Abstract
Skeletal muscles are the most abundant tissues in the human body. They are composed of a heterogeneous collection of muscle fibers that perform various functions. Skeletal muscle troponin (sTn) regulates skeletal muscle contraction and relaxation. sTn consists of 3 subunits, troponin I (TnI), troponin T (TnT), and troponin C (TnC). TnI inhibits the actomyosin Mg2+-ATPase, TnC binds Ca2+, and TnT is the tropomyosin (Tm)-binding subunit. The cardiac and skeletal isoforms of Tn share many similarities but the roles of modifications of Tn in the two muscles may differ. The modifications of cardiac Tn are known to alter muscle contractility and have been well-characterized. However, the modification status of sTn remains unclear. Here, we have employed top-down mass spectrometry (MS) to decipher the modifications of human sTnT and sTnI. We have extensively characterized sTnT and sTnI proteoforms, including alternatively spliced isoforms and post-translationally modified forms, found in human skeletal muscle with high mass accuracy and comprehensive sequence coverage. Moreover, we have localized the phosphorylation site of slow sTnT isoform III to Ser1 by tandem MS with electron capture dissociation. This is the first study to comprehensively characterize human sTn and also the first to identify the basal phosphorylation site for human sTnT by top-down MS.
Keywords: Muscle contraction, Myofilament, Proteomics, Fourier transform mass spectrometry, Electron capture dissociation
Introduction
Skeletal muscles are highly abundant tissues present in mammals, which are known to be heterogeneous in nature and are capable of performing a wide variety of activities (Bottinelli and Reggiani, 2000; Ohlendieck, 2011; Schiaffino and Reggiani, 2011). Structural and functional diversity are essential properties of skeletal muscles that allow them to respond to a broad range of functional demands in the human body. Generally, human skeletal muscle can be classified as either fast or slow twitch, according to the maximum speed at which fibers shorten, myofibrillar protein isoform expression, and energy supply systems (Moss et al., 1995; Schiaffino and Reggiani, 1996).
Muscle fibers are composed of sarcomeres, which are the primary functional units of muscle and contain the contractile proteins myosin and actin as the primary components of the thick and thin filaments, respectively (Moss et al., 1995; Schiaffino and Reggiani, 1996). Muscular contractions are driven by interactions between the myosin thick filament and the actin thin filament (Moss et al., 1995; Schiaffino and Reggiani, 1996). The thin filament also contains the functionally important regulatory protein complex, troponin (Tn) (McComas, 1996; Scott et al., 2001). Tn regulates skeletal and cardiac muscle contraction and consists of three subunits, troponin I (TnI), troponin C (TnC), and troponin T (TnT) (Farah et al., 1994; Scott et al., 2001; Takeda et al., 2003). TnI inhibits the actomyosin Mg2+-ATPase, TnC abolishes TnI-mediated inhibition of actin-myosin cross-bridge formation upon binding to Ca2+, and TnT is the tropomyosin (Tm)-binding subunit. The Tn complex, along with Tm, is responsible for Ca2+-dependent regulation of myofilaments (Farah et al., 1994; Gahlmann et al., 1987; Leavis and Gergely, 1984). When the muscle fiber receives a stimulus in the form of an action potential, Ca2+ released from the sarcoplasmic reticulum is bound by TnC inducing a change in the Tn-Tm conformation that exposes myosin binding sites on the actin filament, thereby allowing for muscular contraction (Scott et al., 2001).
Not surprisingly, due to their critical role in muscle contraction and relaxation, both cardiac Tn (cTn) and skeletal Tn (sTn) have been implicated in a number of muscle-related diseases (Berchtold et al., 2000). cTnI and cTnT are released into the circulation following myocardial injury and, thus, these proteins are now the gold standard serum biomarkers for acute myocardial infarction owing to their cardiac specific sequences (Babuin et al., 2006; Colantonio et al., 2002; Guy et al., 2013). Differential detection of sTnI isoforms in serum has been proposed as a biomarker for muscle injury (Simpson et al., 2002a; Simpson et al., 2002b). cTn subunits are known to undergo extensive post-translational modifications (PTMs) such as phosphorylation proteolytic degradation that have been shown to modulate the regulation of contraction, and both spontaneous and heritable mutations of these subunits have been implicated in the pathogenesis of heart disease (Labugger et al., 2000; Dong et al., 2012; Gregorich et al., 2014; Gregorich and Ge, 2014; Marston and de Tombe, 2008; Marston and Redwood, 2003; Solaro et al., 2008; Wei and Jin, 2011). Most notably, the phosphorylation of cTnI is known to modulate the contractility of cardiac muscles (Dong et al., 2012; Layland et al., 2005; Solaro et al., 2008; Sumandea et al., 2004; Zhang et al., 2011a) and has recently been identified as a candidate biomarker for heart failure (Zhang et al., 2011a). Given the high degree of sequence homology between human Tn in cardiac and skeletal muscles, it is reasonable to hypothesize that modifications of sTn play an important role in the regulation of skeletal muscle contractility. However, little is known about the modification status of sTn. Thus, in this study, we employed top-down mass spectrometry (MS) to identify potential PTM’s in human sTnT and sTnI.
Top-down mass spectrometry (MS) (Ayaz-Guner et al., 2009; Ge et al., 2002; Ge et al., 2009; Han et al., 2006; Jebanathirajah et al., 2005; Kelleher et al., 1999; Kuhn et al., 2009; Ryan et al., 2010; Sze et al., 2002; Zabrouskov et al., 2008; Zhang et al., 2011a; Zhang et al., 2011b; Zhang et al., 2010; Gregorich and Ge, 2014) is a preferred technology for assessing proteoforms, a unified term to “designate all of the different molecular forms in which the protein product of a single gene can be found, including changes due to genetic variations, alternatively spliced RNA transcripts and PTMs” (Smith et al., 2013). Unlike the bottom-up MS method, the top-down approach allows for the detection of all proteoforms without a priori knowledge due to the fact that intact proteins, rather than peptides, are analyzed (Chait, 2006; Siuti and Kelleher, 2007; Zhang and Ge, 2011; Zhang et al., 2011b; Gregorich and Ge, 2014). A specific proteoform of interest can then be directly isolated in the mass spectrometer and subsequently fragmented by tandem MS (MS/MS) techniques, such as collisionally activated dissociation (CAD) and electron capture dissociation (ECD), for highly reliable mapping of modification sites and sequence identification (Ayaz-Guner et al., 2009; Ge et al., 2009; Han et al., 2006; Kelleher et al., 1999; Kuhn et al., 2009; Ryan et al., 2010; Sze et al., 2002; Zhang et al., 2011a; Zhang et al., 2011b; Zhang et al., 2010). ECD is particularly suitable for the localization of labile PTMs since they are well-preserved during the ECD fragmentation process (Ayaz-Guner et al., 2009; Cooper et al., 2005; Ge et al., 2009; Shi et al., 2001; Zabrouskov et al., 2008; Zhang et al., 2011a; Zhang et al., 2011b; Zhang et al., 2010; Zubarev et al., 2000).
Herein, we have utilized top-down MS to comprehensively characterize the complexity of Tn subunit proteoforms in human skeletal muscle. We have comprehensively characterized the sequences of the most abundant proteoforms of sTnT and sTnI, including alternatively spliced isoforms and post-translationally modified forms, with high mass accuracy. Furthermore, the phosphorylation site of sTnT(III) has been mapped to Ser1 (accounting for removal of the N-terminal methionine) by MS/MS. This is the first study to comprehensively characterize human sTnI and sTnT proteoforms by top-down MS.
Experimental Procedures
Chemicals and Reagents
All chemicals and reagents were purchased from Sigma Chemical Co. (St Louis, MO) unless noted otherwise. All solutions were prepared in Milli-Q water (Millipore Corporation, Billerica, MA).
Preparation of Human Skeletal Muscle Troponin
The affinity purified human skeletal muscle troponin T (sTnT) and troponin I (sTnI) were purchased from Hytest Ltd. (Turku, Finland). Approximately 0.1–0.2 mg of human sTnT and sTnI were used per preparation. The sTnT samples were dissolved in 30% methanol in water, and the sTnI samples were dissolved in 50% methanol in water. 1% of acetic acid was added to the sample prior to high-resolution top-down MS analysis.
Percutaneous skeletal muscle biopsies were taken from the tibialis anterior (predominantly slow) and the vastus lateralis (mixed) from two young healthy women. All procedures have been approved by the ethical committee of the Uppsala University and carried out according to the guidelines of the Declaration of Helsinki. The tissues were then snap frozen in liquid nitrogen, and stored in -80 °C freezer before tissue homogenization and myofilament protein extraction. Approximately 5 mg of human skeletal muscle tissues were used per purification of sTn using liquid chromatography (LC).
Tissues were homogenized in HEPES extraction buffer (HEPES, 25 mM, pH 7.5, NaF, 50 mM, Na3VO4, 0.25 mM, PMSF (in isopropanol), 0.25 mM, EDTA, 2.5 mM, and half tablet of commercial protease inhibitor cocktail) using Teflon homogenizer (1.5 mL tube rounded tip, Cienceware, Pequannock, NJ, USA). The homogenate was centrifuged at 16.1×1000 rcf in 4 °C for 40 min (Centrifuge 5415R, Eppendorf, Hamburg, Germany), and supernatant was removed. The pellet was then resuspended and homogenized in trifluoroacetic acid (TFA) extraction buffer (1% TFA, 1 mM TCEP) using Teflon homogenizer similarly as described previously (Peng et al., 2014). The homogenate was then centrifuged again at 16.1 × 1000 rcf in 4 °C for 40 min to collect the supernatant which contains mainly the myofilament proteins for further analysis. The myofilament proteins were separated by a 2D-nano LC system (Eksigent, Dublin, CA) using a home-made polymeric reversed phase spherical (PLRP) column (200 mm ×500 μm, 10 μm, 1000Å, PLRP-S particles purchased from Varian, Lake Forest, CA), which is coupled with a linear ion trap mass spectrometer (LTQ, Thermo Scientific Inc., Bremen, Germany). After separation, about 5% of the LC eluant was ionized by electrospray ionization (ESI) in online LC/MS mode. Meanwhile, the remainder went through the split and was fraction-collected for further offline high-resolution top-down MS analysis.
Top-Down Mass Spectrometry
Fractions containing sTnT and sTnI from human skeletal muscle biopsies and affinity purified Hytest sTn samples were analyzed using a 7T linear trap/Fourier transform ion cyclotron resonance (FTICR) (LTQ FT Ultra) hybrid mass spectrometer (Thermo Scientific Inc., Bremen, Germany) equipped with an automated chip-based nano electrospray ionization (ESI) source (Triversa NanoMate; Advion BioSciences, Ithaca, NY) as described previously (Ayaz-Guner et al., 2009; Dong et al., 2012; Ge et al., 2009; Horn et al., 2000; Peng et al., 2012; Zhang et al., 2011a; Zhang et al., 2011b). The spray voltage was 1.2–1.6 kV versus the inlet of the mass spectrometer, resulting in a flow rate of 50–200 nL/min. Ion transmission into the linear trap and, subsequently, into the FTICR cell, was automatically optimized for the maximal ion signal. The number of accumulated ions for the full scan linear trap (IT), FTICR cell (FT), MSn FTICR cell, and ECD were 3×104, 8×106, 8×106, and 8×106, respectively. The resolving power of the FTICR mass analyzer was typically set at 200,000. For CAD and ECD fragmentation, individual charge states of protein molecular ions were first isolated and then dissociated using 15–20% of normalized collision energy for CAD or 2.8–3.5% electron energy for ECD with a 70–75 ms duration and no delay. Typically, 2000–5000 transient scans were averaged to ensure high quality ECD spectra. All MS and MS/MS data were processed by an in-house developed software, MASH Suite (version 1.2), using the THRASH (Horn et al., 2000) algorithm with S/N threshold of 3 and a fit factor of 60% and manually validated. The fragments in MS/MS spectra were assigned on the basis of the protein sequence of human sTnT and human sTnI obtained from Swiss-Prot protein knowledgebase (Unit-ProtKB/Swiss-Prot). Allowance was made for possible PTMs such as acetylation of the N-terminus, phosphorylation, and two amino acid polymorphisms using 1 Da tolerance for the precursor and fragment ions, respectively. The assigned ions were manually validated to ensure the accuracy of the assignments. For fragment ions containing possible phosphorylation sites, the masses of fragment ions were manually validated for 80 Da mass shifts to confirm or exclude the existence of phosphorylation. All the reported molecular weights (MWs) are the most abundant MWs.
Results
Sequence characterization of sTnT proteoforms
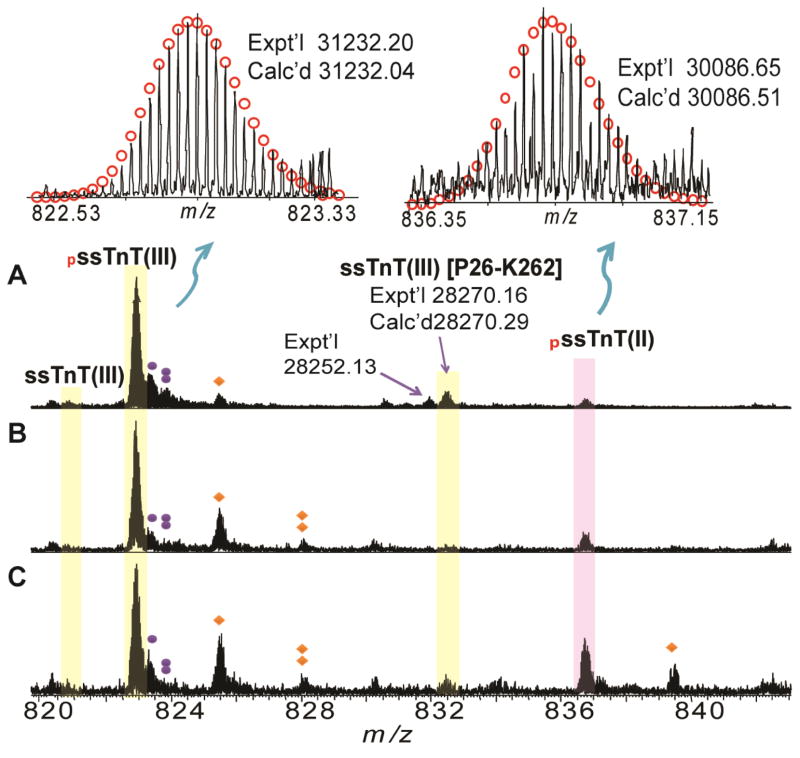
The FTMS spectrum of affinity purified sTnT from Hytest revealed one major peak with a molecular weight (MW) of 31232.20 as well as two minor peaks with MWs of 30086.65 and 31152.24, respectively (Figure 1A, Table 1). Surprisingly, none of the experimentally determined MWs matched with the MWs of human sTnT calculated based on sequences from the Uniprot database. However, the experimental MW of 31152.24 matched with the sequence of human slow sTnT isoform III (ssTnT(III)) reported in the Swiss-Prot database (P13805-3, TNNT1_HUMAN) after considering removal of the N-terminal methionine and acetylation at the new N-terminus (Calc’d: 31152.07 Da). The peaks with MWs of 31152.24 and 31232.20 differed by 79.96 Da, suggesting that the most abundant peak with MW of 31232.20, is likely mono-phosphorylated ssTnT(III) (Figure 1). Although the un-phosphorylated peak was not observed, the peak with MW 30086.65 may be mono-phosphorylated ssTnT isoform II as the MW is close to that calculated based on the sequence of ssTnT isoform II from the database, assuming removal of the N-terminal methionine, acetylation at the new N-terminus, and the addition of a phosphate group (Calc’d: 30086.51 Da). In addition to the major peaks, several minor peaks were also detected. The minor peak with a MW of 28270.16 matches well with the proteolytic degradation product of ssTnT(III) [P26-K262] (Calc’d 28270.29) (Figure 1). Minor peaks corresponding to oxidized forms and those associated with non-covalent phosphate adducts were also detected.
Figure 1. High-resolution FTICR-MS analysis of human sTnT.
A. Affinity purified sTnT purchased from Hytest; B. LC purified sTnT from human vastus lateralis muscle; C. LC purified sTnT from human tibialis anterior muscle. Representative FTICR-MS spectrum of human ssTnT, M38+ for ssTnT(III), M36+ for ssTnT(II), respectively. Red circles represent theoretical isotopic abundance distribution of the isotopomer peaks. Solid circle(s) represents oxidation species. Diamonds represent the phosphate adducts. Calc’d, calculated most abundant mass; Expt’l, experimental most abundant mass. pssTnT (III), and pssTnT (II) represents mono-phosphorylated ssTnT isoform III (P13805-3), and isoform II (P13805-2), respectively.
Table 1. Human sTnI and sTnT proteoforms identified in Hytest Samples by high-resolution FTICR-MS.
Molecular weight (MW) of fsTnI was calculated based on DNA-predicted sequence of human fsTnI (UnitprotKB/Swiss-Prot P48788 (TNNI2_HUMAN)) with the removal of methionine and acetylation of the first amino acid at the N-terminus (+42 Da). The MW of ssTnI was calculated based on DNA-predicted sequence of human ssTnI (UnitprotKB/Swiss-Prot P19237 (TNNI1_HUMAN)) with removal of methionine at the N-terminus. The MW of ssTnT was calculated based on DNA-predicted sequence of human ssTnT (UnitprotKB/Swiss-Prot P13805-3) with the removal of N-terminal Met and acetylation at the new N-terminus.
| Identification | Experimental MW | Calculated MW | Error (ppm) |
|---|---|---|---|
| Human sTn Isoforms | |||
| ssTnT(III) | 31152.24 | 31152.07 | 5.46 |
| fsTnI | 21249.03 | 21248.90 | 6.12 |
| ssTnI | 21560.69 | 21560.55 | 6.49 |
| Phosphorylation Products | |||
| pssTnT(II) | 30086.65 | 30086.51 | 4.65 |
| pssTnT(III) | 31232.20 | 31232.04 | 5.12 |
| Degradation Product | |||
| ssTnT(III) [P26-K262] | 28270.16 | 28270.29 | 4.60 |
In the LC fraction that contains ssTnT, a major peak with MW of 31232.20 [mono-phosphorylated ssTnT (III)] and a minor peak of MW 30086.65 [mono-phosphorylated ssTnT (II)] was also observed in human skeletal muscle biopsies (both vastus lateralis and tibialis anterior) (Figure 1B–C, Table 2–3). The un-phosphorylated forms of these proteins were not detected suggesting that they are relatively low in abundance in this sample. The degradation product of ssTnT(III) [P26-K262] was also detected in the skeletal muscle biopsies although the intensity of this peak was greatly reduced in this sample compared to the sTnT from Hytest.
Table 2.
Human sTnI and sTnT proteoforms identified in human vastus lateralis muscle by high-resolution FTICR-MS.
| Identification | Experimental MW | Calculated MW | Error (ppm) |
|---|---|---|---|
| Human sTn Isoforms | |||
| fsTnT(III) | 30874.59 | 30874.11 | 15.5 |
| fsTnT(VI) | 30260.24 | 30259.85 | 12.9 |
| fsTnT(VII) | 29645.95 | 29645.55 | 13.5 |
| fsTnI | 21249.19 | 21248.90 | 13.6 |
| ssTnI | 21560.68 | 21560.55 | 6.03 |
| Phosphorylation Products | |||
| pfsTnT(III) | 30954.49 | 30954.08 | 13.2 |
| pfsTnT(VI) | 30340.21 | 30339.82 | 12.9 |
| pfsTnT(VII) | 29725.90 | 29725.52 | 12.8 |
| pssTnT(II) | 30086.71 | 30086.51 | 6.65 |
| pssTnT(III) | 31232.22 | 31232.04 | 5.76 |
| pssTnI | 21640.62 | 21640.51 | 5.08 |
| Degradation Product | |||
| ssTnT(III) [P26-K262] | 28270.16 | 28270.29 | 4.60 |
Table 3.
Human sTnI and sTnT proteoforms identified in human tibialis anterior muscle by high-resolution FTICR-MS.
| Identification | Experimental MW | Calculated MW | Error (ppm) |
|---|---|---|---|
| Human sTn Isoforms | |||
| fsTnT(VII) | 29645.95 | 29645.55 | 13.5 |
| fsTnI | 21249.19 | 21248.90 | 13.6 |
| ssTnI | 21560.68 | 21560.55 | 6.03 |
| Phosphorylation Products | |||
| pfsTnT(III) | 30954.49 | 30954.08 | 13.2 |
| pfsTnT(VI) | 30340.21 | 30339.82 | 12.9 |
| pfsTnT(VII) | 29725.90 | 29725.52 | 12.8 |
| pssTnT(II) | 30086.71 | 30086.51 | 6.65 |
| pssTnT(III) | 31232.22 | 31232.04 | 5.76 |
| pssTnI | 21640.62 | 21640.51 | 5.08 |
| Degradation Product | |||
| ssTnT(III) [P26-K262] | 28270.16 | 28270.29 | 4.60 |
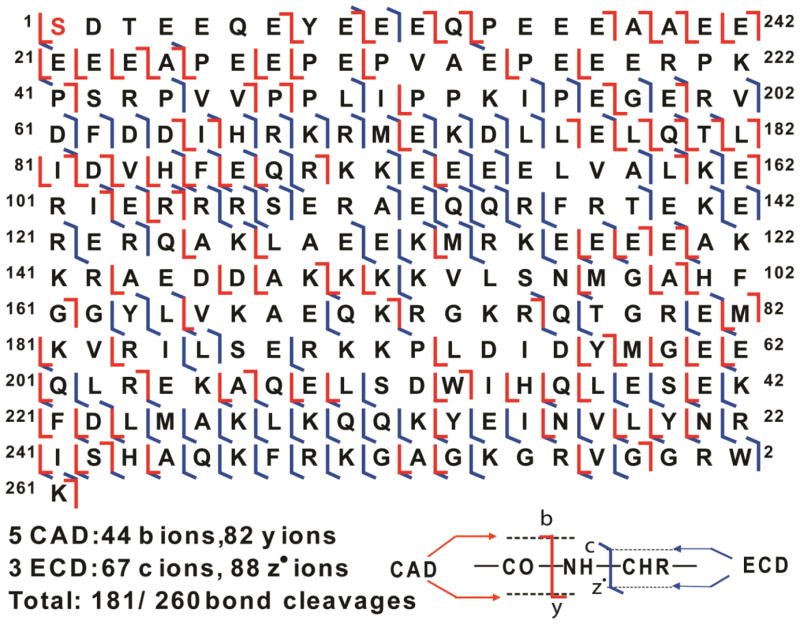
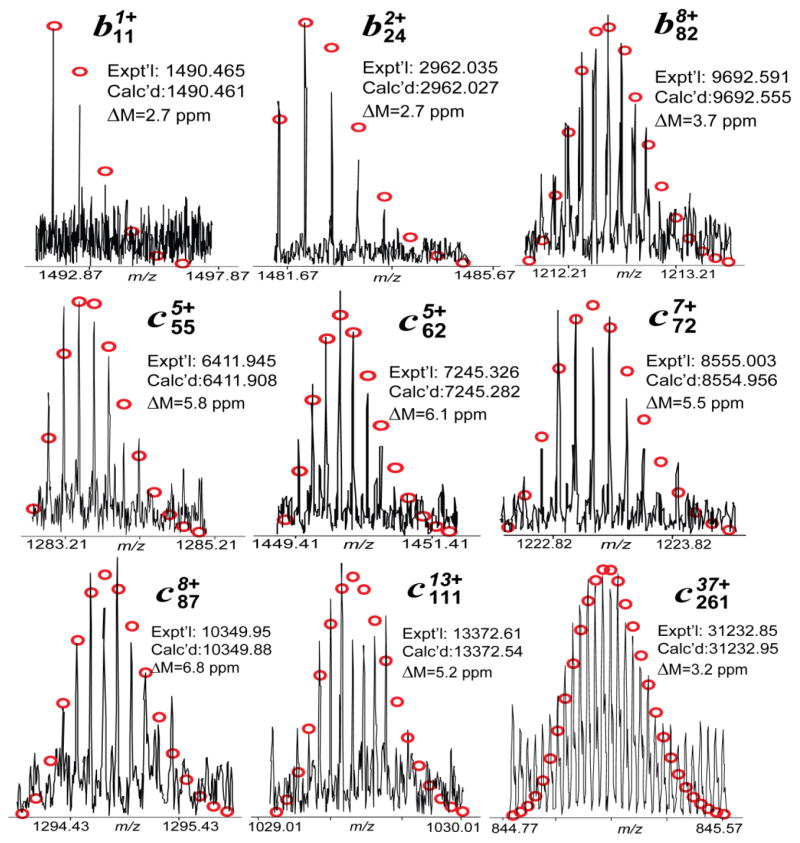
To confirm the identities of the peaks assigned based on the high accuracy MWs and to identify/localize the phosphorylation site of ssTnT(III), a single charge state of mono-phosphorylated ssTnT(III) was isolated in the mass spectrometer (a “gas-phase” purification) and subsequently dissociated using either ECD or CAD. A total of eight sets of MS/MS data including five CAD and three ECD spectra were combined, providing near-complete sequence coverage (Figure 2). Overall, the total number of ions generated for mono-phosphorylated ssTnT(III) was 281, which corresponds to 44 b ions, 82 y ions, 67 c ions, and 88 z· ions assigned to the DNA-predicted sequence of human ssTnT (UnitprotKB/Swiss-Prot P13805-3) considering removal of N-terminal methionine, acetylation at the new N-terminus and the addition of a phosphate group (Figure 2). For the fragmentation map generated by MS/MS experiments, no c ions were detected before adding any modifications, suggesting that the phosphorylation site might be located within the first few amino acids at the N-terminus of ssTnT(III). Based on the sequence, there are three residues in this stretch of amino acids that could potentially be phosphorylated, namely, Ser1, Thr3, and Tyr8. However, the probability of serine being the phosphorylation site is much higher (around 90%) than threonine or tyrosine (around 10% for Thr and less than 0.05% for Tyr) (Mann et al., 2002; Zhang et al., 2011b). In addition, the mono-phosphorylated site for mouse cTnT is also Ser1 (Zhang et al., 2011b). Thus, Ser1 was assigned as the more probable phosphorylation site for ssTnT(III) (with removal of methionine at the N-terminus) by MS/MS experiments. Representative MS/MS product ions of mono-phosphorylated sTnT(III) are shown in Figure 3. Thus, we have confirmed that the most abundant peak, with MW 31232.20, is the mono-phosphorylated form of sTnT(III), and have mapped the site of basal phosphorylation to Ser1. Based on this information, the peak with MW of 31152.24 was assigned as the unphosphorylated ssTnT(III). As mention above, the peak with experimental MW of 30086.65 matches the calculated MW of ssTnT(II) after accounting for removal of the N-terminal methionine, acetylation at the new N-terminus, and the addition of a phosphate group (Cal’d: 30086.78 Da); however, the intensity of this peak was too low to perform MS/MS. It is well known that cTnT and all the isoforms of sTnT sequences were highly conserved as shown in the sequences alignment (Supplementary Figure 1). Consistently, two out of the three ssTnT isoforms (II and III) reported in the database were observed in both commercially available sTnT sample and the ssTnT fraction from human skeletal muscle biopsies. The isoform I of human ssTnT (P13805), which has been chosen as the “canonical” form for ssTnT, was not detected in any of the samples.
Figure 2. MS/MS characterization of the mono-phosphorylated sTnT(III).
Fragment assignments were made to the DNA-predicted sequence of human ssTnT (UnitprotKB/Swiss-Prot P13805-3) with the removal of N-terminal Met, acetylation and phosphorylation at the new N-terminus. A total of eight sets of MS/MS data including five CAD and three ECD spectra were analyzed, and a total of 181 out of 260 bond cleavages are shown. Based on the fragmentation map, the most abundant peak with a molecular mass of 31232.20 Da was confirmed to be the mono-phosphorylated peak of ssTnT isoform III (P13805-3) with the phosphorylation site at the N-terminus.
Figure 3. Representative MS/MS product ions of mono-phosphorylated human sTnT(III) for the identification of phosphorylation site in purified human sTnT sample.
Circles, the theoretical isotopic distribution of isotopic peaks. Calc’d, calculated most abundant molecular weight (MW). Expt’l, experimental most abundant MW.
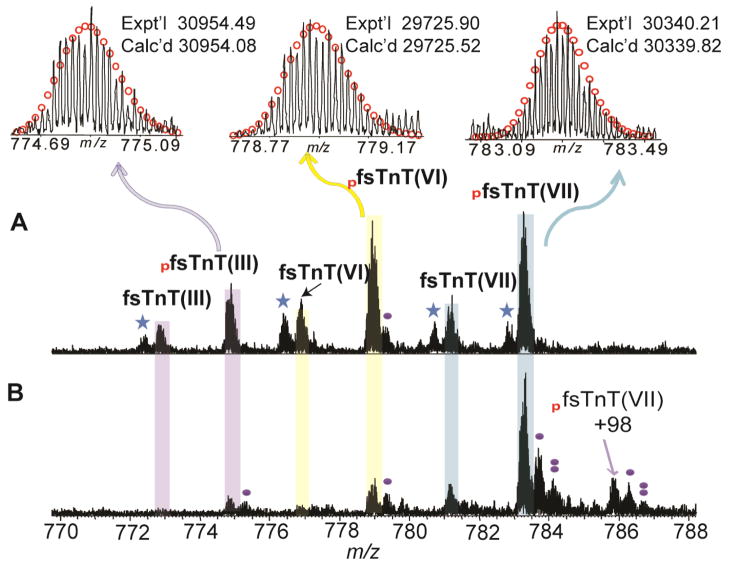
On the other hand, although none of the fast sTnT isoforms (fsTnT) were observed in the spectra from the sTnT sample from Hytest, we have identified fsTnT isoforms in human skeletal muscle biopsies using a top-down LC/MS method. The experimentally obtained MWs with high accuracy matched well with the calculated MWs from the database. The fsTnT proteoforms identified in human vastus lateralis include fsTnT(III), mono-phosphorylated fsTnT(III), fsTnT(VI), mono-phosphorylated fsTnT(VI), fsTnT(VII), mono-phosphorylated fsTnT(VII) (Figure 4A, Table 2). Peaks with MWs 30874.59, 30260.24, and 29645.95 match well with fsTnT(III) (UnitprotKB/Swiss-Prot P45378-3, Calc’d: 30874.11), fsTnT(VI) (UnitprotKB/Swiss-Prot P45378-6, Calc’d: 30259.85), and fsTnT(VII) (UnitprotKB/Swiss-Prot P45378-7, Calc’d: 29645.55), respectively, with removal of N-terminal methionine and acetylation as observed in the vastus lateralis sample. In addition, the peaks with MWs 30954.49, 30340.21, and 29725.90 are 80 Da apart from fsTnT(III), fsTnT(VI), and fsTnT(VII) correspondingly and are thus assigned as the mono-phosphorylated species of these three isoforms. In comparison, fewer proteoforms were detected including mono-phosphorylated fsTnT(III), mono-phosphorylated fsTnT(VI), fsTnT(VII), and mono-phosphorylated fsTnT(VII) in tibialis anterior muscle (Figure 4B, Table 3). The lower abundance of fsTnT isoforms detected in the tibialis anterior muscle compared to the vastus lateralis muscle might due to the composition of muscle types. Vastus lateralis is mixed muscle whereas the tibialis anterior is predominantly slow-twitch muscle fibers.
Figure 4. High-resolution FTICR-MS analysis of fsTnT from human skeletal muscle biopsies.
(A) LC purified sTnT from human vastus lateralis muscle; (B) LC purified sTnT from human tibialis anterior muscle. Representative FTICR-MS spectra of human fsTnT, M40+ for fsTnT(III), M39+ for fsTnT(VI), M38+ for fsTnT(VII), respectively. Red circles represent theoretical isotopic abundance distribution of the isotopomer peaks. Solid circle(s) represents oxidation species. Star represents potential water loss (-18 Da). Calc’d, calculated most abundant mass; Expt’l, experimental most abundant mass. +98 represents the non-covalent phosphate adduct ((H3PO4, +98 Da). pfsTnT (III), pfsTnT (VI), and pfsTnT(VII) represents mono-phosphorylated fsTnT isoform III (P45378-3), isoform VI (P45378-6) and isoform, VII (P45378-7), respectively.
sTnI sequence determination
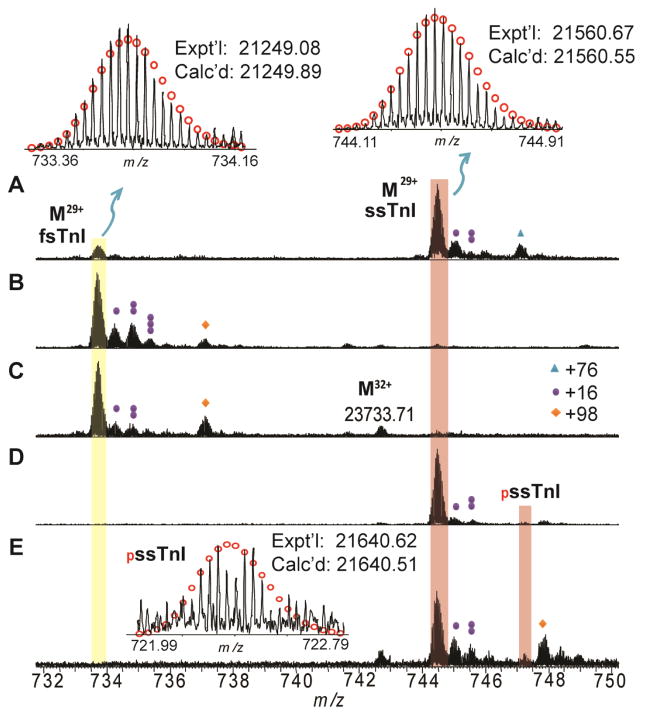
High-resolution ESI/FTMS analysis of human sTnI from Hytest revealed two major peaks corresponding to proteins with MWs of 21249.03 and 21560.69, respectively (Figure 5A). The MW of 21249.08 matches with the DNA-predicted sequence of human fsTnI (UnitprotKB/Swiss-Prot P48788 (TNNI2_HUMAN)) after removing the N-terminal methionine and adding acetylation at the new N-terminus (Calc’d: 21248.90 Da) (Supplementary Figure 2). Similarly, the MW of 21560.69 matches with the DNA-predicted sequence of human slow skeletal muscle TnI (ssTnI) (UnitprotKB/Swiss-Prot P19237 (TNNI1_HUMAN)) after accounting for removal of the N-terminal methionine (Calc’d: 21560.55 Da) (Supplementary Figure 2).
Figure 5. High-resolution FTICR-MS analysis of human sTnI.
A. Affinity purified sTnT purchased from Hytest. B. LC purified fsTnI from human vastus lateralis muscle. C. LC purified fsTnI from human tibialis anterior biopsy. D. LC purified ssTnI from human vastus lateralis muscle. E. LC purified ssTnI from human tibialis anterior muscle. Representative FTICR-MS spectrum (M29+) of human fsTnI and ssTnI were shown. Circles represent theoretical isotopic abundance distribution of the isotopomer peaks. Calc’d, calculated most abundant mass; Expt’l, experimental most abundant mass. MW of fsTnI was calculated based on DNA-predicted sequence of Human fsTnI (UnitprotKB/Swiss-Prot P48788 (TNNI2_HUMAN)) with removal of methionine and acetylation of the first amino acid at the N-terminus (+42 Da), and molecular weight of ssTnI was calculated based on DNA-predicted sequence of human ssTnI (UnitprotKB/Swiss-Prot P19237 (TNNI1_HUMAN)) with removal of methionine at the N-terminus. The peaks marked with one (+16 Da) two (+32Da) or three solid circle(s) (+48 Da) represent single, double or triple oxidation(s), respectively. Triangles (+76 Da), β-mercaptoethanol adduct (β-met), +98 represents non-covalent phosphate adduct (H3PO4, +98 Da).
In the skeletal muscle biopsies, fsTnI (Figure 5B–C) and ssTnI (Figure 5D–E) are well-separated using our newly developed LC method. The major peak of MW 21249.19 was detected in fsTnI and the major peak of MW 21560.68 was observed in ssTnI. The MWs of both fsTnI and ssTnI are consistent with those in Hytest sTnI sample. Nonetheless, the new LC method developed in this study allows separation of fsTnI and ssTnI, compared to the affinity purified Hytest sTnI sample composing a mixture of both fsTnI and ssTnI. In addition, a minor peak with MW 21640.62 is only observed in the human skeletal muscle biopsies, which matches well with mono-phosphorylated ssTnI with removal of N-terminal methionine and addition of a phosphate group (Figure 5D–E).
To confirm the sequence of the hypothetical proteoforms of sTnI, a single charge state of fsTnI or ssTnI was isolated and subjected to MS/MS (ECD and CAD). Before adding any modifications, barely any c ions matched the N-terminal sequence for both fsTnI and ssTnI, which indicates possible modifications at the N-terminus (Supplementary Figures 3 and 4). As shown in the ECD map, c ions started to show up after adding the modifications; one representative ECD experiment generated 51 c ions and 33 z· ions according to the modified sequence of human fsTnI (UnitprotKB/Swiss-Prot P48788 (TNNI2_HUMAN)) (Figure 6A), and the other ECD experiment generated 42 c ions and 42 z· ions according to the modified sequence of human ssTnI (UnitprotKB/Swiss-Prot P19237 (TNNI1_HUMAN)) (Figure 6B). Therefore, the tentative proteoform identifications listed above are confirmed—the peaks with experimental MWs 21249.03 and 21560.69 correspond to fsTnI and ssTnI, respectively.
Figure 6. Characterization of human fsTnI and ssTnI by ECD.
(A) The ECD map of fsTnI assigned based on DNA-predicted sequence of Human fsTnI (UnitprotKB/Swiss-Prot P48788 (TNNI2_HUMAN)) after removal of methione and acetylation at the N-terminus. (B) The ECD map of ssTnI assigned based on based on DNA-predicted sequence of human ssTnI (UnitprotKB/Swiss-Prot P19237 (TNNI1_HUMAN)) after removal of methione.
Interestingly, two peaks were observed to have mass difference of +76 from fsTnI and ssTnI respectively. However, a mass difference of +76 Da does not correspond to any of the common adducts (such as Na+ or K+) we see in high-resolution FTMS spectra. Based on the mass, one potential explanation is that these +76 peaks correspond to beta-mercaptoethanol adducts; however β-mercaptoethanol was not added during any of the steps prior to sample injection. Nevertheless, β-mercaptoethanol is a popular reducing agent used frequently in protein purification to reduce disulfide bonds. Therefore, it is reasonable to hypothesize that β-mercaptoethanol was used during the commercial purification of the sTn sample, which explains the presence of β-mercaptoethanol adducts of fsTnI and ssTnI in the spectra. Since we used TCEP instead of β-mercaptoethanol as a reducing agent in our new purification method, the β-mercaptoethanol adduct is absent in the MS spectra of sTnT and sTnI.
Discussion
Here we present the characterization of human sTnT and sTnI proteoforms by high-resolution top-down MS. It is known that there is a high level of similarity between human cardiac and skeletal isoforms of TnT and TnI as shown in the sequence alignment of TnT and TnI (Supplementary Figure 1 and 2). This agrees with previous findings that the sequences of human TnT and TnI are well conserved across different categories of muscle and moreover, the contraction mechanisms in skeletal muscle and cardiac muscle share many similarities (Jin et al., 2008). Given the fact that modifications of cTn, including PTMs and alternatively splicing, represent a key mechanism in regulating cardiac function, it is reasonable to hypothesize that sTn modifications may also influence muscle contractility; however, until now, the modification status of sTn was uncharacterized.
Top-down MS provides an overview of all possible proteoforms by providing a global view of the full complement of all PTMs and sequence variations (Peng et al., 2013; Roth et al., 2005; Siuti and Kelleher, 2007; Zhang and Ge, 2011; Zhang et al., 2011b). For the human ssTnT samples obtained from Hytest, ssTnT(III), mono-phosphorylated ssTnT(III), and mono-phosphorylated ssTnT(II) were identified, and the phosphorylation site of ssTnT(III) was mapped to Ser1 using ECD and CAD experiments. The sequence of human ssTnT was first reported by Gahlmann et al. in 1987 (Gahlmann et al., 1987). Sequence and hybridization analyses showed that two sTnT variants were produced from a single TnT gene via alternative splicing and that these variants harbor major differences in an important functional domain of the protein (Gahlmann et al., 1987). Variable expression of protein isoforms in muscles is known to be a main determinant of contractility and TnT is known to be regulated by alternative splicing (Jin et al., 2008; Pinto et al., 2012). Previous reports using skinned fibers suggest that varied expression of TnT isoforms leads to significant changes in the Ca2+ sensitivity of tension (Moss et al. 1995). A total of 64 variants can be generated owing to the alternative splicing of five exons at the 5′ end of one exon near the 3′ end of the gene (Bottinelli and Reggiani, 2000; Schiaffino and Reggiani, 2011). Nevertheless, only a limited number of splicing isoforms are detected in mammalian muscles (Schiaffino and Reggiani, 2011). Even though a total of 10 TnT isoforms (produced by alternative splicing) are reported in Supplementary Figure 1, including seven isoforms of fsTnT and three of ssTnT, only one major and one minor isoform of ssTnT were detected in the high-resolution FTMS data from Hytest affinity purified sample (Figure 1), and none of the fsTnT isoforms were observed. In contrast, not only the two ssTnT isoforms but also three fsTnT isoforms including isoform III, VI and VII were detected in the skeletal muscle biopsies (Figure 4). Interestly, a higher abundance of fsTnT isoforms is observed in vastus lateralis, composed of mixed-fast skeletal muscle in comparison with a lower abundance in tibialis anterior, predominantly composed of slow-twitch muscle fibers. The difference in fsTnT abundances is likely due to the difference in composition of the skeletal muscle types. Although more sTnT isoforms were observed from the skeletal muscle biopsies, not all the isoforms were seen. This is probably due to the diversity of skeletal muscle that not all alternatively spliced isoforms are expressed in all skeletal tissues. Previous studies have revealed the presence of four major variants of fsTnT in human limb skeletal muscle whereas two isoforms of ssTnT have been identified in the slow fibers of rat, rabbit, and human muscles (Bottinelli and Reggiani, 2000; Schiaffino and Reggiani, 2011).
Moreover, although the fsTnT isoforms exist in both fetal and adult tissues, fetal tissues generally have a higher level of expression of fsTnT isoforms than adult tissues (http://www.uniprot.org/uniprot/P45378). Briggs et al. identified an exon for human fsTnT at the 5′ alternatively spliced region that plays an important regulatory role during development (Briggs et al., 1994). This exon was only found in fetal skeletal muscle, and not in adult skeletal muscle (Briggs et al., 1994), providing evidence that some isoforms only express in fetal muscle. Isoform I (with accession number P45378) and isoform II (with accession number P45378-2) sequences contained the fetal exon and, therefore, are considered embryonic forms of human fsTnT. In addition, isoform V of fsTnT was reported as a minor isoform that was only detected in an approximate of 1% of cDNA clones (Briggs et al., 1994). Consistently we did not detect these three isoforms of fsTnT purified from human skeletal muscle biopsies (Figure 5). Nevertheless, further investigation of the diversity of sTnT proteoforms in various skeletal muscle fibers is needed.
A proteolytic product with accurate mass of 28270.16 Da was observed for ssTnT, [P26-K262]. Similarly, proteolytic products were also observed consistently in mouse cTnT [P31-K290] (Zhang et al., 2011b) and rat [P30-288] cTnT (Solis et al., 2008). The N-terminal domain of TnT is highly acidic, thus can be disordered and unstable resulting in the proteolytic degradation. Feng et al. previously reported a proteolytic fragment of cTnT with removal of 71 amino acids from the N-terminus during myocardial ischemic reperfusion indicating thin filament functional adaptation under stress condition (Feng et al., 2008). Nevertheless, no such proteolytic product was observed in our previously reported mouse and human cTnT study (Zhang et al. 2011) as well as in this study of human sTnT possibly because of the use of healthy muscle tissues.
Regarding sTnI, both fsTnI and ssTnI were observed in the FTMS spectra of human sTnI, however, ssTnI is more abundant than fsTnI (Figure 5A). Previously, Cummings and Perry analyzed sTnI from freshly excised human gastrocnemius, rectus abdominis, biceps, diaphragm, and soleus using affinity chromatography and gel electrophoresis and identified two forms of human sTn, which could not be readily distinguished by electrophoresis but could be separated after forming complexes with TnC (Cummins and Perry, 1978). Here we have shown that these two forms of sTnI can be easily distinguished by high-resolution MS, and can also be separated by LC (Figure 5). No alternatively spliced isoforms of sTnI were detected in both Hytest samples and the human skeletal muscle biopsies, which is consistent with previous results that no alternative RNA splicing has yet been shown for TnI (Jin et al., 2008). Similarly, it has been shown that only one fsTnI and one ssTnI isoform are expressed in human skeletal muscle tissues, with fsTnI being more abundant in fast fibers and ssTnI more abundant in slow fibers (Bottinelli and Reggiani, 2000; Salviati et al., 1984).
Besides alternatively spliced isoforms, top-down MS can detect all protein PTMs simultaneously within one spectrum (Gregorich and Ge, 2014; Zhang and Ge, 2011). Here, we have detected N-terminal methionine removal, acetylation of the new N-terminus, and the phosphorylation of Ser1 for ssTnT (II) and (III), similarly as we observed previously for cTnT (Zhang, et al. 2011). Thus, phosphorylation of TnT is conserved in various species (human, rat, and mouse) and various muscle tissues (cardiac and skeletal)(Solis et al., 2008; Zhang et al., 2011b). Casein kinase II has been proposed to phosphorylate Ser1, however, the functional significance of Ser1 phosphorylation of TnT remains to be elucidated (Katrukha and Gusev, 2013).
Although we have observed removal of the N-terminal methionines for both fsTnI and ssTnI, acetylation of the new N-terminus has been detected for fsTnI but not for ssTnI. Previously, cTnI has been shown to be actylated consistently in human, non-human primate, swine, and rodent hearts (Zhang et al, 2011, Xu et al. 2011, Zhang, et al. 2010, Ayaz-Guner et al., 2009). N-terminal acetylation is the most common co-translational covalent modifications of proteins in eukaryote and it is known that 85% of all human proteins are N-terminally acetylated. Generally, N-terminal acetylation is known to play an important role in the stability and localization of proteins albeit additional functional roles have been revealed recently (Arnesen et al., 2011). Coulton et al. showed that N-terminally acetylated and un-acetylated tropomyosin bound to distinct actin structure within the cell and thus resulted in profound effect on the shape and integrity of the polymeric actin filament (Coulton et al. 2010). So it is likely that the N-terminal acetylation may be important for understanding the specific properties of fsTnI and ssTnI.
In contrast to cTnI, no phosphorylation has been detected for both ssTnI and fsTnI from Hytest samples and minimal phosphorylation was detected in ssTnI from human skeletal muscle biopsies (Figure 5), likely due to the lack of the cardiac-specific N-terminal sequence which contains Ser22 and Ser23, two well-known protein kinase A (PKA) phosphorylation sites (Layland et al., 2005). Despite a high degree of sequence homology in the C-terminal region, the N-terminal region of TnI is rather divergent (Supplementary Figure 2). Both fsTnI and ssTnI lack the long N-terminal extension in cTnI, which is encoded by a distinct exon (Schiaffino and Reggiani, 1996). Our previous studies have shown that Ser22 and Ser23 are the only basal phosphorylation sites in cTnI purified from healthy animal and human heart tissues (Ayaz-Guner et al., 2009; Zhang et al., 2011a). PKA phosphorylation at Ser22/23 reduces the Ca2+ sensitivity of myofilaments and dephosphorylation of Ser22 and Ser23 has been shown to be associated end stage heart failure (Messer et al., 2006, Zhang, et al. 2011b).
In summary, we have comprehensively analyzed both fsTnI and ssTnI by high-resolution high accuracy top-down MS. For the first time, we have characterized the proteoforms arising from PTMs and alternatively spliced isoforms for human skeletal Tn and mapped the modification sites. As demonstrated, top-down MS is a powerful tool to dissect the molecular complexity of myofilament proteins. Top-down MS can be utilized to further delineate the skeletal muscle diversity and establish the correlation between the modifications of sTn and muscle disorders.
Supplementary Material
Acknowledgments
We would like to thank Ying-Hua (Edith) Chang and Ying (Lynn) Peng for helpful discussions, and Zachery Gregorich for critical reading and editing of the manuscript. We would like to acknowledge National Institutes of Health Grants R01HL096971 and R01HL109810 (to YG). We would also like to thank the Wisconsin Partnership Program for the establishment of UW Human Proteomics Program Mass Spectrometry Facility.
Abbreviations
- sTn
Skeletal muscle troponin
- fsTn
Fast skeletal muscle troponin
- ssTn
Slow skeletal muscle troponin
- TnI
Troponin I
- TnC
Troponin C
- TnT
Troponin T
- Tm
Tropomyosin
- MS
Mass spectrometry
- PTMs
Post-translational modifications
- CAD
Collisionally activated dissociation
- ECD
Electron capture dissociation
- FTICR
Fourier transform ion cyclotron resonance
- ESI
Electrospray ionization
- MWs
Molecular weights
- PKA
Protein kinase A
References
- Arnesen T. Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 2011;9:e1001074. doi: 10.1371/journal.pbio.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz-Guner S, Zhang J, Li L, Walker JW, Ge Y. In Vivo Phosphorylation Site Mapping in Mouse Cardiac Troponin I by High Resolution Top-Down Electron Capture Dissociation Mass Spectrometry: Ser22/23 Are the Only Sites Basally Phosphorylated. Biochemistry. 2009;48:8161–8170. doi: 10.1021/bi900739f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babuin L, Jaffe AS. Troponin: the biomarker of choice for the detection of cardiac injury. (vol 173, pg 1191, 2005) Can Med Assoc J. 2006;174:353–353. doi: 10.1503/cmaj.050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- Bottinelli R, Reggiani C. Human skeletal muscle fibres: molecular and functional diversity. Prog Biophys Mol Biol. 2000;73:195–262. doi: 10.1016/s0079-6107(00)00006-7. [DOI] [PubMed] [Google Scholar]
- Briggs MM, Maready M, Schmidt JM, Schachat F. Identification of a Fetal Exon in the Human Fast Troponin-T Gene. Febs Lett. 1994;350:37–40. doi: 10.1016/0014-5793(94)00729-2. [DOI] [PubMed] [Google Scholar]
- Chait BT. Mass spectrometry: Bottom-up or top-down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- Colantonio DA, Pickett W, Brison RF, Collier CE, Van Eyk JE. Detection of cardiac troponin I early after onset of chest pain in six patients. Clin Chem. 2002;48:668–671. [PubMed] [Google Scholar]
- Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom Rev. 2005;24:201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP. The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J Cell Sci. 2010;123:3235–3243. doi: 10.1242/jcs.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins P, Perry SV. Troponin I from Human Skeletal and Cardiac Muscles. Biochem J. 1978;171:251–259. doi: 10.1042/bj1710251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Sumandea CA, Chen YC, Garcia-Cazarin ML, Zhang J, Balke CW, Sumandea MP, Ge Y. Augmented Phosphorylation of Cardiac Troponin I in Hypertensive Heart Failure. J Biol Chem. 2012;287:848–857. doi: 10.1074/jbc.M111.293258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah CS, Miyamoto CA, Ramos CH, da Silva AC, Quaggio RB, Fujimori K, Smillie LB, Reinach FC. Structural and regulatory functions of the NH2- and COOH-terminal regions of skeletal muscle troponin I. J Biol Chem. 1994;269:5230–5240. [PubMed] [Google Scholar]
- Feng HZ, Biesiadecki BJ, Yu ZB, Hossain MM, Jin JP. Restricted N-terminal truncation of cardiac troponin T: a novel mechanism for functional adaptation to energetic crisis. J Physiol. 2008;586:3537–3550. doi: 10.1113/jphysiol.2008.153577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann R, Troutt AB, Wade RP, Gunning P, Kedes L. Alternative splicing generates variants in important functional domains of human slow skeletal troponin T. J Biol Chem. 1987;262:16122–16126. [PubMed] [Google Scholar]
- Ge Y, Lawhorn BG, ElNaggar M, Strauss E, Park JH, Begley TP, McLafferty FW. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J Am Chem Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- Ge Y, Rybakova IN, Xu QG, Moss RL. Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. P Natl Acad Sci USA. 2009;106:12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorich ZR, Chang YH, Ge Y. Proteomics in heart failure: top-down or bottom-up? Pflugers Arch Eur J Physiol. 2014;466:1199–1209. doi: 10.1007/s00424-014-1471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorich ZR, Ge Y. Top-down proteomics in health and disease: Challenges and opportunities. Proteomics. 2014;14:1195–1210. doi: 10.1002/pmic.201300432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MJ, Chen YC, Clinton L, Zhang H, Zhang J, Dong XT, Xu QG, Ayaz-Guner S, Ge Y. The impact of antibody selection on the detection of cardiac troponin I. Clin Chim Acta. 2013;420:82–88. doi: 10.1016/j.cca.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XM, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectr. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- Jebanathirajah JA, Pittman JL, Thomson BA, Budnik BA, Kaur P, Rape M, Kirschner M, Costello CE, O’Connor PB. Characterization of a new qQq-FTICR mass spectrometer for post-translational modification analysis and top-down tandem mass Spectrometry of whole proteins. J Am Soc Mass Spectr. 2005;16:1985–1999. doi: 10.1016/j.jasms.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Jin JP, Zhang ZL, Bautista JA. Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit Rev Eukar Gene. 2008;18:93–124. doi: 10.1615/critreveukargeneexpr.v18.i2.10. [DOI] [PubMed] [Google Scholar]
- Katrukha IA, Gusev NB. Enigmas of cardiac troponin T phosphorylation. J Mol Cell Cardiol. 2013;65:156–158. doi: 10.1016/j.yjmcc.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J Am Chem Soc. 1999;121:806–812. [Google Scholar]
- Kuhn P, Xu QG, Cline E, Zhang D, Ge Y, Xu W. Delineating Anopheles gambiae coactivator associated arginine methyltransferase 1 automethylation using top-down high resolution tandem mass spectrometry. Protein Sci. 2009;18:1272–1280. doi: 10.1002/pro.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labugger R, Organ L, Collier C, Atar D, Van Eyk JE. Extensive troponin I and T modification detected in serum from patients with acute myocardial infarction. Circulation. 2000;102:1221–1226. doi: 10.1161/01.cir.102.11.1221. [DOI] [PubMed] [Google Scholar]
- Layland J, Solaro RJ, Shah AM. Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc Res. 2005;66:12–21. doi: 10.1016/j.cardiores.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Leavis PC, Gergely J. Thin filament proteins and thin filament-linked regulation of vertebrate muscle contraction. CRC Crit Rev Biochem. 1984;16:235–305. doi: 10.3109/10409238409108717. [DOI] [PubMed] [Google Scholar]
- Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- Messer AE, Jacques AM, Marston SB. Dephosphorylation of Ser23/24 on Troponin I could account for the contractile defect in end-stage failing human heart. J Mol Cell Cardiol. 2006;40:941–941. doi: 10.1016/j.yjmcc.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Marston SB, de Tombe PP. Troponin phosphorylation and myofilament Ca2+-sensitivity in heart failure: Increased or decreased? J Mol Cell Cardiol. 2008;45:603–607. doi: 10.1016/j.yjmcc.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston SB, Redwood CS. Modulation of thin filament activation by breakdown or isoform switching of thin filament proteins - Physiological and pathological implications. Circ Res. 2003;93:1170–1178. doi: 10.1161/01.RES.0000105088.06696.17. [DOI] [PubMed] [Google Scholar]
- McComas AJ. Skeletal Muscle: Form and Function. Champaign, Il: 1996. [Google Scholar]
- Moss RL, Diffee GM, Greaser ML. Contractile properties of skeletal muscle fibers in relation to myofibrillar protein isoforms. Rev Physiol Biochem Pharmacol. 1995;126:1–63. doi: 10.1007/BFb0049775. [DOI] [PubMed] [Google Scholar]
- Ohlendieck K. Skeletal muscle proteomics: current approaches, technical challenges and emerging techniques. Skelet Muscle. 2011;1:6. doi: 10.1186/2044-5040-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Chen X, Sato T, Rankin SA, Tsuji RF, Ge Y. Purification and High-Resolution Top-Down Mass Spectrometric Characterization of Human Salivary alpha-Amylase. Anal Chem. 2012;84:3339–3346. doi: 10.1021/ac300083y. [DOI] [PubMed] [Google Scholar]
- Peng Y, Chen X, Zhang H, Xu QG, Hacker TA, Ge Y. Top-down Targeted Proteomics for Deep Sequencing of Tropomyosin Isoforms. J Proteome Res. 2013;12:187–198. doi: 10.1021/pr301054n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Gregorich ZR, Valeja SG, Zhang H, Cai W, Chen Y, Guner H, Chen AJ, Schwahn DJ, Hacker TA, Liu X, Ge Y. Top-down proteomics reveals concerted reductions in myofilament and Z-disc protein phosphorylation after acute myocardial infarction. Mol Cell Proteomics. 2014;13:2752–2764. doi: 10.1074/mcp.M114.040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto JR, Gomes AV, Jones MA, Liang JS, Nguyen S, Miller T, Parvatiyar MS, Potter JD. The Functional Properties of Human Slow Skeletal Troponin T Isoforms in Cardiac Muscle Regulation. J Biol Chem. 2012;287:37362–37370. doi: 10.1074/jbc.M112.364927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MJ, Forbes AJ, Boyne MT, Kim YB, Robinson DE, Kelleher NL. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol Cell Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- troponin as a potential therapeutic approach for treating neuromuscular diseases. Nat Med. 18:452–455. doi: 10.1038/nm.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping PP, Durazo A, Bowie JU, Hasan SS, et al. Post-translational Modifications of Integral Membrane Proteins Resolved by Top-down Fourier Transform Mass Spectrometry with Collisionally Activated Dissociation. Mol Cell Proteomics. 2010;9:791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salviati G, Betto R, Betto DD, Zeviani M. Myofibrillar-protein isoforms and sarcoplasmic-reticulum Ca2+-transport activity of single human muscle fibres. Biochem J. 1984;224:215–225. doi: 10.1042/bj2240215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Scott W, Stevens J, Binder-Macleod SA. Human skeletal muscle fiber type classifications. Phys Ther. 2001;81:1810–1816. [PubMed] [Google Scholar]
- Shi SDH, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- Simpson JA, Labugger R, Hesketh G, D’Arsigny C, O’Donnell D, Collier C, Iscoe S, Van Eyk JE. Fast and slow skeletal TnI: Potential serum markers of skeletal muscle injury and disease. Clin Chem. 2002a;48:A38–A38. [PubMed] [Google Scholar]
- Simpson JA, Labugger R, Hesketh GG, D’Arsigny C, O’Donnell D, Matsumoto N, Collier CP, Iscoe S, Van Eyk JE. Differential detection of skeletal troponin I isoforms in serum of a patient with rhabdomyolysis: Markers of muscle injury? Clin Chem. 2002b;48:1112–1114. [PubMed] [Google Scholar]
- Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Kelleher NL, Proteomics CTD. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaro RJ, Rosevear P, Kobayashi T. The unique functions of cardiac troponin I in the control of cardiac muscle contraction and relaxation. Biochem Bioph Res Co. 2008;369:82–87. doi: 10.1016/j.bbrc.2007.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis RS, Ge Y, Walker JW. Single amino acid sequence polymorphisms in rat cardiac troponin revealed by top-down tandem mass spectrometry. J Muscle Res Cell Motil. 2008;29:203–212. doi: 10.1007/s10974-009-9168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumandea MP, Burkart EM, Kobayashi T, De Tombe PP, Solaro RJ. Molecular and integrated biology of thin filament protein phosphorylation in heart muscle. Ann Ny Acad Sci. 2004;1015:39–52. doi: 10.1196/annals.1302.004. [DOI] [PubMed] [Google Scholar]
- Sze SK, Ge Y, Oh H, McLafferty FW. Top-down mass spectrometry of a 29-kDa protein for characterization of any posttranslational modification to within one residue. Proc Natl Acad Sci USA. 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- Wei B, Jin JP. Troponin T isoforms and posttranscriptional modifications: Evolution, regulation and function. Arch Biochem Biophys. 2011;505:144–154. doi: 10.1016/j.abb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Xu Q, Dong X, Guy M, Guner H, Hacker TA, Ge Y. Top-down high-resolution electron capture dissociation mass spectrometry for comprehensive characterization of post-translational modifications in rhesus monkey cardiac troponin I. Int J Mass Spectrom. 2011;305:95–102. [Google Scholar]
- Zabrouskov V, Ge Y, Schwartz J, Walker JW. Unraveling Molecular Complexity of Phosphorylated Human Cardiac Troponin I by Top Down Electron Capture Dissociation/Electron Transfer Dissociation Mass Spectrometry. Mol Cell Proteomics. 2008;7:1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ge Y. Comprehensive Analysis of Protein Modifications by Top-Down Mass Spectrometry. Circ-Cardiovasc Gene. 2011;4:711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Guy MJ, Norman HS, Chen YC, Xu Q, Dong X, Guner H, Wang S, Kohmoto T, Young KH, et al. Top-Down Quantitative Proteomics Identified Phosphorylation of Cardiac Troponin I as a Candidate Biomarker for Chronic Heart Failure. J Proteome Res. 2011;10:4054–4065. doi: 10.1021/pr200258m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang H, Ayaz-Guner S, Chen YC, Dong XT, Xu QG, Ge Y. Phosphorylation, but Not Alternative Splicing or Proteolytic Degradation, Is Conserved in Human and Mouse Cardiac Troponin T. Biochemistry. 2011b;50:6081–6092. doi: 10.1021/bi2006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Dong XT, Hacker TA, Ge Y. Deciphering Modifications in Swine Cardiac Troponin I by Top-Down High-Resolution Tandem Mass Spectrometry. J Am Soc Mass Spectr. 2010;21:940–948. doi: 10.1016/j.jasms.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.