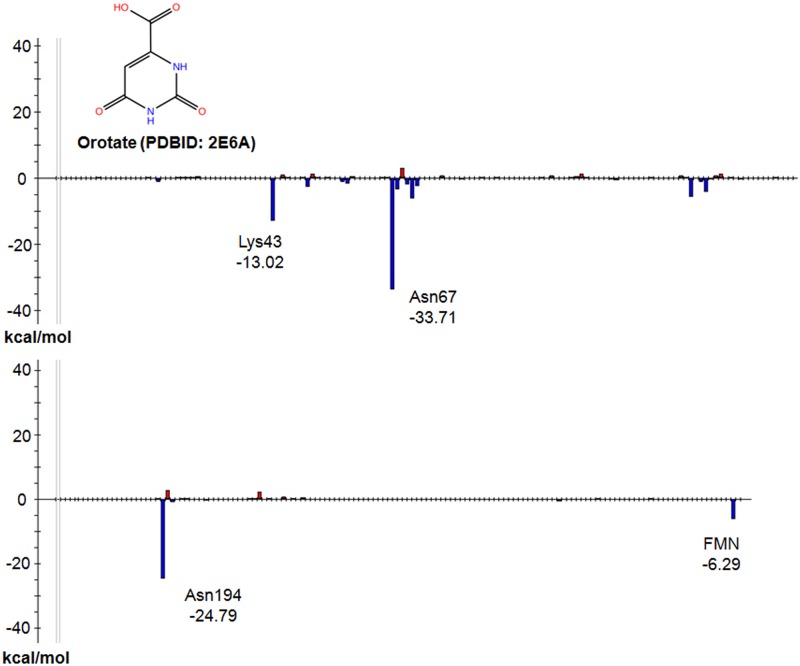
The FMO-based interaction energy analysis indicates that the hydrogen bonds with Lys43, Asn67 and Asn194 residues are important in this interaction (-13.02, -33.71, and -24.79 kcal mol-1, respectively), with interactions with Asn67 being strongest (Fig 4).
Fig 4. Interaction energy analysis of TcDHODH—orotate.
The vertical axis shows the interaction energy between the ligand and each fragment, and the horizontal axis shows the fragment numbers. Fragments are numbered in order from N to C termini, followed by ligands, such as substrate and cofactor, to preserve the order described in the PDB file.
These findings indicate that Asn67 and Asn194 contribute to the specificity of orotate’s pyrimidine ring, and Lys43 contributes to the hydrogen bonding of orotate at the carboxylate moiety at the pyrimidine 6-position. The π-π interaction energy between orotate and FMN was −6.29 kcal mol-1 and contributed significantly to ligand binding (Fig 4). Given that Asn67 and Asn194 residues form two hydrogen bonds with orotate, large interaction energies would be predicted from this binding mode.
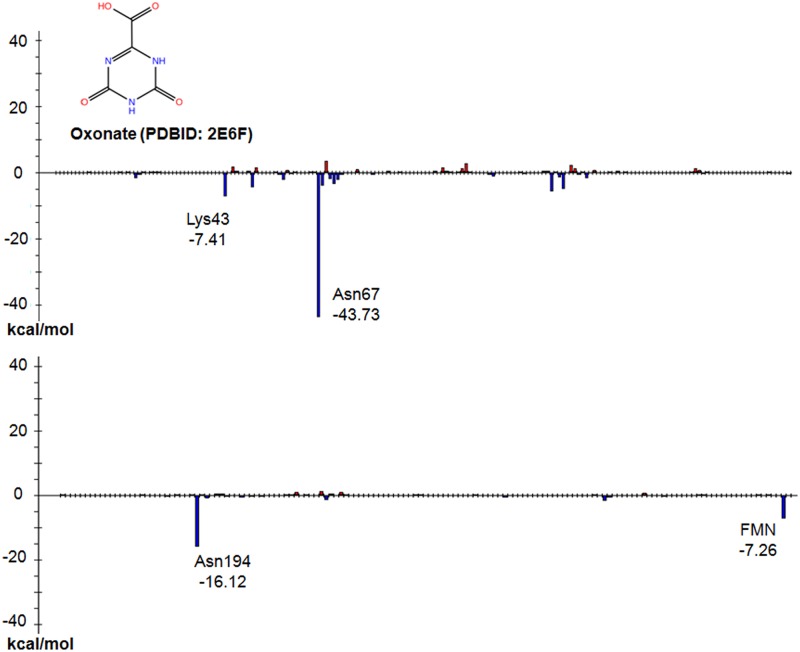
Oxonate, a well-known inhibitor of TcDHODH [15], also binds to the active center of TcDHODH, similarly to orotate. The FMO-based interaction energy analysis confirms the interaction with Lys43, Asn67, Asn194 and FMN as well as that of orotate (Fig 5: -7.41, -43.73, -16.21, and -7.26 kcal mol-1, respectively).
Fig 5. Interaction energy analysis of TcDHODH—oxonate.
The vertical axis and horizontal axis are same as the Fig 4. (PDB ID: 2E6F, A-chain).
These results suggest that inhibitors targeting the active site of TcDHODH should preferentially have an aromatic moiety containing hydrogen bond donors and acceptors.
Analysis of interaction energies between TcDHODH and orotate derivatives
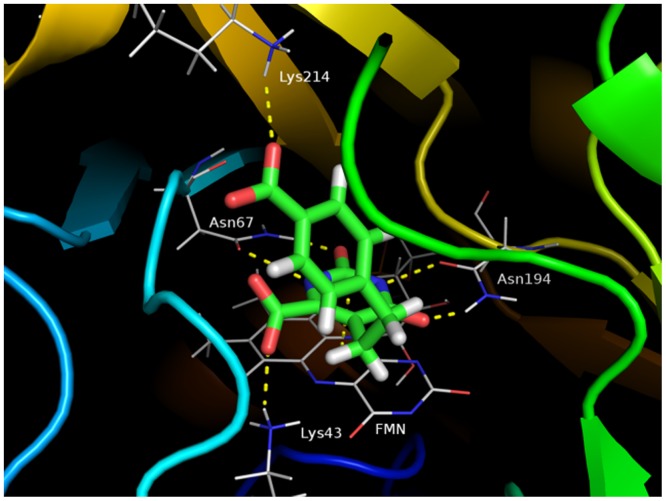
Next, 43 orotate derivatives (Fig 2) predicted to form additional hydrogen bond and hydrophobic interactions were synthesized and their co-crystal structures were determined. These derivatives contain a functional group substituted at the pyrimidine 5-position through an ethylene or butylene linker. The binding mode between the active site of TcDHODH and derivative 27 is shown in Fig 6 as an example. Similarly to orotate, derivative 27 and other derivatives interact with Lys43, Asn67, Asn194 and FMN.
Fig 6. Binding mode of derivative 27.
Crystal structures of the TcDHODH—derivative 27 complex active site. Hydrogen bond distance with the Lys43 amine group: 2.73 Å, with the Asn67 amine group: 2.89 Å, with the Asn67 carbonyl group: 2.96, with the Asn194 amine group: 2.86, with the Asn194 carbonyl group: 2.80, with the Lys214 residue: 2.91 Å.
Furthermore, new interactions were predicted from the crystal structure, such as Lys214 by the introduction of a functional group at the pyrimidine 5-position.
These orotate derivatives were used to analyze the interaction energy by the FMO method. Table 2 shows the results of the interaction energy analyses for some amino acid residues and FMN. Lys43, Asn67, Asn194, and FMN showed strong interactions with the orotate moiety.
Table 2. Interaction energies between TcDHODH and orotate derivatives (kcal mol-1).
| PDB ID | Derivative | Lys43 | Asn67 | Asn194 | FMN | Lys214 |
|---|---|---|---|---|---|---|
| 3W1R | 1 | -11.40 | -36.98 | -25.47 | -9.64 | 1.72 |
| 3W1T | 2 | -10.26 | -35.07 | -23.67 | -8.39 | 3.17 |
| 3W1U | 3 | -11.01 | -36.88 | -30.16 | -8.92 | 0.17 |
| 3W1X | 4 | -12.89 | -38.06 | -24.23 | -9.78 | 2.66 |
| 3W2J | 5 | -7.78 | -40.08 | -25.61 | -10.69 | -11.89 |
| 3W2K | 6 | -9.97 | -39.10 | -26.14 | -9.37 | 0.26 |
| 3W2L | 7 | -13.14 | -35.55 | -24.46 | -8.65 | 4.41 |
| 3W2M | 8 | -8.87 | -39.46 | -26.45 | -10.48 | -7.13 |
| 3W2N | 9 | -13.00 | -39.49 | -27.22 | -8.41 | -2.00 |
| 3W2U | 10 | -5.28 | -37.27 | -24.90 | -9.33 | -7.48 |
| 3W3O | 11 | -14.69 | -36.40 | -28.38 | -7.77 | 2.84 |
| 3W22 | 12 | -12.59 | -37.19 | -27.77 | -8.49 | 0.46 |
| 3W23 | 13 | -8.46 | -37.26 | -24.82 | -9.77 | 2.78 |
| 3W6Y | 14 | -14.43 | -34.32 | -25.60 | -8.09 | 5.73 |
| 3W7C | 15 | -14.39 | -36.21 | -25.72 | -8.73 | 3.13 |
| 3W7D | 16 | -15.93 | -38.93 | -28.17 | -7.48 | 0.58 |
| 3W7E | 17 | -12.63 | -39.04 | -27.31 | -8.50 | 1.59 |
| 3W7G | 18 | -2.77 | -39.69 | -23.95 | -10.71 | 0.69 |
| 3W7H | 19 | -12.81 | -38.30 | -24.42 | -8.36 | 1.51 |
| 3W7I | 20 | -16.99 | -34.75 | -23.45 | -8.51 | 2.58 |
| 3W7J * | 21* | -10.24 | -39.80 | -27.15 | -9.62 | 1.88 |
| 3W7J ** | 21** | -9.52 | -35.73 | -26.05 | -12.73 | -16.50 |
| 3W7K | 22 | -17.90 | -34.85 | -29.04 | -7.66 | -3.67 |
| 3W7L | 23 | -5.54 | -39.55 | -24.21 | -9.74 | 1.06 |
| 3W7M | 24 | -11.72 | -39.73 | -26.86 | -7.00 | 4.99 |
| 3W7N | 25 | -13.58 | -40.59 | -26.87 | -7.37 | 1.93 |
| 3W7O | 26 | -11.13 | -37.11 | -25.37 | -9.26 | 2.05 |
| 3W7P | 27 | -8.50 | -37.28 | -25.71 | -9.76 | -13.55 |
| 3W7Q | 28 | -13.21 | -32.89 | -26.72 | -7.85 | -1.95 |
| 3W70 | 29 | -17.93 | -31.13 | -27.90 | -4.56 | -0.07 |
| 3W71 | 30 | -14.88 | -37.71 | -25.69 | -8.27 | 5.48 |
| 3W72 | 31 | -15.76 | -38.29 | -30.00 | -8.21 | 0.78 |
| 3W73 | 32 | -12.73 | -39.97 | -29.00 | -8.74 | -1.02 |
| 3W74 | 33 | -10.35 | -33.38 | -27.55 | -10.09 | -3.62 |
| 3W75 | 34 | -17.08 | -40.20 | -29.55 | -8.96 | 3.53 |
| 3W76 | 35 | -15.64 | -39.26 | -28.43 | -9.97 | 4.56 |
| 3W83 | 36 | -16.05 | -25.20 | -25.57 | -6.43 | 0.84 |
| 3W84 | 37 | -10.78 | -34.59 | -24.88 | -7.99 | 4.40 |
| 3W85 | 38 | -13.72 | -34.61 | -29.44 | -8.75 | -0.12 |
| 3W86 | 39 | -11.87 | -42.10 | -29.49 | -8.79 | 6.61 |
| 3W87 | 40 | -15.37 | -39.60 | -27.92 | -8.41 | -3.40 |
| 3W88 | 41 | -13.33 | -39.38 | -29.22 | -9.34 | -4.98 |
| 4JD4 | 42 | -16.41 | -38.66 | -29.45 | -8.39 | -1.31 |
| 4JDB | 43 | -14.67 | -36.77 | -26.81 | -8.33 | 1.24 |
* Lys214 alternative conformer A
** Lys214 alternative conformer B
The interaction energies between three amino acid residues and all orotate derivatives were strongly similar to those of orotate. Our analyses indicated that interactions between the orotate moiety and these three amino acid residues as well as FMN were strongly conserved for all of the derivatives. In contrast, some derivatives such as the alternative conformer B of 21 showed the strongest interaction with Lys214, followed by 27, 5, 10, and 8 with interaction energies of -16.50, -13.55, -11.89, -7.48, and -7.13 kcal mol-1, respectively. The carboxylate group introduced in the phenyl moiety of 27 formed a hydrogen bond with the Lys214 residue (Fig 6). Derivatives 40 and 41 interacted with Lys214 with low energy despite the introduction of a carboxylate group on the aromatic ring moiety. Derivatives 40 and 41 were linked to a functional group at the pyrimidine 5-position through a butylene linker. In contrast, derivatives 21 and 27 interacted strongly (>10 kcal mol-1) when coupled with functional groups through an ethylene linker. These results suggest that the ethylene linker is appropriate for mediating interactions with Lys214.
Pharmacophore obtained from calculation
From these results, intermolecular interactions with the Lys43, Asn67 and Asn194 by hydrogen bond and FMN through π-π interaction were obtained as a common pharmacophore in the orotate moiety and intermolecular interactions with the Lys214 by hydrogen bond obtained as particular pharmacophore at derivatives 5, 21 and 27.
Discussion
This study provides a comprehensive FMO-based interaction analysis between TcDHODH residues and 43 orotate derivative inhibitors. Our analysis indicated that the orotate moiety formed hydrogen bonds with the Lys43, Asn67, and Asn194 residues of TcDHODH and a π-π interaction with FMN. These interactions were conserved in oxonate and orotate derivatives, and are also consistent with the magnitude of the interaction energy. From these results, the orotate moiety of orotate derivatives maintained these interactions whether or not functional groups were introduced at the pyrimidine ring 5-position. We accordingly expected a pharmacophore model containing molecules with hydrogen-bond donor/acceptor groups corresponding to Lys43, Asn67, and Asn194, such as amine, carbonyl, and carboxylate groups with an aromatic ring close to FMN.
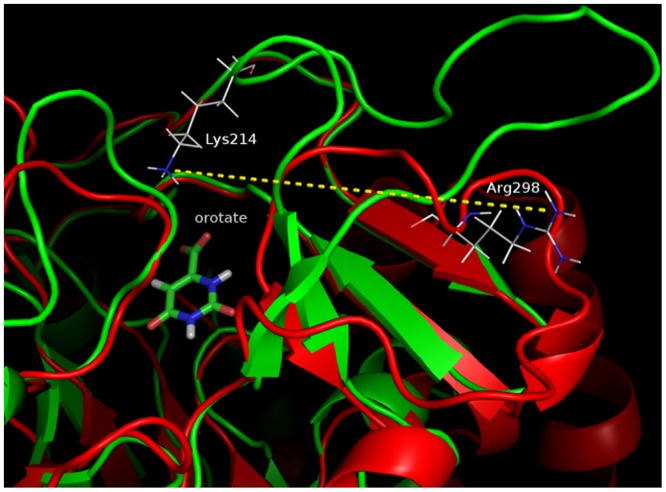
In contrast, some derivatives containing an acceptor at a functional group form a hydrogen bond with the Lys214 residue. In particular, the energies of interaction between Lys214 and derivatives 5, 21, 27 were predicted to be stronger than -10 kcal mol-1. Furthermore, alignment analysis has shown that the TcDHODH Lys214 residue is replaced by Arg298 in human DHODH [15]. In their crystal structures, TcDHODH Lys214 and human DHODH Arg298 are each located in the loop connecting βF–βG and β6–βE [14–15]. These loop regions are poorly conserved within the family of DHODHs, an observation consistent with the difference in the loop structures (Fig 7); whereas, the amino group of TcDHODH Lys214 points towards the active site, the guanidium group of human DHODH Arg298 points to the opposite side, with a distance of 20 Å between the two.
Fig 7. Comparison of loop structures.
Alignment structure based on orotate (green ribbon: 2E6A, blue ribbon: 1D3G, distance between TcDHODH Lys214 and human DHODH Arg298: 19.5 Å). Superimposition was performed by “Align Binding Sites” in Maestro based on orotate.
Thus, even if TcDHODH Lys214 and human DHODH Arg298 are apparently conserved by alignment analysis, they are structurally not conserved. Consequently, orotate derivatives such as derivatives 5, 21, and 27 may specifically inhibit TcDHODH by interacting with the Lys214 residue. Thus, a hydrogen bond acceptor as a particular pharmacophore corresponding to Lys214 is necessary to inhibit TcDHODH selectively.
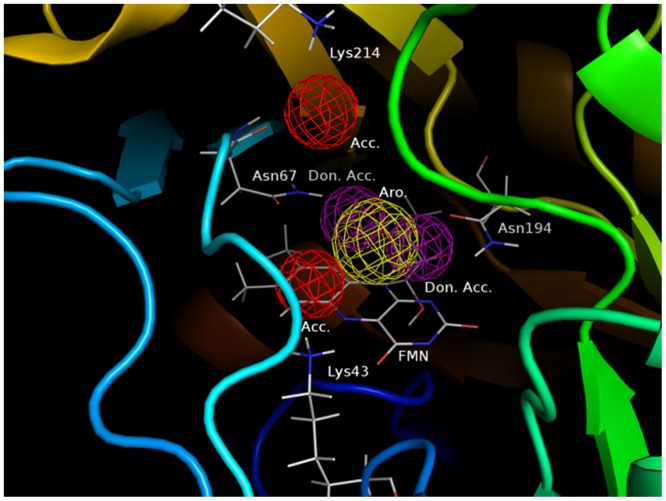
In conclusion, we propose a pharmacophore model inferred from FMO-based interaction analysis. Hydrogen bond acceptor pharmacophores correspond to Lys43 and Lys214, hydrogen bond donor and acceptor pharmacophores correspond to Asn67 and Asn197, and an aromatic ring pharmacophore corresponds to FMN, indicating important characteristics of compounds expected to inhibit Trypanosoma cruzi DHODH (Fig 8).
Fig 8. Pharmacophore inferred from interaction energy analysis.
Acc.: Acceptor, Don.: Donor and Aro.: Aromatic.
These characteristics inferred from single-point FMO calculations for a large number of structures provide insights into the ligand—amino acid residue interactions important for pharmacophore modeling and may facilitate the development of TcDHODH—targeted anti-Chagas drugs.
Acknowledgments
The authors would like to thank the anonymous reviewers for their valuable comments and suggestions to improve the quality of the paper. We also thank all beamline staff members from SPring-8 as well as Photon Factory for their help with X-ray diffraction data collection. This research was partially supported by Japanese Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 25870222 and 24240044 (to M.S.). This work was supported in part by Creative Scientific Research Grant 18GS0314 (to K.K.), Grant-in-aid for Scientific Research 18073004 and 26253025 (to K.K.) and 26870119 (to D.K.I.) from the JSPS, and Targeted Proteins Research Program (to K.K. and S.H.) from the Japanese Ministry of Education, Science, Culture, Sports and Technology (MEXT). We also acknowledge the support from Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (to K.K. and S.H.) and JST/JICA, SATREPS (Science and Technology Research Partnership for Sustainable Development) (10000284) to K.K. Astellas Pharma Inc., provided support in the form of salaries for authors YH, KO and MO.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was partially supported by Japanese Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 25870222 and 24240044 (to M.S.). This work was supported in part by Creative Scientific Research Grant 18GS0314 (to K.K.), Grant-in-aid for Scientific Research 18073004 and 26253025 (to K.K.) and 26870119 (to D.K.I.) from the JSPS, and Targeted Proteins Research Program (to K.K. and S.H.) from the Japanese Ministry of Education, Science, Culture, Sports and Technology (MEXT). The authors also acknowledge the support from Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (to K.K. and S.H.) and JST/JICA, SATREPS (Science and Technology Research Partnership for Sustainable Development) (10000284) to K.K. Astellas Pharma Inc. provided support in the form of salaries for authors YH, KO and MO (YZ1005).
References
- 1. Lockman JW, Hamilton AD. Recent developments in the identification of chemotherapeutics for Chagas disease. Curr Med Chem. 2005;12: 945–959. [DOI] [PubMed] [Google Scholar]
- 2. Schmunis GA, Yadon ZE. Chagas disease: a Latin American health problem becoming a world health problem. Acta Trop. 2010;115: 14–21. 10.1016/j.actatropica.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 3. Roellig DM, Brown EL, Barnabe C, Tibayrenc M, Steurer FJ, Yabsley MJ. Molecular typing of Trypanosoma cruzi isolates. United States. Emerg Infect Dis. 2008;14: 1123–1125. 10.3201/eid1407.080175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marcet PL, Duffy T, Cardinal MV, Burgos JM, Lauricella MA, Levin MJ, et al. PCR-based screening and lineage identification of Trypanosoma cruzi directly from faecal samples of triatomine bugs from northwestern Argentina. Parasitology. 2006;132: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rassi A Jr, Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375: 1388–1402. 10.1016/S0140-6736(10)60061-X [DOI] [PubMed] [Google Scholar]
- 6. Chagas C. Nova trypanozomiaze humana: estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homem. Mem Inst Oswaldo Cruz. 1909;1: 159–218. [Google Scholar]
- 7. Kirchhoff LV. American trypanosomiasis (Chagas’ disease). Gastroenterol Clin North Am. 1996;25: 517–533. [DOI] [PubMed] [Google Scholar]
- 8. Rossi MA, Bestetti RB. The challenge of chagasic cardiomyopathy. Cardiology. 1995;86: 1–7. [DOI] [PubMed] [Google Scholar]
- 9. Hugo VDB. Chemotherapy for parasitic infections. Nature. 1978;273: 626–630. 351413 [Google Scholar]
- 10. Linares GEG, Ravaschino EL, Rodriguez JB. Progresses in the field of drug design to combat tropical protozoan parasitic diseases. Curr Med Chem. 2006;13: 335–360. [DOI] [PubMed] [Google Scholar]
- 11. Maya JD, Cassels BK. Iturriaga-Vasquez P, Ferreira J, Faundez M, Galanti N, et al. Mode of action of natural and synthetic drugs against Trypanosoma cruzi and their interaction with the mammalian host. Comp Biochem Physiol A Mol Integr Physiol. 2007;146: 601–620. [DOI] [PubMed] [Google Scholar]
- 12. Buckner FS, Navabi N. Advances in Chagas disease drug development: 2009–2010. Curr Opin Infect Dis. 2010;23: 609–616. 10.1097/QCO.0b013e3283402956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen KF, Björnberg O. Evolutionary and functional families of dihydroorotate dehydrogenases. Paths Pyrimidines. 1998;6:20–28. [Google Scholar]
- 14. Liu S, Neidhardt EA, Grossman TH, Ocain T, Clardy J. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure. 2000;8: 25–33. [DOI] [PubMed] [Google Scholar]
- 15. Inaoka DK, Sakamoto S, Shimizu H, Shiba T, Kurisu G, Nara T, et al. Structures of Trypanosoma cruzi dihydroorotate dehydrogenase complexed with substrates and products: atomic resolution insights into mechanisms of dihydroorotate oxidation and fumarate reduction. Biochemistry. 2008;47: 10881–10891. 10.1021/bi800413r [DOI] [PubMed] [Google Scholar]
- 16. Annoura T, Nara T, Makiuchi T, Hashimoto T, Aoki T. The origin of dihydroorotate dehydrogenase genes of kinetoplastids, with special reference to their biological significance and adaptation to anaerobic, parasitic conditions. J Mol Evol. 2005;60: 113–127. [DOI] [PubMed] [Google Scholar]
- 17. Davies M, Heikkilä T, McConkey GA, Fishwick CW, Parsons MR, Johnson AP. Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases. J Med Chem. 2009;52: 2683–2693. 10.1021/jm800963t [DOI] [PubMed] [Google Scholar]
- 18. Davis JP, Cain GA, Pitts WJ, Magolda RL, Copeland RA. The immunosuppressive metabolite of leflunomide is a potent inhibitor of human dihydroorotate dehydrogenase. Biochemistry. 1996;35: 1270–1273. [DOI] [PubMed] [Google Scholar]
- 19. Coteron JM, Marco M, Esquivias J, Deng X, White KL, White J, et al. Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J Med Chem. 2011;54: 5540–5561. 10.1021/jm200592f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phillips MA, Rathod PK. Plasmodium dihydroorotate dehydrogenase: a promising target for novel anti-malarial chemotherapy. Infect Disord Drug Targets. 2010;10: 226–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Copeland RA, Marcinkeviciene J, Haque TS, Kopcho LM, Jiang W, Wang K, et al. Helicobacter pylori-selective antibacterials based on inhibition of pyrimidine biosynthesis. J Biol Chem. 2000;275: 33373–33378. [DOI] [PubMed] [Google Scholar]
- 22. Diao Y, Lu W, Jin H, Zhu J, Han L, Xu M, et al. Discovery of Diverse Human Dihydroorotate Dehydrogenase Inhibitors as Immunosuppressive Agents by Structure-Based Virtual Screening. J Med Chem. 2012;55: 8341–8349. 10.1021/jm300630p [DOI] [PubMed] [Google Scholar]
- 23. Marwaha A, White J, El Mazouni F, Creason SA, Kokkonda S, Buckner FS, et al. Bioisosteric Transformations and Permutations in the Triazolopyrimidine Scaffold To Identify the Minimum Pharmacophore Required for Inhibitory Activity against Plasmodium falciparum Dihydroorotate Dehydrogenase. J Med Chem. 2012;55: 7425–7436 10.1021/jm300351w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wermuth CG, Ganellin CR, Lindberg P, Mitscher LA. Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998). Pure and Applied Chemistry. 1998;70: 1129–1143. [Google Scholar]
- 25. Lingling J, Robert CR. Pharmacophore-based similarity scoring method for DOCK. J Phys Chem B. 2015. January 22;119(3):1083–102. 10.1021/jp506555w Epub.2014.10.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kitaura K, Sawai T, Asadaa T, Nakanob T, Uebayasi M. Pair interaction molecular orbital method: an approximate computational method for molecular interactions. Chem Phys Lett. 1999;312: 319–324. [Google Scholar]
- 27. Tomonaga O, Kosuke O. CH/π hydrogen bonds determine the selectivity of the Src homology 2 domain to tyrosine phosphotyrosyl peptides: an ab initio fragment molecular orbital study. Journal of Computational Chemistry. 2008;29: 2656–2666. 10.1002/jcc.20998 [DOI] [PubMed] [Google Scholar]
- 28. Tomonaga O, Eiichi T, Motoyasu O, Cchiaki H, Harunobu M, Tochihiro N,et al. The importance of CH/π hydrogen bonds in rational drug design: an ab initio fragment molecular orbital study to leukocyte-specific protein tyrosine (LCK) kinase. Bioorganic and Medicinal Chemistry. 2008;16: 10311–10318. 10.1016/j.bmc.2008.10.041 [DOI] [PubMed] [Google Scholar]
- 29. Tomonaga O, Kosuke O, Kazuo K. CH/π hydrogen bonds play a role in ligand recognition and equilibrium between active and inactive states of the b2 adrenergic receptor: An ab initio fragment molecular orbital (FMO) study. Bioorganic and Medicinal Chemistry. 2011;19: 5231–5237. 10.1016/j.bmc.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 30. Tomonaga O, Kosuke O, Kazuo K. Importance of CH/π hydrogen bonds in recognition of the core motif in proline-recognition domains: an ab initio fragment molecular orbital study. Journal of Computational Chemistry. 2011;32: 2774–2782. 10.1002/jcc.21857 [DOI] [PubMed] [Google Scholar]
- 31. Kenichi F, Yoshimasa S. The role of fluorine atoms in a fluorinated prostaglandin agonist. Chem Med Chem. 2010;5: 1254–1257. 10.1002/cmdc.201000174 [DOI] [PubMed] [Google Scholar]
- 32. Ohno K., Mori K., Orita M., Takeuchi M.. Computational insights into binding of bisphosphates to farnesyl pyrophosphate synthase. Current Medicinal Chemistry. 2011;18: 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Osamu I, John B, Richard JL, Mark W, Compound design by fragment linking. Molecular Informatics. 2011;30: 298–306. [DOI] [PubMed] [Google Scholar]
- 34. Michael PM, Osamu I, Richard JL, Mark W, Prediction of cyclin dependent kinase 2 inhibitor potency using the fragment molecular orbital method. Journal of Cheminformatics. 2011;3: 2 10.1186/1758-2946-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Björnberg O., Jordan DB, Palfey BA, Jensen KF. Dihydrooxonate is a substrate of dihydroorotate dehydrogenase (DHOD) providing evidence for involvement of cysteine and serine residues in base catalysis. Arch. Biochem. Biophys. 2001;391: 286–94. [DOI] [PubMed] [Google Scholar]
- 36. Zbyszek O and Wladek M. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997; 276: 307–326. [DOI] [PubMed] [Google Scholar]
- 37. Emsley P and Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53: 240–255. [DOI] [PubMed] [Google Scholar]
- 39.http://www1.bbiq.jp/zzzfelis/Facio.html
- 40. Fedorov DG, Kazuo K. Pair interaction energy decomposition analysis. J Comput Chem. 2007;28: 222–237. [DOI] [PubMed] [Google Scholar]
- 41. Ozawa T, Tsuji E, Ozawa M, Handa C, Mukaiyama H, Nishimura T, et al. The importance of CH/pi hydrogen bonds in rational drug design: An ab initio fragment molecular orbital study to leukocyte-specific protein tyrosine (LCK) kinase. Bioorg Med Chem. 2008;16: 10311–10318. 10.1016/j.bmc.2008.10.041 [DOI] [PubMed] [Google Scholar]
- 42. Watanabe C, Fukuzawa K, Okiyama Y, Tsukamoto T, Kato A, Tanaka S, et al. Three- and four-body corrected fragment molecular orbital calculations with a novel subdividing fragmentation method applicable to structure-based drug design. J Mol Graph Model. 2013l;41: 31–42. 10.1016/j.jmgm.2013.01.006 [DOI] [PubMed] [Google Scholar]
- 43.David. AC, Tom D, Thomas EC, Carlos S, Junmei. W, Robert. ED, et al., Amber 11. San Francisco. 2010
- 44. Michael WS, Kim KB, Jerry AB, Steven TE, Mark SG, Jan HJ, et al. General atomic and molecular electronic structure system. J Comput Chem. 1993;14: 1347–1363. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.