Summary
The vertebrate antiviral innate immune system is often considered to consist of two distinct groups of proteins: pattern recognition receptors (PRRs) that detect viral infection and induce the interferon (IFN) signaling, and effectors that directly act against viral replication. Accordingly, previous studies on PRRs, such as RIG-I and MDA5, have primarily focused on their functions in viral double-stranded RNA (dsRNA) detection and consequent antiviral signaling. We here report that both RIG-I and MDA5 efficiently displace viral proteins pre-bound to dsRNA in a manner dependent on their ATP hydrolysis, and that this activity assists a dsRNA-dependent antiviral effector protein, PKR, and allows RIG-I to promote MDA5 signaling. Furthermore, truncated RIG-I/MDA5 lacking the signaling domain, and hence the IFN stimulatory activity, displace viral proteins and suppress replication of certain viruses in an ATP-dependent manner. Thus, this study reveals novel “effector-like” functions of RIG-I and MDA5 that challenge the conventional view of PRRs.
Introduction
The DExD/H motif helicases are nucleic acid-dependent ATP hydrolases and are involved in a broad spectrum of cellular functions, from transcription, splicing and translation, to antiviral immunity (Linder and Jankowsky, 2011). One family of such helicases includes the viral RNA receptors, RIG-I and MDA5, which are ubiquitously expressed cytoplasmic proteins and are responsible for eliciting the type I IFN antiviral response against a broad range of viruses (Goubau et al., 2013; Kato et al., 2011). RIG-I and MDA5 share the same domain architecture, consisting of the N-terminal tandem caspase activation recruitment domain (2CARD), the central DExD/H motif helicase domain, and the C-terminal domain (CTD). 2CARD is responsible for activation of the type I IFN signaling pathway by interacting with the adaptor molecule, MAVS, whereas the helicase domain and CTD together function as an RNA recognition unit. Despite these similarities, RIG-I and MDA5 play non-redundant roles by recognizing largely distinct groups of viral RNAs (Goubau et al., 2013; Kato et al., 2011).
As with other RNA helicases, RIG-I and MDA5 hydrolyze ATP upon binding to their cognate RNA molecules. The consequence of ATP hydrolysis, however, differs between RIG-I and MDA5. RIG-I primarily binds to a dsRNA end containing a 5’-triphosphate (5’ppp) in an ATP-independent manner (Cui et al., 2008), but during ATP hydrolysis, it translocates to the dsRNA interior (Myong et al., 2009) and stacks along the translocation track to form short filamentous oligomers (Patel et al., 2013; Peisley et al., 2013). Filament formation brings together neighboring 2CARDs into proximity and promotes its oligomerization (Peisley et al., 2013), a prerequisite for activation of MAVS (Jiang et al., 2012; Wu et al., 2014). MDA5 also assembles into filaments (Peisley et al., 2011), but unlike RIG-I, the MDA5 filament nucleates in the dsRNA interior and propagates to the exterior in a highly cooperatively manner in the absence of ATP (Peisley et al., 2012). ATP hydrolysis instead triggers end-disassembly of the MDA5 filament, which results in selective accumulation of MDA5 molecules on long dsRNA and robust discrimination between long viral dsRNA and short cellular dsRNAs (Peisley et al., 2012; Peisley et al., 2011). The ATP-driven disassembly also makes the MDA5 filament a highly dynamic entity that undergoes repetitive cycles of assembly and disassembly during ATP hydrolysis (Peisley et al., 2012; Peisley et al., 2011).
Several helicases utilize their conformational dynamics during ATP binding or hydrolysis to remodel protein:nucleic acid complexes (Jankowsky and Bowers, 2006). In many cases, such remodeling activities are key to their functions in transcription, splicing and translation. We asked whether RIG-I and MDA5 also utilize ATP-driven translocation and filament formation (for RIG-I) or repetitive filament dynamics (for MDA5) to remodel viral protein:dsRNA complexes, thereby exerting IFN signaling-independent antiviral functions. We here report data supporting this hypothesis.
Results
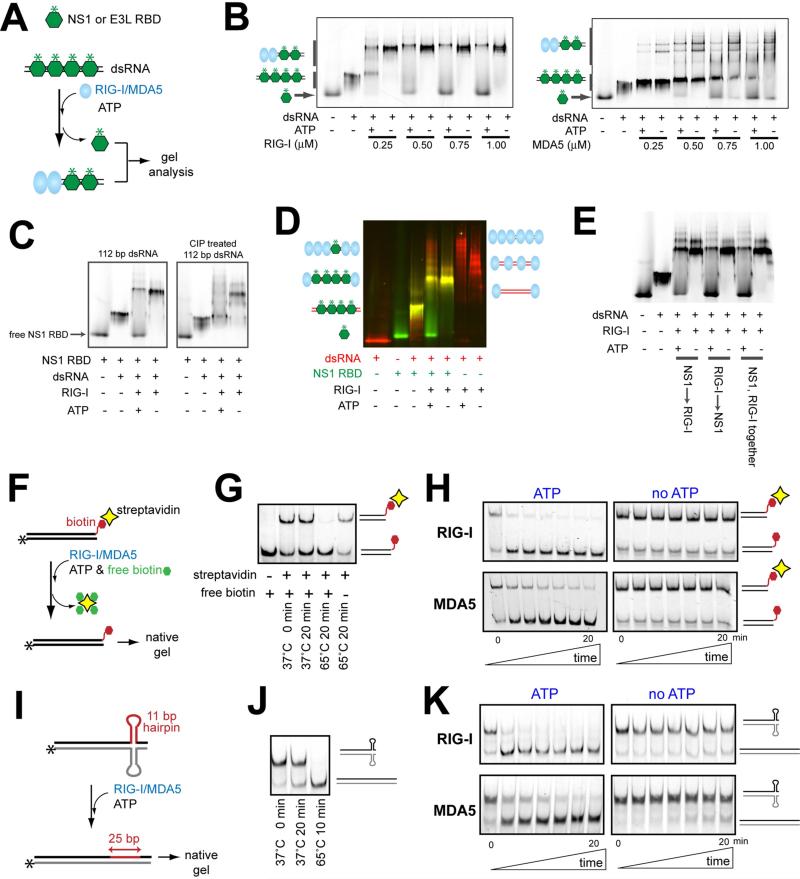
RIG-I and MDA5 can displace viral proteins bound to dsRNA in an ATP-dependent manner
To examine whether RIG-I and MDA5 can remodel viral protein:RNA complexes, we used NS1 of influenza A and E3L of vaccinia virus as model proteins with affinity for dsRNA (Figure S1A) and asked whether RIG-I and MDA5 can displace these viral proteins from dsRNA. For simplicity, we purified the RNA binding domains (RBDs) of NS1 and E3L and fluorescently labeled them to monitor their dsRNA binding and unbinding by native gels. We first saturated an in vitro transcribed 112 bp or 512 bp dsRNA with NS1 or E3L RBDs, added RIG-I/MDA5, and monitored appearance of free RBDs (Figure 1A). In the absence of ATP, addition of RIG-I or MDA5 alone induced dissociation of neither NS1 nor E3L from dsRNA, except at high concentrations of MDA5 (Figure 1B for NS1, see Figure S1B for E3L). Addition of ATP, however, significantly promoted dissociation of viral proteins from dsRNA for both RIG-I and MDA5 (Figures 1B & S1B). Note that displacement by MDA5 was in general less efficient and required longer dsRNA (512 bp) for E3L displacement (Figure S1B), consistent with its reported preference for long dsRNAs (Peisley et al., 2011). The N-terminal 2CARD truncation mutants (ΔNs) of RIG-I and MDA5, which can bind dsRNA and hydrolyze ATP, also displaced viral proteins in an ATP-dependent manner (Figure S1C). However, the ATP hydrolysis-deficient catalytic mutants, RIG-I (K270A) and MDA5 (K335A), did not show a similar displacement activity (Figure S1D). Additionally, use of a non-hydrolyzable ATP analog, 5’-adenylyl-β, γ-imidodiphosphate (ADPNP) did not promote viral protein displacement by wild-type RIG-I and MDA5 (Figure S1E). These results suggest that ATP hydrolysis, not binding, is important for RIG-I and MDA5 to clear dsRNA of viral proteins.
Figure 1.
RIG-I and MDA5 can displace viral proteins bound to dsRNA in an ATP-dependent manner.
A. Schematic of the viral protein displacement assay. The in vitro transcribed 112 bp dsRNA (45 nM) was pre-incubated with a saturating concentration (1 μM) of fluorescently labeled (*) RBDs of NS1 and E3L, and further incubated with RIG-I or MDA5 in the presence or absence of ATP. Unless mentioned otherwise, all dsRNAs in this manuscript contain 5’ppp at both ends.
B. Gel analysis of the protein displacement assay using NS1 RBD as described in (A). Gel images were obtained using fluorescein conjugated to NS1 RBD. For results with E3L RBD, see Figure S1B.
C. The NS1 displacement assay using RIG-I and 112 bp dsRNA before and after CIP treatment.
D. The NS1 displacement assay using RIG-I and 3’-Cy3-labeled 112 bp dsRNA (before CIP). The gel image was obtained by overlaying the images obtained using the Cy3 fluorescence (red) and the fluorescein fluorescence (green).
E. The NS1 displacement assay using RIG-I and 112 bp dsRNA. NS1 RBD and RIG-I were incubated with dsRNA in the indicated order.
F. Schematic of the streptavidin displacement assay. The 3’-biotinylated 112 bp dsRNA was complexed with streptavidin and then with RIG-I/MDA5. The displacement reaction was initiated with ATP and an excess amount of free biotin (streptavidin trap) at 37°C, and quenched with EDTA prior to gel analysis.
G. Stability of the streptavidin:biotinylated dsRNA interaction at 37°C.
H. Gel analysis of the streptavidin displacement as described in (F), using the fluorescence of RNA.
I. Schematic of RNA hairpin displacement assay. The 112 bp dsRNA containing the complementary hairpins (Figure S1F) was complexed with RIG-I/MDA5 in the presence or absence of ATP at 37°C prior to gel analysis.
J. Stability of the hairpin containing duplex at 37°C.
K. Gel analysis of the hairpin displacement as described in (I), using the fluorescence of RNA.
For RIG-I, ATP hydrolysis causes its translocation along dsRNA (Myong et al., 2009) and subsequent filamentous oligomerization (Peisley et al., 2013). Both phenomena are known to be dependent on 5’ppp of dsRNA although slow translocation and less efficient filament formation was also observed even in the absence of 5’ppp (Myong et al., 2009; Peisley et al., 2013). We found that the protein displacement activity of RIG-I was also significantly reduced in the absence of 5’ppp (Figure 1C). Simultaneous visualization of Cy5-labeled dsRNA and fluorescein-labeled NS1 RBD showed that NS1 displacement was accompanied by increased accumulation of RIG-I molecules on dsRNA (Figure 1D). Displacement was more efficient when RIG-I was placed prior to or together with NS1 (Figure 1E), suggesting an additional role of RIG-I filament assembly in blocking NS1 binding, in addition to the active displacement of pre-bound NS1. Relative importance of translocation, filament formation and local conformational change of RIG-I during ATP hydrolysis could not be determined as these characteristics are tightly coupled.
To further examine whether the specific identity of the dsRNA-binding protein is important for the displacement by RIG-I/MDA5, we asked whether RIG-I/MDA5 can displace artificial roadblocks, such as streptavidin bound to 3’-biotinylated dsRNA (Figure 1F) and complementary hairpins (Figures 1I & S1F) embedded within dsRNA. In the absence of RIG-I/MDA5, spontaneous dissociation of streptavidin from biotinylated dsRNA and relaxation of the complementary hairpins were negligible at 37°C over 20 min (Figures 1G & 1J). Addition of RIG-I or MDA5 alone had little impact on these roadblocks, but further addition of ATP significantly accelerated their displacement (Figures 1H & 1K). None of the nonhydrolyzable ATP analogs promoted the hairpin removal, further supporting the role of ATP hydrolysis in roadblock displacement (Figure S1G). Interestingly, unlike NS1 RBD, displacement of the hairpins by RIG-I was independent of 5’ppp (Figure S1H). Since these hairpins are quasi-stable (kinetically trapped) species, this result suggests that slow translocation or less efficient filament formation observed with 5’-dephosphorylated dsRNA may be sufficient to remove weak roadblocks. Altogether, our data show that both RIG-I and MDA5 can remove a wide range of roadblocks placed on dsRNA in a manner dependent on ATP hydrolysis but the requirement for RNA (length for MDA5 and 5’ppp for RIG-I) is dependent on the specific nature of the roadblock.
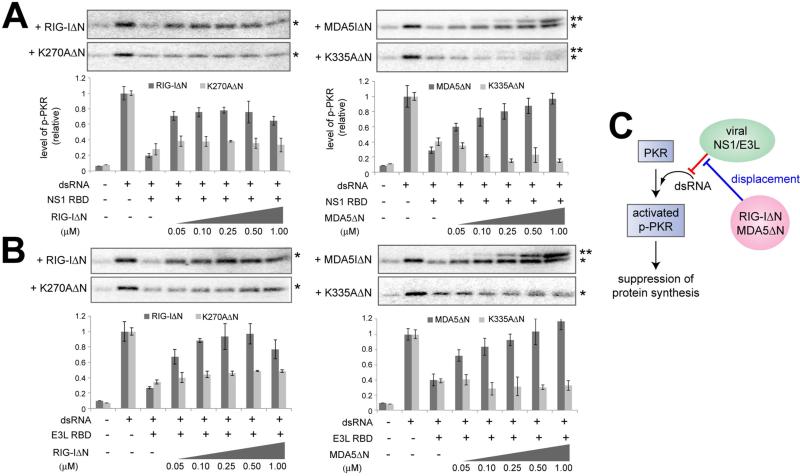
2CARD deletion mutants, RIG-IΔN and MDA5ΔN, relieve the viral suppression of PKR
We next asked whether the ability of RIG-I/MDA5 to displace viral proteins can increase the overall accessibility of dsRNA, thereby promoting functions of other dsRNA-dependent antiviral proteins in the host, such as PKR. PKR is a protein kinase whose activation requires dsRNA binding and subsequent autophosphorylation (McKenna et al., 2007b) (Figure S2A). Upon activation, PKR phosphorylates the translation initiation factor, eIF2α, resulting in the global suppression of cap-dependent protein-synthesis and inhibition of viral replication (Garcia et al., 2007). Viral proteins, such as NS1 and E3L, are known to suppress PKR by sequestering dsRNA (Garcia et al., 2007), although dsRNA-independent mechanisms have been also proposed (Li et al., 2006). Consistent with the previous reports, the dsRNA-induced PKR activity, as measured by the level of autophosphorylation, was suppressed upon addition of NS1 and E3L RBDs (Figure S2B). The suppression was relieved when supplemented with an excess amount of dsRNA (Figure S2C), suggesting that dsRNA sequestration is the primary mechanism by which NS1 and E3L RBDs inhibit PKR. It is worth noting that at high concentration of NS1 RBD (~1 μM), the PKR inhibition persisted regardless of dsRNA supplementation (Figure S2D), suggesting that a direct, RNA-independent inhibition of PKR also occurs at high concentrations of NS1 RBD (Figure S2E).
To examine whether the protein displacement activity of RIG-I/MDA5 could assist the activity of PKR by removing NS1/E3L from dsRNA, we first examined in vitro the effect of RIG-IΔN and MDA5ΔN on the autophosphorylation activity of PKR. In the absence of NS1/E3L RBDs, RIG-IΔN and MDA5ΔN neither suppressed nor activated PKR (Figure S2F). The lack of suppression of PKR by RIG-I and MDA5 may reflect the dynamic nature of the interactions between RIG-I/MDA5 and dsRNA (due to the dynamic instability of the MDA5 filament and the translocation activity of RIG-I), and the sufficiency of transient contact with dsRNA for activation of PKR (McKenna et al., 2007a). In the presence of NS1 RBD (0.2 μM) or E3L RBD (1 μM), RIG-IΔN or MDA5ΔN increased the PKR activity to the level comparable to that without NS1/E3L (Figures 2A-B). Catalytic mutants of RIG-IΔN and MDA5ΔN did not restore the PKR activity (Figures 2A-B). Consistent with the notion that RIG-IΔN and MDA5ΔN increase the PKR activity by removing NS1/E3L from dsRNA, the RNA-independent inhibition by NS1 RBD at 1 μM could be not relieved by RIG-IΔN and MDA5ΔN (Figure S2G).
Figure 2.
2CARD deletion mutants, RIG-IΔN and MDA5ΔN, relieve the viral suppression of PKR. A. Impact of RIG-IΔN or MDA5ΔN on the PKR autophosphorylation activity in the presence of NS1 RBD. PKR (0.2 μM) was stimulated with 112 bp dsRNA (5 nM) in the presence or absence of NS1 RBD (0.2 μM) and indicated amount of RIG-IΔN or MDA5ΔN (or their catalytic mutants). * and ** indicate phosphorylated PKR and MDA5ΔN, respectively. Data are shown in mean ± SD (n=3).
B. Same as (A) with E3L RBD (1 μM) in place of NS1 RBD. Data are shown in mean ± SD (n=3).
C. Model in which RIG-IΔN and MDA5ΔN indirectly promote PKR by displacing NS1/E3L from dsRNA.
To confirm that RIG-I and MDA5 can promote the function of PKR in cells, we measured the effect of RIG-IΔN and MDA5ΔN on the autophosphorylation level of PKR in human embryonic kidney 293T cells. We used RIG-IΔN and MDA5ΔN instead of full-length RIG-I and MDA5 to avoid the confounding effect of the IFN signaling and subsequent induction of PKR by the full-length receptors. For NS1 and E3L, we used full-length proteins instead of RBDs because neither expression nor PKR inhibition could be detected with isolated RBDs of NS1 and E3L (Figures S2H & S2I). Similarly to our biochemical assay, PKR was inhibited by NS1 and E3L, but this inhibition was relieved by wild-type RIGIΔN/MDA5ΔN (Figures S2J). The limited relief of the NS1-mediated suppression, particularly by MDA5ΔN, may reflect the presence of RNA-independent suppression of PKR by NS1 at a high concentration (Figure S2E). For both NS1 and E3L, no relief of viral inhibition was observed with the catalytic mutants of RIG-IΔN/MDA5ΔN (Figures S2K), further supporting the notion that RIG-IΔN and MDA5ΔN displace NS1/E3L from dsRNA, thereby promoting the PKR activity (Figure 2C).
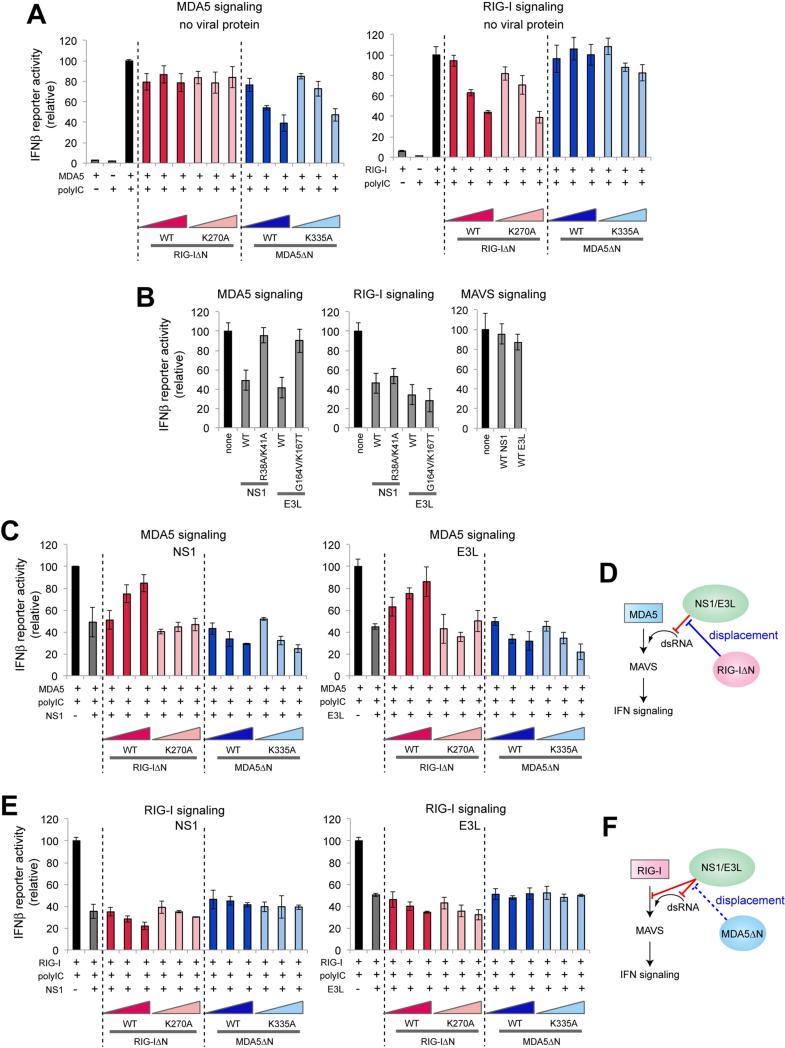
RIG-IΔN relieves the viral suppression of the MDA5 signaling activity
The ability of RIG-IΔN/MDA5ΔN to clear dsRNA of the viral proteins further prompted the question whether RIG-I and MDA5 could indirectly assist each other through a similar ATP-dependent, signaling-independent mechanism as for PKR. To address this question, we examined the impact of RIG-IΔN and MDA5ΔN on the signaling activities of full-length RIG-I and MDA5 in the presence or absence of NS1/E3L as measured by the IFNβ reporter activity and the mRNA levels of IFNβ and RANTES. In the absence of NS1 and E3L, ectopically expressed RIG-I and MDA5, but not their ΔN truncation mutants, robustly stimulated IFN signaling in response to a dsRNA mimic, polyinosinic-polycytidylic acid (polyIC) (Figures S3A & S3B). When ΔNs were co-expressed with full-length receptors, signaling activities of RIG-I and MDA5 were diminished by respective ΔN, but not by ΔN of the other receptor (Figures 3A and S3C). This suggests that the competition for RNA exists between RIG-I and RIG-IΔN, and between MDA5 and MDA5ΔN, but not between RIG-I(ΔN) and MDA5(ΔN), in agreement with their known preference for the distinct sites on RNA (Yoneyama and Fujita, 2008).
Figure 3.
RIG-IΔN relieves the viral suppression of the MDA5 signaling activity.
A. Impact of RIG-IΔN or MDA5ΔN on the IFNβ reporter activity of full-length MDA5 or RIG-I in the absence of viral proteins. Wild-type (WT) and their catalytic mutants, K270A (for RIG-I) and K335A (for MDA5), were compared. Data are shown in mean ± SD (n=3).
B. Impact of full-length NS1, E3L and their RNA-binding defective mutants on the signaling activities of MDA5, RIG-I and MAVS. See Figure S3E for western blot analysis.
C. Impact of RIG-IΔN or MDA5ΔN on the IFNβ reporter activity of full-length MDA5 in the presence of NS1 or E3L. Data are shown in mean ± SD (n=3).
D. Model in which RIG-I indirectly promotes the MDA5 signaling activity by displacing NS1/E3L from dsRNA.
E. Impact of RIG-IΔN or MDA5ΔN on the IFNβ reporter activity of full-length RIG-I in the presence of NS1 or E3L. Data are shown in mean ± SD (n=3).
F. Model in which the RIG-I signaling activity is affected by NS1/E3L and MDA5ΔN. NS1/E3L suppress RIG-I through both RNA-dependent and -independent mechanisms. Accordingly, the inhibitory effect of NS1/E3L on RIG-I could not be relieved by the protein displacement activity of MDA5.
In the presence of NS1 or E3L, polyIC-mediated signaling activities of both RIG-I and MDA5 were suppressed (Figure 3B). No suppression was observed for polyIC-indepdent MAVS-mediated signaling (Figure S3B). For MDA5, this suppression is due to sequestration of polyIC by NS1/E3L, as the dsRNA-binding defective mutants of NS1 (R38A/K41A) (Donelan et al., 2003) and E3L (G164V/K167T) (Chang and Jacobs, 1993) failed to suppress MDA5 signaling (Figure 3B). Notably, co-expression of MDA5 with RIG-IΔN, but not with K270AΔN, relieved the suppression by NS1/E3L (Figures 3C & S3D). As with PKR, this result is consistent with the notion that RIG-IΔN displaces NS1/E3L, thereby increasing the dsRNA accessibility for MDA5 (Figure 3D). In contrast to MDA5, RIG-I was suppressed by NS1/E3L through an RNA-independent mechanism (Gack et al., 2009) (Figure 3B), and the suppression was not relieved by either RIG-IΔN or MDA5ΔN (Figures 3E & S3D). These results suggest that displacement of NS1/E3L by RIG-I (and potentially MDA5) could indirectly increase their signaling activities, but this activity of RIG-I/MDA5 can be countered by viral proteins through RNA-independent inhibitory mechanisms (Figure 3F).
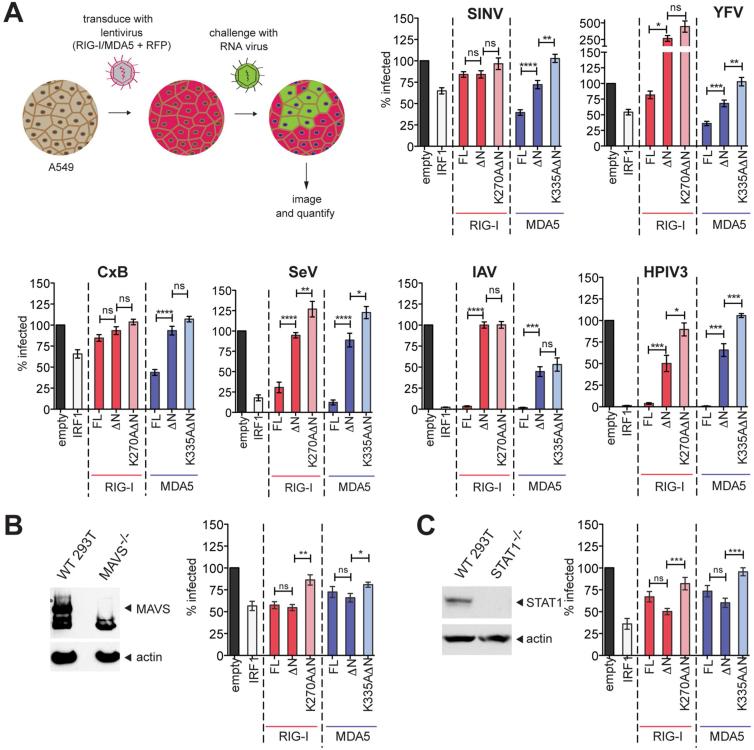
RIG-IΔN and MDA5ΔN suppress replication of certain viruses in an ATP-dependent manner
To examine whether these signaling-independent activities of RIG-I and MDA5 could confer an antiviral function, we next examined the antiviral potency of RIG-IΔN and MDA5ΔN using a panel of RNA viruses: Sindbis (SINV), yellow fever (YFV), Coxsackie B (CxB), Sendai (SeV), influenza A (IAV), and human parainfluenza 3 (HPIV3). Human lung carcinoma A549 cells were transduced with the lentivirus expressing RIG-I, MDA5 or their variants and challenged with the viruses engineered to express a reporter gene, green fluorescent protein (GFP), or with wild-type non-reporter IAV. Efficiency of virus spread, as measured by the number of GFP- or anti-IAV-NP -positive cells, was assessed (Figure 4A). Consistent with their reported divergence in viral specificity (Goubau et al., 2013; Kato et al., 2011), full-length RIG-I and MDA5 suppressed different families of RNA viruses (Figure 4A). Notably, RIGIΔN and MDA5ΔN also displayed antiviral activities against several viruses (SINV, YFV and HPIV3 for MDA5, and HPIV3 for RIG-I) (Figure 4A), albeit less efficiently than full-length receptors. Introduction of the ATP-hydrolysis impairing catalytic mutations, however, completely abrogated the antiviral activities of RIG-IΔN and MDA5ΔN. In full-length receptors, catalytic mutations induced cytotoxicity for MDA5 (Figure S4A) and reduced antiviral potency of RIG-I against most but not all viruses (Figure S4B), suggesting a more complex role of ATP-hydrolysis in signaling-dependent activities of these receptors.
Figure 4.
RIG-IΔN and MDA5ΔN suppress replication of certain viruses in an ATP-dependent manner.
A. Impact of RIG-IΔN and MDA5ΔN on viral spread efficiencies. A549 cells were transduced to express full-length (FL) RIG-I, MDA5 or their ΔN variants, together with RFP as the transduction control, and challenged with indicated viruses at MOI=0.01. Samples were fixed after several rounds of replication and the fraction of infected cells was normalized against the empty control. IRF1, a broadly acting antiviral ISG (Schoggins et al., 2011), served as the positive control. Data are represented as mean ± SEM from n=3 independent experiments with 4 technical replicates each. Indicated pairs were analyzed by Mann Whitney test. ns = not significant. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.1.
B. Viral spread assay on 293T MAVS−/− cells. MAVS knock-out was confirmed by Western Blot (left panel). Spread data (right panel) are represented as mean ± SEM from n=4 independent experiments with 4 technical replicates each. Indicated pairs were analyzed by Mann Whitney test. ns = not significant. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.1
C. Same as (B) using 293T STAT1−/− cells instead of MAVS−/−.
To exclude the possibility that RIG-IΔN and MDA5ΔN exert antiviral activities through endogenous RIG-I/MDA5 and their IFN signaling activities, we examined the mRNA levels of IFN-stimulated genes (ISGs) in A549 cells. In parallel, we also measured viral spread in 293T cells deficient in MAVS, the common signaling adaptor for RIG-I and MDA5, and cells deficient in STAT1, the transcription factor required for ISG induction. We chose to examine HPIV3 as this virus exhibited robust sensitivity to both RIG-IΔN and MDA5ΔN (Figure 4A). We found that RIG-IΔN and MDA5ΔN induced little or no ISG expression above the empty control (Figure S4C) and that RIG-IΔN and MDA5ΔN, but not their catalytic mutants, suppressed HPIV3 in MAVS−/− (Figures 4B & S4D) and STAT−/− cells (Figure 4C). These results suggest that the suppression of HPIV3 by RIG-IΔN and MDA5ΔN is independent of the canonical RIGI/MDA5 signaling pathway involving MAVS and type I IFN, but dependent on their ATP hydrolysis.
Discussion
We here demonstrate that RIG-I and MDA5 utilize the ATP-driven receptor dynamics to remodel ligands and their accessibility, and can utilize this activity to perform signaling-independent antiviral functions. This finding challenges the conventional views that pattern recognition receptors (PRRs) passively bind pre-exposed ligands and that their antiviral activities are solely dependent on their ability to induce immune signaling. Thus, our finding of signaling-independent antiviral activities of RIG-I and MDA5 re-defines these proteins as a novel class of PRRs with “effector-like” functions.
Our findings also reveal a new dimension of complexity in host-virus and host-host interactions. To evade detection by the host innate immune system, some viruses encode dsRNA-binding proteins, such as NS1 of influenza A, whereas RIG-I and MDA5 work to counteract this viral evasion mechanism by stripping away the viral proteins from dsRNA. While it is possible that RIG-I/MDA5 can also displace host dsRNA-binding proteins, our results suggest unexpected cooperative, rather than competitive, relationships among the host proteins, as shown by the positive action of RIG-I/MDA5 on PKR as well as that of RIG-I on MDA5. This may reflect the efficient detection of transiently exposed dsRNA by PKR and MDA5 (McKenna et al., 2007a), and perhaps their ability to then amplify the signal.
As with many antiviral effector molecules, high protein concentration is likely required for the robust effector-like functions of RIG-I and MDA5. As RIG-I and MDA5 are themselves ISGs, one plausible model is that their canonical PRR function dominates at baseline, whereas their effector-like function emerges when sufficient levels of RIG-I and MDA5 accumulate through the IFN response (Figure S4E). This would enable temporal coordination with other ISGs, such as PKR, which RIG-I and MDA5 assist by exposing dsRNA. Are the protein-displacement activities of RIG-I/MDA5 limited to assisting other host dsRNA sensors? The answer to this question will likely vary with different viruses, but the wide range of roadblocks that can be displaced by RIG-I and MDA5 (Figure 1) raise the possibility that they may also interfere directly with core viral life cycle steps. For example, this could be mediated by displacing viral transcription or replication complexes, or by disrupting capsid interactions required for packaging and infectious virus production. In any case, this work provides the foundation and strong justification to begin addressing these interesting questions.
Experimental Procedures
Detailed experimental procedures are provided in Supplemental Information.
Protein displacement, streptavidin displacement and hairpin removal assays
For protein displacement assay, dsRNA was pre-incubated with fluorescently labeled viral RBDs in buffer A (20 mM Hepes, pH 7.5, 100 mM NaCl, 1.5 mM MgCl2 and 2 mM DTT) and then with an indicated amount of RIG-I/MDA5 in the presence or absence of ATP, quenched with EDTA and immediately analyzed by Bis-Tris native PAGE (Life). For streptavidin displacement, 3’-biotinylated dsRNA was incubated with streptavidin and subsequently with RIG-I or MDA5 in buffer A. The reaction was initiated by the addition of ATP and an excess amount of free biotin at 37°C, quenched with EDTA at indicated time points, and immediately analyzed on native TBE gels. For hairpin removal assay, the hairpin-containing dsRNA was incubated with RIG-I/MDA5 with or without ATP at 37°C for indicated time period, quenched with the loading buffer (see Supplemental Information) and immediately analyzed on native TBE gels.
PKR activity assay
112 bp dsRNA was pre-incubated with unlabeled NS1 or E3L RBDs in buffer A at 37°C for 5 min, and was additionally incubated with a mixture of PKR, ATP and RIG-IΔN (or MDA5ΔN) at 37°C for 15 min. The auto-phosphorylation reaction was quenched with EDTA, and analyzed by 10% SDS-PAGE.
IFN-β promoter reporter assay
293T cells were transfected with pFLAG-CMV4 plasmids encoding full-length NS1/E3L, full-length RIG-I/MDA5 and RIG-IΔN/MDA5ΔN, along with IFNβ promoter driven firefly luciferase reporter plasmid and CMV promoter driven Renilla luciferase reporter plasmid. Cells were stimulated with high molecular weight polyIC (Invivogen), and harvested 20 hr post-stimulation prior to Dual Luciferase Reporter assay (Promega). Firefly luciferase activity was normalized against Renilla luciferase activity.
Image-based virus spread assays
A549 cells or 293T-based knock-out cells were transduced with lentiviral stocks expressing RIG-I and MDA5 (along with the transduction control, RFP) for 2 days, and challenged with viruses at 100 PFU/well (MOI 0.01). Cells were fixed after multiple rounds of replication, permeabilized, stained with DAPI (for all viruses) or IAV NP (Millipore)(for IAV), and imaged using the ImageXPressMICRO System (Molecular Devices) for analysis of the virus infected population.
Supplementary Material
Acknowledgement
This work was supported by the Chinese Scholarship Council joint PhD program (H.Y.), a fellowship from the Deutsche Forschungsgemeinschaft (M.D.), anonymous donors, and R01 grants AI106912 (S.H.), AI111784 (S.H.) and AI091707 (C.M.R.). We thank Jihong Han for supporting the graduate program of H.Y., Yoandris del Toro Duany for helping protein purification and Melody Li, Hachung Chung and Kierstin Bell for generously providing virus stocks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.Y. and S.H. conceived the study. H.Y., M.D., C.R. and S.H. designed the experiments and analyzed the data. H.Y. performed biochemical and cellular assays, and M.D. performed viral assays. A.P. purified proteins for biochemical assays; H.H and R.G. assisted with viral assays; and T.S., J.S-B, and V.H. generated the MAVS and STAT1 knock-out cell lines. H.Y. M.D. and S.H. wrote the manuscript.
References
- Chang H-W, Jacobs BL. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Donelan NR, Basler CF, Garcia-Sastre A. A Recombinant Influenza A Virus Expressing an RNA-Binding-Defective NS1 Protein Induces High Levels of Beta Interferon and Is Attenuated in Mice. J. Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn K-S, Huang I-C, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A Virus NS1 Targets the Ubiquitin Ligase TRIM25 to Evade Recognition by the Host Viral RNA Sensor RIG-I. Cell Host & Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Goubau D, Deddouche S, Reis e Sousa C. Cytosolic Sensing of Viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Research. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immuno Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Li S, Min J-Y, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping- the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular Framework for the Activation of RNA-dependent Protein Kinase. J Biol Chem. 2007a;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- McKenna SA, Lindhout DA, Shimoike T, Puglisi JD. Biophysical and Biochemical Investigations of dsRNA-Activated Kinase PKR. Methods in Enz. 2007b;430:373–396. doi: 10.1016/S0076-6879(07)30014-1. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner K-P, Ha T. Cytosolic Viral Sensor RIG-I Is a 5′-Triphosphate–Dependent Translocase on Double-Stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Jo M, Lin C, Wu B, Orme-Johnson M, Walz T, Hohng S, Hur S. Kinetic Mechanism for Viral dsRNA Length Discrimination by MDA5 Filament. Proc Natl Acad Sci U S. 2012;A109:E3340–3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Bin W, Orme-Johnson M, Liu M, Walz T, Hur S. Cooperative Assembly and Dynamic Disassembly of MDA5 Filaments for Viral dsRNA Recognition. Proc Natl Acad Sci U S A. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I Forms Signaling-Competent Filaments in an ATP-Dependent, Ubiquitin-Independent Manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular imprinting as a signal activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.