SUMMARY
IRGM, encoded by a uniquely human gene conferring risk for inflammatory diseases, affects autophagy through a hitherto unknown mechanism. Here we show how IRGM controls autophagy. IRGM interacts with ULK1 and Beclin 1 and promotes their co-assembly thus governing the formation of autophagy initiation complexes. We further show that IRGM interacts with pattern recognition receptors including NOD2. IRGM, NOD2 and ATG16L1, all of which are Crohn’s disease risk factors, form a molecular complex to modulate autophagic responses to microbial products. NOD2 enhances K63-linked polyubiquitination of IRGM, which is required for interactions of IRGM with the core autophagy factors and for microbial clearance. Thus, IRGM plays a direct role in organizing the core autophagy machinery to endow it with antimicrobial and anti-inflammatory functions.
INTRODUCTION
Autophagy is a cellular homeostatic mechanism with broad roles in human health and disease (Mizushima et al., 2008). Autophagy is at the intersection of metabolic (Mizushima et al., 2008) and anti-microbial processes (Deretic et al., 2015; Levine et al., 2011). Thus, the system responds to a range of inputs such as starvation (Mizushima et al., 2008), and endogenous danger associated molecular patterns and microbial products commonly referred to as pathogen-associated molecular patterns (PAMPS) (Deretic et al., 2015). Autophagic responses to PAMPS lead to direct antimicrobial action through a process termed xenophagy and control of inflammation and other immune processes (Deretic et al., 2015; Levine et al., 2011).
Among the better-established links between autophagy and human diseases are the genetic polymorphisms in ATG16L1 and IRGM conferring risk for Crohn’s disease (CD), an intestinal inflammatory disorder (Consortium, 2007; Craddock et al., 2010). The human population polymorphisms in IRGM have been linked to autophagy (Consortium, 2007; Craddock et al., 2010) and to its effector outputs including direct antimicrobial defense (Brest et al., 2011; McCarroll et al., 2008). In keeping with its autophagy-mediated antimicrobial role, IRGM is additionally a genetic risk factor for tuberculosis in different human populations (Intemann et al., 2009; Song et al., 2014) and may afford protection in leprosy (Yang et al., 2014). However, the molecular mechanism of IRGM’s function in autophagy has remained a mystery.
IRGM has no homologs among the Atg genes in yeast, which makes it difficult to assign to it an autophagy-specific function; instead, IRGM has been considered to affect autophagy indirectly (Singh et al., 2006; Singh et al., 2010). A complicating factor in understanding the exact function of IRGM is that it is distinctly a human gene (Bekpen et al., 2009). Its orthologs are present only in African great apes and Homo sapiens but active alleles are absent in ancestral evolutionary lineages leading up to them (Bekpen et al., 2009). The mouse genome encodes a large family of immunity related GTPase (21 IRG genes) compared to a single gene (IRGM) in humans; furthermore, all murine IRGs encode ~40-kDa proteins that are much larger than the human IRGM (21 kDa). The prevailing view of the murine IRGs is that they have predominantly non-autophagy functions (Choi et al., 2014). Thus the significant information gathered in the murine systems may have limited import on how the human IRGM works.
Given the significance of IRGM in human populations and the notoriously high prevalence of diseases such as CD and tuberculosis, it is surprising that IRGM’s mechanism of action in autophagy remains unknown. Here we report that IRGM physically interacts with key autophagy regulators, ULK1, Beclin 1, ATG14L and ATG16L1. We also show that IRGM links inputs from PAMP sensors by making molecular complexes with NOD2, another genetic risk factor in CD (Hugot et al., 2001; Ogura et al., 2001). The formation of NOD2-IRGM complex is stimulated in response to PAMPs, whereas increased association of NOD2 with IRGM promotes IRGM-directed assembly of autophagy regulators.
RESULTS
IRGM activates the core regulators of autophagy
Prior work has indicated that IRGM affects autophagy through processes influencing mitochondrial function, including mitochondrial fission and membrane potential collapse (Singh et al., 2010). Similar changes in mitochondrial function often lead to AMPK activation (Romanello et al., 2010; Turkieh et al., 2014). Thus, we tested the activation status of AMPK. A knockdown of IRGM reduced the total amounts of AMPK in both control or starved cells (Figure 1A) and decreased the levels of the activated form of AMPK phosphorylated at Thr-172 (Figure 1A). Overexpression of IRGM increased levels of Thr-172 phosphorylated AMPK (Figure 1B).
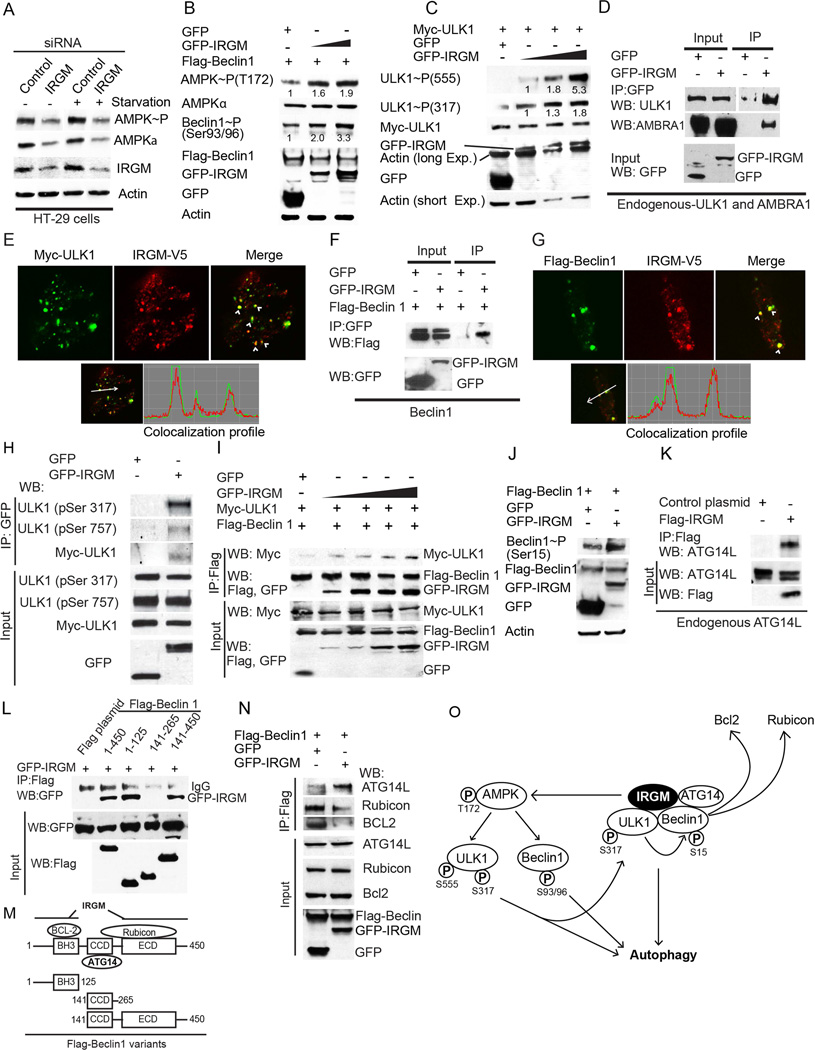
Figure 1. IRGM activates AMPK signaling and interacts with core autophagy machinery.
(A) Lysates from HT-29 colon epithelial cells transfected with control and IRGM siRNA were subjected to Western blotting with antibodies to phospho-AMPK (Thr-172), AMPK, IRGM and actin. (B) Levels of phospho-AMPK (Thr-172) and phospho-Beclin 1 (Ser-93/96) in lysates from HEK293T cells co-expressing Flag-Beclin 1 and GFP or GFP-IRGM. (C) Levels of active phospho-ULK1 (Ser-555 and Ser-317) in lysates of HEK293T cells co-expressing Myc-ULK1 and either GFP or GFP-IRGM. Numbers beneath bands in B, C, quantification of phosphorylated proteins relative to the total abundance of the same protein. (D) Co-immunoprecipitation (Co-IP) analysis of interaction between IRGM and endogenous ULK1 and AMBRA1 in HEK293T lysates of cells expressing GFP or GFP-IRGM. (E) Top, confocal microscopy images of HEK293T cells expressing IRGM-V5 and Myc-ULK1 subjected to starvation for 2 h. Arrowheads, co-localization. Bottom, fluorescence intensity line tracing. (F) Co-IP analysis in lysates of HEK293T cells expressing indicated proteins. (G) Confocal microscopy images of HEK293T cells transiently expressing V5-IRGM and Flag-Beclin1 subjected to starvation for 2 h. Details as for panel E. (H) Lysates of HEK293T cells expressing GFP or GFP-IRGM with Myc-ULK1 subjected to immunoprecipitation with anti-GFP and blots probed with phospho-ULK1 Ser-317 or Ser-757 antibodies. (I) Lysates of cells expressing Myc-ULK1, Flag-Beclin-1 and increasing concentrations of GFP-IRGM subjected to immunoprecipitation with anti-Flag; blots probed as indicated. (J) HEK293T cell lysates co-expressing GFP-IRGM and Flag-Beclin 1 subjected to Western blotting with antibody to phospho-Beclin 1 (Ser-15) and antibodies as indicated. (K) Co-IP analysis of Flag-IRGM and endogenous ATG14. (L, M) Mapping of Beclin 1 regions interacting with IRGM. (L) Lysates of HEK293T cells co-expressing GFP-IRGM and Flag-Beclin 1 variants in panel M were subjected to immunoprecipitation with anti-Flag and blots probed as indicated. (M) Beclin 1 domain organization indicating its interacting proteins along with deletion constructs used in Co-IP analysis in panel L. (N) Co-IP analysis of the effects of IRGM overexpression on the interaction of Beclin 1 with its regulatory proteins. Lysates of HEK293T cells co-expressing GFP-IRGM and Flag-Beclin 1 were subjected to immunoprecipitation with anti-Flag and blots probed as indicated. (O) Model of IRGM-dependent autophagy induction based on the results obtained in Figure 1 and Figure S1. See also Figure S1.
AMPK has been previously shown to induce autophagy by directly phosphorylating ULK1 (Egan et al., 2011; Kim et al., 2011) and Beclin 1(Kim et al., 2013). When we tested the phosphorylation status of ULK1 and Beclin 1, we observed that the expression of IRGM, which caused induction of autophagy (Figure S1A), enhanced phosphorylation at activating sites of Beclin 1 at Ser93/96 (Kim et al., 2013), and ULK1 at Ser-555 (Egan et al., 2011) and at Ser-317 (Kim et al., 2011) (Figure 1B,C).
IRGM assembles the core regulatory machinery for autophagy
The entire signaling cascade described above could explain how IRGM induces autophagy, e.g. by its effects on AMPK and activation of downstream autophagy regulators. However, IRGM showed a further, more direct role by interacting with the key regulators of autophagy. We found that IRGM co-immunoprecipitated and co-localized with both endogenous and overexpressed ULK1 and Beclin 1 (Figure 1D–G and S1B–C) but not with AMPK (Figure S1D). IRGM complexes with ULK1 were enriched for the activated, AMPK-dependent Ser-317, form of ULK1 relative to the inhibitory, mTOR-dependent, Ser-757 form (Figure 1H). Furthermore, expression of IRGM enriched ULK1 in the immunoprecipitated Beclin 1 complexes (Figure 1I and S1G). In keeping with this, cells overexpressing IRGM also showed increased Beclin 1 Ser-15 phosphorylation, the phosphorylated form of Beclin 1 dependent on ULK1 activity (Kim et al., 2013) (Figure 1J).
IRGM determines the composition of the Beclin 1 complex
We found that IRGM complexes also included autophagy-enhancing Beclin1 interactors, AMBRA1 (Figure 1D and S1E), ATG14L (Figure 1K) and UVRAG (Figure S1F) but not the autophagy inhibitory factor Rubicon (Figure S1F) (Fimia et al., 2007; Itakura et al., 2008; Matsunaga et al., 2009). Next, we mapped Beclin 1 regions required for interaction with IRGM (Figure 1M). IRGM interacted with two Beclin 1 regions: (i) BH3-containing 1–125 N-terminal portion, and (ii) a segment encompassing CCD and ECD, whereas it did not bind to the intervening CCD domain alone (Figure 1L,M).
Incidentally, two Beclin1 negative regulators Bcl-2 and Rubicon bind respectively to the regions spanning Beclin 1’s BH3 domain and Beclin 1’s CCD and ECD domains, whereas ATG14L, a factor enabling Beclin 1 to activate the initiation complex (Kim et al., 2013), binds to the CCD domain of Beclin 1(Sun et al., 2008). This domain occupancy on Beclin1 is compatible with simultaneous binding of IRGM and ATG14L and exclusion of autophagy negative regulators. When IRGM was overexpressed, it dis-enriched Rubicon and Bcl-2 from Beclin 1 and enriched ATG14L in Beclin 1 complexes (Figure 1N).
The above data indicate that IRGM forms protein complexes with the central regulators of autophagy and activates Beclin 1 by displacing its negative regulators (Figure 1O, Right). This, taken together with IRGM’s ability to sponsor the phosphorylation cascade that activates ULK1 and Beclin 1, shows how IRGM promotes autophagy (Figure 1O, Left).
IRGM affects levels of autophagy regulators
As observed with AMPK (Figure 1A), IRGM affected the levels of a number of other autophagy regulators. IRGM knockdown in U937 monocytic cells (Figure S2A) reduced total amount of ULK1 (Figure 2A,B, Figure S2B), ATG14L (Figure 2C, Figure S2B), and AMBRA1 (Figure 2C, Figure S2B). In contrast to the above suite of autophagy regulators, Beclin 1 was not affected (Figure S2C). In addition to Beclin 1, IRGM did not alter cytoplasmic levels of ATG5-ATG12 conjugates (Figure 2C). However, the physical organization of ATG5-ATG12 was affected, since the numbers of its puncta, revealed by ATG5 immunofluorescence, were reduced upon IRGM knockdown (Figure 2D).
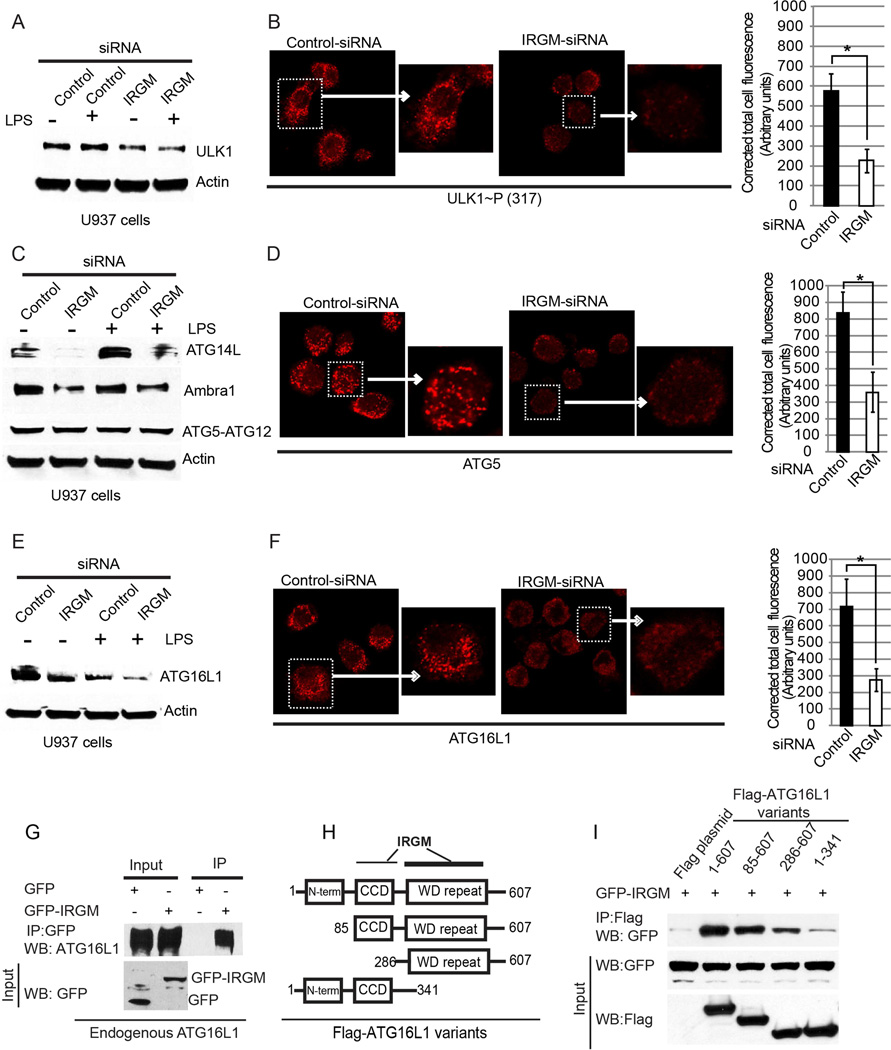
Figure 2. IRGM is required for stable levels of the autophagy initiation proteins.
(A,C,E) U937 cells transfected with control or IRGM siRNAs, untreated or treated with LPS (500 ng/ml for 4 h) were lysed and subjected to Western blotting with antibody to (A) ULK1, (C) ATG14L, AMBRA1 and ATG5, and (E) ATG16L1. IRGM knock down efficiency and quantifications are shown in Supplementary Figure S2A,B. (B,D,F) Left, confocal images of U937 cells transfected with control or IRGM siRNA treated with LPS (500 ng/ml for 4 h), Immunofluorescence analysis was performed with (B) phopho-ULK1 (Ser-317), (D) ATG5, and (F) ATG16L1. Graphs, means ± SD (corrected total cell fluorescence of cells; > 30 cells from 5 fields measured using Image J). *, p < 0.05 (Student’s unpaired t test). (G) Lysates from HEK293T cells expressing GFP or GFP-IRGM were subjected to immunoprecipitation with anti-GFP and blot probed with indicated antibodies. (H) Schematic of ATG16L1 domain structure indicating IRGM interacting regions mapped in panels I. (I) Lysates of HEK293T cells co-expressing GFP-IRGM and the indicated Flag-ATG16L1 variants in panel H were subjected to immunoprecipitation with anti-Flag and blots probed as indicated. Results, representative of three independent experiments. See also Figure S2.
ATG5 puncta formation is governed by ATG16L1 (Mizushima, 2003). We thus looked at the effects of IRGM on ATG16L1 levels and observed that they were reduced in IRGM knockdown cells (Figure 2E,F). This prompted us to test whether IRGM might interact with ATG16L1. IRGM was in complexes with endogenous Atg16L1 (Figure 2G). Further domain mapping showed that IRGM primarily interacted with the WD repeats region of ATG16L1 (Figure 2H,I). The residual weak interaction between IRGM and ATG16L1 outside of the WD repeats (construct ATG16L1(1–341)) was not due to FIP200, previously shown to bridge ATG16L1 with ULK1 (Gammoh et al., 2013) since interaction was not reduced upon FIP200 knockdown, and if anything was slightly increased (Figure S2D). In summary, in addition to directing the assembly of key autophagy-specific regulators, IRGM also stabilizes them. Furthermore IRGM interacts with and stabilizes ATG16L1, a component of the ATG5-Atg12/ATG16L1 E3 complex, which governs LC3 conjugation and autophagosome formation (Mizushima, 2003).
Expression of IRGM and its assembly with autophagy factors responds to microbial signals
Infection with CD-associated adhesive invasive Escherichia coli (AIEC) LF82 (Lapaquette et al., 2010) or treatment with LPS or muramyl dipeptide (MDP) induced IRGM expression in U937 cells (Figure 3A–C). The induction of IRGM was similar to other physiological inducers of autophagy: starvation and IFN-γ (Gutierrez et al., 2004) which acted in a cell type-dependent manner, and, in the case of starvation, showed AMPK dependence (Figure 3D, Figure S3A–I). When autophagy was induced by LPS (Figure S3J) or MDP (Figure S3K) (Cooney et al., 2010), a knockdown of IRGM (Figure S3L) precluded LC3B-II conversion and LC3B puncta formation in response to these stimuli (Figure 3E–H). Thus, IRGM is required for autophagy elicited by microbial products.
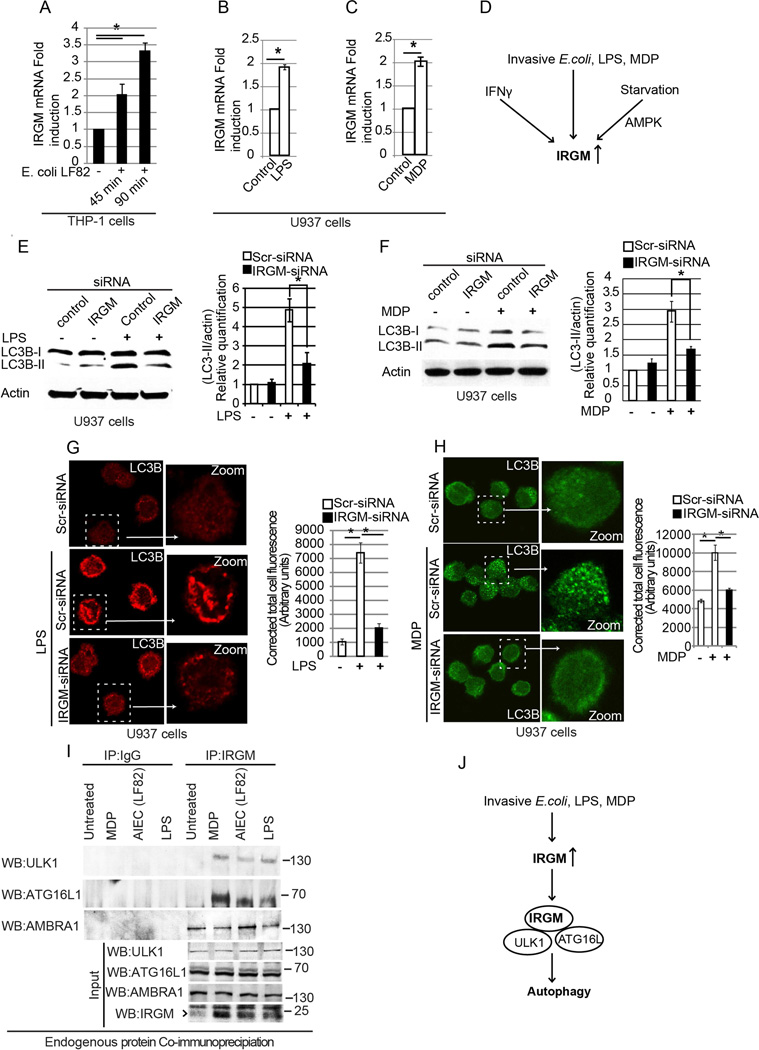
Figure 3. IRGM is required for PAMPs induced autophagy.
(A) Abundance of IRGM mRNA (relative to GAPDH) in THP-1 cells (control or infected with invasive E. coli LF82) determined by quantitative real-time PCR (qRT-PCR). (B) Effect of LPS (30 min) or (C) MDP exposure (16 h) on IRGM mRNA levels in U937 cells. Gene expression (qRT-PCR) was normalized relative to GAPDH. Data, means ± SD (n>3); *, p< 0.05 (Student’s unpaired t test). (D) Schematic summary of the physiological signals activating IRGM expression based on data in panels A–C and in Figure S3A–H. (E, F) Left, Western blot analysis of LC3-II abundance in U937 cells transfected with control or IRGM siRNA: (E) treated or not with LPS (500 ng/ml; 4 h); (F) treated or not with MDP (5 µg/ml for 8 h). Right, densitometric analysis of Western blots using ImageJ software. (G, H) Left, confocal images of LC3 puncta in LPS treated (500 ng/ml; 4 h) (G) or MDP-treated (5 µg/ml; 8 h), (H) U937cells transfected with control or IRGM siRNA. Graphs (right of panels G and H), represent mean corrected total cell fluorescence ± SE (25–35 cells from 10–15 fields measured using ImageJ. *, p<0.05 (ANOVA). (I) Analysis of endogenous interactions (Co-IP) using THP-1 lysates infected with invasive E. coli LF82 (1 h) or stimulated with LPS (2 µg/ml, 2 h) or MDP (10 µg/ml, 2 h). Lysates were subjected to immunoprecipitation with IRGM antibody or control IgG and probed as indicated. (J) Schematic summary of the results obtained in Figure 3E–I. See also Figure S3.
In experiments with endogenous proteins, we could not detect interactions of IRGM with ULK1 and ATG16L1 under basal conditions (Figure 3I, untreated lane). However, when a monocytic cell line (THP-1) was infected with E. coli LF82, immunoprecipitates of endogenous IRGM contained ULK1 and ATG16L1 (Figure 3I, AIEC lane). Similar effects were observed with MDP and LPS (Figure 3I). Of note, MDP (a NOD2-cogante ligand) was a stronger promoter of these effects than LPS. In contrast to ULK1 and ATG16L1, which showed interactions with endogenous IRGM only in samples from cells infected or treated with MDP or LPS, AMBRA1 showed association with endogenous IRGM even under basal conditions (Figure 3I). Thus, exposure of cells to microbes or their products affects IRGM expression and also influences interactions with the autophagic apparatus (Figure 3J).
Three Crohn’s disease risk factors, NOD2, IRGM, and ATG16L1 interact
A known receptor for MDP is NOD2, a risk factor for familial CD (Ogura et al., 2001). Furthermore, ATG16L1, harboring an important CD-associated polymorphism (Consortium, 2007), interacts with NOD2 (Cooney et al., 2010; Travassos et al., 2010). Hence, we wondered whether IRGM, a third genetic CD risk factor (incidentally co-discovered with ATG16L1) (Consortium, 2007), is a part of this complex. Endogenous and overexpressed IRGM immunoprecipitates contained both NOD2 and ATG16L1 (Figure 4A, B). IRGM increased interactions between NOD2 and ATG16L1 (Figure S4A). In contrast, co-expression of NOD2 did not affect IRGM-ATG16L1 interactions (Figure S4B), suggesting that IRGM is important for promoting the assembly of the tri-partite complex. Morphologically, NOD2 co-expression changed IRGM intracellular distribution from diffuse cytosolic to punctate (Figure S4C). A subset of these profiles colocalized with mitochondrial markers (Tom20; Figure S4D, E), in keeping with a partial NOD2 colocalization with mitochondrial antiviral signaling protein MAVS (Sabbah et al., 2009), and the previously reported partial IRGM localization to mitochondria (Singh et al., 2010).
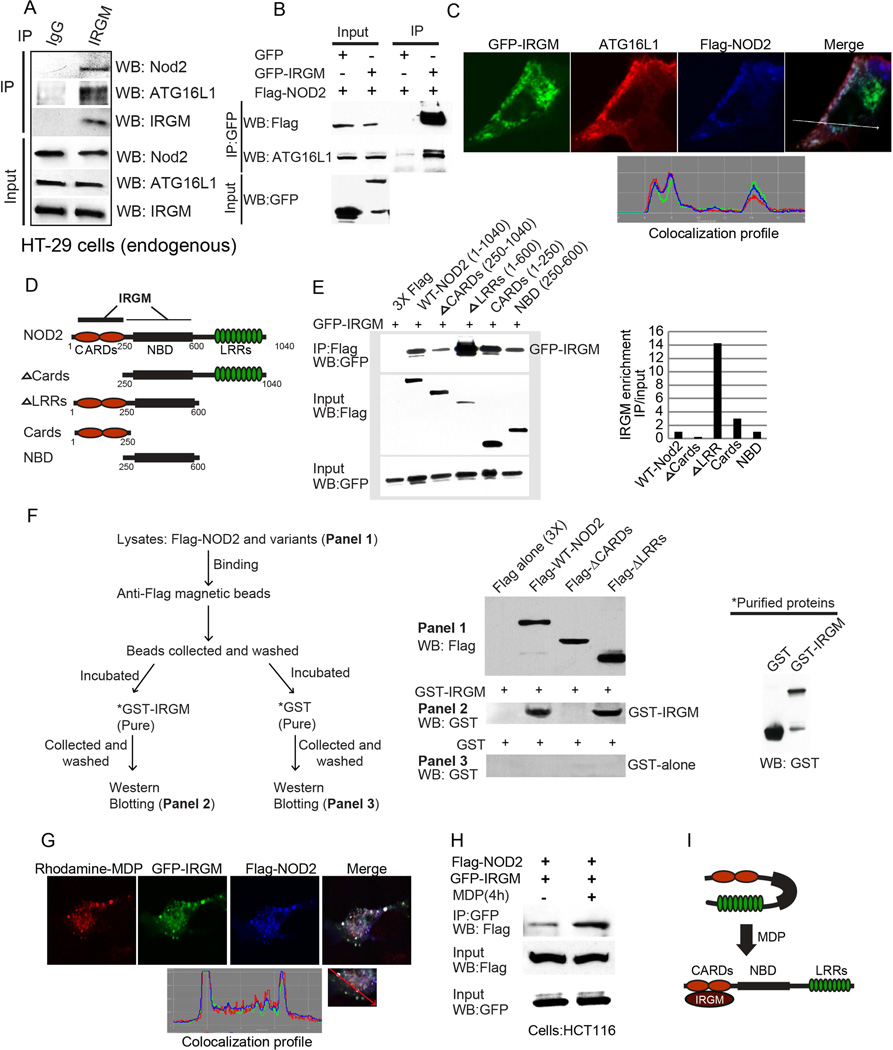
Figure 4. IRGM interacts and co-localizes with ATG16L1 and NOD2.
(A, B) Co-IP analysis of endogenous (A) or overexpressed (B) IRGM, with NOD2 and ATG16L1 in (A) starved HT29 cells and (B) HEK293T cells. (C) Top, confocal microscopy images of HEK293T cells transiently expressing GFP-IRGM and Flag-NOD2. Bottom, fluorescence intensity line tracing corresponding to dashed line. (D) Schematic of NOD2 domain organization along with deletion constructs used in Co-IP analysis in panel E. (E) Left panel, lysates of HEK293T cells coexpressing GFP-IRGM and the Flag-NOD2 variants shown in panel D subjected to immunoprecipitation with anti-Flag and blot probed with antibodies as indicated. Right panel, densitometric analysis of Western blots (IP blot/Input blot). (F) Flag tag pull-down assays performed with affinity purified NOD2 variants from 293T cell lysates and purified recombinant GST-IRGM shown in the schematic (left panel). (G) Top, confocal microscopy images showing co-localization of GFP-IRGM and Flag-NOD2 and Rhodamine-MDP in HEK293T cells. Bottom, fluorescence intensity line tracing corresponding to red line. (H) Effect of MDP (10 µg/ml, 8 h) on GFP-IRGM and Flag-NOD2 interactions in HCT116 cells. (I) Model of IRGM-NOD2 interactions. See also Figure S4.
All three factors, IRGM, ATG16L1, and NOD2 co-localized in co-transfected cells (Figure 4C). Mapping of interaction domains revealed that association of IRGM with NOD2 is likely a regulated event. A region containing the two CARD domains of NOD2 was required for IRGM interaction (Figure 4D,E). A deletion of the LRR domains in NOD2 enhanced interactions between IRGM and NOD2 (Figure 4D,E). The LRR domain region is known to be inhibitory to the previously established NOD2 activities (Tanabe et al., 2004) by keeping NOD2 in a closed conformation until it is activated through stimuli such as MDP (Tanabe et al., 2004). IRGM and NOD2 interaction was confirmed by proximity ligation assay (PLA; Figure S4F), which reports direct protein-protein interactions in situ. Positive PLA readouts of direct in situ interactions between proteins appear as fluorescent dots, the products of in situ PCR that generates a fluorescent product physically attached to antibodies against the two proteins that are <16 nm (FRET distance) apart. A deletion of the CARD domains in NOD2 reduced the NOD2-IRGM PLA signal (Figure S4F), in keeping with the importance of CARDs for the interactions between IRGM and NOD2. We carried out additional interaction experiments with purified GST-IRGM protein (Singh et al., 2010), prepared from insect cells (Figure S4G, isoform d, used in all experiments in this work), and Flag-NOD2 (full length and its variants ΔCARD and ΔLRR) prepared from 293T overexpressing cells. The results show that IRGM interacts with full length NOD2 and ΔLRR NOD2, but not with ΔCARD NOD2 (Figure 4F). These findings demonstrate that the NOD2 CARD domain is key for interactions with IRGM.
Fluorescently labeled MDP co-localized with NOD2 and IRGM in the cells (Figure 4G). In the presence of MDP, interactions between IRGM and NOD2 were enhanced (Figure 4H). These findings are consistent with the inhibitory action of LRRs in the resting state of NOD2, and with the observation that following activation with MDP, NOD2 becomes available for interactions with IRGM (Figure 4I). In summary, IRGM, NOD2, and ATG16L1 form a complex, with IRGM-NOD2 assembly being controlled by MDP, thus rendering the IRGM autophagy-promoting system responsive to microbial products.
NOD2 enhances IRGM interactions with ULK1 and Beclin 1
NOD2 affected IRGM quaternary structure. Co-expression of NOD2 and IRGM induced IRGM oligomerization within protein complexes (Figure 5A). NOD2 furthermore promoted interactions between IRGM and ULK1 as well as between IRGM and Beclin 1 (Figure 5B,C). Incidentally, NOD2 was also found in complexes with ULK1 (Figure S5A). IRGM co-expression increased ULK1-NOD2 complexes (Figure S5A). Thus, NOD2 modulates IRGM interactions with ULK1 and Beclin 1, in contrast to the above-described (Figure S4B) absence of NOD2 effects on IRGM-ATG16L1 complex formation. Based on these and above observations, IRGM is a pivotal organizer of the core parts of the autophagy initiation machinery (ULK1/Beclin1 and ATG16L1) along with NOD2.
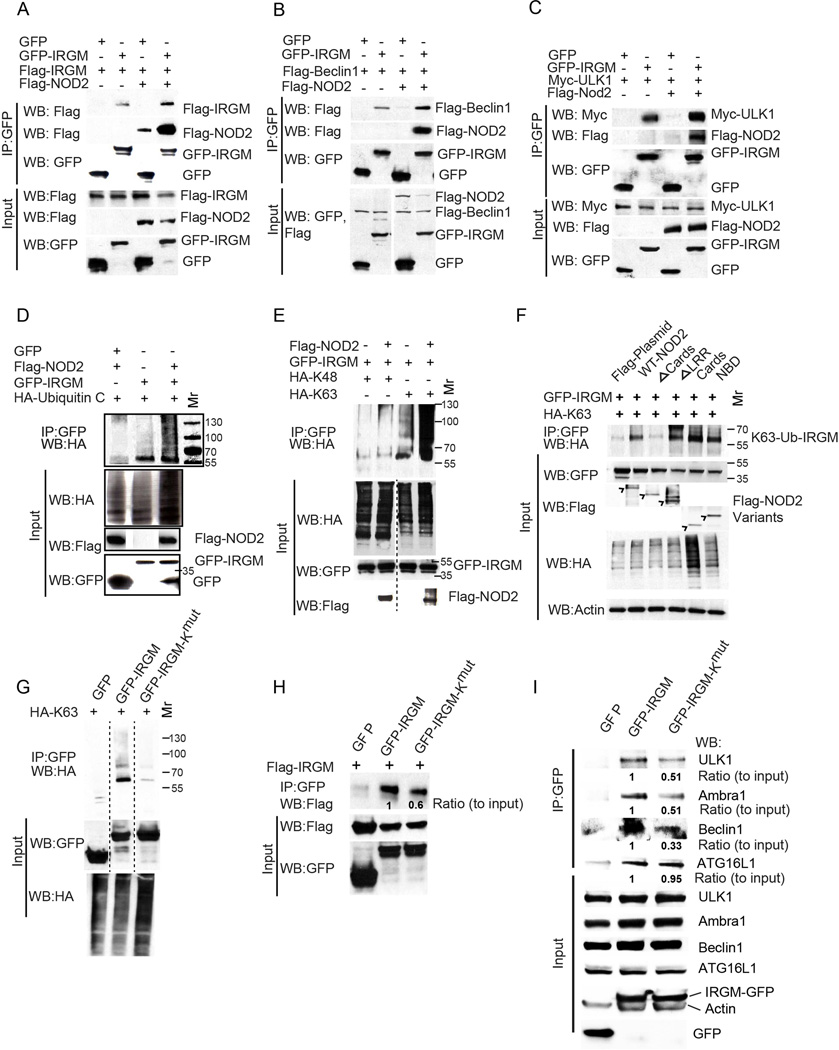
Figure 5. NOD2 promotes K63-linked polyubiquitination of IRGM, enhancing its interactions with autophagy initiation factors.
(A–C) Effects of NOD2 expression on IRGM self-association (A), and IRGM’s interaction with Beclin 1 (B) or with ULK1 (C) in HEK293T cells. (D, E) Analysis of IRGM ubiquitination in HEK293T cells. Cells co-expressing GFP or GFP-IRGM and (D) HA-tagged Ubiquitin C or (E) HA-tagged Ubiquitin C mutated for all lysines except lysine 48 (HA-K48) or Lysine 63 (HA-K63) and Flag-NOD2 were subjected to immunoprecipitation with GFP antibody and blots probed with indicated antibodies. Blot in (E) was processed to remove irrelevant lanes (dashed vertical line). (F) Cells co-expressing GFP-IRGM, HA-K63 and Flag-NOD2 deletion variants as in Figure 4D were subjected to immunoprecipitation analysis with anti-GFP and blot probed with indicated antibodies. (G) Cells co-expressing GFP or GFP-IRGM or GFP-IRGM-Kmut (IRGM variant with all lysine residues mutated to alanine) and HA-K63 were subjected to immunoprecipitation analysis with anti-GFP and blot was probed with indicated antibodies. Blot was processed (dashed vertical line) to remove irrelevant lanes. (H) Lysates of cells co-expressing GFP or GFP-IRGM or GFP-IRGM-Kmut and Flag-IRGM were subjected to immunoprecipitation with anti-GFP and blot probed with indicated antibodies. (I) Lysates of cells expressing GFP or GFP-IRGM or GFP-IRGM-Kmut were subjected to immunoprecipitation with anti-GFP and blots probed with indicated antibodies. Results representative of three independent experiments. See also Figure S5.
Polyubiquitination of IRGM promotes its assembly with ULK1 and Beclin 1
In the co-immunoprecipitation experiments of NOD2 with IRGM, we observed the presence of multiple GFP-IRGM bands (Figure S5B). NOD2 is known to promote ubiquitination of several target proteins (Abbott et al., 2007; Hasegawa et al., 2008). We tested whether IRGM was ubiquitinated and observed that it can be polyubiquitinated whereas NOD2 enhanced IRGM ubiquitination (Figure 5D). To determine which ubiquitination linkage was involved, we co-expressed GFP-IRGM with two HA-tagged ubiquitin variants, one that can be ubiquitinated only at K63 and another one that can be ubiquitinated only at K48. The IRGM ubiquitination showed a much stronger signal with the HA-Ub-K63 (Figure 5E). Endogenous IRGM as well as a construct with a tag smaller than GFP (V5 tag; IRGM-V5) were K63 polyubiquitinated (Figure S5C, S5D). The K63 ubiquitination of IRGM was strongly enhanced in the presence of NOD2 (Figure 5E). Overexpression or downregulation of TRAF6, an E3 ligase known to work in concert within the NOD2 pathway (Abbott et al., 2007; Yang et al., 2007) increased or decreased IRGM ubiquitination (Figure S5E and S5F) suggesting a role for TRAF6 in IRGM ubiquitination. However, TRAF6 knockdown destabilized NOD2, so it was not possible to conclude that TRAF6 was the only E3 ligase responsible for IRGM ubiquitination. Next, we mapped which of the NOD2 domains are necessary for effective ubiquitination of IRGM, and found that deletion of the CARD domain in NOD2 prevented IRGM ubiquitination, consistent with IRGM’s ability to bind to that region of NOD2 (Figure 5F). Moreover, when the CARD domain of NOD2 was expressed alone, it enhanced IRGM K63 ubiquitination (Figure 5F).
Mutation of either individual or small clusters of K (Lys) residues in IRGM did not prevent K-63 linkage ubiquitination of IRGM in the presence of NOD2 (Figure S5G). In the absence of NOD2, the low level ubiquitination (see Figure 5E) of the same series of K mutants of IRGM also persisted (Figure S5H). A similar phenomenon, i.e. an absence of a dominant ubiquitination residue, has been described for several proteins including p53 (Chan et al., 2006) and cyclins (Fung et al., 2005). Paradoxically, mutation of the K-23/K-27 cluster in IRGM, enhanced K-63 linkage ubiquitination (Figure S5H); it nevertheless reduced K-48 linked ubiquitination (Figure S5I) suggesting that K-23/K-27 cluster may be a dominant K-48 ubiquitination site, and that its elimination enhances K-63 ubiquitination of IRGM. Thus, multiple K residues in IRGM are K63-ubiquitinated. When we mutated all twelve lysine residues in IRGM (IRGM-Kmut; K residues converted to R), the GFP-IRGM fusion lost ubiquitination capacity (Figure 5G). Nevertheless, GFP-IRGM-Kmut still bound ATG16L1 equally well as the wild type IRGM (Figure 5I). In contrast to its unaltered association with ATG16L1, GFP-IRGM-Kmut showed a reduced ability to oligomerize within protein complexes (revealed by using IRGM with two different tags; Figure 5H) and displayed diminished capacity for interactions with ULK1, Beclin 1 and AMBRA1 (Figure 5I). In addition, NOD2 could not increase Beclin 1-IRGM-Kmut interactions, although NOD2 increased Beclin 1 interactions with wild type IRGM (Figure S5J). Thus, polyubiquitination of IRGM is important for the assembly of the core regulatory machinery centered on ULK1 and Beclin 1, and this modification of IRGM is under the control by NOD2.
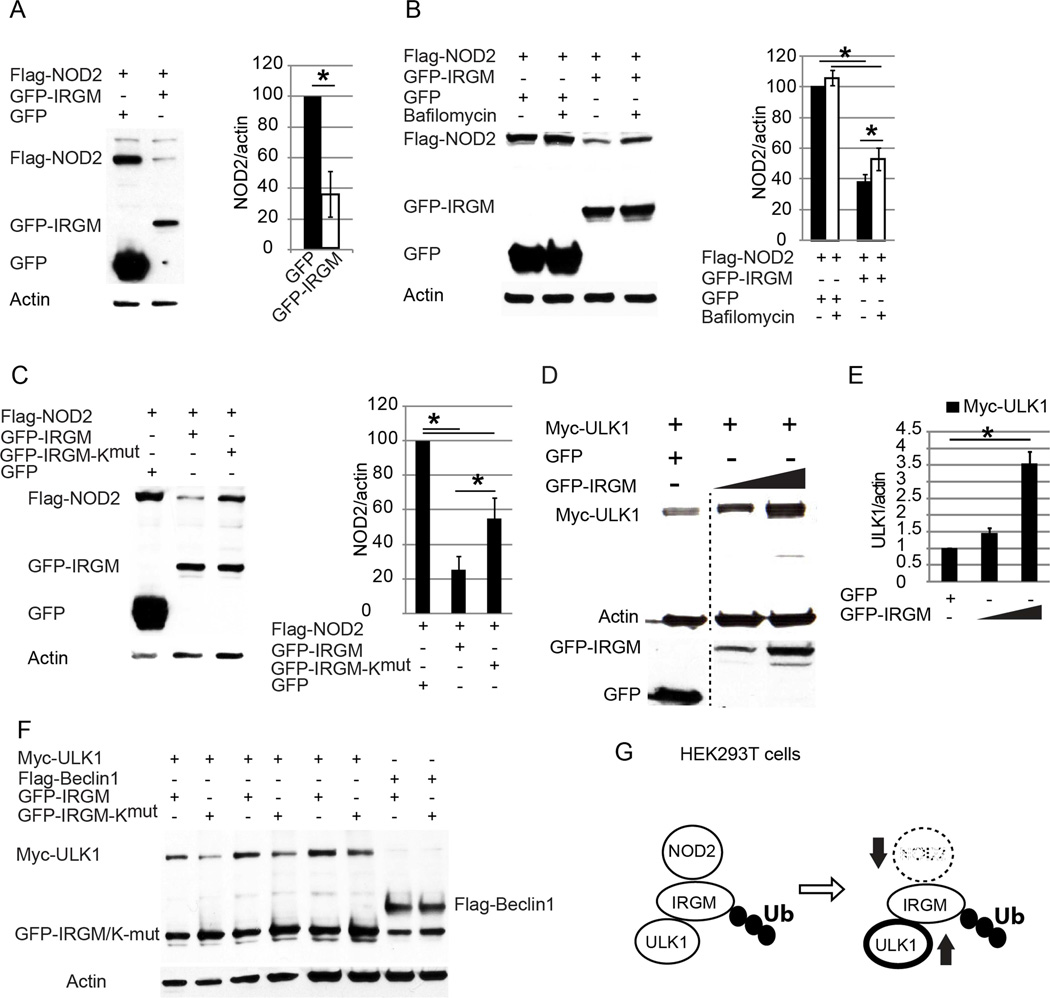
Polyubiquitinated IRGM inversely controls NOD2 and ULK1 protein levels
We observed that co-expression of GFP-IRGM had an effect on NOD2 protein amount, by diminishing its levels relative to control (Figure 6A). IRGM promoted NOD2 degradation, which was partially blocked by bafilomycin A1, commonly used to inhibit autolysosomal degradation (Figure 6B). The IRGM-Kmut variant of IRGM displayed a decreased ability to commit NOD2 for degradation (Figure 6C). In contrast to the destabilizing effects of IRGM on NOD2, expression of IRGM increased co-expressed myc-ULK1 in a dose-dependent manner (Figure 6D). The total amount of ULK1 was not increased when the IRGM-Kmut variant was co-expressed (Figure 6E). This effect was ULK1-specific, since Beclin 1 levels were not affected when IRGM vs IRGM-Kmut were compared, corroborating with a related finding that IRGM did not affect Beclin 1 stability (Figure S3B). Thus, polyubiquitinated IRGM protects ULK1 and promotes degradation of NOD2 (Figure 6F). This represents a negative feedback regulatory loop, which induces autophagy but at the same time limits NOD2’s ability to continue unabated stimulation of this process (Figure 6G).
Figure 6. Ubiquitination of IRGM is required for NOD2 degradation and ULK1 stability.
(A) Effects of IRGM expression on NOD2 levels in transfected HEK293T cells. Data, means ± SE; *, p< 0.05 (Student’s unpaired t test). (B) Lysates of HEK293T cell co-expressing GFP or GFP-IRGM and Flag-NOD2, untreated/treated with Bafilomycin A1 (100 nM for 8 h) were subjected to Western blotting. (C) Lysates of cells co-expressing Flag-NOD2 and GFP, GFP-IRGM, or GFP-IRGM-Kmut were subjected to Western blotting. (D, E) Lysates from HEK293T cells co-expressing Myc-ULK1 and either GFP or increasing amounts of GFP-IRGM were subjected to Western blotting as in (D) with the relative abundance of Myc-ULK1 shown in (E). Blot was processed (dashed vertical line) to remove irrelevant lanes. (F) HEK293T cells transfected with plasmids encoding GFP, GFP-IRGM, or GFP-IRGM-Kmut and either Myc-ULK1 or Flag-Beclin 1 were lysed and subjected to Western blotting. Data from densitometric analyses of Western blots (B, C, E), means ± SE, n=3 *, p< 0.05 (ANOVA). (G) Depiction of the role of IRGM ubiquitination in NOD2 degradation and ULK1 stabilization. See also Figure S6.
IRGM affects antimicrobial and inflammatory outputs and interfaces with several innate immunity systems
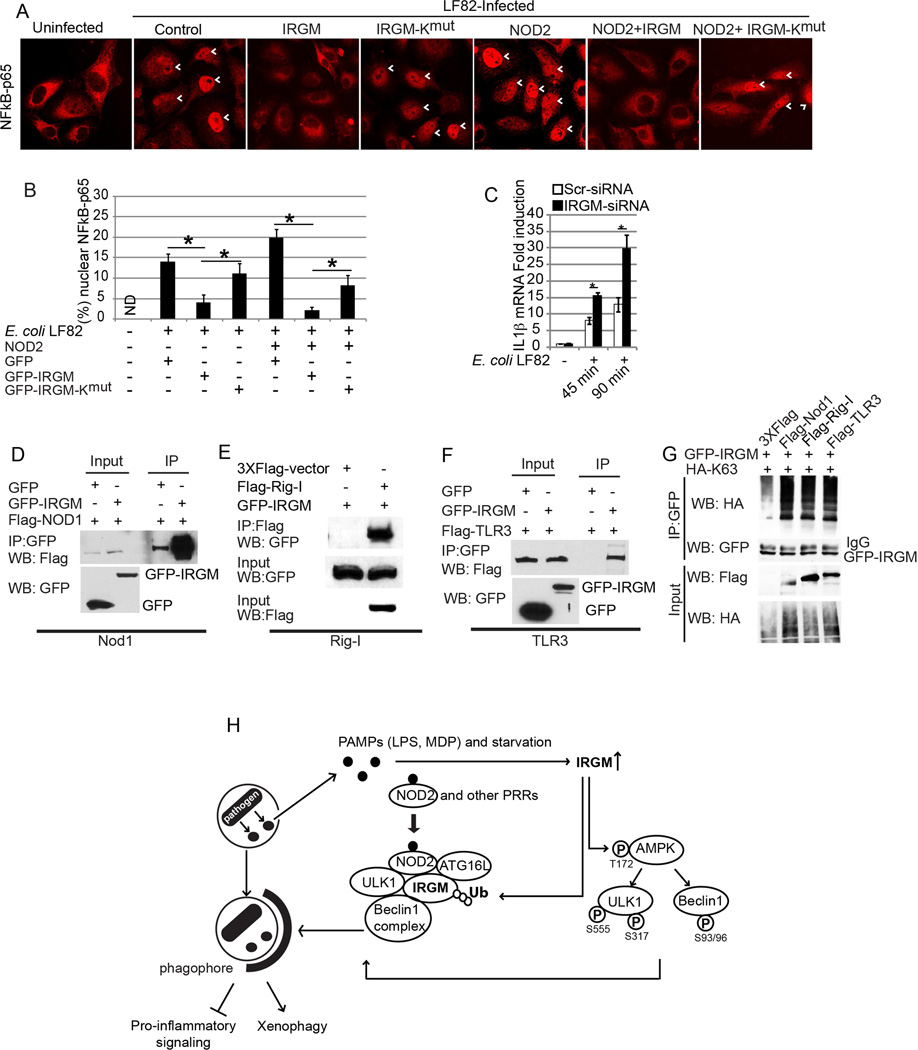
IRGM has been shown to control intracellular bacteria (Brest et al., 2011; McCarroll et al., 2008](Singh et al., 2006). Using a model system of transfected epithelial cells previously developed by others (Brest et al., 2011; Lapaquette et al., 2010) for monitoring autophagic handling of invasive bacteria, we tested how IRGM-Kmut, the mutant form of IRGM disabled for ubiquitination and examined for its effects in molecular relationships above, affected a subset of IRGM’s immune outputs. Co-expression of NOD2 with IRGM-Kmut resulted in increased NF-κB p65 nuclear translocation in response to E. coli LF82 (a CD isolate of adherent invasive E. coli) (Lapaquette et al., 2010) relative to NOD2 co-expression with IRGM wild type (Figure 7A,B, Figure S6A). Consistent with this observation, a monocytic cell line THP-1 infected with E. coli LF82 showed elevated pro-inflammatory response (increased IL-1β transcription) when IRGM was knocked down (Figure 7C, Figure S6B). The increased NF-κB response with IRGM-Kmut (Figure 7A,B) was mirrored in the effects of expressing IRGM or IRGM-Kmut on bacterial survival, reflected in the diminished ability of IRGM-Kmut to control E. coli LF82 (Figure S6C). Although IRGM expression on its own enhanced bacterial elimination, this was increased by co-expression with NOD2, an effect that was diminished when IRGM-Kmut was employed (Figure S6C). Although the overall magnitude of the effects on bacterial killing was subtle, it was in keeping with the known limitations of the system (Brest et al., 2011; Lapaquette et al., 2010) as reflected in its maximum output (upon starvation induction) of bacterial control by autophagy (Figure S6D). Based on the above experiments with IRGM-Kmut, the properties of IRGM that are essential for the assembly of the core autophagy machinery affect its antimicrobial and inflammatory outputs.
Figure 7. Ubiquitination of IRGM is important for preventing inflammation.
(A) Effect of IRGM (WT and Kmut) expression with and without NOD2 on the nuclear localization of NF-κB-p65 in HeLa cells upon E. coli LF82 infection. (B) Graph, mean % cells with NFκB-p65 nuclear localization (from 10 microscopic fields) ± SD; *, p< 0.05 (ANOVA). (C) Effect of E. coli infection on IL-1β mRNA expression in THP-1 cells subjected to IRGM knockdown (qRT-PCR normalized to GAPDH). Data, means ± SD (n>3); *, p< 0.05 (ANOVA). (D, E, F) Lysates of cells co-expressing either GFP or GFP-IRGM and (D) Flag-NOD1, (E) Flag-Rig-I, or (F) Flag-TLR3, subjected to immunoprecipitation with anti-GFP (D, E) or anti-Flag (F); blots were probed with indicated antibodies. (G) Effect of FLAG-tagged NOD1, RIG-I, or TLR3 expression on IRGM ubiquitination (K63-linked) in HEK293T cells. (H) Model of IRGM-mediated xenophagy. IRGM expression is induced by physiological cues including starvation, microbes, or microbial products (PAMPs). IRGM protein increases the abundance of active AMPK, which subsequently promotes autophagy by activating ULK1 and Beclin 1. Not only does IRGM amplify this fundamental autophagy signaling but it also assembles the core autophagy machinery. Association of IRGM with NOD2, which is enhanced in the presence of MDP, promotes IRGM ubiquitination and the assembly of autophagy initiation factors. Together, these molecular events promote antimicrobial autophagy and suppress excessive inflammatory responses. See also Figure S7.
We also tested localization of IRGM relative to the CD isolate E. coli LF82 (Lapaquette et al., 2010). We observed that without the co-expression of NOD2, IRGM had a diffuse cytosolic localization even when the cells were infected with bacteria (Figure S7A). However, when NOD2 was co-expressed with GFP-IRGM, IRGM was recruited to the invading bacteria (Figure S7B), in keeping the previously observed recruitment of ATG16L1 and NOD2 to bacterial entry sites (Travassos et al., 2010). While studying IRGM interacting partners, we observed a further ability of IRGM to engage other pattern recognition receptors (PRRs), such as NOD1, RIG-I, and TLR3 (Figure 7D–F). In contrast, IRGM did not interact with TLR4 (Figure S7C). Similarly to NOD2, NOD1, RIG-I, and TLR3 induced IRGM ubiquitination (Figure 7G). In conclusion, not only does IRGM assemble the core autophagy machinery to control innate immune responses to NOD2 agonists, but IRGM potentially has a broader repertoire of interactors among the PRR systems.
DISCUSSION
In this study we have shown that human IRGM, hitherto believed to have indirect effects on autophagy, directly governs the assembly of the principal autophagy regulators. Furthermore, it physically links the microbial sensors, including NOD2, to the core autophagic apparatus. This solves the long-standing puzzle regarding how IRGM works, and places it mechanistically at the center of action in autophagic responses to microbes.
IRGM assembles ULK1 and Beclin 1 in their activated forms to promote autophagy. Of relevance for how these proteins become activated is that IRGM also stimulates AMPK by stabilizing it in its Thr-172 phosphorylated form, which is required for AMPK activation (Mihaylova and Shaw, 2011). This is likely due to effects of IRGM on mitochondria (Singh et al., 2010), which activates AMPK (Romanello et al., 2010; Turkieh et al., 2014), and may involve specific kinases upstream of AMPK including TAK1 (Criollo et al., 2011) and CAMKKβ (Hoyer-Hansen et al., 2007) that have been shown to phosphorylate AMPK at Thr-172 (Mihaylova and Shaw, 2011) and activate autophagy (Criollo et al., 2011; Hoyer-Hansen et al., 2007). The stabilization of phospho-Thr-172 AMPK likely contributes to AMPK-dependent phosphorylation and activation of ULK1 (Egan et al., 2011; Kim et al., 2011) and Beclin 1 (Kim et al., 2013). Consistent with this, IRGM increases total activated ULK1 phosphorylated at Ser-317 and Ser-555 by AMPK (Egan et al., 2011; Kim et al., 2011), and the activated form of Beclin 1 that is phosphorylated at Ser-15 by ULK1 (Kim et al., 2013) and at Ser-93 and Ser-96 by AMPK (Kim et al., 2013). IRGM has a second effect on autophagic regulators by assembling the activated ULK1 with Beclin 1. Thus, IRGM promotes phosphorylation cascade of key autophagy regulators and assembles them into autophagy initiation complexes (Figure 7J).
Of interest is that IRGM increases levels of a number of autophagy regulators (ULK1, ATG14L, AMBRA1, and ATGL1) but does not affect the stability of others (Beclin 1 and the ATG5-ATG12 complex). The apparent absence of effects on Beclin 1 stability may be explained by the bulk of Beclin 1 being predominantly in non-autophagy related hVPS34 complexes whereas ATG14L-associated Beclin 1 represents a minority of Beclin 1 species in the cell (Kim et al., 2013). IRGM also has an effect on NOD2 levels. However, IRGM reduces NOD2 levels, in contrast to IRGM-dependent stabilization of autophagy regulators. We interpret this dichotomy as a part of the well tuned circuitry in response to microbial challenge: whereas autophagy is activated as an antimicrobial effector mechanism, the stimulatory inputs into the system mediated by NOD2 are downregulated lest the system overcommits, which in turn may result in detrimental consequences for the host. PAMP (e.g. MDP) tolerance is an important mechanism to avoid septic shock, which is in part achieved by NOD2 degradation (Zurek et al., 2012).
It has been previously shown that ATG16L1 and NOD2 interact (Cooney et al., 2010; Travassos et al., 2010). This has placed two of the Crohn’s disease-genetic risk factors together, but has left the role of IRGM unexplained. The data presented here show that IRGM is in complexes with ATG16L1 and NOD2 and that IRGM enhances assembly of Atg16L1 with NOD2. Moreover, IRGM affects the stability of each of the components of this complex. Although bringing ATG16L1 to the bacterial entry site marked by NOD2 is a previously known important step (Travassos et al., 2010), how this links up with the core autophagy regulators including ULK1 and Beclin 1 has not been addressed in prior studies. In this work we show that IRGM plays that bridging role by stimulating phosphorylation and activation of key autophagy regulators and placing them together with ATG16L1 (Figure 7J). This point is not trivial, as for example it has not been easy to connect the two seemingly separate systems of autophagy initiation: ULK1-Beclin1 complexes vs. LC3-II conjugation and localized autophagosomal membrane build up. Only recently a part of this key issue has been solved for conventional (non-immunological) autophagy by showing that ATG16L1 and WIPI2 directly interact (Dooley et al., 2014), with WIPI2 recognizing the lipid modification products of the Beclin 1-directed hVPS34 activity. We propose here that IRGM acts with a similar purpose by bridging ULK1-Beclin 1 complexes with the autophagy conjugation machinery, as shown here for ATG16L1. This can additionally explain why ATG5 is found in IRGM complexes (Gregoire et al., 2011).
Ubiquitination has been implicated in autophagy in several ways primarily in targeting of substrates for autophagic elimination (Stolz et al., 2014). However, the role of K63-linked polyubiquitination has also begun to be appreciated as a mechanism for stabilization of large autophagy-initiating complexes (Nazio et al., 2013; Shi and Kehrl, 2010). Polyubiquitination of IRGM and its role in autophagy (Figure 7J) does not play a role in targeting substrates for autophagy; instead, it stabilizes multi-protein autophagy initiation complexes. The ubiquitination of IRGM is under the control by NOD2. NOD2 enhances association of ubiquitination-competent IRGM with ULK1 and Beclin 1, whereas NOD2 has no similar effect on the ubiquitination-null mutant of IRGM (IRGM-Kmut). Importantly, IRGM-Kmut retains certain activities: it maintains the ability to bind ATG16L1 equally well as the ubiquitination-competent IRGM.
IRGM gene expression is cell-type dependent and responds to both starvation and microbial products. IRGM is particularly inducible in cells (intestinal epithelial cells and macrophages) derived from tissues affected in diseases where IRGM has been implicated as a genetic risk factor: CD and tuberculosis (Consortium, 2007; Craddock et al., 2010; Intemann et al., 2009). PAMPs induce autophagy in macrophages through IRGM linking the PAMP detection by NOD2 with the autophagic machinery activation (Figure 7J). IRGM controls not just initiation of autophagy but may also affect its maturation. IRGM complexes include UVRAG, a regulator of autophagic maturation (Itakura et al., 2008). IRGM displaces Rubicon, known to inhibit maturation complexes (Matsunaga et al., 2009). Thus, IRGM controls several points along the autophagy pathway and contributes to efficient xenophagy.
In conclusion, IRGM orchestrates antimicrobial autophagic responses. We have shown here how IRGM does that and what are the exact molecular processes that IRGM controls. This explains the hitherto mysterious role of IRGM in autophagy, places it at the center of molecular complexes controlling and executing autophagy, and molecularly connects biological inputs with autophagic outputs. Finally, our findings indicate that IRGM links up not only with NOD2 but also with several other PRRs, such as NOD1, RIG-I and TLR3. Thus, IRGM and possibly its distant IRG homologs in other vertebrates may act as transmission modules between a selective sub-repertoire of innate immune responses and the autophagy machinery.
EXPERIMENTAL PROCEDURES
Antibodies, plasmids, and siRNA
Antibodies were from Cell Signaling (AMPK, AMPK Thr-172, ULK1, ULK1 p-Ser 317, p-Ser 757, p-Ser555, NOD2, Beclin 1 p-Ser-93/96 and ATG5), MBL international corp. (ATG16L1, ATG14L, Rubicon and UVRAG), Abcam (GFP, IRGM, LPS, TRAF6 and BCL2), Sigma (LC3B, Flag), Millipore (V5 tag and HA tag), Abbiotec (Beclin 1 p-Ser15) and Novus biological (AMBRA1). GFP-tagged IRGM expression plasmid (GFP-IRGMd) was described previously (Singh et al., 2010). GFP-IRGM-Kmut was generated from GFP-IRGMd plasmid by replacing wild type IRGMd gene with synthetic mutated IRGMd gene (GeneScript) with all lysine residues mutated to arginine. Flag-IRGM and IRGM-V5 were generated by Gateway cloning (Life technologies). HA-UbiquitinC, HA-UbiquitinC-K63 (all lysine mutated except K63, Plasmid 17606), HA-UbiquitinC-K48 (all lysine mutated except K48, Plasmid 17605), Flag-TLR3 (Plasmid 13084) and YFP-TLR4 (Plasmid 13018) were from Addgene. Flag-NOD2 and variants were from Dr. Thomas Kufer (University of Cologne, Germany). Flag-ATG16L1 and variants were from Dr. Ramnik Xavier (Massachusetts General Hospital, Boston). Flag-TRAF6 was from Dr. Edward Harhaj (Johns Hopkins School of Medicine, US). IRGM siRNA, TRAF6 siRNA, AMPKα2 siRNA were from Dharmacon (siGENOME SMART pool).
Autophagy induction
U937 cells were treated with LPS (500 ng/ml) for 4 h or by transfecting MDP (5 µg/ml) with calcium phosphate for 8 h. For induction of autophagy by starvation, cells were cultured in EBSS.
Protein interactions analyses
For co-immunoprecipitation assays, the cells were lysed using NP-40 buffer containing protease inhibitor cocktail and PMSF. Lysates were incubated with antibody for 2 h followed by incubation with proteinG Dynabeads (Life technologies) for 2 h. Beads were washed for four times with 1XPBS and then boiled with SDS-PAGE buffer for analysis of interacting protein by Immunoblotting. Immunoblots were quantified using Image J software.
Supplementary Material
Acknowledgments
We thank R. Xavier, T. Kufer and E. Harhaj for Atg16L1, NOD2 and TRAF6 constructs. This work was supported by grants AI042999 and AI111935 from NIH. We acknowledge contributions by D. Bhattacharya and M. Mudd.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary information
Supplementary information includes Extended Experimental Procedures and seven figures.
REFERENCES
- Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, Ventura M, Kidd JM, Siswara P, Howard JC, Eichler EE. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- Chan WM, Mak MC, Fung TK, Lau A, Siu WY, Poon RY. Ubiquitination of p53 at multiple sites in the DNA-binding domain. Mol Cancer Res. 2006;4:15–25. doi: 10.1158/1541-7786.MCR-05-0097. [DOI] [PubMed] [Google Scholar]
- Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, Fujita N, Saitoh T, Akira S, Yoshimori T, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40:924–935. doi: 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, et al. Inhibition of autophagy by TAB2 and TAB3. The EMBO journal. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Kimura T, Timmins G, Moseley P, Chauhan S, Mandell M. Immunologic manifestations of autophagy. The Journal of Clinical Investigation. 2015;125:75–84. doi: 10.1172/JCI73945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12-5-16L1. Molecular cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Fung TK, Yam CH, Poon RY. The N-terminal regulatory domain of cyclin A contains redundant ubiquitination targeting sequences and acceptor sites. Cell Cycle. 2005;4:1411–1420. doi: 10.4161/cc.4.10.2046. [DOI] [PubMed] [Google Scholar]
- Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol. 2013;20:144–149. doi: 10.1038/nsmb.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS pathogens. 2011;7:e1002422. doi: 10.1371/journal.ppat.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, Inohara N. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Molecular cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Intemann CD, Thye T, Niemann S, Browne EN, Amanua Chinbuah M, Enimil A, Gyapong J, Osei I, Owusu-Dabo E, Helm S, et al. Autophagy gene variant IRGM-261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. Journal of cell science. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nature cell biology. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Kim SY, Chung KS, Moon CM, Kim SW, Kim EY, Jung JY, Park MS, Kim YS, Kim SK, et al. Association between genetic variants in the IRGM gene and tuberculosis in a Korean population. Infection. 2014;42:655–660. doi: 10.1007/s15010-014-0604-6. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature immunology. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Turkieh A, Caubere C, Barutaut M, Desmoulin F, Harmancey R, Galinier M, Berry M, Dambrin C, Polidori C, Casteilla L, et al. Apolipoprotein O is mitochondrial and promotes lipotoxicity in heart. J Clin Invest. 2014;124:2277–2286. doi: 10.1172/JCI74668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Chen J, Zhang L, Cha Z, Han S, Shi W, Ding R, Ma L, Xiao H, Shi C, et al. Mycobacterium leprae Upregulates IRGM Expression in Monocytes and Monocyte-Derived Macrophages. Inflammation. 2014;37:1028–1034. doi: 10.1007/s10753-014-9825-1. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, Kelliher MA. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. The Journal of biological chemistry. 2007;282:36223–36229. doi: 10.1074/jbc.M703079200. [DOI] [PubMed] [Google Scholar]
- Zurek B, Schoultz I, Neerincx A, Napolitano LM, Birkner K, Bennek E, Sellge G, Lerm M, Meroni G, Soderholm JD, et al. TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PloS one. 2012;7:e41255. doi: 10.1371/journal.pone.0041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.