Summary
In mammalian cells, DNA methylation on the 5th position of cytosine (5mC) plays an important role as an epigenetic mark. However, DNA methylation was considered to be absent in C. elegans because of the lack of detectable 5mC as well as homologs of the cytosine DNA methyltransferases. Here, using multiple approaches, we demonstrate the presence of adenine N6-methylation (6mA) in C. elegans DNA. We further demonstrate that this modification increases trans-generationally in a paradigm of epigenetic inheritance. Importantly, we identify a DNA demethylase, NMAD-1, and a potential DNA methyltransferase, DAMT-1, which regulate 6mA levels and crosstalk between methylation of histone H3K4me2 and 6mA, and control the epigenetic inheritance of phenotypes associated with the loss of the H3K4me2 demethylase spr-5. Together, these data identify a DNA modification in C. elegans and raise the exciting possibility that 6mA may be a carrier of heritable epigenetic information in eukaryotes.
Introduction
An increasing number of complex phenotypes, including physical appearance (Cavalli and Paro, 1998; Morgan et al., 1999), energy metabolism (Benyshek et al., 2006), behavioral state (Dias and Ressler, 2014), and longevity (Greer et al., 2011; Rechavi et al., 2014) have been shown to be regulated in part by non-genetic information. The molecular nature of the epigenetic information that is transmitted from generation to generation is still incompletely understood. It has been postulated that anything in the zygote that is not the DNA sequence itself could carry this non-genetic information. This includes proteins, non-coding RNA, and modifications to both proteins and DNA in chromatin (Greer and Shi, 2012; Martin and Zhang, 2007; Moazed, 2011). Arguments have been made for each of these modes of epigenetic inheritance, and it is possible that a given mode of inheritance may play a larger role than others depending on the paradigm of inheritance. One paradigm of epigenetic inheritance in C. elegans involves mutation of the histone H3 lysine 4 dimethyl (H3K4me2) demethylase spr-5 (Katz et al., 2009), which is an ortholog of the mammalian LSD1/KDM1A (Shi et al., 2004). The spr-5 mutant worms initially do not exhibit phenotypes; however, after successive generations lacking this demethylase, they display a progressively increased infertility. This fertility decline is concomitant with a global increase in the activating histone mark H3K4me2 and decline in the repressive histone mark H3K9me3 (Greer et al., 2014; Katz et al., 2009; Kerr et al., 2014; Nottke et al., 2011). Despite the fact that early and late generation spr-5 mutant worms should be genetically identical, late generation spr-5 mutant worms display altered phenotypes, most likely because of the inheritance of non-genetic information.
Previous studies searched for DNA modifications that carry epigenetic information in C. elegans. An early report performed high-performance liquid chromatography (HPLC) on C. elegans as they age and suggested that C. elegans have 5-methylcytosine (5mC) and that it accumulates with age (Klass et al., 1983). Other nematode species have also been reported to have 5mC (Gao et al., 2012), however, subsequent studies in C. elegans were unable to replicate this finding (Simpson et al., 1986). This lack of reproducibility, coupled with the fact that C. elegans do not contain homologs of the enzymes that add methyl moieties to cytosine - DNA (cytosine-5-)-methyltransferase 1 (DNMT1) or DNMT3, has led to the prevailing view that C. elegans do not possess DNA methylation (Wenzel et al., 2011). However, DNA is not only methylated at the 5th position of the pyrimidine ring of cytosines. Other DNA methylation events have been reported, including methylation of the exocyclic NH2 groups at the 6th position of the purine ring in adenines (6mA) and at the 4th position of the pyrimidine ring in cytosines (4mC) (Iyer et al., 2011). In prokaryotes, 4mC and 6mA are primarily used for distinguishing self from foreign DNA (Iyer et al., 2011). These modifications are considered to be signaling or epigenetic modifications because they are predicted not to disrupt DNA base pairing (Iyer et al., 2011). Conversely, methylation of the 1st position of the purine ring in adenines (1mA) and the 3rd position of the pyrimidine ring in cytosines (3mC) are considered DNA damage methylation events since they disrupt the hydrogen bonding with their base pairs. Additional DNA modifications have also been discovered or predicted in bacteria and eukaryotes (Iyer et al., 2011; Iyer et al., 2013), but it remains to be seen whether they are conserved across all species. Studies in eukaryotic organisms typically focus on 5mC and its role as an epigenetic modification (Koh and Rao, 2013; Martin and Zhang, 2007). However, it remains unknown whether DNA modifications such as 6mA and 4mC can also be used as epigenetic marks in eukaryotes and potentially even perpetuated through cell divisions and generations via the semi-conservative nature of DNA replication.
Here, we demonstrate that 6mA occurs in C. elegans DNA, is broadly distributed across the genome, and increases trans-generationally in spr-5 mutant worms. We identify a 6mA DNA demethylase, NMAD-1, and show that deletion of nmad-1 accelerates the progressive fertility defect phenotype of spr-5 mutant worms. Conversely, deletion of damt-1, a potential 6mA DNA methyltransferase, reduces 6mA levels in worms and suppresses the progressive fertility defect of spr-5 mutant worms. Additionally, we also identify reciprocal regulation between DNA 6mA and histone methylation. Our study identifies a new DNA modification in C. elegans, as well as regulators that control the dynamics of this modification, and advances 6mA as a potential carrier of non-genetic information across generations.
Results
6mA occurs in C. elegans and increases trans-generationally in spr-5 mutant worms
To investigate whether any forms of DNA methylation are present in C. elegans and could be potential carriers of epigenetic memory in worms lacking spr-5, we extracted genomic DNA (gDNA) from whole worms and performed dot blot analysis on wildtype (WT) and late generation spr-5(by101) mutant worms using a number of DNA modification-specific antibodies. Excitingly, we found that: 1) 6mA, but not 5mC or 5hmC, was detectable in gDNA from WT worms; and 2), the level of 6mA appears to be elevated in spr-5 mutant worms (Figure S1A). To exclude the possibility that the detected 6mA is due to contamination from bacterial DNA, which contains 6mA, we used a bacterial food source that was deficient in the DNA adenine methyltransferase (dam) and DNA cytosine methyltransferase (dcm) enzymes, which are responsible for 6mA and 5mC modifications in bacteria, respectively (we confirmed that this mutant bacterial strain does not contain 6mA (Figure S1B)). To exclude the possibility that the detected 6mA was due to contaminating methylated RNA, we treated purified gDNA with enzymes targeting all major forms of RNA, including RNase A, RNase T1, and RNase H. We found that gDNA extracted from wildtype and late generation spr-5 mutant worms fed with dam−dcm− bacteria and treated with several RNases still exhibited detectable 6mA (Figure S1B). Furthermore, 6mA antibodies only detected very low signals from worm RNA dot blots, confirming that the observed 6mA DNA dot blot signals were not derived from potentially contaminating RNA in our genomic DNA preparations (Figures 1A and S1C). Lastly, we detected 6mA in worm gDNA samples using 6mA antibodies from two independent sources (Figures S1A and S1B).
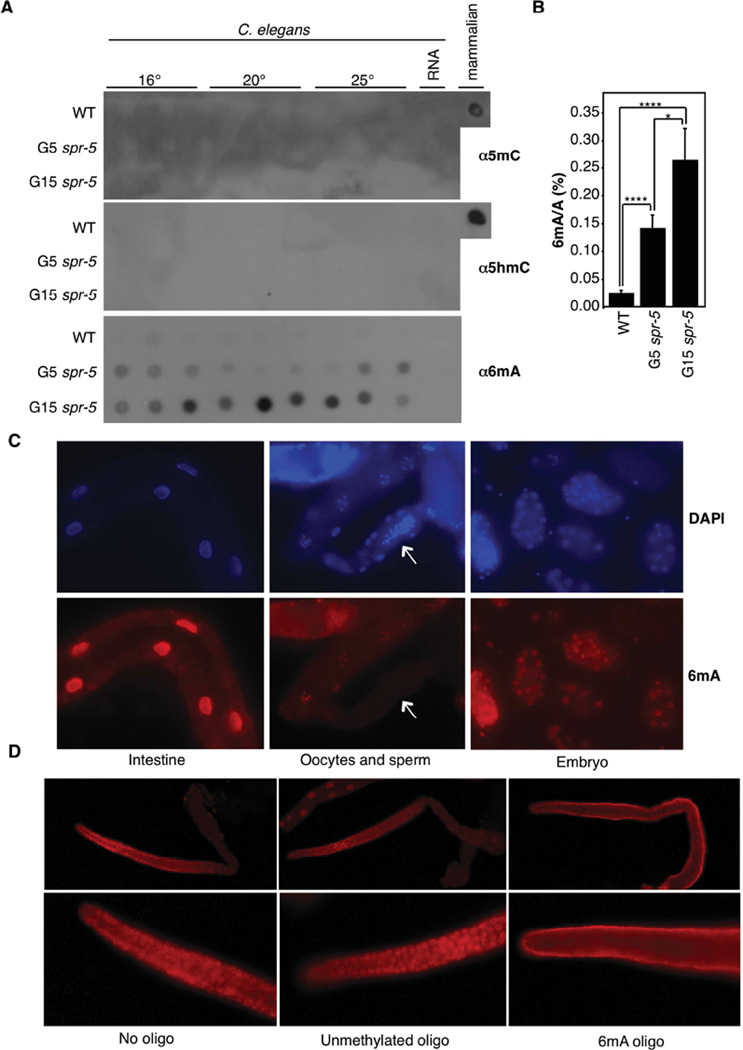
Figure 1. 6mA occurs in C. elegans DNA and increases across spr-5 generations.
A) Dot blots of 3 biological replicates of WT, generation 5, and generation 15 spr-5(by101) mutant worms grown at 16°, 20°, or 25° all show progressively elevated 6mA and lack detectable 5mC and 5hmC. 250 ng of gDNA are loaded per dot. Mammalian gDNA is used as a control for 5mC and 5hmC antibody strength. B) 6mA levels increase across generations of spr-5(by101) mutant worms as assessed by UHPLC-MS/MS. Each column represents the mean and standard deviation of 3–5 biological replicates per group. * p < 0.05, **** p <0.0001. C) Immunofluorescence displays 6mA staining in the intestine, oocytes, and every cell of the embryo. Arrow indicates sperm. D) Immunofluorescence of wildtype extracted germlines show 6mA in every nuclei. This staining was competed by a 6mA premethylated oligo but not by unmethylated oligos. See also Figures S1, S2, and S3.
We confirmed the specificity of the antibodies used in our dot blot analysis using a panel of unmethylated and premethylated DNA oligos (Figure S1D). Two 6mA antibodies (Synaptic Systems and Megabase) recognized either single- or double-stranded 6mA- but not 3mC- containing oligos. The 6mA antibodies also recognized the non-denatured (double-stranded, ds), but not denatured (single-stranded, ss) 1mA (Figure S1D). Because the worm gDNA was denatured before being loaded onto blots, the 6mA antibody-detected signal was likely N6 adenine methylated DNA.
The elevation of 6mA in late generation spr-5 mutant worms raises the possibility that 6mA might potentially play a role in transmitting heritable epigenetic information. Therefore, we next investigated whether the 6mA level changes in a trans-generational manner, as spr-5 mutant worms have been shown to display a trans-generational increase in H3K4me2 level concomitant with trans-generational fertility defects (Greer et al., 2014; Katz et al., 2009). We found that 6mA increased in a trans-generational manner in spr-5 mutant worms, regardless of worm culturing temperatures (Figure 1A). The magnitude of the increase in 6mA was variable across biological replicates but the trend towards more 6mA in spr-5 mutant worms was consistent.
To confirm that we were detecting 6mA, we turned to an antibody-independent approach, i.e, ultra high performance liquid chromatography coupled with a triple-quadrupole tandem mass spectrometry (UHPLC-MS/MS) analysis. We found that 6mA levels were variable from experiment to experiment in wildtype worms (occurring on between 0.01–0.4% of adenines). However, 6mA levels were invariably elevated in the spr-5(by101) mutant worms, though the degree of up-regulation differs from experiment to experiment (between 1.5 to 17-fold) and depending on the generation of worms assayed (Figure 1B and data not shown).
We initially noted that 1mA appeared to also increase in spr-5 mutant worms as detected by the 1mA antibody (Figure S1C). However, the 1mA antibody recognizes both 1mA and 6mA oligos and therefore cannot distinguish the two modifications (Figure S1D), whereas UHPLC-MS/MS readily separates 1mA and 6mA (Figure S2). UHPLC-MS/MS analysis of WT and spr-5 mutant worms gDNA typically failed to detect any 1mA in either strain (Figure S2B), indicating that the changes observed with our 1mA antibody likely reflected recognition of the elevated 6mA levels. On one occasion (out of more than 10 trials) in which 1mA was detected by UHPLC-MS/MS, it was observed to be at similarly low levels in WT and spr-5 mutant worms (Figure S1E).
We next investigated tissue distributions of 6mA by performing immunofluorescence (IF) on extracted germlines, embryos, and whole worms (Figures 1C, 1D and S3A), which had been treated with RNase to remove potential RNA 6mA signal. We found 6mA present ubiquitously throughout the worm except for sperm nuclei (Figures 1C and S3A) and in every other cell in the worms’ germline (Figure 1D). The absence of 6mA in sperm (Figure 1C) could reflect the high compaction of sperm chromatin (which might hamper the antibody accessibility) or could be indicative of a paternal erasure of 6mA. The IF signal likely represents 6mA, as pre-incubation of the antibodies with premethylated 6mA oligos, but not unmethylated oligos, abrogated the nuclear signal and resulted in a diffused, non-specific staining (Figure 1D). We also detected 6mA signal ubiquitously throughout the embryo (Figure 1C). While 6mA was elevated in spr-5 mutant worms (Figure S3A), 3mC and 1mA signals were essentially undetectable in germlines extracted from generation 20 (G20) spr-5(by101) mutant and wildtype worms (Figure S3B).
To determine whether 6mA might be associated with DNA damage, we performed dot blot analysis and stained gonads extracted from WT and DNA damage deficient mutant strains. We found that deletion of the DNA damage genes, xpa-1 (UV damage), ercc-1 (nucleotide excision repair), and sod-2 and sod-3 (oxidative stress) did not lead to appreciably altered levels of 6mA in the germline (Figures S4A and S4B), nor did treatment with lethal doses of the DNA damaging agent methyl methanesulfonate (MMS) (Figure S4C). Together, these results suggest that 6mA is not a DNA damage-induced modification.
6mA genomic locations
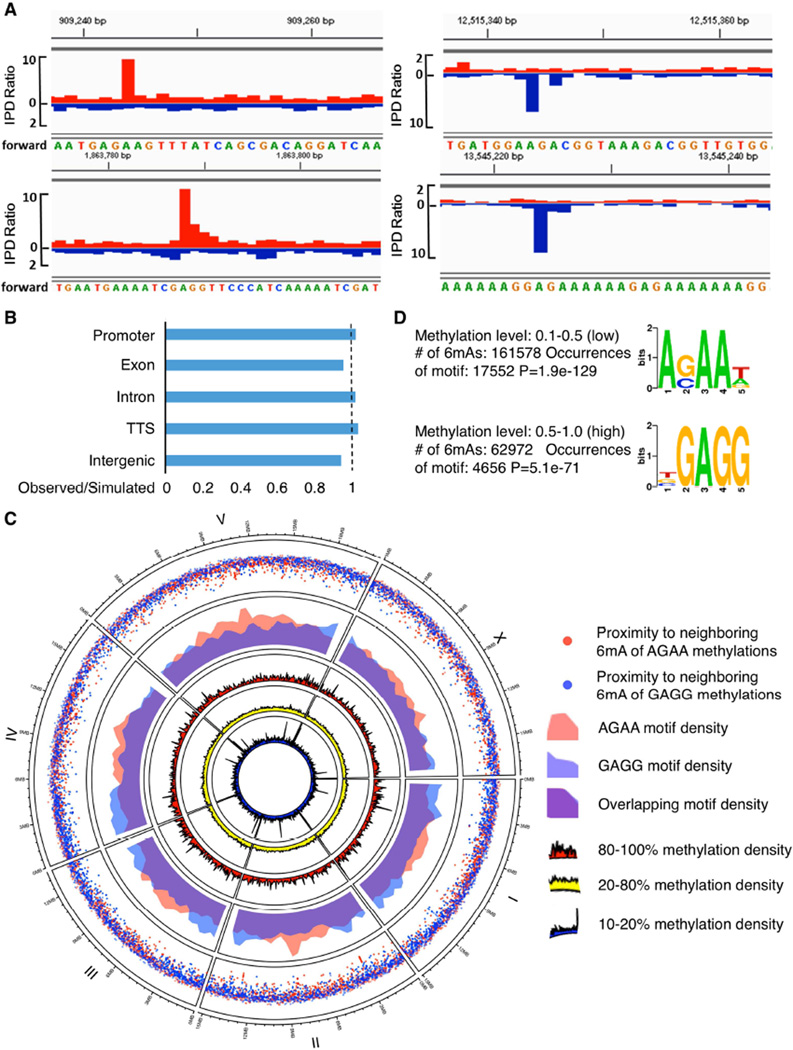
For an initial investigation of 6mA genomic localization, we performed 6mA methylated DNA immunoprecipitation (Figure S5A) followed by sequencing (MeDIP-seq) on mixed stage WT worms. MeDIP-seq identified 766 6mA peaks broadly distributed throughout the genome and evenly represented across major genomic features, except for a modest depletion in introns (Figure S5B). The most prevalent motif, AAGAAGGAAGAA, was present in 314 of the peaks identified (p=1e-42, Figure S5C).
To more directly interrogate 6mA localization using an antibody-independent, base pair resolution approach, we carried out Single Molecule Real Time sequencing (SMRT sequencing), which not only identifies individual bases but also their modifications (Flusberg et al., 2010). We generated a SMRT sequencing dataset, using gDNA from mixed stage, wildtype worms. To increase our read density we merged our dataset with the publicly available C. elegans SMRT sequencing data generated by Pacific Biosciences (http://datasets.pacb.com.s3.amazonaws.com/2014/c_elegans/list.html). In this analysis, SMRT sequencing detected 6mA on 225,586 adenines, about ~0.7% of the total adenines in the worm genome, which is equivalent to 0.3% bulk adenine methylation as some adenines were methylated 10% of the time while others were methylated 90% of the time. This value (0.3%) is comparable to some of the UHPLC-MS/MS results (Figure 5E). SMRT sequencing does not distinguish 6mA versus 1mA, but 1mA is rarely above the level of detectability by UHPLC-MS/MS in worm gDNA. This suggests that the signals detected through SMRT sequencing were 6mA (Figure 2A), although we cannot completely exclude the possibility that rare occurrences of 1mA could have been detected as 6mA in our SMRT sequencing analysis. Similar to the MeDIP-seq results, the SMRT sequencing analysis identified a broad distribution of 6mA across all chromosomes of the worm genome, with no one genomic feature being significantly enriched or depleted for 6mA (Figures 2B and 2C). Since lowly methylated regions usually include functional elements in mammalian cells (Stadler et al., 2011), we examined 6mA distribution (Figure 2C) by separating it into low (10–20%, dark blue circle), middle (20–80%, yellow circle), and high (80–100%, red circle) categories and presenting the data in circos plot format in which concentric rings represent the density distributions of 6mA across the six worm chromosomes in the given category. We found that some lowly methylated regions appeared in dense clusters similar to lowly methylated 5mC (Figure 2C, innermost concentric circle). Notably, two sequence motifs were significantly associated with the presence of 6mA (Figure 2D): AGAA (p=1.9e-129) and GAGG (p=5.1e-71). Importantly, the AGAA motif identified by SMRT sequencing was also identified by MeDIP-seq (Figure S5C). Interestingly, the GAGG motif was most prevalent in sites that were frequently 6mA methylated (50–100% methylation level), whereas the AGAA motif was most prevalent in infrequently 6mA methylated sites (10–50% methylation level). The two 6mA motifs did not significantly differ in chromosomal distribution (Figure 2C, fourth concentric circle), though there were some regions that showed increased clustering density for each of the 6mA motifs (Figure 2C, outer rainfall plot). Notably, both motifs indicate that methylation at these sites occurs only on one of the strands, unlike the strong propensity for 5mC to occur in the context of CG doublets in various eukaryotes. Both SMRT sequencing and MeDIP-seq – which have been performed on mixed tissues and mixed stage worms – confirmed the presence of 6mA in worm DNA across the genome and at similar sequence motifs.
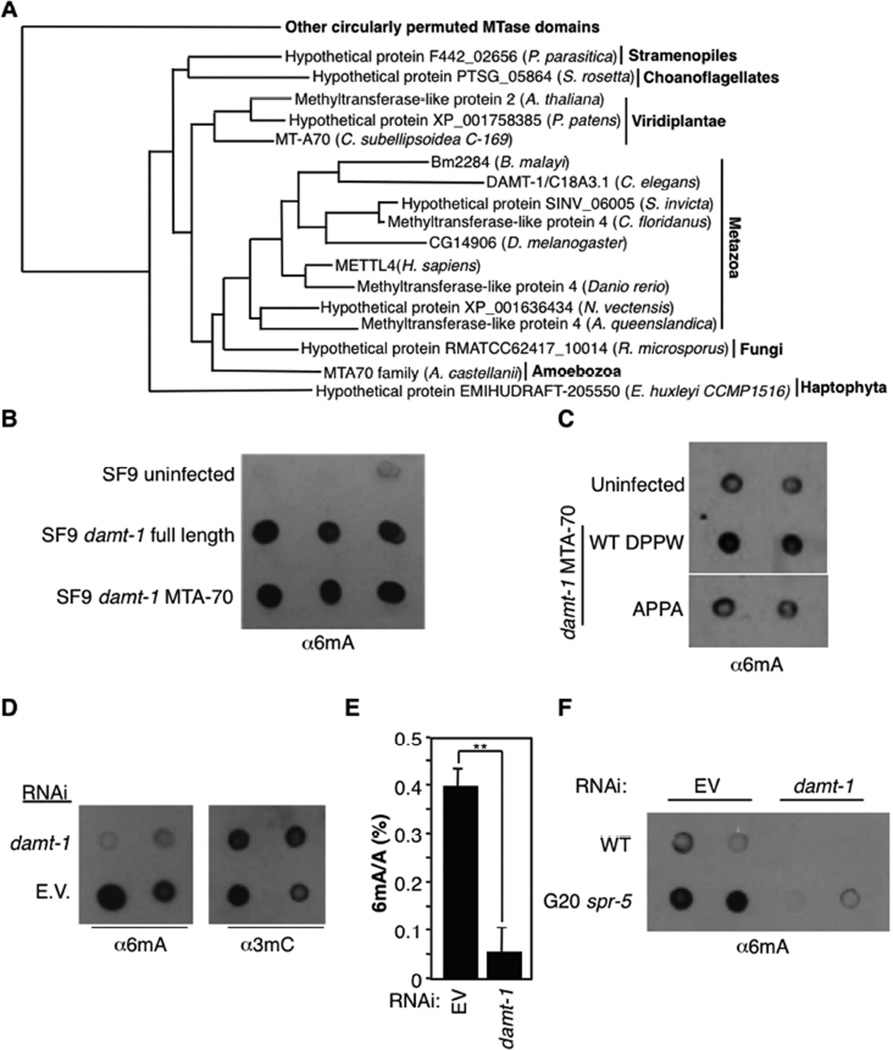
Figure 5. DAMT-1 regulates 6mA levels.
A) Phylogeny tree shows conservation of DAMT-1 in other eukaryotic species. Full tree and details of related clades are presented in Figure S6B. B) gDNA extracted from SF9 cells infected with full length or the catalytic domain of damt-1 show elevated levels of 6mA by dot blot. C) Mutation of the catalytic domain of DAMT-1 (DPPW to APPA) limits the increase in 6mA levels of infected SF9 cells. DAMT-1 expression is presented in Figure S6C. D) damt-1 knockdown decreases 6mA without affecting detectable 3mC levels. E) damt-1 mutants have decreased levels of 6mA as assessed by LC-MS/MS. Each bar represents the mean and standard error of the mean of 3 independent experiments of 3 biological replicates each measured in duplicate. ** p < 0.01. F) Generation 20 (G20) spr-5 mutant worms show elevated 6mA levels compared with WT worms and damt-1 knockdown suppresses the elevated 6mA in spr-5 mutant worms. Each dot represents 250 ng of gDNA of independent biological replicates.
Figure 2. 6mA genomic location.
A) Representative interpulse duration (IPD) ratios, of SMRT sequencing data of mixed stage wildtype worms. IPD ratio is defined as the change in IPD distribution in the sample compared to unmodified bases. Red: positive strand; Blue: negative strand. B) Comparison of observed vs. simulated distributions of 6mA across the C. elegans genome indicates that 6mA is not enriched or depleted in any major genomic feature. A permutation was used to calculate the average of 10,000 simulations for comparison to the observed data. C) Circos plots of 6mA and motif distributions; three inner rings: 6mA density normalized to adenines in each bin of 6mAs within different methylation fractions. Red, yellow, and blue represent highly methylated (80–100%), intermediate (20–80%), and lowly methylated (10–20%) 6mA, respectively. The middle ring shows AGAA (red) and GAGG (blue) motif densities, with purple indicating the overlap. The outer ring (rainfall plot) shows the distribution of inter-distance between each two adjacent 6mAs in the same motif. Red dots represent 6mAs in AGAA motif and blue dots represent 6mAs in GAGG motif; increasing vertical distance towards the center of the circle indicates increasing local density of 6mA occurrences. D) SMRT sequencing identified two motifs associated with 6mA. AGAA and GAGG are associated with low and high percentage 6mA, respectively. Methylation level refers to the percentage of times (1.0 = 100%) a given A in the sample population was read as methylated by SMRT sequencing. See also Figure S5 for 6mA MeDIPseq.
Deletion of potential dealkylating enzyme, nmad-1, accelerates the progressive fertility defect of spr-5 mutant worms
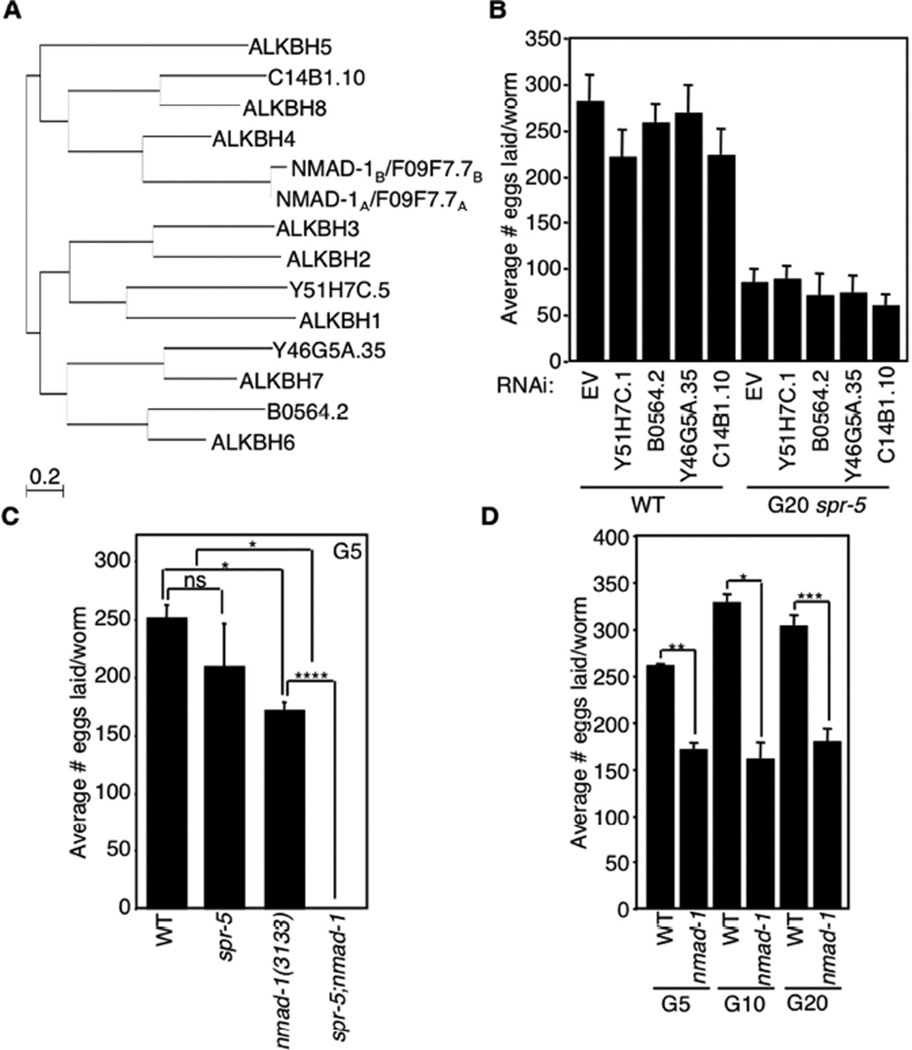
To identify the enzymes responsible for the addition and removal of 6mA in C. elegans, we first examined the ALKB family of dealkylating enzymes, which have been shown in other species to remove methyl groups from DNA and RNA oxidatively, utilizing 2- oxoglutarate as a cofactor (Yi and He, 2013). Since 6mA levels increased across generations of spr-5 mutant worms, we hypothesized that deletion of a 6mA demethylase would accelerate the trans-generational fertility defect of spr-5 mutant worms. To determine whether any of the five C. elegans ALKB family members (Figure 3A) regulates 6mA, we first investigated whether knockdown or deletion of the family members had any effect on the progressive fertility defect of spr-5 mutant worms. We found that knockdown of Y51H7C.1, B0564.2, Y46G5A.35, and C14B1.10 had no effect on the fertility of WT or spr-5 mutant worms (Figures 3B and S6A). Although we were unable to efficiently knockdown the fifth ALKB family member, F09F7.7 (Figure S6A), we obtained a worm strain carrying deletion of F09F7.7(ok3133) and found that loss of F09F7.7 accelerated progressive fertility defect of spr-5 mutant worms such that the spr-5;F09F7.7 double mutant worms became completely sterile by generation 4 (Figure 3C). As a control, we found that at a similar generation, the spr-5 mutant worms did not display a significant fertility defect (Figure 3A, (Greer et al., 2014; Katz et al., 2009)). As a further control, we examined and found that F09F7.7 mutants laid fewer eggs than WT worms (Figure 3C), but, importantly, this phenotype was not progressive (Figure 3D), suggesting that the acceleration of the progressive fertility defect of spr-5 mutant worms is a result of a specific genetic interaction between F09F7.7 and spr-5. These findings suggested that F09F7.7 may act as a 6mA demethylase in vivo, which is further supported by the biochemical experiments discussed below. We thus renamed F09F7.7 N6-methyl adenine demethylase 1 (nmad-1) to reflect this newly identified function.
Figure 3. Deletion of nmad-1 accelerates the progressive fertility defect of spr-5 mutant worms.
A) Phylogeny tree of human and C. elegans ALKBH family members. B) Knockdown of 4 of the ALKBH family members has no effect on egg laying of WT and spr-5 mutant worms treated for 20 generations with bacteria expressing the specific dsRNAs. Knockdown efficiency was tested by real-time RT PCR (Figure S6A). C) Early generation (G5) spr-5 mutant worms do not display significant fertility defects, but when combined with nmad-1 deletion these worms become sterile by generation 4. Each bar represents the mean ± SEM of 3 independent experiments. D) nmad-1 mutants lay fewer eggs than WT worms but do not display a progressive fertility decline. Each bar represents the mean ± SEM of 2–6 independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns not significant.
NMAD-1 demethylates 6mA in vitro and in vivo
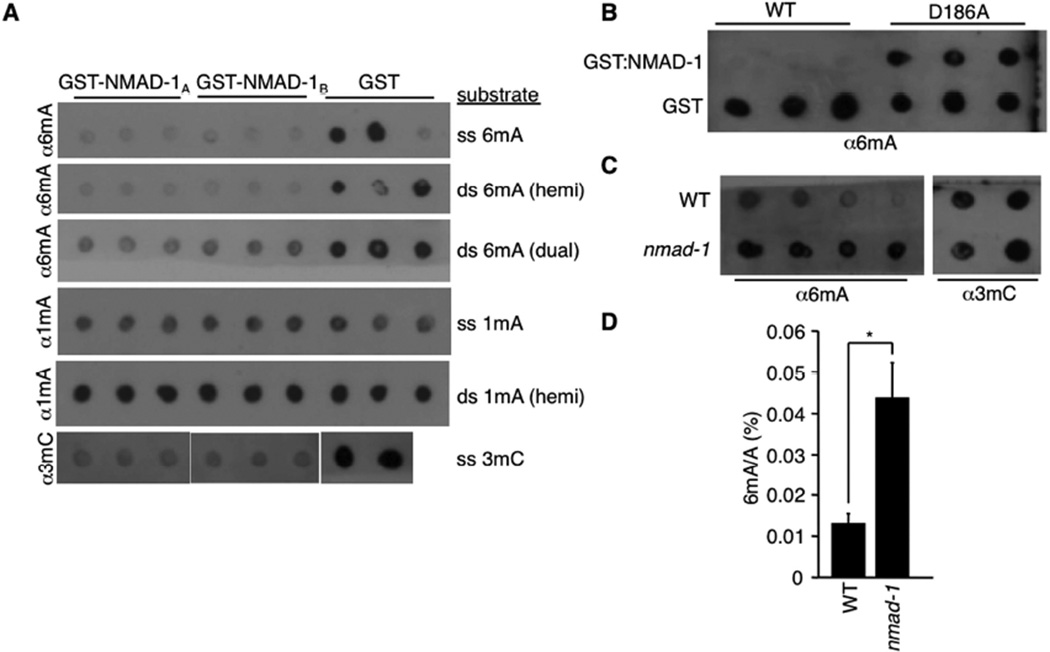
To biochemically determine whether NMAD-1 was a 6mA demethylase, we GST tagged and purified the protein and tested its demethylating activity in vitro. We found that two different isoforms of NMAD-1 were able to demethylate 6mA and 3mC oligos but not 1mA oligos in vitro (Figure 4A). To determine whether this demethylating activity was intrinsic to NMAD-1, we mutated the iron-chelating aspartic acid 186 in the catalytic domain of NMAD-1 to an alanine (D186A) and found that this mutation abrogated the ability of NMAD-1 to demethylate 6mA oligos (Figure 4B), suggesting that NMAD-1 possesses 6mA demethylase activity in vitro. We next investigated whether nmad-1 mediates demethylation of both 6mA and 3mC in vivo. As shown in Figure 4C, nmad-1 mutant worms showed elevated levels of 6mA, but not 3mC. This elevated 6mA was further confirmed by UHPLC-MS/MS (Figure 4D). Together, these results suggest that NMAD-1 is primarily a 6mA demethylase in vivo, although recombinant NMAD-1 protein can demethylate both 6mA and 3mC in vitro.
Figure 4. NMAD-1 demethylates 6mA in vitro and in vivo.
A) Two different isoforms of NMAD-1 demethylate single stranded denatured (ss) and double stranded non-denatured (ds, hemi or dual methylated) oligos premethylated at 6mA and 3mC but not 1mA. B) Mutation of the catalytic domain of NMAD-1 abrogates the ability of NMAD-1 to demethylate 6mA premethylated oligos. C) nmad-1 mutants have elevated levels of 6mA without detectable changes in 3mC levels. Each dot represents 250 ng of DNA of independent biological replicates. D) nmad-1 mutants have elevated levels of 6mA as assessed by UHPLC-MS/MS. Each bar represents the mean and standard error of the mean of 2 independent biological replicates measured in duplicate. * p < 0.05.
Deletion and over-expression of the potential methyltransferase damt-1 decreases and increases 6mA levels in vivo and in tissue culture, respectively
We next sought to identify enzymes that mediate adenine N6-methylation in C. elegans. Although candidate 6mA DNA methyltransferases have been identified in chlorophyte algae, ciliates, some fungi, and certain other eukaryotic lineages (Iyer et al., 2011; Iyer et al., 2014), none has been identified in Metazoa thus far. While the eukaryotic candidate 6mA methyltransferases belong to multiple distinct methylase lineages (Iyer et al., 2011), the most widespread versions belong to the MTA-70 family exemplified by the yeast mRNA adenine methylase complex Ime4/Kar4 (Anantharaman et al., 2002; Clancy et al., 2002). These enzymes have evolved from m.MunI-like 6mA DNA methyltransferases of bacterial restriction-modification systems (Iyer et al., 2011) and are typified by a C-terminal circularly permuted methyltransferase domain fused to a distinctive N-terminal, predicted α-helical domain with a strongly positively charged segment. C. elegans has one representative of this family - the gene C18A3.1, which is conserved across eukaryotes including humans, plants, basal fungi, certain amoebozoans and stamenopiles, and can be distinguished by phylogenetic analysis from Ime4 and Kar4 that are absent in C. elegans (Figures 5A and S6B). The orthologs of C18A3.1 form a distinct clade, separated from the mRNA methylase complex clade, within the primary eukaryotic radiation of the MTA-70 family. C. elegans also lack the transposon-encoded 6mA DNA methylase domains, which are found in related nematodes like C. remanei. These observations suggest that C18A3.1 is the primary 6mA DNA methylase candidate in C. elegans.
We investigated whether C18A3.1 could methylate the 6th position of adenines, but due to its high hydrophobicity we were unable to purify this protein from bacterial or insect cells in sufficient quantities to study its activity in vitro. However, when we analyzed the gDNA isolated from SF9 cells expressing full length C18A3.1 or the catalytic domain of C18A3.1 alone, we found that 6mA levels were elevated compared to DNA from insect cells that do not express C18A3.1 (Figure 5B). To determine whether this potential methylating activity was intrinsic to C18A3.1, we mutated amino acids in the N6A methyltransferase signature (DPPW) important for substrate recognition and catalytic activity (Iyer et al., 2011) and found that mutation of DPPW to APPA in the catalytic domain ablated the 6mA induction in SF9 gDNA (Figures 5C and S6C). This result suggests that C18A3.1 (renamed damt-1 for DNA N6 adenine methyltransferase 1) is itself a 6mA methyltransferase, although we cannot rule out the less likely possibility that C18A3.1 expression in insect cells coincidentally activated an endogenous insect cell enzyme that is responsible for the observed 6mA. To determine whether DAMT-1 was a 6mA methyltransferase in vivo, we knocked down damt-1 in WT worms and found decreased 6mA but not 3mC levels in the extracted gDNA (Figures 5D and 5E). damt-1 knockdown also decreased 6mA levels in spr-5(by101) mutant worms to similar levels as in WT worms (Figure 5F). Taken together, these data suggest that DAMT-1 is a 6mA methyltransferase in C. elegans.
Deletion of damt-1 suppresses the trans-generational phenotypes of spr-5 mutant worm s
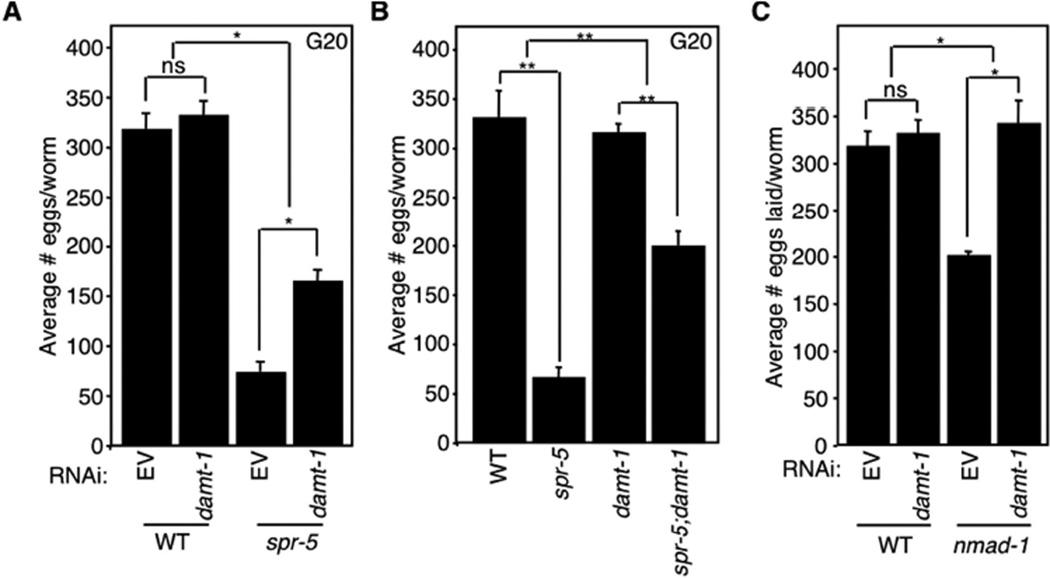
If DAMT-1 is a 6mA methyltransferase, then we would expect that its knockdown or deletion would suppress the trans-generational phenotypes of spr-5 mutant worms. Indeed, knockdown of damt-1 for 20 generations partially suppressed the progressive fertility defect of spr-5(by101) mutant worms without affecting the fertility of WT worms (Figure 6A). Specifically, late generation spr-5 mutant worms on damt-1 RNAi laid 2–3 times more eggs than late generation spr-5 mutant worms on bacteria containing an empty RNAi vector (Figure 6A). Similarly, a genetic deletion (gk961032) that removes the entirety of damt-1 and a portion of the nearby Ras GTPase superfamily gene rab-3, had no effect on egg laying by itself but suppressed the progressive fertility defect of spr-5(by134) mutant worms at generations 10, 17, 20, and 26 (Figure 6B and data not shown). damt-1 knockdown also eliminated the fertility defect of the nmad-1 mutant worms, suggesting that DAMT-1 functions to counteract the activity of the 6mA demethylase, NMAD-1, in vivo (Figure 6C). Collectively, these data suggest that DAMT-1 is a 6mA methyltransferase that suppresses the trans-generational phenotypes of spr-5 mutant worms.
Figure 6. Deletion of damt-1 suppresses the trans-generational phenotypes of spr-5 mutant worms.
A) damt-1 knockdown has no effect on WT egg laying but partially suppresses the progressive fertility defect of spr-5(by101) mutant worms. B) damt-1 deletion has no effect on WT egg laying but partially suppresses the progressive fertility defect of spr-5(by134) mutant worms. C) damt-1 knockdown reverts the egg laying defect of nmad-1 mutant worms. All assays were performed at generation 20. Each bar represents the mean ± SEM of 3 independent experiments. * p < 0.05, ** p < 0.01, ns not significant.
Crosstalk between H3K4me2 and 6mA
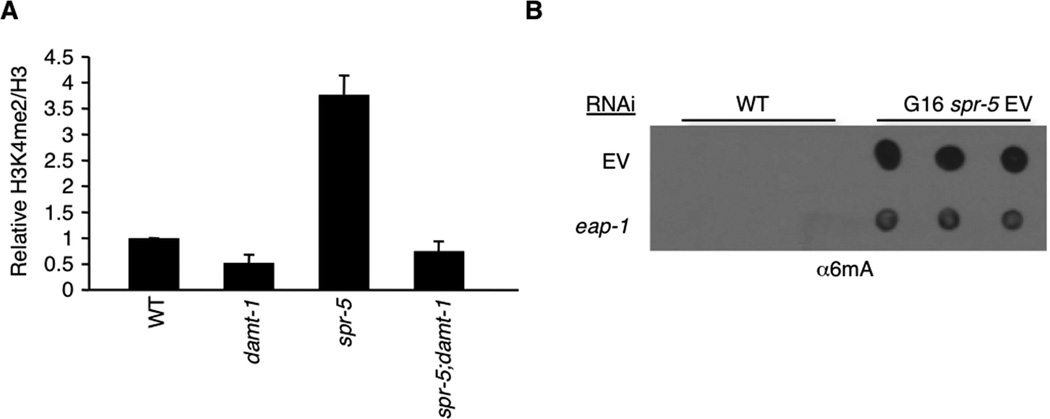
As discussed earlier, we initially observed an increase in 6mA levels in the histone H3K4me1/me2 demethylase mutant spr-5. Conversely, we found that deletion of the potential 6mA methyltransferase, damt-1, reduced the elevated H3K4me2 levels of spr-5 mutant worms (Figures 7A, S7A, and S7B). Furthermore, we found that knockdown of the H3K9me binding protein eap-1, which reduces H3K4me2 levels in spr-5 mutant worms (Greer et al., 2014), also reduced the levels of 6mA in spr-5 mutant worms (Figure 7B and S7C). Collectively, these findings suggest reciprocal regulation of H3K4 and adenine N6 methylation and crosstalk between regulators that control adenine and histone methylation.
Figure 7. DNA methylation and histone methylation crosstalk.
A) Deletion of damt-1 suppresses the elevated H3K4me2 levels of late generation spr-5(by134) mutant worms. Each bar represents the mean ± SEM of 3 independent experiments performed in biological duplicate. Image J was used to analyze the relative intensity of H3K4me2 compared to Histone H3. Western blots corresponding to two of these experiments are shown in Figures S7A and S7B. B) Knockdown of H3K9me binding protein, eap-1, suppresses the elevated 6mA level detected in spr-5 mutant worms as assessed by dot blots. A longer exposure showing 6mA levels in WT worms is shown in Figure S7C.
Discussion
To date, 6mA has primarily been studied in prokaryotes, where it has been shown as a mark to discriminate invasive DNA (Arber and Dussoix, 1962; Meselson and Yuan, 1968). However, prokaryotic 6mA also functions as a binding platform and influences gene expression (Braun and Wright, 1986; Han et al., 2004). 6mA has also been reported in more ancient eukaryotes such as ciliates, in which it is observed in the macro (somatic) and not in the micro (germline) nucleus, highlighting its potential function in a broad range of biological contexts (Gutierrez et al., 2000). Both fungi and animals are known to undergo methylation of adenosine in mRNA, with 6mA influencing mRNA stability (Fu et al., 2014) and RNA splicing (Dominissini et al., 2012; Liu et al., 2015). However, whether 6mA is present in DNA of Metazoa has been unclear, and it has been widely assumed that 5mC, rather than 6mA, plays a primary role as the key carrier of epigenetic information on DNA in these organisms (Wion and Casadesus, 2006). Importantly, this study not only identifies the presence of 6mA in C. elegans, but also raises the exciting possibility that this modification may play a role in carrying and transmitting epigenetic information across generations, and that in addition to 5mC, 6mA may also be used across eukaryotes as a potential epigenetic modification.
Our conclusion that 6mA is present in the C. elegans genome is supported by multiple lines of evidence. First, 6mA was detected by two independently developed 6mA-specific antibodies (Figures 1A and S1A). Second, 6mA was detected on the DNA of most cells throughout the worm by immunofluorescence (Figures 1C, 1D, S3, and S4A). Third, the presence of 6mA was also identified by an antibody-independent means, i.e., UHPLC- MS/MS, which showed that C. elegans genome possesses 6mA (Figure 1B). Fourth, two independent sequencing methods - direct, antibody-independent DNA sequencing using SMRT sequencing and the antibody-dependent MeDIPseq - both detected 6mA on C. elegans DNA (Figures 2 and S5). Although both sequencing methods have caveats about distinguishing between 1mA and 6mA, the DNA samples subjected to sequencing had undetectable 1mA (as determined by UHPLC-MS/MS), suggesting that the majority of the methylation events detected by SMRT sequencing likely represent 6mA. Finally, we also identified potential enzymatic machineries that mediate addition and removal of 6mA (Figures 4 and 5). Importantly, manipulation of these enzymes in vivo not only affects 6mA levels, but also impacts trans-generational epigenetic inheritance in C. elegans (Figures 3 and 6), raising the exciting and attractive possibility that 6mA may indeed carry epigenetic information.
Both SMRT sequencing and MeDIP-seq identified a broad 6mA genomic distribution with a common sequence motif, but without a clear enrichment pattern; in contrast, 5mC distributions in mammals are highly tissue-specific (Smith and Meissner, 2013). Given that worms of mixed developmental stages were used for sequencing, the possibility that 6mA may be enriched in specific genomic locations in a tissue-, cell type-, or developmental stage-specific manner remains, and such enrichment patterns may only emerge when DNA samples from specific cell types or developmental stages are analyzed.
While DNA methylation may be a more efficient carrier of epigenetic information, it remains to be seen whether 6mA, H3K4me2, or some as of yet unidentified mark carry the epigenetic information on their own or collaborate to transmit epigenetic information across generations in C. elegans. A recent study provided evidence that both the histone modification mark (H3K27me3) and the PRC2 machinery are transmitted across generations epigenetically (Gaydos et al., 2014), implicating chromatin modifications as possible carriers of heritable non-genetic information. Interestingly, our study identified robust genetic interactions between the H3K4me1/2-specific demethylase SPR-5 and machineries that regulate 6mA, i.e., NMAD-1 and DAMT-1, in the regulation of trans-generational epigenetic inheritance. These results suggest crosstalk between 6mA and histone methylation and possible collaboration of these modifications in transmitting epigenetic information. Further evidence for this crosstalk was provided by the finding that knockdown of the H3K9me binding protein, eap-1, which reduces H3K4me2 levels in spr-5 mutant worms (Greer et al., 2014), also decreases 6mA levels in spr-5 mutant worms (Figure 7B). Conversely, deletion of the potential 6mA methyltransferase, damt-1, decreases H3K4me2 levels in spr-5 mutant worms (Figure 7A). Consistent with the possibility of crosstalk between H3K4 and adenine N6 methylation regulation, analysis of the domain architectures of DNA N6A methyltransferases in eukaryotes, such as chlorophytes and fungi, showed that the DNA-modifying catalytic domain is fused to histone-recognition domains (Iyer et al., 2011; Iyer et al., 2014).
At the present time, the molecular function of 6mA is still unclear. DNA methylation systems such as 6mA and 5mC are proposed to serve various functions, including protection of host genomes (Arber and Dussoix, 1962; Meselson and Yuan, 1968), silencing of transposable elements (Kato et al., 2003; Zemach and Zilberman, 2010), transcriptional silencing (Csankovszki et al., 2001; Sado et al., 2000; Stein et al., 1982), prevention of cryptic transcription in intragenic regions (Zemach et al., 2010), and heterochromatin state transitions (Saksouk et al., 2014). A study conducted in Chlamydomonas reinhardtii (Fu, Y. and He, C., personal communications) shows a correlation of 6mA modification with active gene transcription, suggesting a possible role in gene expression regulation. We observed that the absolute 6mA levels were variable from experiment to experiment and found that some environmental manipulations altered 6mA levels (data not shown). This raises the possibility that this modification could integrate environmental stimuli to regulate biological processes. Future studies will be required to fully explore the molecular function of 6mA in worms.
Finally, it will be informative to place 6mA regulation within a cellular pathway(s). In Arabidopsis, for example, the RNAi pathway feeds into 5mC regulation and heterochromatin formation and propagation (Law and Jacobsen, 2010; Teixeira et al., 2009; Wassenegger et al., 1994). Whether molecular pathways governing the trans-generational epigenetic inheritance of fertility and other phenotypes feed into 6mA regulation in C. elegans remains to be determined. It will be of significant interest to understand whether 6mA contributes to regulating the epigenome landscape that governs trans-generational epigenetic inheritance. Furthermore, given that orthologs of damt-1 are widely conserved across eukaryotes, including mammals and other vertebrates, it will now be of great interest to investigate which other eukaryotic species might also have 6mA in their DNA, and in which biological contexts this modification is regulated and plays a biological function. If other eukaryotes are found to have 6mA, it raises the exciting possibility that 6mA could carry epigenetic information in multiple paradigms of epigenetic inheritance.
Experimental Procedures
Worm Strains
The N2 Bristol strain was used as the wildtype background. The following mutations were used in this study: LG1: spr-5(by101), spr-5(by134), ercc-1(tm1981), xpa-1(mn157), sod-2(gk257); LGII: damt-1(gk961032); LGIII: nmad-1(ok3133), LGX: sod-3(tm760). In this paper mutant worms were backcrossed: damt-1: 5-7 times, nmad-1: 5–9 times. Worms were grown on dam−dcm− bacteria (NEB C2925) in all experiments except for Figure S1A where they were grown on OP50-1 bacteria.
Fertility assays
From day 3 to day 8 post-hatching, 10 worms were placed on NGM plates with bacteria in triplicate (30 worms total per condition). Worms were grown at 20°C. After 24 h, the adult worms were removed from each plate and placed on new plate. The numbers of eggs and hatched worms on the plate were counted. Statistical analyses of fertility were performed using two-way ANOVA tests with Bonferroni post-tests, or t-tests using mean and standard error values.
Worm gDNA extraction
Worms were washed 2 times with M9 buffer. 250 µl of worm genomic DNA lysis buffer (200 mM NaCl, 100 mM Tris-HCl pH 8.5, 50 mM EDTA pH 8.0, 0.5% SDS) + proteinase K (0.1 mg/ml) was added. Worms were incubated at 65°C for 1 hour with occasional vortexing and then incubated at 95°C for 20 minutes. RNase A was added (0.1 mg/ml) and incubated at 37°C for 1 hour. 250 µl of phenol;chloroform;isoamylic acid was added. Samples were mixed and then spun @ 13,000 rpm at room temperature for 15 minutes. The aqueous phase was removed to a new tube and phenol;chloroform;isoamylic acid extraction was repeated. To the aqueous phase 25 µl of 3M sodium acetate and 750 µl of 100% EtOH were added and samples were placed at −80°C for at least 1 hour. Samples were spun at 13,000 rpm at 4°C for 30 minutes. The supernatant was removed. 350 µl of cold 75% EtOH was added and samples were again spun at 13,000 rpm for 10 minutes. The supernatant was discarded and pellet was allowed to dry before being resuspended in TE (10 mM Tris-HCl, 1 mM EDTA pH 8.0 final pH 7.5). For samples presented in Figures 7B, S1B, S4C, and S7C purified gDNA was then treated with RNase A/T1 mix (Thermo Scientific) at a 1:20 dilution and RNaseH (NEB) at a 1:50 dilution for 1 hour at 37°C for 1 hour prior to subsequent re-purification starting with proteinase K digestion.
Dot blot
Samples were diluted to 100 ng/µl and heated at 95°C for 10 minutes to denature DNA. Samples were immediately placed on ice for 5 minutes and 250 ng were loaded per dot on Hybond + membranes. Membranes were allowed to air dry and placed in boxes with damp paper towels. DNA was then autocrosslinked in a UV stratalinker 2400 (stratagene) 2 times. The membrane was allowed to dry then blocked for 1 hour in 5% milk TBS. Membranes were probed for 1 hour at room temperature or overnight at 4°C with primary antibody in 5% milk TBS. Blots were washed 3 times for 10 minutes with TTBS then probed with secondary antibody in 5% milk for 1 hour at room temperature. Blots were washed 3 times for 10 minutes with TTBS and ECL was applied and film was developed.
SMRT sequencing
The raw data is from two parts: (1) our own data, uploaded into GEO (accession no. GSE66504); (2) from PacBio public database (http://datasets.pacb.com.s3.amazonaws.com/2014/c_elegans/list.html). Each of the raw data in bax.h5 format was first aligned to ce10 genome using pbalign with the option --forQuiver. The polymerase kinetics information was further loaded after alignment by loadChemistry.py and loadPulses scripts. Then two post-aligned datasets were merged and sorted by using cmph5tools. Finally, the 6mA was identified using ipdSummary.py script. We then further filtered 6mAs with less than 50× coverage. For motif identification, we first separated the whole 6mAs into 10 groups based on their methylation fraction (methylation fraction: 0–0.1; 0.1–0.2…0.9–1.0). For each 6mA, we then extracted 2bp from the upstream and downstream sequences. MEME-ChIP (Machanick and Bailey, 2011) was then used to identify motifs in each group. The genome-wide 6mA and motif profiles are generated from circlize (Gu et al., 2014) Part of the analysis was done by customized scripts in R, Python and Perl.
Antibodies
The following antibodies were used: α6mA (Synaptic Systems, 202 003), α6mA (Megabase Research), α5mC (Active Motif, 39649), α5hmC (Active Motif 39769), α3mC (Active Motif, 61111 and 61179), and α1mA (Active Motif, custom). α6mA (Megabase Research) was only used in Figure S1A.
Supplementary Material
Acknowledgments
We thank members of the Shi lab for helpful discussions, Elizabeth Pollina for feedback on the manuscript, and Madeline Schuck and LaVondea Elow for technical and administrative support. We thank the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40OD010440) for C. elegans strains, and the Tufts University Core Facility Genomics Core and the University of Massachusetts Medical School Deep Sequencing Core for MeDIP-seq and SMRT sequencing, respectively. E.L.G. was supported by a Helen Hay Whitney postdoctoral fellowship and a National Institute on Aging of the NIH grant (K99AG043550). M.A.B. was supported by an NIH NRSA postdoctoral fellowship (1F32CA180450-01) and is currently supported by a Special Fellow award from the Leukemia & Lymphoma Society (3353-15). L.A. was supported by funds of the Intramural Research Program of the National Library of Health, NIH, US department of Health and Human Services. C.H. is a Howard Hughes Medical Institute investigator. This work was supported by NIH grants to Y.S. (GM058012, CA118487, and MH096066) and E.L.G. (K99AG043550) and by an Ellison Foundation Senior Scholar Award to Y.S. Y.S. is an American Cancer Society Research Professor. Y.S. is also a cofounder of Constellation Pharmaceuticals Inc. and a member of its scientific advisory board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
E.L.G., M.A.B., and Y.S. conceived and planned the study and wrote the paper. E.L.G. produced Figures 1A, 3, 5A, 5D, 5F, 6, 7A, S1C, S3A, S4B, S4C, S6A, S7A and S7B. M.A.B. produced Figures 1A, 4A, 4B, 4C, S1A, S1B, S1D, and S5A. L.G. performed bioinformatics analysis presented in Figures 2, S5B, and S5C. E.S. produced Figures 5B, 5C, 7B, S6C, and S7C. J.L. performed UHPLC-MS/MS experiments shown in Figure 1B, 4D, 5E, S1E, and S2 and was advised by C.H. D.A.C. produced Figures 1C, 1D, S3B, and S4A. C-H.H. performed protein purifications and DNA methylation assays. L.A. identified damt-1 bioinformatically and produced Figures 5A and S6B.
References
- Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J Mol Biol. 1962;5:18–36. doi: 10.1016/s0022-2836(62)80058-8. [DOI] [PubMed] [Google Scholar]
- Benyshek DC, Johnston CS, Martin JF. Glucose metabolism is altered in the adequately-nourished grand-offspring (F3 generation) of rats malnourished during gestation and perinatal life. Diabetologia. 2006;49:1117–1119. doi: 10.1007/s00125-006-0196-5. [DOI] [PubMed] [Google Scholar]
- Braun RE, Wright A. DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol Gen Genet. 1986;202:246–250. doi: 10.1007/BF00331644. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A–seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nature reviews Genetics. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Gao F, Liu X, Wu XP, Wang XL, Gong D, Lu H, Xia Y, Song Y, Wang J, Du J, et al. Differential DNA methylation in discrete developmental stages of the parasitic nematode Trichinella spiralis. Genome Biol. 2012;13:R100. doi: 10.1186/gb-2012-13-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos LJ, Wang W, Strome S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science (New York, NY. 2014;345:1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Beese-Sims SE, Brookes E, Spadafora R, Zhu Y, Rothbart SB, Aristizabal-Corrales D, Chen S, Badeaux AI, Jin Q, et al. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Reports. 2014;7:113–126. doi: 10.1016/j.celrep.2014.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature reviews Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- Gutierrez JC, Callejas S, Borniquel S, Martin-Gonzalez A. DNA methylation in ciliates: implications in differentiation processes. Int Microbiol. 2000;3:139–146. [PubMed] [Google Scholar]
- Han JS, Kang S, Kim SH, Ko MJ, Hwang DS. Binding of SeqA protein to hemi-methylated GATC sequences enhances their interaction and aggregation properties. The Journal of biological chemistry. 2004;279:30236–30243. doi: 10.1074/jbc.M402612200. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, Burroughs AM, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, de Souza RF, Pukkila PJ, Rao A, Aravind L. Lineage-specific expansions of TET/JBP genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1676–1683. doi: 10.1073/pnas.1321818111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Current biology : CB. 2003;13:421–426. doi: 10.1016/s0960-9822(03)00106-4. [DOI] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SC, Ruppersburg CC, Francis JW, Katz DJ. SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state during passage through the germ line. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9509–9514. doi: 10.1073/pnas.1321843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass M, Nguyen PN, Dechavigny A. Age-correlated changes in the DNA template in the nematode Caenorhabditis elegans. Mechanisms of ageing and development. 1983;22:253–263. doi: 10.1016/0047-6374(83)90080-5. [DOI] [PubMed] [Google Scholar]
- Koh KP, Rao A. DNA methylation and methylcytosine oxidation in cell fate decisions. Current opinion in cell biology. 2013;25:152–161. doi: 10.1016/j.ceb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature reviews Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Moazed D. Mechanisms for the inheritance of chromatin States. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature genetics. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Nottke AC, Beese-Sims SE, Pantalena LF, Reinke V, Shi Y, Colaiacovo MP. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12805–12810. doi: 10.1073/pnas.1102298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Houri-Ze'evi L, Anava S, Goh WS, Kerk SY, Hannon GJ, Hobert O. Starvation-Induced Transgenerational Inheritance of Small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Developmental biology. 2000;225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla ME, et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Molecular cell. 2014;56:580–594. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Simpson VJ, Johnson TE, Hammen RF. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711–6719. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. A role for RNAi in the selective correction of DNA methylation defects. Science (New York, NY. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Wenzel D, Palladino F, Jedrusik-Bode M. Epigenetics in C. elegans: facts and challenges. Genesis. 2011;49:647–661. doi: 10.1002/dvg.20762. [DOI] [PubMed] [Google Scholar]
- Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, He C. DNA repair by reversal of DNA damage. Cold Spring Harb Perspect Biol. 2013;5:a012575. doi: 10.1101/cshperspect.a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science (New York, NY. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zemach A, Zilberman D. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Current biology : CB. 2010;20:R780–R785. doi: 10.1016/j.cub.2010.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.