Abstract
Memory impairment is the cardinal early feature of Alzheimer's disease (AD), a highly prevalent disorder whose causes remain only partially understood. To identify novel genetic predictors, we used an integrative genomics approach to perform the largest study to date of human memory (n=14,781). Using a genome-wide screen, we discovered a novel association of a polymorphism in the pro-apoptotic gene FASTKD2 (fas-activated serine/threonine kinase domains 2; rs7594645-G) with better memory performance and replicated this finding in independent samples. Consistent with a neuroprotective effect, rs7594645-G carriers exhibited increased hippocampal volume and gray matter density and decreased cerebrospinal fluid levels of apoptotic mediators. The MTOR (mechanistic target of rapamycin) gene and pathways related to endocytosis, cholinergic neurotransmission, EGFR signaling, and immune regulation, among others, also displayed association with memory. These findings nominate FASTKD2 as a target for modulating neurodegeneration and suggest potential mechanisms for therapies to combat memory loss in normal cognitive aging and dementia.
Keywords: genome-wide association study (GWAS), Alzheimer's disease (AD), dementia, cognitive aging, memory, magnetic resonance imaging (MRI), apoptosis, neurodegeneration
INTRODUCTION
Human memory is a complex, dynamic trait with significant roles in development, aging, and disease. Impairment in episodic memory – involving the encoding and conscious recollection of experiences – is an early hallmark of AD, the most common cause of dementia.1 Declines in episodic and other memory domains are also found in normal cognitive aging and many age-related disorders, including Parkinson's disease, diabetes, and cancer.2 With the rising incidence and burdens of dementia, a better understanding of its causes is crucial for risk stratification and the development of memory-sparing lifestyle and drug therapies.3, 4
At present, the molecular mechanisms underlying memory performance are not fully understood. Epidemiological studies have linked many factors to memory, including the presence of vascular and metabolic disease, mental and physical activity, and educational and occupational attainment.5, 6 Episodic memory is estimated to have substantial heritability (30-60%) based on twin studies and is thought to be influenced by common and rare genetic variation in multiple pathways.6 Although genetic studies have implicated numerous single nucleotide polymorphisms (SNPs) in episodic memory performance,7 significant heritability remains unexplained.
Recently, there has been great interest in harnessing the power of genome-wide association studies (GWAS) to gain additional mechanistic insight into complex neurological traits and diseases.8 Extant approaches for maximizing the translational impact of GWAS findings include using large samples with adequate statistical power for discovery,6 performing complementary analyses to detect broader trends of association within genes and pathways,9 and assessing for convergence of genetic associations across phenotypic modalities to understand their biological context.10, 11 Here, we integrated GWAS with molecular and neuroimaging data to identify and characterize novel predictors of episodic memory function in older adults.
MATERIALS AND METHODS
Study participants
Launched in 1992, the Health and Retirement Study (HRS) is a nationally representative longitudinal study of more than 26,000 Americans over 50 years old.12 Biennial interviews were used to collect detailed information on the health, social, and economic status of participants.
The AddNeuroMed Consortium,13 Alzheimer's Disease Neuroimaging Initiative (ADNI),14 Indiana Memory and Aging Study (IMAS),15 Rush Memory and Aging Project (MAP),16 and Religious Orders Study (ROS)17 include older adults representing clinical stages along the continuum from normal aging to AD who received longitudinal clinical evaluations.
All participants provided written informed consent and study protocols were approved by each site's institutional review board.
Clinical and cognitive assessment data
We obtained the RAND Corporation's public release HRS data files (version L) which integrated information across study waves.18 For discovery, we analyzed HRS wave 3 since it contained the largest cross-sectional sample with complete data for cognitive functioning variables. To maximize statistical power for discovery, we analyzed the full available sample including multiple ethnicities and included principal component eigenvectors to control for population stratification.19 Episodic memory performance was assessed through immediate free recall of a 10-word list.20 Age, gender, and years of education were used as covariates for GWAS.
For replication, we used independent samples from HRS wave 7, AddNeuroMed, ADNI, IMAS, MAP, and ROS. With the exception of HRS, word list immediate recall tasks included multiple trials and therefore the sum of those trials at the baseline visit was used as the outcome. Immediate recall tasks used in these cohorts included the Rey Auditory Verbal Learning Test (RAVLT) in ADNI, the California Verbal Learning Test (CVLT) in IMAS, and the Consortium to Establish a Registry for Alzheimer's Disease (CERAD) word list test in AddNeuroMed, MAP, and ROS. Age, gender, and education were included as covariates in all replication analyses. Diagnosis (as a categorical variable) was also included as a covariate for analyses in AddNeuroMed, ADNI, IMAS, MAP, and ROS to account for the clinical stages represented among participants.
Structural magnetic resonance imaging (MRI) data
Baseline visit T1-weighted cranial MRI scans were analyzed for 315 healthy control participants from ADNI who were not included in the memory analyses. Scan protocols, quality control, image processing, and extraction of volume and gray matter density for brain regions of interest were performed as described previously.21
Cerebrospinal fluid (CSF) proteomic data
Baseline visit CSF samples were analyzed for 82 healthy control participants from ADNI who were not included in the memory analyses. Sample acquisition and quality control procedures were performed as described previously.22 Levels for 159 proteins were measured by multiplex immunoassay on the Myriad Genetics (Austin, TX) Rules Based Medicine Human Discovery MAP panel. Stringent quality control for protein level measurements was performed as described previously.23 Additional technical details are available in the ADNI Biomarker Consortium CSF Proteomics Data Primer (http://adni.loni.usc.edu/wp-content/uploads/2012/01/2011Dec28-Biomarkers-Consortium-Data-Primer-FINAL1.pdf). Based on literature search, we performed a pathway-guided analysis of CSF levels of four proteins (adiponectin, fas ligand, TNFR2, and TRAIL-R3) involved in fas-mediated apoptosis. Protein levels were log10− transformed to ensure a normal distribution of each phenotype.
Genotyping and imputation
Genomic DNA for 12,507 participants was obtained during HRS waves 8-9 via buccal swab or saliva sample. Genotyping was performed at the Center for Inherited Disease Research (CIDR; Johns Hopkins University) using the Illumina (San Diego, CA) HumanOmni2.5-4v1 array. Genotype data underwent stringent quality control including identity checks, sample exclusion for call rate<98%, and SNP exclusion for call rate<98%, Hardy-Weinberg p<10−4, minor allele frequency (MAF)<1%, or failure of other technical and quality filters.24 Identity by descent analysis in PLINK25 (http://pngu.mgh.harvard.edu/~purcell/plink/) revealed 80 pairs and 4 trios with significant relatedness; therefore one participant from each unit was randomly selected for inclusion. SNPRelate26 was used to identify population group outliers and generate principal components to control for population substructure. As described previously, on scree plot the fraction of variance explained was minimal and level after the first two principal components.24 The final dataset included 1,681,327 SNPs for 12,419 participants.
We used Minimac27 and the 1000 Genomes Project to impute the SNPs characterizing the APOE ε4 allele (rs429358, rs7412) in HRS and the specific SNPs required for replication studies in AddNeuroMed, ADNI, IMAS, MAP, and ROS. Imputation and quality control were performed as described previously.28, 29 Due to the control for population substructure required for imputation quality, imputation was restricted to participants with non-Hispanic Caucasian ancestry as determined by multidimensional clustering in PLINK.28
Statistical analysis
Basic statistical analyses were performed using IBM SPSS Statistics for Windows, Version 22.0 (Armonk, NY). Genetic associations were tested using linear regression under an additive genetic model in PLINK. In the discovery sample GWAS, the first three principal components were included as covariates consistent with prior studies of AD endophenotypes,30, 31 and a conservative significance threshold (p<5×10−8) was employed based on a Bonferroni correction of one million independent tests.32 For analysis of the HRS replication sample (wave 7), the first three principal components were included as covariates as in the discovery GWAS. For all other replication analyses, the potential effects of population stratification were minimized through restricting analyses to participants with non-Hispanic Caucasian ancestry as determined by multidimensional clustering in PLINK.28 Manhattan and Q-Q plots were generated with Haploview33 and regional association and linkage disequilibrium (LD) plots were generated with LocusZoom34 and SNAP.35
Complementary approaches were used to validate and extend the GWAS findings. We used HYST36 to calculate a summary p-value for each gene accounting for its size, LD structure, and constituent GWAS SNP associations. We then utilized these summary p-values and GSA-SNP37 to identify pathways enriched with significant association to memory. Pathway definitions, consisting of canonical representations of biological processes compiled by expert reviewers, were downloaded from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp; canonical pathways set, version 4.0) and the false discovery rate (FDR) was used to correct for pathway-level multiple comparisons.
Genome-wide significant associations were further investigated in independent samples through inverse-variance weighted meta-analysis using METAL38 and through association testing with MRI and CSF biomarkers using multivariate analyses of covariance with Wilks’ lambda as the test statistic. For meta-analysis, effect sizes were denoted by standardized β coefficients from linear regression. Cohen's d statistics were also calculated to facilitate comparison of effect sizes across phenotypes.
RESULTS
Study participants
This study involved a total of 14,781 participants from six independent cohorts (Table 1). For discovery, we analyzed data for 6,705 participants from HRS wave 3. In the HRS, immediate recall was approximately normally distributed (Supplementary Figure 1) and was highly-correlated with delayed recall (r=0.74). Using linear regression, younger age (p=3.73×10−56), female gender (p=2.70×10−41), and more years of education (p=4.74×10−170) were associated with better episodic memory performance and collectively explained 16.4% of the variance in immediate recall (omnibus F(3, 6701)=437.88, p=8.77×10−260).
Table 1.
Selected characteristics (number or mean (standard deviation)) of participants in the discovery* and replication cohorts.
| HRS* (wave 3) | HRS (wave 7) | Add-Neuro-Med | ADNI | IMAS | MAP | ROS | |
|---|---|---|---|---|---|---|---|
| Participants | 6,705 | 5,116 | 64 | 1,147 | 68 | 878 | 803 |
| Gender | |||||||
| Male | 2,529 (38%) | 2,177 (43%) | 27 (42%) | 669 (58%) | 25 (37%) | 244 (28%) | 276 (34%) |
| Female | 4,176 (62%) | 2,939 (57%) | 37 (58%) | 478 (42%) | 43 (63%) | 634 (72%) | 527 (66%) |
| Age (y) | 61.8 (8.2) | 60.4 (10.5) | 70.9 (6.8) | 74.3 (7.1) | 71.9 (8.1) | 81.0 (6.6) | 75.7 (7.3) |
| Education (y) | 12.4 (3.1) | 13.0 (3.1) | 10.2 (5.0) | 15.8 (2.9) | 16.5 (2.5) | 14.7 (2.9) | 18.2 (3.4) |
| Ethnicity | |||||||
| NHC | 5,283 | 4,042 | 64 | 1,147 | 68 | 878 | 803 |
| AA | 817 | 711 | 0 | 0 | 0 | 0 | 0 |
| Other | 605 | 363 | 0 | 0 | 0 | 0 | 0 |
Abbreviations: NHC = non-Hispanic Caucasian; AA = African-American
GWAS of episodic memory
To identify novel genetic predictors of episodic memory functioning, we performed a GWAS (n=6,705) controlling for age, gender, education, and the first three principal components to address population substructure. The genomic inflation factor (λ) was 1.007, indicating no substantial inflation of association (Supplementary Figure 2).
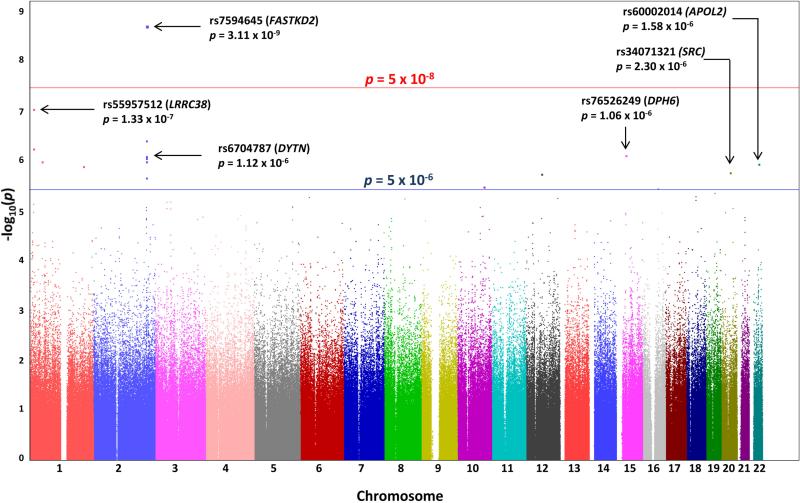
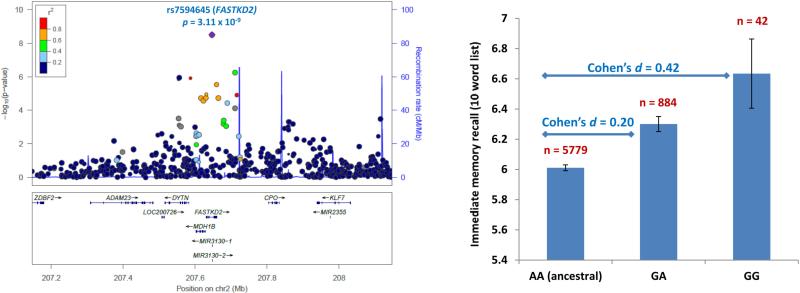
A genome-wide significant association with immediate recall was identified on chromosome 2 for rs7594645 (p=3.11×10−9; Figure 1). This SNP resides in an intron of FASTKD2 that is overlapped by MIR3130-1 and MIR3130-2 (Figure 2a). The rs7594645-G allele exhibited a modest additive effect associated with better episodic memory performance and explained an additional 0.5% of the phenotypic variance (Figure 2b).
Figure 1. Manhattan plot for the HRS discovery GWAS of immediate recall.
Observed -log10 p-values (y-axis) are displayed for all tested SNPs on each chromosome (x-axis). A genome-wide significant association (p<5×10−8) to immediate recall was identified on chromosome 2 within FASTKD2. Suggestive associations (p<5×10−6) were identified on additional chromosomes. Selected top associations are labeled on the plot.
Figure 2. Association and effect of rs7594645-G (FASTKD2) on immediate recall in the HRS discovery GWAS.
a) All SNPs within 500 kb of FASTKD2 are plotted based on their GWAS -log10 p-values, NCBI build 37 genomic position, and recombination rates calculated from the 1000 Genomes Project reference data. The color scale of r2 values is used to label SNPs based on their degree of LD with rs7594645. Genes in the region are labeled with arrows denoting 5’-to-3’ orientation. b) Mean immediate recall scores (adjusted for clinical covariates and population structure) +/- standard errors are displayed based on rs7594645 genotype. Presence of the minor allele (G) of rs7594645 imparted a modest additive effect of improving memory performance and accounted for 0.5% of the phenotypic variance.
Suggestive associations with immediate recall (p<5×10−6) were also identified (Figure 1; Supplementary Table 1). These included SNPs near FASTKD2 within DYTN (dystrotelin) as well as SNPs on additional chromosomes within LRRC38 (leucine rich repeat containing 38), SRC (v-Src tyrosine kinase), and APOL2 (apolipoprotein L2). We also observed nominal associations with immediate recall (p<0.05) for 14 of 25 memory- and AD-related candidate genes identified through literature review (Supplementary Table 2). These genes included BCHE (butyrylcholinesterase), BDNF (brain-derived neurotrophic factor), CR1 (complement receptor 1), and TREM2 (triggering receptor expressed on myeloid cells 2), among others. Significant association after Bonferroni correction for 25 genes (p<0.002) was observed for three of these genes, CAMTA1 (calmodulin binding transcription activator 1), DISC1 (disrupted in schizophrenia 1), and WWC1 (WW and C2 domain containing 1; also known as KIBRA).
Due to its well-known association with AD,39 we further investigated the effect of the APOE (apolipoprotein E) ε4 allele in the GWAS sample. Since the SNPs characterizing APOE ε4 (rs429358, rs7412) failed initial genotyping quality control, to perform additional analyses we imputed these SNPs in non-Hispanic Caucasian participants (n=5,288). We did not observe an association of APOE ε4 with immediate (p=0.904) or delayed (p=0.466) recall in this sample. However, APOE ε4 was associated with increased odds of self-reported diagnosis of AD by a doctor (p=5.96×10−13, OR 5.59, 95% CI 3.50-8.91) among participants re-evaluated 14 years later (n=4,491).
Gene- and pathway-based analyses of episodic memory
To extend the GWAS findings, we performed complementary analyses at the gene and pathway levels. Using HYST to calculate summary gene-based p-values, we identified additional genome-wide significant associations with episodic memory based on a Bonferroni correction for 23,672 genes (p<2.11×10−6; Supplementary Table 3). Top genome-wide significant associations included the genes LARS2 (leucyltRNA synthetase 2, mitochondrial; p=5.56×10−11) and MTOR (p=2.50×10−9), which along with FASTKD240 encode proteins with critical roles in mitochondrial function.41, 42
Numerous pathways exhibited enrichment of association (FDR-corrected p<0.05) with episodic memory performance (Table 2). These included multiple mechanisms related to immune regulation and cell adhesion as well as pathways related to cholinergic neurotransmission, endocytosis, and epidermal growth factor receptor (EGFR; ErbB1) signaling, among others.
Table 2.
Biological pathways associated with immediate recall in the HRS discovery GWAS (n=6,705).
| Pathway Description (Source Database) | Size* | Uncorrected p** |
|---|---|---|
| Valine, leucine, and isoleucine biosynthesis (KEGG) | 11 | 3.78 × 10−9 |
| IFNγ pathway (Protein Interaction Database) | 40 | 1.21 × 10−6 |
| GA12 pathway (Signal Transduction Knowledge Environment) | 23 | 3.24 × 10−6 |
| MTORC1-mediated signaling (Reactome) | 11 | 3.28 × 10−6 |
| CDC42 pathway (Protein Interaction Database) | 70 | 5.48 × 10−6 |
| Mitochondrial tRNA aminoacylation (Reactome) | 21 | 1.45 × 10−5 |
| B-cell survival pathway (Biocarta) | 16 | 2.27 × 10−5 |
| CD28 co-stimulation pathway (Reactome) | 32 | 2.33 × 10−5 |
| CTCF pathway (Biocarta) | 23 | 4.12 × 10−5 |
| ErbB2/ErbB3 pathway (Protein Interaction Database) | 44 | 4.27 × 10−5 |
| Highly calcium-permeable postsynaptic nicotinic acetylcholine receptors (Reactome) | 13 | 6.05 × 10−5 |
| ErbB1 downstream pathway (Protein Interaction Database) | 105 | 8.48 × 10−5 |
| Creation of C4 and C2 activators (Reactome) | 10 | 1.16 × 10−4 |
| G1 phase (Reactome) | 38 | 1.48 × 10−4 |
| L1CAM interactions (Reactome) | 86 | 2.08 × 10−4 |
| Glioma (KEGG) | 65 | 2.48 × 10−4 |
| Acetylcholine binding and downstream events (Reactome) | 16 | 4.80 × 10−4 |
| Protein kinase B mediated events (Reactome) | 29 | 5.04 × 10−4 |
| Endocytosis (KEGG) | 183 | 5.85 × 10−4 |
| LKB1 pathway (Protein Interaction Database) | 47 | 6.27 × 10−4 |
| Nectin pathway (Protein Interaction Database) | 30 | 6.83 × 10−4 |
| Prion diseases (KEGG) | 35 | 6.83 × 10−4 |
Number of genes in the pathway
All pathways displayed are significant at FDR-corrected p<0.05
Replication of FASTKD2 association with episodic memory
For replication of our major SNP-based genome-wide significant finding, we analyzed independent samples from the HRS, AddNeuroMed, ADNI, IMAS, MAP, and ROS cohorts. Due to heterogeneity of memory instruments and clinical populations, we initially analyzed within cohorts and then performed a replication meta-analysis involving 7,761 participants which validated the association of rs7594645-G with higher immediate recall (p=0.013; Supplementary Table 4 and Supplementary Figure 3). In all replication cohorts except ROS, rs7594645-G exhibited a positive direction of effect on episodic memory performance consistent with the discovery sample. The heterogeneity statistic (I2) p-value was 0.71, indicating no significant heterogeneity across the replication studies.43 Overall, a summary meta-analysis of rs7594645-G across the discovery and replication samples combined displayed genome-wide significant association (p=3.73×10−9, total n=14,466).
Association of rs7594645 with FASTKD2 mRNA expression
We assessed the functional impact of rs7594645 on FASTKD2 mRNA expression using two expression quantitative locus (eQTL) databases. Using Genevar,44 we analyzed published eQTL data for 856 healthy female twins from the MuTHER (Multiple Tissue Human Expression Resource) project.45 Although rs7594645 was not available in this dataset, we analyzed rs10490541 as a proxy SNP since its minor allele (T) is in complete LD (D’=1, r2=1) with rs7594645-G in the CEU reference population from the 1000 Genomes Project. We observed that rs10490541-T was associated with decreased FASTKD2 mRNA expression in skin cells (p=0.035), controlling for participant age, experimental batch, and sample processing.45 Using GeneNetwork,46 we next analyzed published brain eQTL data for 187 healthy older adults.47, 48 In this analysis, we identified a SNP (rs4578841) in strong LD with rs7594645 (D’=1, r2=0.13) that displayed significant association to neocortical FASTKD2 expression (p=9.16×10−4).
Association of rs7594645 with cerebrospinal fluid (CSF) levels of pro-apoptotic factors
FASTKD2 encodes a pro-apoptotic protein that is predominantly localized to mitochondria.40, 49 Given the association of rs7594645-G with higher memory performance and lower FASTKD2 mRNA expression, we hypothesized that rs7594645-G would also be associated with decreased activation of apoptosis in the central nervous system. To test this hypothesis, we analyzed CSF levels of four proteins involved in fas-mediated apoptosis in 82 healthy control participants from ADNI (Supplementary Figure 4). Controlling for age and gender, rs7594645-G carriers displayed decreased CSF levels of adiponectin (p=5.85×10−5) and fas ligand (p=0.012) and trended towards decreased levels of TNFR2 (tumor necrosis factor receptor 2; p=0.057) and TRAIL-R3 (tumor necrosis factor-related apoptosis inducing ligand receptor 3; p=0.083), with a significant multivariate effect of genotype (p=1.07×10−4).
Association of FASTKD2 with hippocampal structure
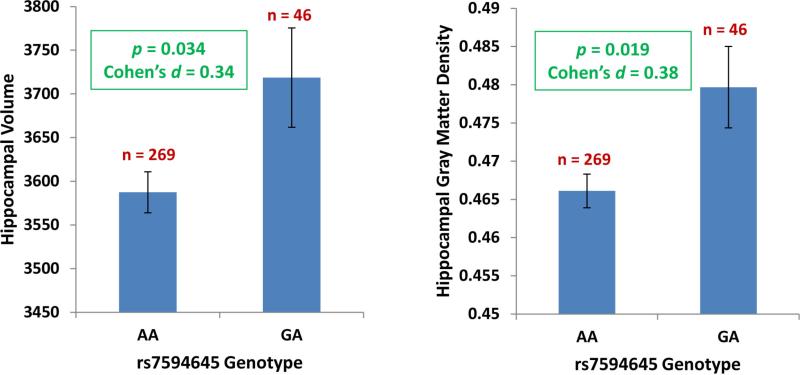
Given the association of rs7594645-G with higher memory performance and the evidence of a functional molecular effect of this SNP, we hypothesized that FASTKD2 would also be associated with structural changes in episodic memory-related brain regions. To test this hypothesis, we assessed hippocampal volume and gray matter density in 315 healthy control participants from ADNI who were genotyped and received high resolution T1-weighted structural MRI (Figure 3). Controlling for age, gender, education, and intracranial volume, rs7594645-G was associated with hippocampal structure (multivariate p=0.036), including higher volume (p=0.034) and gray matter density (p=0.019).
Figure 3. Effect of FASTKD2 rs7594645-G on hippocampal volume and gray matter density in 315 older healthy control participants from the ADNI.
Using high resolution T1-weighted structural MRI, mean volumes and gray matter densities in the hippocampus (adjusted for age, gender, education, and intracranial volume) ± standard errors are displayed based on rs7594645 genotype. Participants with the minor allele (G) of rs7594645 displayed increased hippocampal volume and gray matter density, with a significant multivariate effect of genotype on these MRI measures (p=0.036).
DISCUSSION
Using GWAS in a large, population-based sample of older adults, this study discovered a genome-wide significant association of FASTKD2 rs7594645-G with better episodic memory performance. This novel association was replicated in independent samples and was further validated by converging evidence from structural MRI and gene and protein expression studies consistent with a neuroprotective effect in the brain.
FASTKD2 encodes a pro-apoptotic protein40, 49 and has not been previously associated with cognition. Signaling through its upstream activator and death receptor fas/CD95 is known to mediate neuronal responses to traumatic brain injury50 and may have roles in neurodegeneration related to amyloid-β,51 methamphetamines,52 and frontotemporal lobar degeneration.53 Rare FASTKD2 mutations have also been linked to infantile encephalopathy associated with electron transport chain complex IV deficiency, suggesting a possible secondary role in cellular energetics.40
rs7594645 is an intronic variant in FASTKD2 that exhibited an effect size comparable to most SNP associations for complex behavioral traits.54 There is substantial precedent for non-coding intronic or intergenic variants to exert functional effects through regulation of chromatin structure, transcription factor binding, and splicing component recognition, among other mechanisms.55-57 In addition, GWAS-implicated SNPs with modest effect sizes have highlighted druggable targets for type 2 diabetes, cancer, and other diseases, demonstrating the potential biological importance of replicable and convergent associations.57, 58
rs7594645 is located in a gene-dense region with extensive LD structure (Supplementary Figure 5). In particular, rs7594645 is overlapped by two genes encoding microRNA molecules. MicroRNAs are small, non-coding RNAs that base pair with complementary sequences in protein-coding mRNAs to direct their degradation or translational repression.59 The location of rs7594645 suggests that allelic variation in this region may regulate FASTKD2 expression through altering mRNA-microRNA interactions. Based on the UCSC Genome Browser, rs7594645 is also 3 base pairs downstream from a binding sequence for CTCF (CCCTC-binding factor), a highly conserved transcription factor required for neuronal development and synapse formation.60 Genetic variation at rs7594645 or a nearby tagging SNP may alter CTCF-mediated transcriptional regulation of FASTKD2 to impact neuronal viability. Molecular characterization in brain tissue, cell cultures, and animal models will be important to assess and clarify these hypothesized mechanisms and the broader functional architecture around FASTKD2 and its neighboring genes DYTN, MDH1B, and MIR3130-1 and -2.
Based on the Human Brain Transcriptome, a database of genome-wide transcriptome data from postmortem human brains, FASTKD2 is highly-expressed in the hippocampus throughout adulthood.61 Behavioral, imaging, and pathological studies have revealed the hippocampus to be crucial for episodic memory storage and retrieval.62 We identified a protective effect of rs7594645-G on hippocampal volume and gray matter density in older adults. Consistent with an anti-apoptotic, neuroprotective effect, we also identified an association of rs7594645-G with decreased FASTKD2 mRNA expression and decreased CSF levels of proteins involved in apoptosis. These convergent findings at the molecular and neural systems levels further support the observed effect of rs7594645-G on memory function and suggest that modulation of FASTKD2 and its functional pathways may be candidates for nootropic and/or neuroprotective drug development.
The overall allelic frequency of rs7594645-G in the discovery sample was 0.072. Among non-Hispanic Caucasian participants, which comprised 79% of this sample, the frequency of rs7594645-G was 0.064. Although rs7594645-G was relatively more common in African-American participants (MAF=0.139), this difference did not appear to exhibit a confounding effect on the association of rs7594645 with episodic memory performance (Supplementary Table 5). In the replication samples, rs7594645-G exhibited an overall frequency of 0.068 and displayed a positive effect on episodic memory performance in all cohorts except ROS. Greater levels of education, among other potential differences, may have modified the effect of rs7594645-G on episodic memory performance in the ROS sample.
Nominal associations for several memory- and AD-related candidate genes were observed in the discovery sample, including WWC163, DISC1,64 and the neurotransmission- or neurotrophic-related genes BCHE,28 BDNF,65 and CAMTA1.66 Although in the discovery sample the APOE ε4 allele was associated with increased odds of self-reported diagnosis of AD by a doctor, no SNPs in APOE were associated with immediate recall. The APOE ε4 allele is the strongest known genetic risk factor for sporadic AD,39 but its relationship to non-pathological cognitive aging is less clear.67, 68 Alternative study designs might have detected effects of APOE on cognition, including cross-sectional analyses of older samples or other cognitive domains as well as longitudinal analyses to identify relative declines within individuals over time.67, 69
Gene- and pathway-based analyses provide additional vehicles for characterizing complex genetic architectures, since variants of modest individual effect can collectively influence susceptibility through action within shared mechanisms.9, 70, 71 Through a gene-based approach accounting for effects of multiple SNPs, we identified genome-wide significant associations of LARS2 and MTOR with episodic memory that were otherwise concealed through standard SNP-based analysis. Mutations in LARS2 have been implicated in Perrault Syndrome, a rare disease characterized by hearing loss and premature ovarian failure and proposed to be mediated by mitochondrial and apoptotic dysfunction.42 The MTOR gene represents a major drug target for AD and other neurodegenerative disorders due to well-studied roles in amyloid and tau pathology.72, 73 We also observed enrichment of association with episodic memory within the EGFR pathway, which has been proposed as a target for treating amyloid-induced memory loss,74 as well as other mechanisms related to cholinergic neurotransmission, endocytosis, and the immune response.75, 76
Although this represents the largest known GWAS of memory performance in older adults, with a bigger sample the suggestive loci we highlighted might have reached genome-wide significance. These included DYTN which encodes a member of a protein family associated with schizophrenia and cognitive aging7 and APOL2 which functions with APOE in lipid pathways.77 In addition, although the confirmatory and novel findings from this study validate its analytical design, alternative assessments of memory performance and clinical status might have yielded more sensitive phenotypic measures for GWAS.
Future extensions of this work using next-generation sequencing and related methods may reveal other associations with episodic memory, as novel SNPs, rare, structural, and epigenetic variants, and dynamic molecular changes in the transcriptome may all play a role in memory.78 Studies of longitudinal change in episodic memory functioning may also elucidate additional mechanistic influences. Further, although it was beyond the scope of this study, characterizing gene-environment interactions will likely guide the optimized application of drug and lifestyle modifiers to preserve or enhance episodic memory.
In conclusion, this study discovered and validated a novel association of FASTKD2 with episodic memory function and hippocampal structure. Overall, these new findings identify potential targets to help improve risk stratification and therapeutic development in normal cognitive aging and dementia.
Supplementary Material
ACKNOWLEDGEMENTS
The HRS is sponsored by the National Institute on Aging (grants U01AG009740, RC2AG036495, and RC4AG039029) and is conducted by the University of Michigan. Further information can be found at http://hrsonline.isr.umich.edu/index.php.
Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Additional ADNI support comes from the NIH grants P30AG010129, K01AG030514, and U24AG21886. Funding for whole genome sequencing in ADNI participants was provided by the Alzheimer's Association and the Brin Wojcicki Foundation. For IMAS, we acknowledge the support of the Indiana CTSI (NIH grants U54 RR025761, RR027710-01, and RR020128). AddNeuroMed was funded through the EU FP6 Programme. Data management and the specific analyses reported here were supported by NIH R01AG19771, P30AG10133, R01LM011360, and R00LM011384, as well as NSF IIS-1117335.
Footnotes
AUTHOR CONTRIBUTIONS
All authors contributed substantively to this report. VKR, LS, and AJS were involved in study conception and design. HS, IK, PM, MT, BV, and SL were involved in coordination and data collection and processing for AddNeuroMed. PSA, RCP, CRJ, LMS, JQT, MWW, RCG, AWT, and AJS were involved in coordination and data collection and processing for ADNI. BCM, MRF, TMF, SG, and AJS were involved in coordination and data collection and processing for IMAS. PLDJ, LY, and DAB were involved in coordination and data collection and processing for ROS and MAP. VKR, KN, LS, SLR, SK, TMF, SG, and AJS were involved in data organization and planning and execution of statistical analyses. VKR and AJS drafted the report and prepared all figures and tables. All authors were involved in reviewing and editing of the report.
POTENTIAL CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer's disease. Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 2.Sweatt JD. Mechanisms of Memory. 2nd Edition Academic Press; 2009. [Google Scholar]
- 3.Hurd MD, Martorell P, Delavande A, Mullen KJ, Langa KM. Monetary costs of dementia in the United States. The New England journal of medicine. 2013;368(14):1326–1334. doi: 10.1056/NEJMsa1204629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stranahan AM, Mattson MP. Recruiting adaptive cellular stress responses for successful brain ageing. Nat Rev Neurosci. 2012;13(3):209–216. doi: 10.1038/nrn3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reitz C, Brayne C, Mayeux R. Epidemiology of Alzheimer disease. Nature reviews Neurology. 2011;7(3):137–152. doi: 10.1038/nrneurol.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papassotiropoulos A, de Quervain DJ. Genetics of human episodic memory: dealing with complexity. Trends Cogn Sci. 2011;15(9):381–387. doi: 10.1016/j.tics.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Harris SE, Deary IJ. The genetics of cognitive ability and cognitive ageing in healthy older people. Trends Cogn Sci. 2011;15(9):388–394. doi: 10.1016/j.tics.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Gandhi S, Wood NW. Genome-wide association studies: the key to unlocking neurodegeneration? Nat Neurosci. 2010;13(7):789–794. doi: 10.1038/nn.2584. [DOI] [PubMed] [Google Scholar]
- 9.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends in genetics : TIG. 2012;28(7):323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461(7261):218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 11.Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, et al. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8(2):183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juster FT, Suzman R. An Overview of the Health and Retirement Study. The Journal of Human Resources. 1995;30:S7. [Google Scholar]
- 13.Lovestone S, Francis P, Kloszewska I, Mecocci P, Simmons A, Soininen H, et al. AddNeuroMed--the European collaboration for the discovery of novel biomarkers for Alzheimer's disease. Annals of the New York Academy of Sciences. 2009;1180:36–46. doi: 10.1111/j.1749-6632.2009.05064.x. [DOI] [PubMed] [Google Scholar]
- 14.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9(5):e111–194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18(7):781–787. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the rush Memory and Aging Project. Curr Alzheimer Res. 2012;9(6):646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Curr Alzheimer Res. 2012;9(6):628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.St. Clair P, Bugliari D, Campbell N, Chien S, Hayden O, Hurd MD, et al. RAND HRS Data Documentation, Version L. RAND Center for the Study of Aging. 2011 Accessed via http://www.rand.org/content/dam/rand/www/external/labor/aging/dataprod/randhrsL.pdf.
- 19.Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol. 2012;8(12):e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace RB, Herzog AR, Weir DR, Ofstedal MB, Langa KM, Fisher GG, et al. Documentation of cognitive functioning measures in the Health and Retirement Study. HRS Documentation Report DR-006. 2005 Accessed via http://hrsonline.isr.umich.edu/sitedocs/userg/dr-006.pdf.
- 21.Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC, et al. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6(4):347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, et al. Update on the biomarker core of the Alzheimer's Disease Neuroimaging Initiative subjects. Alzheimers Dement. 2010;6(3):230–238. doi: 10.1016/j.jalz.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Swaminathan S, Inlow M, Risacher SL, Nho K, Shen L, et al. Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PLoS One. 2013;8(7):e70269. doi: 10.1371/journal.pone.0070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weir DR, Faul JD, Kardia SL, Smith JD, Doheny KF, Romm J, et al. Quality control report for genotypic data. Health and Retirement Study. 2012 accessed via http://hrsonline.isr.umich.edu/sitedocs/genetics/HRS_QC_REPORT_MAR2012.pdf.
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28(24):3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19(3):351–357. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69(3):560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruchaga C, Kauwe JSK, Harari O, Jin SC, Cai YF, Karch CM, et al. GWAS of Cerebrospinal Fluid Tau Levels Identifies Risk Variants for Alzheimer's Disease. Neuron. 2013;78(2):256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee S, Kim S, Gibbons LE, Nho K, Risacher SL, Glymour MM, et al. Genetic architecture of resilience of executive functioning. Brain Imaging Behav. 2012;6(4):621–633. doi: 10.1007/s11682-012-9184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li MX, Kwan JS, Sham PC. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012;91(3):478–488. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam D, Kim J, Kim S-Y, Kim S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Research. 2010;38(suppl 2):W749–W754. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 40.Ghezzi D, Saada A, D'Adamo P, Fernandez-Vizarra E, Gasparini P, Tiranti V, et al. FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am J Hum Genet. 2008;83(3):415–423. doi: 10.1016/j.ajhg.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramanathan A, Schreiber SL. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci U S A. 2009;106(52):22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierce SB, Gersak K, Michaelson-Cohen R, Walsh T, Lee MK, Malach D, et al. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am J Hum Genet. 2013;92(4):614–620. doi: 10.1016/j.ajhg.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26(19):2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44(10):1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams RW, Mulligan MK. Genetic and molecular network analysis of behavior. International review of neurobiology. 2012;104:135–157. doi: 10.1016/B978-0-12-398323-7.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84(4):445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39(12):1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 49.Yeung KT, Das S, Zhang J, Lomniczi A, Ojeda SR, Xu CF, et al. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Molecular and cellular biology. 2011;31(11):2287–2298. doi: 10.1128/MCB.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beier CP, Kolbl M, Beier D, Woertgen C, Bogdahn U, Brawanski A. CD95/Fas mediates cognitive improvement after traumatic brain injury. Cell Res. 2007;17(8):732–734. doi: 10.1038/cr.2007.60. [DOI] [PubMed] [Google Scholar]
- 51.Su JH, Anderson AJ, Cribbs DH, Tu C, Tong L, Kesslack P, et al. Fas and Fas ligand are associated with neuritic degeneration in the AD brain and participate in beta-amyloid-induced neuronal death. Neurobiology of disease. 2003;12(3):182–193. doi: 10.1016/s0969-9961(02)00019-0. [DOI] [PubMed] [Google Scholar]
- 52.Jayanthi S, Deng X, Ladenheim B, McCoy MT, Cluster A, Cai NS, et al. Calcineurin/NFAT-induced up-regulation of the Fas ligand/Fas death pathway is involved in methamphetamine-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2005;102(3):868–873. doi: 10.1073/pnas.0404990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu WT, Chen-Plotkin A, Grossman M, Arnold SE, Clark CM, Shaw LM, et al. Novel CSF biomarkers for frontotemporal lobar degenerations. Neurology. 2010;75(23):2079–2086. doi: 10.1212/WNL.0b013e318200d78d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papassotiropoulos A, Stefanova E, Vogler C, Gschwind L, Ackermann S, Spalek K, et al. A genome-wide survey and functional brain imaging study identify CTNNBL1 as a memory-related gene. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 56.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis A, Tomlinson I. Cancer. The utility of mouse models in post-GWAS research. Science. 2012;338(6112):1301–1302. doi: 10.1126/science.1231733. [DOI] [PubMed] [Google Scholar]
- 58.Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. The New England journal of medicine. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 59.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2(2):345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 61.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frankland PW, Bontempi B. The organization of recent and remote memories. Nat Rev Neurosci. 2005;6(2):119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- 63.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314(5798):475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 64.Thomson PA, Parla JS, McRae AF, Kramer M, Ramakrishnan K, Yao J, et al. 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: analysis for association with psychiatric disorder and cognitive traits. Mol Psychiatry. 2014;19(6):668–675. doi: 10.1038/mp.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nature reviews Neurology. 2009;5(6):311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- 66.Huentelman MJ, Papassotiropoulos A, Craig DW, Hoerndli FJ, Pearson JV, Huynh KD, et al. Calmodulin-binding transcription activator 1 (CAMTA1) alleles predispose human episodic memory performance. Hum Mol Genet. 2007;16(12):1469–1477. doi: 10.1093/hmg/ddm097. [DOI] [PubMed] [Google Scholar]
- 67.Schiepers OJ, Harris SE, Gow AJ, Pattie A, Brett CE, Starr JM, et al. APOE E4 status predicts age-related cognitive decline in the ninth decade: longitudinal follow-up of the Lothian Birth Cohort 1921. Mol Psychiatry. 2012;17(3):315–324. doi: 10.1038/mp.2010.137. [DOI] [PubMed] [Google Scholar]
- 68.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature reviews Neurology. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, et al. Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. The New England journal of medicine. 2009;361(3):255–263. doi: 10.1056/NEJMoa0809437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramanan VK, Saykin AJ. Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer's disease, Parkinson's disease, and related disorders. American journal of neurodegenerative disease. 2013;2(3):145–175. [PMC free article] [PubMed] [Google Scholar]
- 71.Papassotiropoulos A, Gerhards C, Heck A, Ackermann S, Aerni A, Schicktanz N, et al. Human genome-guided identification of memory-modulating drugs. Proc Natl Acad Sci U S A. 2013;110(46):E4369–4374. doi: 10.1073/pnas.1314478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer's disease. PLoS One. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maiese K, Chong ZZ, Shang YC, Wang S. mTOR: on target for novel therapeutic strategies in the nervous system. Trends in molecular medicine. 2013;19(1):51–60. doi: 10.1016/j.molmed.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, Chiang HC, Wu W, Liang B, Xie Z, Yao X, et al. Epidermal growth factor receptor is a preferred target for treating amyloid-beta-induced memory loss. Proc Natl Acad Sci U S A. 2012;109(41):16743–16748. doi: 10.1073/pnas.1208011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramanan VK, Kim S, Holohan K, Shen L, Nho K, Risacher SL, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6(4):634–648. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lucin KM, Wyss-Coray T. Immune activation in brain aging and neurodegeneration: too much or too little? Neuron. 2009;64(1):110–122. doi: 10.1016/j.neuron.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martins IJ, Hone E, Foster JK, Sunram-Lea SI, Gnjec A, Fuller SJ, et al. Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer's disease and cardiovascular disease. Mol Psychiatry. 2006;11(8):721–736. doi: 10.1038/sj.mp.4001854. [DOI] [PubMed] [Google Scholar]
- 78.Bras J, Guerreiro R, Hardy J. Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nature Reviews Neuroscience. 2012;13(7):453–464. doi: 10.1038/nrn3271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.