SUMMARY
The mammalian target of rapamycin complex 1 (mTORC1) is regulated, in part, by the endogenous inhibitor DEPTOR. However, the mechanism of DEPTOR regulation with regard to rapid mTORC1 activation remains unknown. We report that DEPTOR is rapidly and temporarily dissociated from mTORC1 upon mitogenic stimulation, suggesting a mechanism underlying acute mTORC1 activation. This mitogen-stimulated DEPTOR dissociation is blocked by inhibition or depletion of the mTORC1 regulator, phospholipase D (PLD), and recapitulated with the addition of the PLD product phosphatidic acid (PA). Our mass spectrometry analysis has independently identified DEPTOR as an mTOR binding partner dissociated by PA. Interestingly, only PA species with unsaturated fatty acid chains, such as those produced by PLD, are capable of displacing DEPTOR and activating mTORC1, with high affinity for the FRB domain of mTOR. Our findings reveal a novel mechanism of mTOR regulation and provide a molecular explanation for the exquisite specificity of PA function.
INTRODUCTION
Mammalian target of rapamycin (mTOR) is a Ser/Thr kinase that responds to a variety of intra- and extra-cellular signals such as amino acids and mitogens in order to coordinate a multitude of cellular processes with the appropriate resources and demand. As a master regulator, mTOR nucleates two biochemically and functionally distinct complexes, namely mTORC1 and mTORC2, which lie at the center of an extensive signaling network (Laplante and Sabatini, 2012). In its active form, mTORC1 stimulates protein synthesis and cell growth through the phosphorylation of the ribosomal S6 kinase (S6K1) and other substrates (Ma, 2009). Catalytically active mTORC2, on the other hand, phosphorylates a different set of substrates, with Akt being the best characterized (Laplante and Sabatini, 2012; Oh and Jacinto, 2011). Downstream signaling of both mTOR complexes is activated in response to mitogens. In the case of mTORC1, this process is dependent on amino acids, which trigger the recruitment of mTORC1 to lysosomal membranes through the Rag small GTPases (Sancak et al., 2010; Sancak et al., 2008), where it is activated by another small GTPase, Rheb (Menon et al., 2014), and phospholipase D1 (PLD1) (Yoon et al., 2011a).
PLD1 and PLD2 catalyze the hydrolysis of phosphatidylcholine (PC) to phosphatidic acid (PA). Both PLD enzymes preferentially hydrolyze mono- or di-unsaturated PC, generating PA species with one or two degrees of unsaturation (Pettitt et al., 2001). Phosphatidic acid produced by PLD serves, primarily, as a second messenger to regulate a range of signaling proteins (Jenkins and Frohman, 2005). Previously, we identified that monounsaturated 16:0-18:1 PA binds with high affinity to the FKBP12 rapamycin binding (FRB) domain of mTOR and that this interaction is in direct competition with the mTOR-specific inhibitor, rapamycin (Fang et al., 2001). Furthermore, we have shown that PLD1 and PA are critical mediators of mTORC1 activation by mitogens as well as amino acid signals (Fang et al., 2003; Fang et al., 2001; Sun et al., 2008; Yoon et al., 2011a), and that PA binding directly stimulates mTORC1 kinase activity (Yoon et al., 2011b). Such regulation of mTOR interactions and, thereby, mTOR activity is crucial for maintaining tight control of cell growth and proliferation.
Indeed, while mutations in mTOR itself are rare, dysregulation of mTOR signaling has been suggested to be a common contributor in cancer (Guertin and Sabatini, 2007). DEPTOR is one such protein that normally binds and inhibits both mTOR complexes but, when overexpressed, alleviates mTORC1 inhibition of mTORC2 signaling, and thereby promotes cancer cell survival (Peterson et al., 2009). In the absence of mitogens, DEPTOR mRNA and protein levels rise, giving way to increased interaction with and inhibition of both mTOR complexes (Peterson et al., 2009). Following the addition of mitogens, DEPTOR is phosphorylated by mTOR, triggering its ubiquitination and subsequent degradation over the course of several hours (Duan et al., 2011; Gao et al., 2011; Peterson et al., 2009; Zhao et al., 2011). While this gradual degradation of DEPTOR may underlie prolonged or basal mTOR activation, it cannot explain how acute stimulation of mTOR triggers maximal signaling activity long before DEPTOR protein levels are affected. Herein, we report that PA produced by PLD specifically binds to mTOR and displaces DEPTOR, revealing, for the first time, a mechanism of acute mTOR regulation involving DEPTOR.
RESULTS
Acute Mitogenic Stimulation Disrupts DEPTOR-mTORC1 Interaction
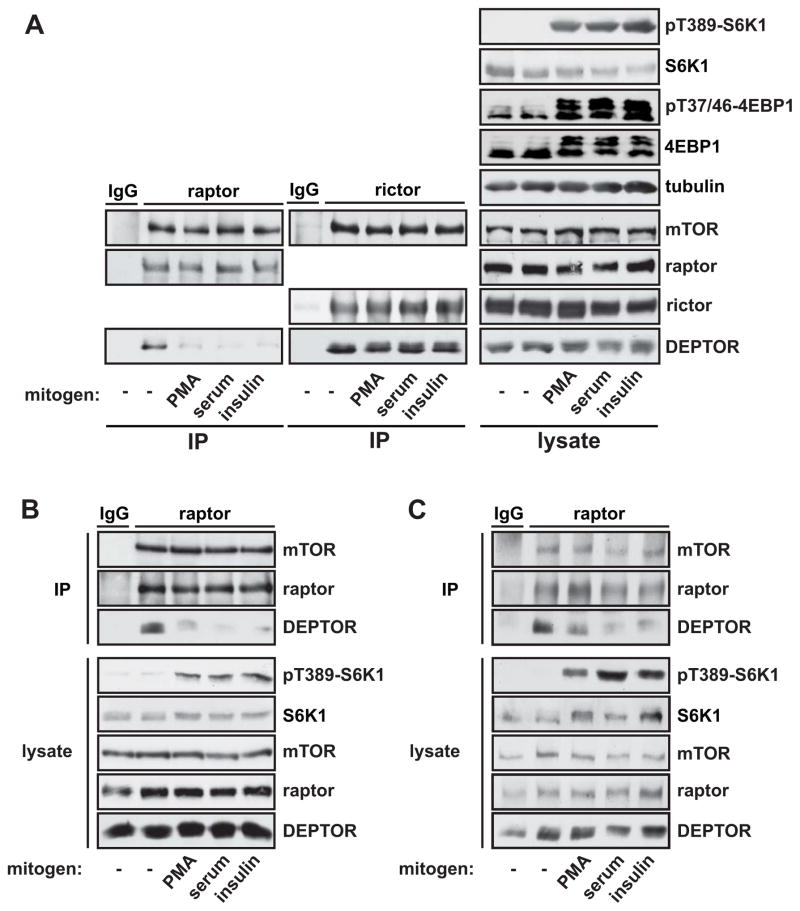
Aware that mitogenic stimulation triggers rapid activation of mTORC1 and mTORC2, we wondered if any effect on the endogenous inhibitor, DEPTOR, could be detected within the same time frame. As shown in Figure 1A, phosphorylation of the mTORC1 substrates S6K1 and 4EBP1 was robustly induced by 30 min stimulation with phorbol 12 myristate 13-acetate (PMA), serum, or insulin. As expected, mTORC1 and mTORC2 isolated from serum-starved cells by raptor and rictor immunoprecipitation, respectively, were bound by DEPTOR. Intriguingly, all three stimuli reduced the amount of DEPTOR associated with mTORC1, but not mTORC2, within the same time window as mTORC1 substrate phosphorylation (Figure 1A).
Figure 1. Acute Mitogenic Stimulation Disrupts DEPTOR-mTORC1 Interaction.
(A) HEK293 cells were serum starved overnight and stimulated for 30 min with 100 nM PMA, 10% serum, or 100 nM insulin. mTORC1 and mTORC2 were isolated by immunoprecipitation of raptor and rictor, respectively. Cell lysates and immunoprecipitates were analyzed by western blotting. C2C12 cells (B) and Swiss 3T3 cells (C) were serum starved overnight and stimulated and analyzed as in (A). Western data shown throughout the paper are representative of three or more independent experiments with similar results. See also Figure S1.
Extending our analysis to C2C12 (Figure 1B) and Swiss 3T3 cells (Figure 1C) revealed that mitogen-stimulated DEPTOR displacement from mTORC1 is conserved across cell types. In addition, we found that lysophosphatidic acid (LPA), which activates PLD1 and thereby mTORC1 (Kam and Exton, 2004), also induced DEPTOR dissociation from mTORC1 in Swiss 3T3 cells (Figure S1A and S1B). We observed that stimulation with amino acids, another mTORC1 stimulus, also caused DEPTOR displacement (Figure S1C), though this effect was not seen in the absence of mitogens (data not shown). Taken together, our results suggest, for the first time, that rapid mitogenic activation of mTORC1 may involve dissociation of the endogenous inhibitor, DEPTOR.
Mitogen-Induced DEPTOR Dissociation is Mediated by PLD1 and its Product, Phosphatidic Acid
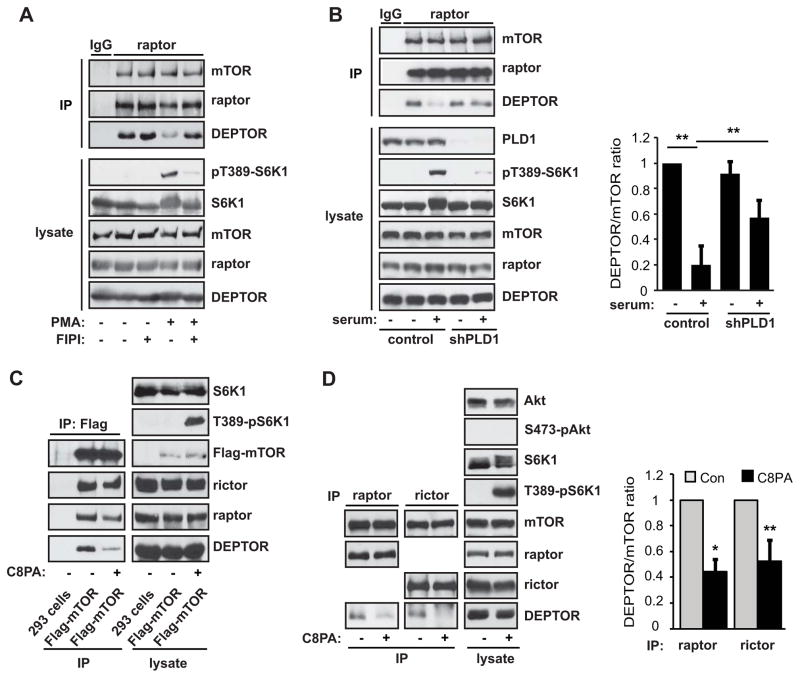
Because all the stimuli shown above to induce DEPTOR dissociation from mTORC1 are also known to stimulate PLD1, we set out to investigate the possibility of a connection between PLD1 and DEPTOR dissociation. Treatment of cells with FIPI (5-Fluoro-2-indolyl des-chlorohalopemide), an inhibitor of PLD, inhibited mTORC1 activity as confirmed by a drastic reduction in PMA-stimulated S6K1 phosphorylation (Figure 2A). At the same time, FIPI blocked PMA-stimulated DEPTOR displacement from mTORC1 (Figure 2A), and the effect of FIPI was dose-dependent (Figure S2A). Although FIPI at 5 μM inhibited the majority of PMA-stimulated PLD activity, an above-basal activity persisted until the cells were treated by 30 μM FIPI (Figure S2B). Importantly, PA production at the lysosome, as demonstrated by a PA sensor concentrating at the lysosomal area and colocalizing with the lysosomal marker LAMP1, was abolished by 30 μM and not 5 μM FIPI (Figure S2C). This is in full agreement with the requirement of PLD1 lysosomal translocation for mTORC1 activation (Yoon et al., 2011a). Additionally, FIPI increased the DEPTOR-mTORC1 association in starved cells (Figure 2A), which likely reflects a robust FIPI inhibition of basal PLD activity otherwise present in starved cells.
Figure 2. Mitogen-Induced DEPTOR Displacement is Mediated by PLD1 and PA.
(A) HEK293 cells were serum starved overnight and stimulated with 200 nM PMA with or without 50 μM FIPI for 30 min. Cell lysates and raptor immunoprecipitates were analyzed by western blotting. (B) HEK293 cells were infected with a negative control- or PLD1-shRNA and selected with puromycin for 3 days, followed by serum starvation overnight, and 10% serum stimulation for 30 min. Cell lysates and raptor immunoprecipitates were analyzed as in (A). DEPTOR:mTOR ratios were calculated based on band intensities measured by densitometry. Data are mean ± SD, with paired t-tests performed as indicated. (C) HEK293 cells stably expressing Flag-mTOR were serum starved overnight. Where indicated, cells were treated for 30 min with 300 μM C8PA vesicles. Flag-IP was performed and analyzed by western blotting. (D) mTORC1 and mTORC2 were isolated as raptor and rictor immunoprecipitates, respectively, from cells starved or stimulated and analyzed as in (B). Data are mean ± SD, analyzed by one-sample t-tests. See also Figure S2 and Figure S3.
To further validate the specific involvement of PLD1, we delivered a previously reported shRNA against PLD1 (Sun et al., 2008) via lentiviral infection of the cells. As shown in Figure 2B, PLD1 knockdown significantly reduced the degree of DEPTOR dissociation seen in serum-stimulated cells, as compared to cells that received control shRNAs. The less complete blockage of dissociation by RNAi compared to FIPI is likely due to the incomplete removal of PLD1 by the knockdown. Collectively, these observations strongly suggest that DEPTOR dissociation from mTORC1 is mediated by PLD1.
Previously, we identified that the PLD1 product PA binds the FRB domain in mTOR and showed that PA binding is necessary for mTOR kinase activity and signaling (Fang et al., 2001; Yoon et al., 2011b). However, the mechanism of mTOR activation remains incompletely understood. In a parallel investigation into the mechanism of mTOR regulation, we utilized nano-liquid chromatography (nanoLC) high resolution Fourier-transform mass spectrometry (FTMS) to analyze protein interactions with stably expressed Flag-tagged mTOR under different conditions. For the purpose of studying exogenous PA effects on mTOR in the cell, we routinely used a short-chain PA (di-8:0 PA, or C8PA) in order to preclude potential signaling from long-chain PA-derived LPA (Yoon et al., 2011b). The nanoLC-FTMS analysis revealed that delivery of C8PA to the cells prior to lysis led to reduced mTOR interaction with DEPTOR (Figure S3A) among other proteins (not shown). Co-immunoprecipitation experiments confirmed that, indeed, DEPTOR association with Flag-mTOR was decreased following treatment of HEK293 cells with PA (Figure 2C). These findings were consistent with our observation that mitogen-stimulated DEPTOR displacement is mediated by PLD1, and suggested that the process likely involves PA production.
Next, we examined the effect of PA on the association of endogenous mTOR with DEPTOR. Indeed, PA treatment led to a reduced level of DEPTOR in the raptor immunoprecipitates (Figure 2D). It is noteworthy that this effect was specific to PA and not recapitulated by rapamycin (Figure S3B), even though PA and rapamycin compete with each other to bind the FRB domain in mTOR (Fang et al., 2001). Surprisingly, the association of DEPTOR with mTORC2, as examined by rictor immunoprecipitation, was also impaired by PA treatment (Figure 2D). Nevertheless, whereas the PA-induced reduction in DEPTOR incorporation correlated with an increase in mTORC1 activity as measured by the phosphorylation of S6K1, mTORC2 activity toward Akt was not stimulated by PA treatment (Figure 2D), as previously described (Yoon et al., 2011b). These data suggest that mTORC1 activity, alone, is affected by the reduced DEPTOR binding triggered by PA treatment.
Phosphatidic Acid Displaces DEPTOR from mTORC1 to Activate Signaling
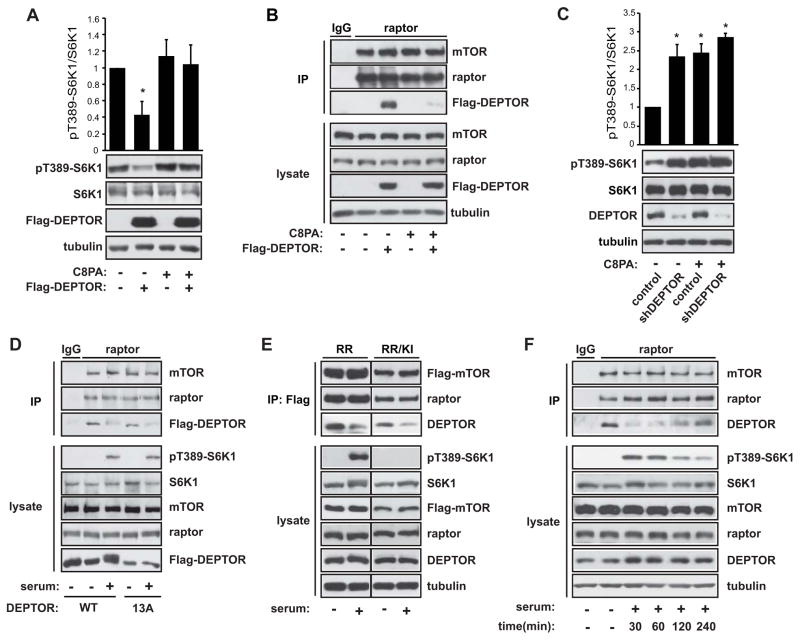
Consistent with the reported role of DEPTOR in inhibiting mTORC1, we found that DEPTOR overexpression inhibited S6K1 phosphorylation in cells (Figure 3A). Importantly, exogenous PA was sufficient to rescue mTORC1 signaling from the effect of DEPTOR overexpression (Figure 3A) while displacing overexpressed DEPTOR from mTORC1 (Figure 3B). Furthermore, DEPTOR knockdown led to an increase in phospho-S6K1 in serum-starved HeLa cells as reported (Peterson et al., 2009), but it had no effect on phospho-S6K1 stimulated by PA (Figure 3C) or mitogens (Figure S4A), validating the role of PA in removing DEPTOR inhibition of mTORC1. The capacity of PA to displace DEPTOR was confirmed in the same cells (Figure S4B).
Figure 3. PA Displaces DEPTOR from mTORC1 to Activate Signaling.
A) HEK293 cells were transiently transfected with Flag-DEPTOR, serum starved overnight, and stimulated with 300 μM C8PA vesicles for 30 min. Cell lysates were analyzed by western blotting, and the ratio of pT389-S6K1 versus total S6K1 was calculated based on band intensities measured by densitometry. (B) HEK293 cells stably expressing Flag-DEPTOR were starved overnight and then stimulated with 300 μM C8PA for 30 min. Cell lysates and raptor immunoprecipitates were analyzed by western blotting. (C) HeLa cells were infected with lentivirus expressing a negative control- or DEPTOR-shRNA and selected with puromycin for 4 days. The cells were then serum starved overnight and stimulated with 300 μM C8PA for 30 min. Cells lysates were analyzed by western blotting, and the ratio of pT389-S6K1 versus total S6K1 was calculated based on band intensities measured by densitometry. Data are mean ± SD. Statistical significance was found when comparing other samples to control (control shRNA and no C8PA), but not when comparing those samples with each other. (D) HEK293 cells stably expressing Flag-DEPTOR-WT or Flag-DEPTOR-13A were serum starved overnight and stimulated with 10% serum for 30 min. Cell lysates and raptor immunoprecipitates were analyzed by western blotting. (E) HEK293 cells stably expressing Flag-mTOR-RR or Flag-mTOR-RR/KI were serum starved overnight, and then stimulated with 10% serum in the presence of 100 nM rapamycin for 30 min. Cell lysates and Flag immunoprecipitates were analyzed by western blotting. (F) HEK293 cells were serum starved overnight and stimulated with 10% serum for the times indicated, followed by raptor IP and analysis by western blotting. See also Figure S4.
While these observations are consistent with a model where PA acts by binding to mTOR and subsequently dissociating DEPTOR, an equally plausible mechanism could be that PA simply activates mTORC1, which leads to phosphorylation (Peterson et al., 2009) and subsequent dissociation of DEPTOR. To examine this alternative possibility, we asked if phosphorylation by mTOR is required for DEPTOR dissociation. To that end, we utilized a non-phosphorylatable mutant of DEPTOR (DEPTOR-13A), which contains alanine substitutions at all 13 identified sites phosphorylated by mTOR (Peterson et al., 2009). As shown in Figure 3D, like the wild-type protein, DEPTOR-13A dissociated from mTORC1 in response to 30-min serum stimulation. To further examine the potential role of mTOR activity in DEPTOR dissociation, we took advantage of cells stably expressing a rapamycin-resistant mTOR in a wild-type (RR) or kinase-inactive (RR/KI) form. In the presence of rapamycin, serum-stimulated DEPTOR dissociation continued to occur even in RR/KI cells that lacked mTORC1 kinase activity (Figure 3E). Therefore, DEPTOR displacement from mTORC1 is independent of mTORC1 activity and DEPTOR’s mTOR-mediated phosphorylation state.
In response to mitogenic stimulation, PA levels quickly rise and fall back toward basal levels within 1 hour of stimulation (Cano et al., 1992; Fang et al., 2001; Fukami and Takenawa, 1992). Given this transient PA presence, we wondered if DEPTOR re-associated with mTORC1 over time. As shown in Figure 3F, DEPTOR indeed began to re-associate with mTORC1 within 2 hours of stimulation. The re-associated DEPTOR exhibited a diffuse resolution by western blotting, most likely reflecting its phosphorylation by mTOR over time as reported. In further support of the functional relevance of DEPTOR displacement, the re-association was accompanied by a reduction in S6K1 phosphorylation (Figure 3F), often observed within 1–2 hours of mitogenic stimulation in non-cancerous cells (e.g., (Chung et al., 1992).. This data is consistent with the idea that as mitogen-induced pools of PA drop to basal levels, DEPTOR-mTOR interaction returns and inhibits mTORC1 signaling.
PA Species Containing Unsaturated Acyl Chains Displace DEPTOR and Activate mTORC1
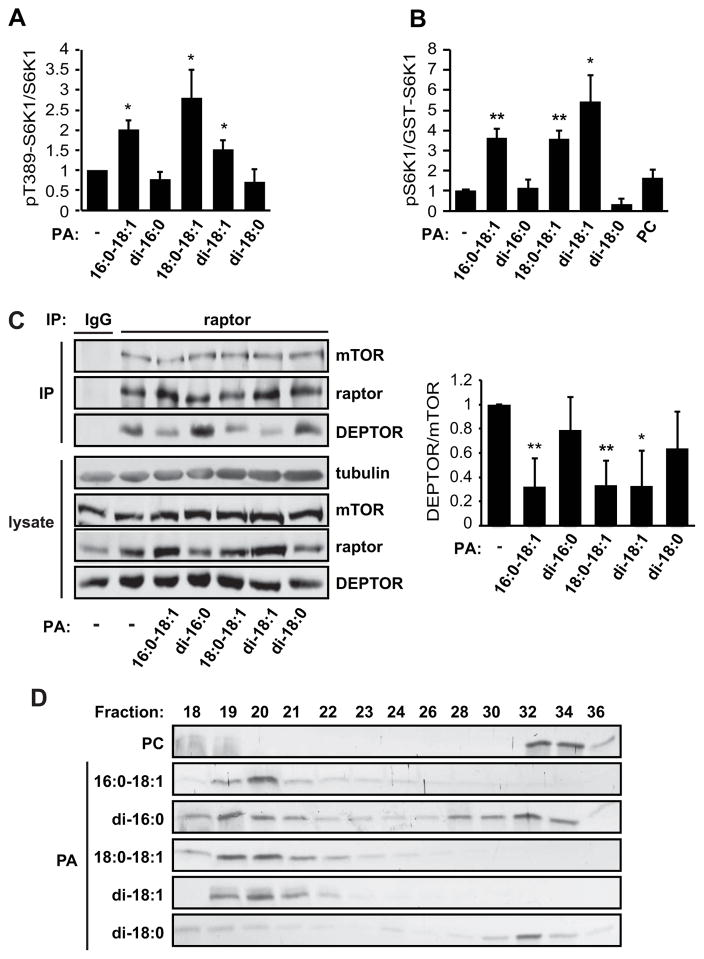
A recent report suggested that not all PA species are created equal but, rather, PA produced by different biosynthetic pathways may have distinct signaling capabilities (Zhang et al., 2012). Such a possibility prompted us to examine the effect of various PA species on mTORC1. PLD primarily generates PA species containing one or two partially unsaturated fatty acid chains, including 16:0-18:1, 18:0-18:1, and di-18:1 (Pettitt et al., 2001). On the other hand, GPAT1 (glyerol-3-phosphate acyltransferase 1)- and AGPAT2 (1-acyl-glyerol-3-phosphate acyltransferase 2)-driven PA production generates mainly PA species with saturated acyl chains, such as di-16:0 (Zhang et al., 2012). Given the necessity of PLD activity for PA-mediated DEPTOR displacement, we reasoned that this mechanism of mTORC1 activation might be specific to the class of PA species produced by PLD. Indeed, when we assessed the ability of different unsaturated or saturated PA species to stimulate mTORC1 activity, we found that all three unsaturated PA species tested (16:0-18:1, 18:0-18:1, and di-18:1) activated mTORC1 both in cells (Figure 4A) and in vitro (Figure 4B), while the saturated species (di-16:0 or di-18:0) had no significant effect on mTORC1 signaling or kinase activity. Furthermore, the ability of the unsaturated PA species to activate mTORC1 correlated with their ability to displace DEPTOR from mTORC1 (Figure 4C). These data suggest that the fatty acid chains confer functional specificity to PA, and that PA species produced by PLD may have a unique ability to displace DEPTOR from mTORC1 and activate the kinase.
Figure 4. Unsaturated PA Species Displace DEPTOR and Activate mTORC1.
(A) HEK293 cells were serum starved overnight and stimulated with 300 μM vesicles of the indicated PA species for 30 min. Cell lysates were analyzed by western blotting, and the ratio of pT389-S6K1 versus total S6K1 was calculated based on band intensities measured by densitometry. (B) mTORC1 was isolated by raptor IP from serum-starved cells and subjected to in vitro kinase assays using a GST-S6K1 peptide as substrate. Vesicles of various PA species were added to the kinase reactions at 300 μM. pT389-S6K1:GST-S6K1 ratios were calculated based on band intensities measured by densitometry. (C) HEK293 cells were treated as in (A), and raptor immunoprecipitates were analyzed by western blotting. DEPTOR:mTOR ratios were calculated based on band intensities measured by densitometry. Data in (A–C) are mean ± SD, and one-sample t-tests were performed to compare each data point to control (no PA). (D) Bacterially purified FRB protein was combined with SUVs of the indicated PA species for 30 min. The reaction mixture was then run on a Sephracryl-300 column to separate vesicle-bound (Fractions 19–22) and unbound (Fractions 30–36) FRB. Protein levels in collected fractions were analyzed by silver staining.
Given the importance of PA-mTOR association for mTORC1 activation (Fang et al., 2001; Yoon et al., 2011b), we reasoned that fatty acid chain unsaturation might influence PA affinity for mTOR, thereby constituting a mechanism of species selectivity. To test this theory, we performed in vitro FRB-lipid binding assays with vesicles composed of different PA species. As previously described (Fang et al., 2001), purified FRB protein bound to small unilamellar vesicles (SUVs) containing 16:0-18:1 PA, as analyzed by size exclusion chromatography. As shown in Figure 4D, 18:0-18:1 and di-18:1 PA vesicles were also completely retained by the FRB protein, whereas vesicles containing the saturated di-16:0 and di-18:0 PA exhibited partial binding to FRB. Because these binding assays were performed with identical protein and vesicle concentrations, the binding patterns reflected affinity of the various lipids for FRB. The strong interaction between FRB and the unsaturated PA species is consistent with the model that PA generated by PLD1 specifically interacts with mTOR to activate mTORC1 by displacing the endogenous inhibitor, DEPTOR.
DISCUSSION
Identification of the endogenous mTOR inhibitor, DEPTOR, prompted several studies into the mechanism and functional consequence of the DEPTOR degradation that occurs several hours after mTOR activation (Duan et al., 2011; Gao et al., 2011; Laplante et al., 2012; Peterson et al., 2009; Zhao et al., 2011). However, there has been little indication of how mTOR rapidly achieves maximal signaling activity long before DEPTOR is degraded. Collectively, the results we describe herein indicate that upon mitogenic stimulation DEPTOR is rapidly displaced from mTORC1 by PLD-generated PA, allowing mTORC1 activation. To our knowledge, this is the first description of a molecular mechanism by which mTORC1 is rapidly and robustly relieved of inhibition by DEPTOR and by which mTORC1 regulation is achieved independently of DEPTOR phosphorylation, the only event previously known to initiate regulation of DEPTOR. Furthermore, we demonstrate that this regulation is dependent on the composition of the PA species interacting with mTOR, revealing a preference of mTORC1 for unsaturated PA species. Fully in line with the well-known transient kinetics of cellular PA production, we have observed re-association of DEPTOR with mTORC1 following prolonged mitogenic stimulation, at which time DEPTOR degradation may take over as the primary means of removing this inhibitor.
It is interesting to note that while exogenously supplied PA displaces DEPTOR from both mTORC1 and mTORC2, this displacement only has functional consequences for mTORC1, as exogenous PA is sufficient to activate mTORC1 but not mTORC2, both in cells and in vitro (Yoon et al., 2011b). This is consistent with the fact that rapamycin, which is a competitive inhibitor of PA-mTOR interaction (Fang et al., 2001; Veverka et al., 2008), specifically inhibits mTORC1 long before it has any effect on mTORC2 (Sarbassov et al., 2006). The regulation of mTORC2 may require additional inputs. Perhaps for the same reason, mTORC2 appears less sensitive to regulation by DEPTOR, as knockdown of DEPTOR is sufficient to activate mTORC1 but not mTORC2, even though overexpression of DEPTOR inhibits both complexes (Peterson et al., 2009).
The reason DEPTOR interaction with mTORC2 is affected by exogenous PA but not by mitogenic stimulation may have to do with compartmentalization and subcellular concentrations of PA. Both mTORC1 and PLD1 are localized to the lysosome in the presence of amino acids (Sancak et al., 2010; Yoon et al., 2011a). mTORC2, on the other hand, has primarily been described to reside at the plasma membrane (Partovian et al., 2008) and the endoplasmic reticulum (ER) (Boulbés et al., 2011), and therefore is unlikely to encounter concentrated pools of PA produced by PLD1. PLD2 also produces PA and is broadly localized to the plasma membrane (Zhang et al., 2014), but it has never been shown to colocalize with mTORC2. The two may well reside in different membrane microdomains.
In addition to the PLD enzymes, glycerol phosphate acyltransferases (GPATs)/acylglycerolphosphate acyltransferases (AGPATs) and diacylglycerol kinases (DGKs) also generate PA. There are many different AGPAT and DGK isoforms; those that are well-characterized localize predominantly to the ER (Takeuchi and Reue, 2009), plasma membrane, and nucleus (Topham and Epand, 2009). Therefore, AGPATs and DGKs are unlikely to closely associate with mTORC1 in cells under physiological conditions. When overexpressed, AGPAT10 (Tang et al., 2006) and DGKζ (Ávila-Flores et al., 2005) were both reported to activate mTORC1, which could be a result of a broadened distribution throughout the cell and an increased proximity to mTORC1. In addition to close proximity with mTORC1, our results indicate that a propensity to produce unsaturated PA is another important dimension of a PA-producing enzyme’s involvement in regulating mTORC1. While AGPAT and DGK isoforms are certainly capable of producing unsaturated PA, none of them have been found to share the strong preference of the PLD enzymes for generating mono- and di-unsaturated PA species (Hollenback et al., 2009; Pettitt et al., 2001; Sukumaran et al., 2009; Takeuchi and Reue, 2009; Topham and Epand, 2009; Zhang et al., 2012). Nevertheless, it has been proposed that PA production by these two pathways could become significantly more important for mTOR regulation when production by PLD is compromised (Foster et al., 2014).
The acyl chain requirement for mTOR binding, DEPTOR displacement, and activation of mTORC1 described herein indicates the importance of subtle species variation within lipid signaling. Together with another recent report that acyl-chain derived specificity may functionally differentiate PA from different sources (Zhang et al., 2012), these findings contribute an additional degree of sophistication to our understanding of mTOR regulation. How the FRB domain of mTOR differentiates between saturated and unsaturated species of PA is still unclear. NMR, crystallography, and mutagenesis studies have provided significant insights into the structure of the FKBP12-rapamcin/PA binding pocket within FRB (Choi et al., 1996; Fang et al., 2001; Leone et al., 2006; Veverka et al., 2008). In the past, emphasis has been placed on the interaction between the lipid head group and FRB; significant additional insights may be gleaned from future studies addressing the involvement of acyl chains in PA-FRB binding.
EXPERIMENTAL PROCEDURES
Cell culture, cell lysis, immunoprecipitation, western blotting, lipid vesicle preparation, PLD assay, and lentivirus-mediated RNAi were all performed following standard procedures or as previously reported, with detailed information described in the Supplemental Experimental Procedures. Additional information regarding antibodies, reagents, plasmids, cell lysis, and mass spectrometry analysis also appear in the Supplemental Experimental Procedures.
mTOR Pulldown and Mass Spectrometry
HEK293 cells stably expressing Flag-mTOR were grown to confluency in 15-cm plates and serum starved overnight. Cells on half of the plates were exposed to di-8:0 PA for 30 min, and then lysed. Lysates from each set of 8 plates were combined as one sample and subjected to anti-Flag IP. As a background control, anti-Flag IP was also performed with lysates of serum-starved plain HEK293 cells. Flag beads-associated proteins were subjected to trypsin digest, and the resulting peptides were resolved by nano-liquid chromatography and analyzed by mass spectrometry as previously reported (Wu et al., 2012). Further details are described in Supplemental Experimental Procedures.
In Vitro mTORC1 Kinase Assay
mTORC1 was immunoprecipitated and the kinase assays were performed as previously described (Yoon et al., 2011b), with 100 ng GST-S6K1 as the substrate. Where applicable, PA or PC vesicles were added to the washed immunocomplexes 15 min before initiation of the kinase assay by the addition of ATP.
FRB Binding Assay
The FRB protein was expressed and purified as we previously reported (Vilella-Bach et al., 1999). Microprobe sonication-generated SUVs containing PA and PC (1:9) or PC alone were tested for FRB binding, following procedures we previously described (Fang et al., 2001). Briefly, SUVs were incubated with purified FRB at room temperature for 30 min, and then analyzed by size-exclusion on a Sephracryl-300 column. Elutions were analyzed on 15% SDS gels by silver-staining.
Statistical analysis
All quantitative data are presented as mean ± standard deviation (SD) of at least three independent experiments. Whenever necessary, statistical significance of the data was analyzed by performing a one-sample or paired Student’s t test as indicated in figure legends. *P ≤ 0.05. **P ≤ 0.01.
Supplementary Material
Acknowledgments
We thank all members of the Chen lab for helpful discussions. This work was supported by grants from the National Institutes of Health (R01 AR048914 & GM089771 to JC; P30 DA018310 to JVS).
Footnotes
AUTHOR CONTRIBUTIONS
MSY and CLR designed the study, performed experiments, analyzed the data, and wrote the manuscript. CW and NT performed experiments and analyzed data. JVS helped with mass spectrometry experiments. JC designed the study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ávila-Flores A, Santos T, Rincón E, Mérida I. Modulation of the mammalian target of rapamycin pathway by diacylglycerol kinase-produced phosphatidic acid. The Journal of biological chemistry. 2005;280:10091–10099. doi: 10.1074/jbc.M412296200. [DOI] [PubMed] [Google Scholar]
- Boulbés DR, Shaiken T, Sarbassov D. Endoplasmic reticulum is a main localization site of mTORC2. Biochemical and biophysical research communications. 2011;413:46–52. doi: 10.1016/j.bbrc.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano E, Munoz-Fernandez MA, Fresno M. Regulation of interleukin-2 responses by phosphatidic acid. Eur J Immunol. 1992;22:1883–1889. doi: 10.1002/eji.1830220731. [DOI] [PubMed] [Google Scholar]
- Choi JW, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Park IH, Wu AL, Du G, Huang P, Frohman MA, Walker SJ, Brown HA, Chen J. PLD1 Regulates mTOR Signaling and Mediates Cdc42 Activation of S6K1. Current Biology. 2003;13:2037–2044. doi: 10.1016/j.cub.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Foster D, Salloum D, Menon D, Frias M. Phospholipase D and the maintenance of phosphatidic acid levels for regulation of mTOR. The Journal of Biological Chemsitry. 2014 doi: 10.1074/jbc.R114.566091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami K, Takenawa T. Phosphatidic acid that accumulates in platelet-derived growth factor-stimulated Balb/c 3T3 cells is a potential mitogenic signal. J Biol Chem. 1992;267:10988–10993. [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, et al. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44:290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hollenback D, Bonham L, Law L, Rossnagle E, Romero L, Carew H, Tompkins CK, Leung DW, Singer JW, White T. Substrate specificity of lysophosphatidic acid acyltransferase β - evidence from membrane and whole cell assays. Journal of Lipid Research. 2009;47:593–604. doi: 10.1194/jlr.M500435-JLR200. [DOI] [PubMed] [Google Scholar]
- Jenkins G, Frohman M. Phospholipase D: a lipid centric review. Cellular and Molecular Life Sciences. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y, Exton JH. Role of phospholipase D1 in the regulation of mTOR activity by lysophosphatidic acid. FASEB J. 2004;18:311–319. doi: 10.1096/fj.03-0731com. [DOI] [PubMed] [Google Scholar]
- Laplante M, Horvat S, Festuccia William T, Birsoy K, Prevorsek Z, Efeyan A, Sabatini David M. DEPTOR Cell-Autonomously Promotes Adipogenesis, and Its Expression Is Associated with Obesity. Cell Metabolism. 2012;16:202–212. doi: 10.1016/j.cmet.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Crowell KJ, Chen J, Jung D, Chiang GG, Sareth S, Abraham RT, Pellecchia M. The FRB domain of mTOR: NMR solution structure and inhibitor design. Biochemistry. 2006;45:10294–10302. doi: 10.1021/bi060976+. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis John. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:1471–0072. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partovian C, Ju R, Zhuang ZW, Martin KA, Simons M. Syndecan-4 regulates subcellular localization of mTOR Complex2 and Akt activation in a PKCalpha-dependent manner in endothelial cells. Mol Cell. 2008;32:140–149. doi: 10.1016/j.molcel.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt TR, McDermott M, Saqib KM, Shimwell N, Wakelam MJ. Phospholipase D1b and D2a generate structurally identical phosphatidic acid species in mammalian cells. Biochem J. 2001;360:707–715. doi: 10.1042/0264-6021:3600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard A, Nada S, Sabatini D. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson T, Shaul Y, Lindquist R, Thoreen C, Bar-Peled L, Sabitini D. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D, Ali S, Sengupta S, Sheen JH, Hsu P, Bagley A, Markhard A, Sabatini D. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sukumaran S, Barnes RI, Garg A, Agarwal AK. Functional characterization of the human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 10/glycerol-3-phosphate acyltransferase isoform 3. Journal of Molecular Endocrinology. 2009;42:469–478. doi: 10.1677/JME-09-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fang Y, Yoon M, Zhang C, Roccio M, Zwartkruis F, Armstrong M, Brown H, JC Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proceedings of the National Academy of Science. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab. 2009;296:E1195–E1209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan J, Chen X, Gu X, Luo K, Li J, Wan B, Wang Y, Yu L. Identificiation of novel human lysophosphatidic acid acyltransferase, LPAAT-theta, which activates mTOR pathway. Journal of Biochemistry and Molecular Biology. 2006;39:626–635. doi: 10.5483/bmbrep.2006.39.5.626. [DOI] [PubMed] [Google Scholar]
- Topham MK, Epand RM. Mammalian diacyglycerol kinases: Molecular interactions and biological functions of selected isoforms. Biochimica et Biophysica Acta. 2009;1790:416–424. doi: 10.1016/j.bbagen.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veverka V, Crabbe T, Bird I, Lennie G, Muskett FW, Taylor RJ, Carr MD. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene. 2008;27:585–595. doi: 10.1038/sj.onc.1210693. [DOI] [PubMed] [Google Scholar]
- Vilella-Bach M, Nuzzi P, Fang Y, Chen J. The FKBP12-Rapamycin-binding Domain Is Required for FKBP12-Rapamycin-associated Protein Kinase Activity and G1 Progression. Journal of Biological Chemistry. 1999;274:4266–4272. doi: 10.1074/jbc.274.7.4266. [DOI] [PubMed] [Google Scholar]
- Wu C, Tran JC, Zamdborg L, Durbin KR, Li M, Ahlf DR, Early BP, Thomas PM, Sweedler JV, Kelleher NL. A protease for ‘middle-down’ proteomics. Nature methods. 2012;9:822–824. doi: 10.1038/nmeth.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Du G, Backer JM, Frohman MA, Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid–sensing mTORC1 pathway. The Journal of Cell Biology. 2011a;195:435–447. doi: 10.1083/jcb.201107033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Sun Y, Arauz E, Jiang Y, Chen J. Phosphatidic acid activates mammalian target of rapamycin complex 1 (mTORC1) kinase by displacing FK506 binding protein 38 (FKBP38) and exerting an allosteric effect. The Journal of biological chemistry. 2011b;286:29568–29574. doi: 10.1074/jbc.M111.262816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wendel AA, Keogh MR, Harris TE, Chen J, Coleman RA. Glycerolipid signals alter mTOR complex 2 (mTORC2) to diminish insulin signaling. Proc Natl Acad Sci U S A. 2012;109:1667–1672. doi: 10.1073/pnas.1110730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang Z, Lu M, Yonekubo Y, Liang X, Zhang Y, Wu P, Zhou Y, Grinstein S, Hancock JF, et al. Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Molecular and Cellular Biology. 2014;34:84–95. doi: 10.1128/MCB.00987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44:304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.