Summary
Cell metabolism is adaptive to extrinsic demands, however the intrinsic metabolic demands that drive the induced pluripotent stem cell (iPSC) program remain unclear. While glycolysis increases throughout the reprogramming process, we show that the estrogen related nuclear receptors (ERRα and γ) and their partnered co-factors PGC-1α and β, are transiently induced at an early stage resulting in a burst of oxidative phosphorylation (OXPHOS) activity. Up-regulation of ERRα or γ is required for both the OXPHOS burst in human and mouse cells, respectively, as well as iPSC generation itself. Failure to induce this metabolic switch collapses the reprogramming process. Furthermore, we identify a rare pool of Sca1−/CD34− sortable cells that is highly enriched in bona fide reprogramming progenitors. Transcriptional profiling confirmed that these progenitors are ERRγ and PGC-1β positive and have undergone extensive metabolic reprogramming. These studies characterize a previously unrecognized, ERR-dependent metabolic gate prior to establishment of induced pluripotency.
Keywords: iPSCs, ERR checkpoint
Introduction
An understanding of the molecular mechanisms that influence the generation, maintenance, and differentiation of human pluripotent stem cells is key to advancing their use in a therapeutic setting. Whereas the transcriptional and epigenetic dynamics have been extensively documented (Buganim et al., 2012; O’Malley et al., 2013; Polo et al., 2012; Theunissen and Jaenisch, 2014), temporal changes in metabolic states during the induction of pluripotency remain largely unknown. Distinct from somatic cells, pluripotent stem cells have unique metabolic pathways (Zhang et al., 2012), which influence their cellular behavior and epigenetic status (Lu and Thompson, 2012; Shyh-Chang et al., 2013a; Shyh-Chang et al., 2013b). Indeed, factors involved in metabolic functions such as mitochondrial proteins are among the first to be up-regulated in cells undergoing reprogramming (Hansson et al., 2012). Therefore, delineating the molecular mechanisms governing the dynamic regulation of cellular metabolism is crucial to understanding the connections between metabolic and epigenetic reprogramming.
Nuclear receptors (NRs) are pleiotropic regulators of organ physiology controlling broad aspects of glucose and fatty acid metabolism and overall energy homeostasis (Mangelsdorf et al., 1995; Yang et al., 2006). While orphan receptors such as the Estrogen Related Receptors (ERRs) are ligand-independent, they nonetheless are capable of directing dramatic changes in both glycolytic and oxidative metabolism in tissues with high energy demands (Giguere et al., 1988). ERRs switch between various oxidative states by associating preferentially with their co-activators PGC-1α/β (Dufour et al., 2011; Schreiber et al., 2003). The ERR family member ERRβ (also known as Esrrb) is glycolytic in the absence of PGC-1α and plays a key role in establishing pluripotency (Buganim et al., 2012; Feng et al., 2009; Festuccia et al., 2012; Martello et al., 2012). In contrast, ERRα and ERRγ, which are expressed in oxidative tissues such as skeletal muscle and heart (Narkar et al., 2011), have not previously been linked to iPSC generation. In this study, we identify transient up-regulation of ERRα and γ in the early stages of reprogramming that induce a unique energetic state. Furthermore, we show that the transient OXPHOS burst and increased glycolysis initiated by this metabolic switch are essential for epigenetic reprogramming. Mechanistically, ERRα and γ are enriched in bona fide reprogramming progenitors and induce widespread changes in metabolic gene networks. Our results suggest that an ERR-mediated metabolic transition is required for induced pluripotency.
Results
ERRα/γ are essential for somatic cell reprogramming
Temporal gene expression studies in mouse embryonic fibroblasts (MEFs) after reprogramming with Oct4, Sox2, Klf4 and cMyc (OSKM) or OSK revealed transient increases in the expression of ERRγ, PGC-1α, PGC-1β, and to a lesser extent, ERRα, 3 days after infection (Figure S1A–D and data not shown). Furthermore, depletion of ERRγ, PGC-1α or PGC-1β by shRNA knockdown coincident with OSKM induction significantly reduced reprogramming efficiency in MEFs (Figure 1A), whereas ERRγ depletion later in reprogramming had little effect (Figure S1E). To further explore the timing of gene induction in early reprogramming, OSKM expression was induced in MEFs isolated from ERRγlox/lox and ERRγlox/loxCreERT mice via doxycycline-inducible lentiviruses (Wei et al., 2009). While tamoxifen-treated ERRγlox/lox MEFs (ERRγ control cells) exhibited multiple foci of reprogramming cells 5 days after doxycycline-induced OSKM expression, ERRγlox/loxCreERT MEFs treated with tamoxifen at day 3 (ERRγ iKO cells) displayed fibroblast-like morphology (Figure 1B). Consistent with a failure of the ERRγ iKO cells to reprogram, few alkaline phosphatase (AP) or Nanog-positive colonies were observed after 3 weeks of OSKM infection, whereas control cells showed normal reprogramming efficiency (Figure 1 C–F). As depletion of ERRγ or ERRα in reprogramming cells leads to a reduction in cell proliferation (Figure S1F), we also compared the reprogramming efficiencies of immortalized MEFs generated from ERRγ knockout (ERRγ−/−) or wildtype (ERRγ+/+) mouse embryos. No Nanog-positive cells were detected in ERRγ−/− cells after OSKM infection (Figure S1G). Together, these findings suggest that the induction of ERRγ early in reprogramming is essential for iPSC generation from MEFs.
Figure 1. ERRα/γ and PGC1α/β are required for induced pluripotency in both mouse and human cells.
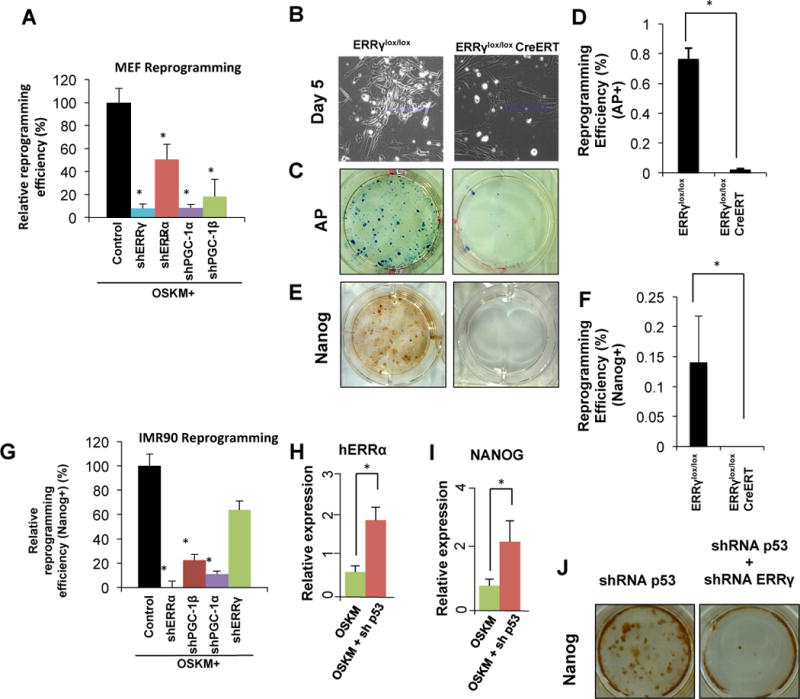
A. MEFs undergoing retroviral reprogramming with OSKM were transduced with control, ERRα, ERRγ, PGC-1α or PGC-1β shRNA. Depletion of ERRα/γ and PGC-1α/β significantly reduce reprogramming efficiency. (n=3, error bars show s.d.) B–F. ERRγlox/lox and ERRγlox/loxCreERT MEFs infected with a doxycycline-inducible OSKM lentivirus were treated with 4-OHT 3 days after OSKM induction. Bright field image showing that ERRγ depletion reduced the clusters of early reprogramming cells (B), significantly reduced AP colonies (C and D), and reduced Nanog-positive colonies (E and F) (n=3, *p<0.01, error bars show s.d.). G. ERRα and PGC-1α/β are required for reprogramming of human lung fibroblast IMR90 (n=3, *p<0.01, error bars show s.d). H, I. qPCR results show that depletion of p53 leads to increased expression of human ERRα during reprogramming of IMR90 cells. (n=3, *p<0.01, error bars show s.e.m) J. Nanog staining of retroviral OSKM-infected MEFs with p53 (left), or p53 and ERRγ (right) shRNA vectors, demonstrating that loss of ERRγ results in complete collapse of reprogramming even with p53 depletion.
Similar gene expression patterns were observed during the reprogramming of human lung fibroblast IMR90 cells and adipose-derived stem cells (ADSCs), with the distinction that ERRα, rather than ERRγ, was up-regulated (Figure S1 H–J). Parallel shRNA knockdown studies in the human IMR90 cells revealed a strong dependence on ERRα expression, alongside PGC-1α and β expression, whereas depletion of ERRγ was partially tolerated (~40% reduction in Nanog+ colonies, Figure 1G), further indicating that ERRα rather than ERRγ is required for iPSC generation in human fibroblasts. Furthermore, knockdown of p53, previously shown to increase iPSC generation (Kawamura et al., 2009), resulted in the hyper-induction of ERRα and Nanog during IMR90 cell reprogramming (Figure 1H and 1I). Notably, the coincident knockdown of ERRγ and p53 essentially blocked iPSC generation in MEFs (Figure 1J), indicating that the ERR signaling pathway is epistatic to p53-induced senescence in iPSC reprogramming.
To decipher the molecular mechanisms driving ERR/PGC-1 induction, IMR90 cells were infected with each of the 4 factors individually. Distinctive expression patterns for ERRα, PGC-1α and -1β were observed 5 days after infection. Klf4, c-Myc and Sox2 were each able to efficiently induce ERRα, Oct3/4 and Klf4 both induced the expression of PGC-1α, while c-Myc efficiently induced PGC-1β expression (Figure S1K–M). These patterns of gene induction correlate well with previous ChIP-Seq data (Chen et al., 2008) and indicate that all four reprogramming factors contribute in complementary ways to produce the operational ERRα transcriptional complex at day 5 (Figure S1N).
ERRs direct a transient hyper-energetic state required for reprogramming
The increased expression of ERRs and their co-activators led us to explore whether acutely altered energy flux in the mitochondria may be fueling reprogramming. MEFs from the reprogramming factor doxycycline-inducible mouse (Carey et al., 2010) reached an OXPHOS peak around days 2–4 after induction (Figure 2A). Importantly, the maximal OXPHOS capacity was also significantly increased in early reprogramming MEFs (Figure 2B and S2A). A similar bioenergetics time course recorded on days 3 to 10 after OSKM infection in human IMR90 cells revealed a transient increase in mitochondrial OXPHOS that peaked 5 days after infection (2.5–5.0 fold increase in oxygen consumption rates (OCR)) accompanied by a sustained increase in glycolysis (2.5–3.5 fold increase in the extra-cellular acidification rates (ECAR)) (Figure S2B–C). Corresponding with the increased expression of energy regulators, the levels of both nicotinamide adenine dinucleotide (NADH) and cellular ATP were increased in IMR90 cells 5 days after infection, while the NAD+/NADH ratio decreased (Figure S2D–F). Together, these results indicate that early reprogramming cells are in a hyper-energetic state. Closer examination of IMR90 cells revealed remarkably coincident temporal expression patterns of ERRα, PGC-1α and β during the early stages of reprogramming that are consistent with the known role of PCG1α/β as an ERR cofactor (days 3 to 8, Figure 2C). ERRs and PGC-1s directly regulate an extensive network of genes controlling energy homeostasis including proteins involved in fatty acid oxidation, the TCA cycle and OXPHOS (Alaynick et al., 2007; Dufour et al., 2007). Therefore, we examined the temporal expression pattern of various known regulators of cellular energy homeostasis during the reprogramming of IMR90 cells. Remarkably, multiple key players in energy metabolism, including ATP synthase in mitochondria (ATP5G1), succinate dehydrogenase (SDHB), isocitrate dehydrogenase (IDH3A) and NADH dehydrogenase (NDUFA2), reach peak expression at day 5 (Figure 2D and S2G). In addition, the induction of superoxide dismutase 2 (SOD2), NADPH oxidase 4 (NOX4) and catalase (CAT) by OSKM infection (Figure S2H), indicates that the antioxidant program is being triggered coordinately with the ERRα-PGC-1 surge.
Figure 2. ERRα/γ induces a metabolic transition in early reprogramming, which is essential to induced pluripotency.
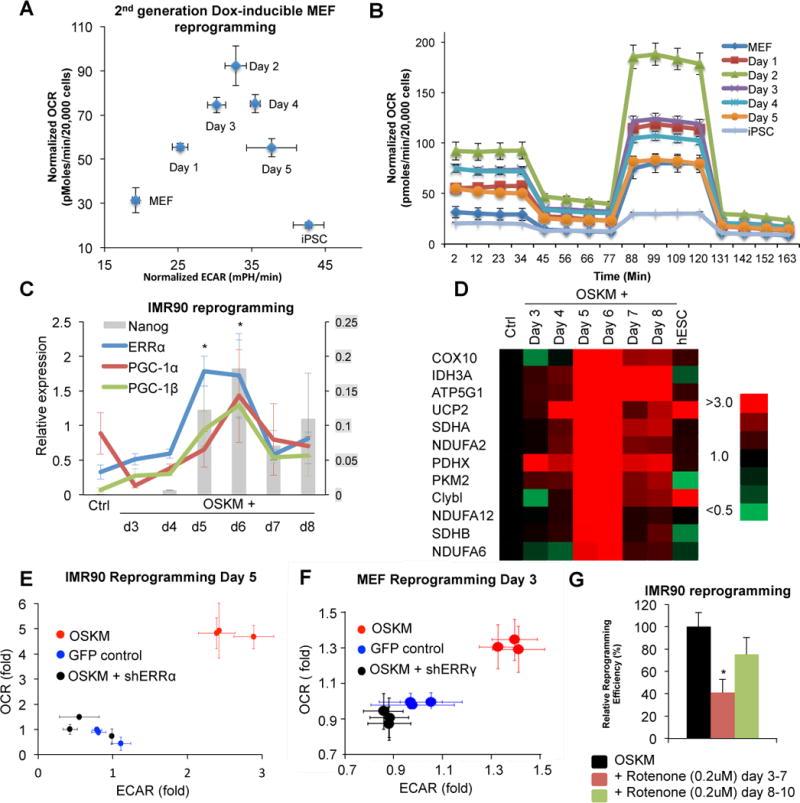
A. Time course of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in Dox-induced reprogramming MEFs, isolated from the single gene transgenic mouse, revealed that the reprogramming population experiences an early OXPHOS burst. B. Mitostress test of early reprogramming MEFs in (A) shows increased basal OCR and maximal OXPHOS capacity. C. Relative gene expression of ERRα, co-activators PGC-1α and PGC-1β, and Nanog after retroviral OSKM infection of IMR90 cells, measured by qPCR, suggests that the expression pattern of ERRs and their co-factors coincide with the metabolic switch in early reprogramming (n=3, *p<0.01, error bars show s.e.m.). D. Temporal expression of metabolic genes during retroviral OSKM-induced IMR90 reprogramming. E. OCR and ECAR measurements of control and ERRα knockdown retroviral reprogramming IMR90 cells demonstrate that ERRα is required for the early OXPHOS burst in human cells. F. OCR and ECAR measurements of control and ERRγ knockdown retroviral reprogramming MEF cells demonstrate that ERRγ is required for the early OXPHOS burst in mouse cells. G. Rotenone treatment, which inhibits the OXPHOS burst, results in significant reduction of retroviral reprogramming efficiency in IMR90, suggesting that the metabolic switch is essential. (n=3, *p<0.05, error bars show s.d.)
Pluripotent stem cells are known to mainly rely on glycolysis to produce energy (Folmes et al., 2012; Shyh-Chang et al., 2013a; Zhang et al., 2012). Previous studies have focused on the changes in glycolytic activity during reprogramming, as elevated glycolysis is linked to a faster cell cycle and iPSC generation (Folmes et al., 2011; Panopoulos et al., 2012; Shyh-Chang et al., 2013b). However, our findings suggest that iPSC precursors undergo a transient increase in oxidative phosphorylation activity. The dynamics of ECAR support previous work showing that the glycolytic activity of the cells is gradually enhanced and maintained during reprogramming to a level similar to iPSCs (Figure 2A and S2C). In contrast, the transient burst of OXPHOS during reprogramming of both human and mouse cells has not been previously documented (Figure 2A, B and S2B). This led us to investigate the potential influence of the ERRα/γ surge on cell plasticity during reprogramming.
To examine a potential causal relationship between ERR expression and the induction of the hyper-energetic state, we compared the metabolic activities of partially reprogrammed cells before and after targeted shRNA knockdowns. Notably, the increase in OXPHOS and glycolysis was completely abrogated in cells depleted of ERRs (ERRα in IMR90 cells at day 5, and ERRγ in MEFs at day 3; Figure 2E and 2F). Furthermore, the mitochondrial inhibitor Rotenone significantly reduced iPSC generation, though only when treatment was coincident with the observed hyper-energetic state, consistent with the OXPHOS burst being necessary for reprogramming (Figure 2G). Additionally, over-expression of ERRγ in MEFs 1 day after induction of OSKM increases reprogramming efficiency ~1.8 fold (Figure S2I). Together these data indicate that ERRα and γ regulate iPSC generation through the induction of a transient enhanced metabolic state that is required for somatic cell reprogramming.
Bona fide iPSC progenitors are enriched for ERRγ expression
Under standard conditions, only a small percentage of cells are successfully reprogrammed into iPSCs (Takahashi and Yamanaka, 2006; Yu et al., 2007). Given our observation of a metabolic switch in the heterogeneous cell populations present in the early stages of reprogramming, we hypothesized that the sub-population of bona fide iPSC progenitors might be enriched for the ERR-mediated hyper-energetic burst. Analysis of cell surface markers differentially expressed during MEF reprogramming revealed that early clusters of reprogramming cells lack the expression of stem cell antigen 1 (Sca1) and cluster of differentiation gene 34 (CD34) expression (Figure 3A and 3B). Upon OSKM induction, CD34 expression is promptly up-regulated, resulting in three distinct cell sub-populations in early reprogramming cells; Sca1−CD34− double negative (DN), Sca1+CD34+ double positive (DP), and Sca1+CD34− single positive (SP) (Figure S3A). Correlating with immunofluorescence staining (Figure 3A), only a minor fraction (~3–5%) of early reprogramming cells are Sca1−CD34− (Figure S3A). Strikingly, ERRγ and PGC-1β expression were ~10- and ~7-fold higher, respectively, in the early reprogramming DN cells compared to DP or SP cells, as determined by qPCR analysis (Figure 3C and 3D). Importantly, these early reprogramming DN cells exhibited significantly elevated ECAR and OCR compared to DP or SP populations (Figure 3E and 3F), consistent with Sca1−CD34− labeling a subpopulation of hyper-energetic cells. Notably, Sca1−CD34− cells present in non-infected MEFs do not show elevated reprogramming efficiency (Figure S3B). To test our hypothesis that this hyper-energetic state is required for reprogramming, we compared the number of iPS colonies generated from isolated DN, SP and DP cells. While DN cells comprise only ~5% of the infected cells, they were approximately 50-fold more efficient at generating iPSCs than the DP or SP populations, based on Nanog staining (Figure 3G; 35.5% (DN) vs 0.6% (DP) or 0.8% (SP)). The iPSCs derived from the DN population showed ESC-like morphology and expressed high levels of alkaline phosphatase activity as well as pluripotency markers (Figure S3C–E). In addition, embryoid body differentiation of the DN-derived iPSCs produced markers from each of the three germ layers (Figure S3F). Moreover, iPSCs generated from DN cells contributed to the formation of chimeric mice with subsequent crosses demonstrating germline-competency (Figure S3G and S3H). Collectively, these data indicate that the hyper-energetic cells identified in early reprogramming represented by the DN population, are bona fide reprogramming precursors that generate iPSCs at high efficiency.
Figure 3. ERRγ enriched sub-population in early reprogramming represent bona fide reprogramming cells with significantly enhanced reprogramming efficiency.
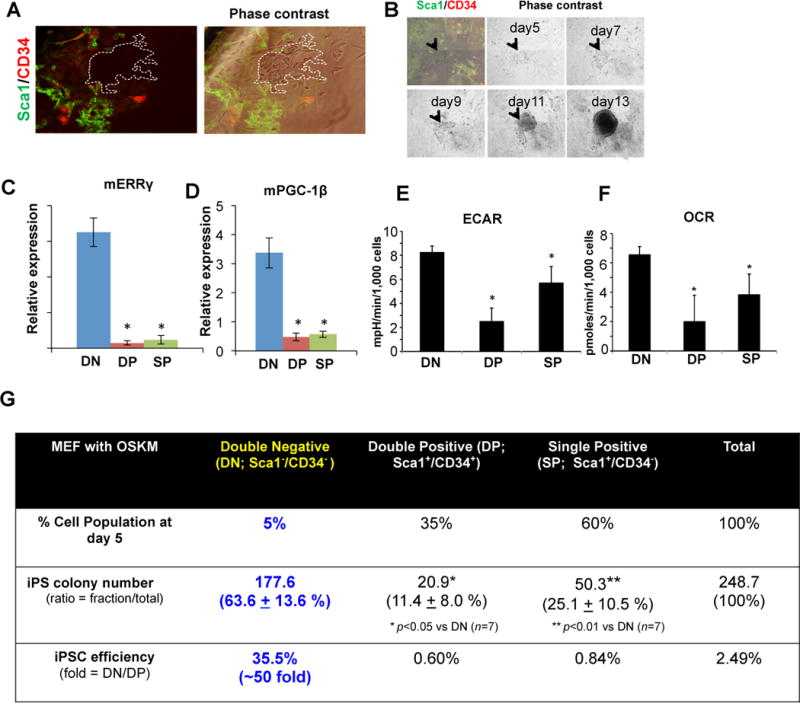
A. Sca1 and CD34 label bona fide reprogramming cells. Retroviral OSKM-infected MEFs stained for Sca1 (green) and CD34 (red) expression, and phase contrast image (right). Sca1−CD34− double negative (DN) cells are demarcated by white dashed lines from phase contrast images. B. Representative phase contrast images of Sca1−CD34− cells during retroviral reprogramming. Arrowheads indicate a representative DN colony. C, D. qPCR demonstrating that ERRγ and PGC-1β are enriched in the DN population (n=3, error bars show s.e.m. *p<0.01). E, F. FACS-isolated DN population exhibits higher ECAR (E) and OCR (F) than DP or SP population (n=4, *p<0.05, error bars show s.d.). G. DN cells demonstrate significantly higher reprogramming efficiency (n=7, *p<0.05, **p<0.01).
Reprogramming cells undergo an ERR-mediated OXPHOS burst
To better understand the molecular underpinnings of cell reprogramming and cell fate determination, we compared the complete transcriptomes, determined by RNA-Seq, of somatic fibroblasts (non-infected MEFs, mock infected MEFs at day 5), intermediate reprogramming cell populations (DN, DP, SP, unsorted day 5 cells) and pluripotent stem cells (iPSCs generated from the DN population and mESCs). Not unexpectedly, distance matrix and clustering analyses grouped the cell types into the above 3 categories (Figure 4A and 4B). The clear separation of the DN population from the pluripotent stem cells indicates that these transitional cells have yet to adopt a durable pluripotency fate. Furthermore, the more subtle separation of the DN population from the other intermediate reprogramming cells in the cluster analysis suggests that they should express a unique gene signature associated with enhanced reprogramming efficiency (Figure 4B). Indeed, the expression of selected pluripotency markers and key cell cycle genes in the DN population more closely resembles that observed in ESCs and iPSCs than found in the DP and SP populations (Figure 4C and 4D). However, a majority of other stem cell markers including ERRβ and Nanog are not enriched in the DN population (data not shown). Thus, the DN cell population is in a definable transcriptional and metabolic state that appears to facilitate efficient progression toward pluripotency.
Figure 4. Transcriptome analysis reveals that ERRs orchestrate the up-regulation of a panel of OXPHOS related genes and promote the metabolic switch during early reprogramming.
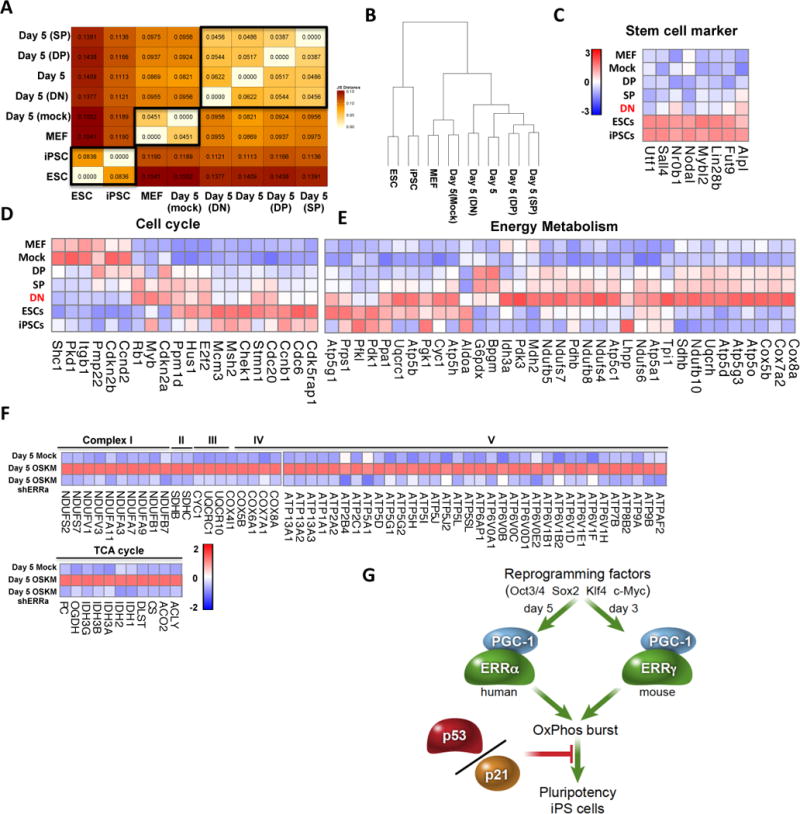
A–B. RNA-Seq analysis reveals that the genome-wide expression pattern of various cell types can be grouped into pluripotent stem cells, MEFs and intermediate retroviral reprogramming cells, demonstrated by distance matrix (A) and clustering analysis (B). C–D. The RNA-Seq patterns of a subset of key pluripotency markers (C) and cell cycle genes (D) reveal similarity between DN cells and ESCs, suggesting that the DN population represent bona fide early reprogramming cells which are in the process of adopting induced pluripotency. E. Expression heat map from RNA-Seq data shows that DN cells have a unique pattern in metabolic genes that represents a hyper-energetic state. F. Heatmap of gene expression from microarray in IMR90 cells after ERRα depletion, showing that a significant portion of the OXPHOS program is directly influenced by ERRα in human fibroblast reprogramming. G. Schematic representation of the role of ERRs and PGC1α/β in inducing the early OXPHOS burst and transition to induced pluripotency. The OXPHOS burst is required for somatic cell reprogramming and transient activation of ERRs and their co-factors are epistatic to the roadblock of p53/p21-induced senescence in reprogramming.
Pivotal pathways controlling the enhanced reprogramming efficiency of DN cells were identified by comparing transcriptomes between DN, DP or SP populations. Interestingly, KEGG pathway analysis of the differentially regulated genes identified oxidative phosphorylation (OXPHOS) as the most significantly altered pathway in DN cells (Figure S4A and S4B). Furthermore, a comparison of the expression levels of genes involved in cellular energy metabolism revealed that the majority are up-regulated in the DN population (Figure 4E), consistent with the DN population comprising the most hyper-energetic cells. This supports the idea that a key feature of bona fide reprogramming is directing progenitors to enter a hyper-energetic state.
Finally, to determine if a causal association exists between the ERR surge and the increased expression of energy metabolism genes, the transcriptional consequences of ERRα knockdown in reprogramming IMR90s were examined. The expression of a large number (1061) of metabolic genes was significantly affected by ERRα depletion (Figure S4C). In particular, dramatic decreases in the expression of regulators of cellular energy homeostasis including NADH dehydrogenases (NDUF), succinate dehydrogenases (SDH), mitochondrial respiratory chains (COX), ATPase, and ATP synthases in mitochondria were seen (Figure 4F). The fact that ERRα depletion influences the expression of a plethora of mitochondrial genes, including a variety of genes in Complex I–V, and the TCA cycle (Figure 4F), further support the conclusion that transient ERRα/γ expression induces an equally transient OXPHOS burst, facilitating reprogramming and enabling the transition from the somatic to pluripotent state (Figure 4G).
Discussion
Recent single-cell expression analyses revealed a requirement for early expression of ERRβ (Buganim et al., 2012), previously demonstrated by Feng et al. to be a ‘Myc substitute’ (Feng et al., 2009). In this model, Sox2 and ERRβ mutually enhance each other’s expression and initiate the reprogramming process, presumably in all transfected cells (Buganim et al., 2012). Here we reveal a downstream requirement for other ERR family members, ERRα and ERRγ, together with their co-activators PGC-1α/β, that define a distinct sub-population of cells with dramatically enhanced efficiency for iPSC generation. A transient surge in ERRα/γ and PGC1α/β, expression during reprogramming induces an early metabolic switch epitomized by a transient OXPHOS burst and sustained enhanced glycolysis. These findings complement a recent study demonstrating stage-specific roles for HIF1α and HIF2α in the early increase in glycolytic metabolism (Mathieu et al., 2014). The surprising functional divergence between ERRα/γ and ERRβ adds a new dimension to the model for reprogramming, in which transient ERRα/γ expression is required to drive an early hyper-energetic metabolic state characterized by increased OXPHOS and glycolysis, whereas ERRβ is required for establishing induced pluripotency at later reprogramming stages (Chen et al., 2008; Martello et al., 2012; Zhang et al., 2008). The fact that metabolic reprogramming is a prerequisite of induced pluripotency reveals the functional relevance of a unique metabolic state to achieving cell plasticity. Furthermore, via cell sorting of Sca1/CD34 double negative cells we demonstrate that ERRγ and PGC-1β are early markers of a newly defined sub-group of reprogramming progenitors. In summary, these studies characterize a previously unrecognized, ERR/PGC-1 dependent metabolic switch prior to establishment of induced pluripotency in both human and mouse cells (Figure 4G).
Methods Summary
MEFs were isolated from embryonic day (E) 13.5 embryos obtained from wild-type and ERRγ-deficient mice (Alaynick et al., 2007). Retroviruses and lentiviruses were produced in HEK293T cells, and 12 to 14 days after infection MEFs were fixed for staining. Reprogramming of MEFs and IMR90 cells were done as previously described (Kawamura et al., 2009; Sugii et al., 2010; Takahashi et al., 2007; Wei et al., 2013; Yu et al., 2007). Illumina BeadChip microarrays on OSKM-induced IMR90 cells infected with shERRα or GFP were performed as previously described (Narkar et al., 2011). RNA-Seq libraries were prepared with TruSeq RNA Sample Preparation Kit v2 (Illumina) and sequenced on an Illumina HiSeq 2000 using multiplexed bar-coding and a 100bp read length. Description of read analysis methods are provided in the Supplemental Information. Bioenergetic assays were performed on a Seahorse XF instrument to measure OXPHOS capacity. OCR and ECAR values were normalized by measuring the cell number in each well using Hoechst 33342 staining followed by quantification of fluorescence at 355 excitation and 460 emission. Intracellular NAD+ and NADH levels were measured by NAD+/NADH Assay Kit (Abcam, San Francisco, CA) as per manufacturer’s instructions. Intracellular ATP was measured by ATP assay kit (Sigma-Aldrich) according to manufacturer’s directions. shRNA constructs were purchased from Openbiosystems. Lentiviral shRNA were produced in 293T cells and polybrene (6μg/ml) was used in transduction. For reprogramming experiments, cells were transduced with lentiviral shRNA at day 0 of reprogramming. Detailed methods can be found in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank Drs. S Fang, J Jonker and W Fan for useful discussions and reagents; L Ong, and C Brondos for administrative assistance. RME is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. YSK was supported by the Japan Society for the Promotion of Science. ZW is supported by a CIRM training grant. This work was supported by grants from CIRM, National Institutes of Health Grants HD105278, DK057978, DK062434, and DK063491, the Leona M. and Harry B. Helmsley Charitable Trust, and the Howard Hughes Medical Institute (R.M.E.), and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, Special Coordination Fund for Promoting Science and Technology of MEXT (TK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Y.S.K., T.K., Z.W., M.D. and R.M.E. designed and supervised the research. Y.S.K., T.K., Z.W., T.S., S.J., A.S-K, and E.Y. performed research. Y.S.K., T.K., Z.W, R.T.Y., C.L., J.R.E., A.R.A., M.D. and R.M.E analyzed data. Y.S.K., T.K., Z.W., A.R.A., R.T.Y., M.D. and R.M.E. wrote the manuscript.
Author Information: Microarray data sets have been deposited in the NCBI Gene Expression Omnibus, accession number GSE31704. Sequence data for RNA-Seq experiments are available in the NCBI SRA database under the accession number SRA023829.2 (ESC and iPSC) and SRP048537 (d5 SP, DP, DN, control and mock populations).
The authors declare no competing financial interests.
References
- In brief. Nature reviews. 12:456. [Google Scholar]
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nature methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Levasseur MP, Pham NH, Eichner LJ, Wilson BJ, Charest-Marcotte A, Duguay D, Poirier-Heon JF, Cermakian N, Giguere V. Genomic convergence among ERRalpha, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS genetics. 2011;7:e1002143. doi: 10.1371/journal.pgen.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell metabolism. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nature cell biology. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell stem cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell stem cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell metabolism. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell reports. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell metabolism. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Gottgens B, Niwa H, Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell stem cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Zhou W, Xing Y, Sperber H, Ferreccio A, Agoston Z, Kuppusamy KT, Moon RT, Ruohola-Baker H. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell stem cell. 2014;14:592–605. doi: 10.1016/j.stem.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Fan W, Downes M, Yu RT, Jonker JW, Alaynick WA, Banayo E, Karunasiri MS, Lorca S, Evans RM. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley J, Skylaki S, Iwabuchi KA, Chantzoura E, Ruetz T, Johnsson A, Tomlinson SR, Linnarsson S, Kaji K. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature. 2013 doi: 10.1038/nature12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerias A, Batchelder EM, Plongthongkum N, Lutz M, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell research. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development (Cambridge, England) 2013a;140:2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science (New York, NY. 2013b;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugii S, Kida Y, Kawamura T, Suzuki J, Vassena R, Yin YQ, Lutz MK, Berggren WT, Izpisua Belmonte JC, Evans RM. Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3558–3563. doi: 10.1073/pnas.0910172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Jaenisch R. Molecular control of induced pluripotency. Cell stem cell. 2014;14:720–734. doi: 10.1016/j.stem.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 Organizes Long-Range Chromosomal Interactions with the Oct4 Locus inReprogramming andPluripotency. Cell stem cell. 2013 doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Wei Z, Yang Y, Zhang P, Andrianakos R, Hasegawa K, Lyu J, Chen X, Bai G, Liu C, Pera M, et al. Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem cells (Dayton, Ohio) 2009;27:2969–2978. doi: 10.1002/stem.231. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, NY) 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic Regulation in Pluripotent Stem Cells during Reprogramming and Self-Renewal. Cell stem cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Wang T, Esteban MA, Pei D. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. The Journal of biological chemistry. 2008;283:35825–35833. doi: 10.1074/jbc.M803481200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


