SUMMARY
The altered metabolism of tumor cells confers a selective advantage for survival and proliferation, and studies have shown that targeting such metabolic shifts may be a useful therapeutic strategy. We developed an intensely fluorescent, rapidly responsive, pH-resistant, genetically encoded sensor of wide dynamic range, denoted SoNar, for tracking cytosolic NAD+ and NADH redox states in living cells and in vivo. SoNar responds to subtle perturbations of various pathways of energy metabolism in real-time, and allowed high-throughput screening for new agents targeting tumor metabolism. Among > 5,500 unique compounds, we identified KP372-1 as a potent NQO1-mediated redox cycling agent that produced extreme oxidative stress, selectively induced cancer cell apoptosis and effectively decreased tumor growth in vivo. This study demonstrates that genetically encoded sensor-based metabolic screening could serve as a valuable approach for drug discovery.
INTRODUCTION
In recent years, there is renewed attention paid to Otto Warburg’s observation in 1924 that cancer cells shift energy metabolism from mitochondrial oxidative phosphorylation to aerobic glycolysis (Warburg, 1956). Cancer cell metabolism is dramatically reprogrammed to support energy demand and increased biosynthesis of macromolecules for rapid cell proliferation, which has attracted many efforts to target it as a selective anticancer strategy (Cairns et al., 2011; Vander Heiden, 2011; Vander Heiden et al., 2009). Biochemical methods, including enzymatic cycling assays, chromatography, mass spectrometry, and nuclear magnetic resonance spectroscopy, can be used to assess cellular metabolism and search for new cancer targeting agents. Most of these methods require cell lysis, are time-intensive, and, therefore, are not suitable for quantitative, real-time high-throughput screening. High-throughput screening of such agents in cells or in vivo is much more difficult and rarely reported. The use of genetically encoded fluorescent sensors may be a more suitable approach for metabolic chemical screening. To date, these sensors have been developed for single-cell real-time monitoring of several key metabolites(Zhao and Yang, 2015). Among these metabolites, NAD+ and its reduced form, NADH, are the most important cofactors involved in energy metabolism. Recently, Yellen’s group and our group independently developed genetically encoded NADH sensors that allow specific monitoring of dynamic changes in NADH levels in cytosol or mitochondria as affected by different metabolic states (Hung et al., 2011; Zhao et al., 2011). These Frex sensors (Zhao et al., 2011) specifically report NADH levels over a large dynamic range; however, they do not adapt an optimal tertiary structure in some cells and their fluorescence is pH sensitive. Peredox sensors (Hung et al., 2011) are much more pH resistant and partially reflect the more physiologically relevant NAD+ /NADH ratio; however, they have a limited dynamic range and their affinity appears too high to be useful under physiological conditions. Importantly, neither Frex nor Peredox sensors show obvious fluorescence response to NAD+. Such limitations make it difficult to use these sensors for measuring metabolic states in vivo and in high-throughput screening.
Herein, we report the development of an intensely fluorescent, rapidly responsive, pH-resistant, genetically encoded sensor of wide dynamic range, denoted SoNar, for the detection of cytosolic NAD+ and NADH redox states in living cells and in vivo. This sensor of superior properties allowed high-throughput metabolic screening of > 5,500 unique compounds in cancer cell lines. We found that many compounds that increased the NAD+/NADH ratio exhibited marked cancer cell cytotoxicity. Among them, KP372-1, the most potent in increasing NAD+/NADH ratio and cancer cell killing, also effectively decreases tumor growth in vivo. Further studies revealed that KP372-1 is a NQO1-catalyzed, NAD(P)H-consuming redox cycling agent that produces extreme oxidative stress in cancer cells.
RESULTS
Generation of a genetically encoded fluorescent sensor for detection of NAD+, NADH, and their ratio
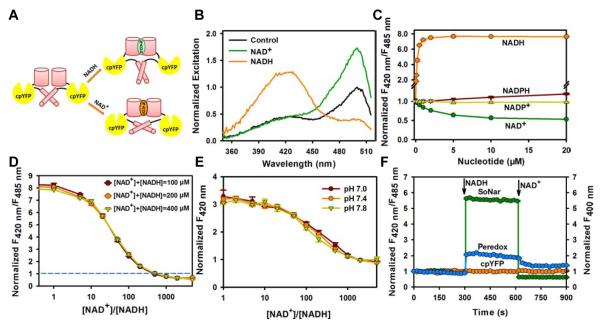
Fifteen chimeric proteins were designed in which cpYFP was inserted between two complete or truncated subunits of Rex protein from Thermus aquaticus (T-Rex), or between amino acid residues located on surface loops of T-Rex (Figure S1A). Among them, the chimera with cpYFP inserted after Phe189 of T-Rex showed a 300% increase in the ratio of fluorescence when excited at 420 nm and 485 nm upon NADH addition (Figure S1B). We then created a series of truncated variants of this protein, either with or without the DNA-binding domain of T-Rex, targeting residues involved in the linker between Rex and cpYFP (Figures S1C and S1D), and found the D2-C2N0 variant to manifest the most dramatic increase in the fluorescence ratio when excited at 420 and 485 nm in the presence of NADH (Figures 1A, 1B, S1D-S1G). Intriguingly, in the presence of saturating NAD+, D2-C2N0 exhibited marked increase in fluorescence when excited at 485 nm (Figures 1B and S1G).
Figure 1. Genetically encoded sensor for NAD+, NADH, and their ratio.
(A) Design of SoNar, which is a fusion of cpYFP and the NADH-binding domain of T-Rex. Binding of NAD+ or NADH both induces changes in protein conformation and fluorescence. (B) Excitation spectra of purified SoNar in the control condition (black), and after addition of 20 μM NAD+ (green) or 20 μM NADH (orange), normalized to the peak intensity in the control condition. Emission was measured at 530 nm. (C) Normalized ratio of fluorescence intensities excited at 420 nm and 485 nm (F420 nm/F485 nm) in the presence of different concentrations of NADH and its analogs. (D) Fluorescence ratios plotted against the NAD+/NADH ratio at the indicated total nicotinamide adenine dinucleotide concentration. Fluorescence ratios were normalized to the control condition in the absence of nucleotides. (E) Fluorescence excited at 420 nm plotted against the NAD+/NADH ratio at the indicated pH. Fluorescence was normalized to the control condition in the absence of pyridine nucleotides at pH 7.4. (F) Kinetics of fluorescence response of purified SoNar, Peredox, and cpYFP protein to sequential addition of 0.2 μM NADH and 2 mM NAD+. (C-F), Error bars represent SEM. See also Figure S1 and Table S1.
Fluorescence titration studies showed that D2-C2N0 had an apparent Kd ≈5.0 μM and ≈ 0.2 μM, respectively, for NAD+ and NADH, at pH 7.4 (Figure 1C), far bellowing the total intracellular pool of NAD+ and NADH in the range of hundreds micromolar (Yamada et al., 2006; Yang et al., 2007). Intracellularly, the sensor would be occupied by either NAD+ or NADH molecules, and its steady-state fluorescence would report the NAD+/NADH ratio rather than the absolute concentrations of the two nucleotides (Figure 1D). We find that D2-C2N0 has an apparent KNAD+/NADH of NAD /NADH of ≈40, the ratio of NAD and NADH at which the response is half-maximal, and is analogous to the dissociation constant (Kd) of a ‘receptor’ for the redox couple. The sensor has high selectivity toward the NAD+/NADH ratio, showing no apparent fluorescence changes towards or in the presence of analogs like NADP+, NADPH, ATP, or ADP (Figures 1C and S1H-S1J). The opposing directional changes of D2-C2N0 fluorescence in the presence of NAD+ and NADH rendered a 15-fold dynamic range, making the sensor among the most responsive genetically encoded sensors available to date (Figure 1D). D2-C2N0 fluorescence excited at 420 nm was much less sensitive to pH, allowing the measurement of NADH levels when modest pH fluctuations occur (Figure 1E). By contrast, D2-C2N0 fluorescence when excited at 485 nm was sensitive to pH, but its dynamic range and KR are more pH-resistant (Figures S1K and S1L). Thus, the pH effects can be alternatively corrected by measuring D2-C2N0 and cpYFP’s fluorescence in parallel, owing to their very similar pH responses (Figures S1L and S1M). Furthermore, kinetic studies showed that the fluorescence of D2-C2N0 responded immediately to sequential addition of NADH and NAD+(Figure 1F), suggesting its usefulness in real-time measurements. Collectively, these data show that D2-C2N0 is highly sensitive and selective for NAD+ and NADH, has rapid responses and a large dynamic range, and is an effective ratiometric sensor for NAD+, NADH, and their ratio with its uniquely designed structure. We, therefore, termed it SoNar (Senor of NAD(H) redox ), and used it for all subsequent experiments.
Select tumor cell lines exhibit decreased NAD+/NADH redox state
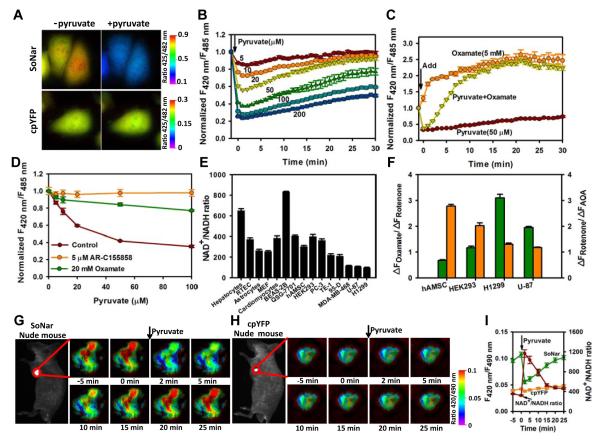
In H1299 human lung cancer cells, SoNar demonstrated much more intense fluorescence compared to the Frex sensor and to a palette of other genetically encoded sensors (Figure S2A), owning to its robust folding and short coding sequence. In cytosol, pyruvate and lactate are generally believed to be in equilibrium with free NAD+ and NADH through the catalytic action of lactate dehydrogenase (LDH). In cells stably expressing SoNar, exogenous pyruvate induced an instantaneous, up to three-fold decrease in the ratio of fluorescence with excitation at 420 and 485 nm, which then recovered slowly (Figures 2A, 2B and S2B). Such effects of pyruvate were almost completely inhibited by the monocarboxylate transporter inhibitor AR-C155858 or lactate dehydrogenase (LDH) inhibitor oxamate (Figure 2D). In control experiments, fluorescence changes were significantly smaller for Frex and Peredox sensors, and occurred much more slowly for the Peredox sensor (Figures S2B and S2C). The more rapid responses of SoNar were due to the faster dissociation rate of the SoNar-NADH complex (Figure 1F). The LDH inhibitor oxamate increased the ratio of SoNar’s fluorescence more effectively than lactate (Figures 2C, S2D and S2E). When cells were treated with pyruvate and oxamate simultaneously, the ratio of SoNar’s fluorescence rapidly decreased initially, and then gradually increased similarly to that caused by oxamate alone (Figure 2C). These data suggest that pyruvate immediately increases the cellular NAD+/NADH ratio, while inhibition of LDH blocked NAD+ recycling and gradually decreased the cellular NAD+/ NADH ratio, consistent with previous reports (Le et al., 2010).
Figure 2. Imaging and quantifying the NAD+/NADH ratio in living cells and in vivo.
(A) Fluorescence images of SoNar or cpYFP-expressing cells before and immediately after incubation with 1 mM pyruvate. Images were pseudocolored with the ratio of fluorescence excited at 425 nm and 482 nm. Scale bar, 10 μm. (B) Kinetics of SoNar fluorescence responses in H1299 cells treated with exogenous pyruvate. (C) Kinetics of SoNar fluorescence responses in H1299 cells treated with pyruvate, oxamate, or pyruvate and oxamate. Data were obtained with a fluorescence plate reader. (D) SoNar fluorescence responses in H1299 cells treated with pyruvate were blocked by AR-C155858 or oxamate. Data were normalized to samples without pyruvate addition. (E) NAD+/NADH ratio in different primary cell cultures and cell lines quantified according to Sonar fluorescence. (F) Effect of oxamate, rotenone, and AOA on fluorescence ratios of different cells expressing SoNar. Cells were treated with inhibitors for 10 min. Green, ΔFOxamate/ΔFRotenone; Orange, ΔFRotenone/ΔFAOA. (G and H) In vivo fluorescence imaging of H1299 xenografts expressing SoNar or cpYFP in response to intravenously administered pyruvate. Sequential frames of fluorescence images of SoNar (G) or cpYFP (H) pseudocolored to the fluorescence ratio. Scale bar, 2 mm. (I) Quantification of the NAD+/NADH ratio in H1299 xenografts according to the SoNar fluorescence data in G and H; error bars represent SD. All experiments performed in H1299 cells and error bars represent SEM unless otherwise indicated. See also Figure S2.
We then quantified the cytosolic NAD+/NADH ratio utilizing SoNar’s fluorescence (Figures 2E, S2F and S2G). The results showed heterogeneity among both cancer cells and non-cancer cells. H1299 and several other cancer cell lines showed significantly lower NAD+/NADH ratios. In these cells, the LDH inhibitor was more potent than mitochondrial NADH oxidase inhibition in reducing the NAD+/NADH ratio, whereas in non-cancer cells, a malate-aspartate shuttle inhibitor was less potent than the mitochondrial NADH oxidase inhibitor in decreasing the NAD+/NADH ratio (Figure 2F). These results suggest that H1299 cells are highly dependent on glycolysis; mitochondrial respiration is sufficiently functional to oxidize NADH derived from glycolysis, but not that derived from the TCA cycle. Further studies showed that inhibition of glycolysis with 3-bromopyruvate (3-BrPA) markedly increased, whereas glucose supplementation decreased, the intracellular NAD+/NADH ratio (Figures S2H-S2J). The glucose concentration for the half-maximal SoNar fluorescence response is ~0.26 mM in H1299 cells and ~1.4 mM in HEK293 cells (Figure S2J), again suggesting that these cancer cells had higher glycolytic activity utilizing lower concentrations of glucose and thereby maintaining their cytosolic NAD+/NADH ratio.
The intense fluorescence of the SoNar sensor allows imaging of cellular redox states in vivo. To this end, nude mice were subcutaneously inoculated with H1299 cells stably expressing SoNar or cpYFP, and imaged when the tumor reached 0.6-1.2 cm. When the mice received an intravenous injection of 110 mg/kg sodium pyruvate, with excitation at 420 and 490 nm the ratio of SoNar fluorescence in the tumor decreased immediately and then gradually recovered over 25 min (Figures 2G and 2I), suggesting that pyruvate in the blood stream significantly affected the NAD(H) redox state in the tumor tissue. In contrast, tumor tissue expressing cpYFP instead of SoNar showed no obvious fluorescence changes upon pyruvate injection, thereby excluding the possibility of interference of the sensor’s fluorescence due to pH variations (Figures 2H and 2I). Taken together, these data indicate that the SoNar sensor displays a superior response, fluorescence intensity, and large dynamic range, and is very useful for the real-time tracking of subtle differences in cellular metabolic and redox states in vitro and in vivo.
SoNar-based high-throughput screening for compounds affecting cancer cell metabolism
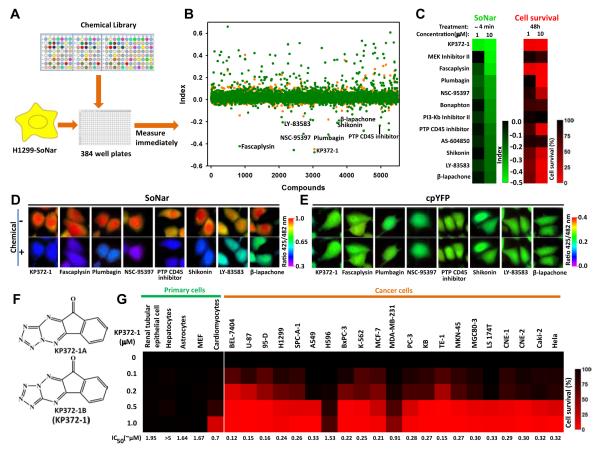
To identify drugs that selectively target cancer cell metabolism, we screened 23 libraries of > 5,500 unique compounds from commercial libraries according to their effects on the intracellular NAD+/NADH redox state of H1299 cells reported by the SoNar sensor (Figures 3A, 3B, and S3A; Table S2). An index was established to describe the intracellular metabolic variation induced by different compounds. While the majority of these compounds have close to zero index and, therefore, minimal impact on SoNar fluorescence and the cellular NAD+/NADH redox state, we did identify 78 compounds that significantly decreased the NAD+/NADH ratio in H1299 cells (index >0.2, ~1.4% of total library) and 12 compounds that significantly increased the NAD+/NADH ratio in H1299 cells (Figures 3B, 3C, and S3B; Table S2). The majority of the compounds that increased (8 of 12) the NAD+/NADH ratio exhibited obvious H1299 cell toxicity (Figures 3C-3E, S3D and S3E), while only a small fraction of the compounds that decreased (9 of 78) the NAD+/NADH ratio exhibited obvious cell toxicity (Figures S3B and S3C). It is interesting that 3 of 8 of these compounds – β-lapachone, shikonin, and fascaplysin -- were potent, widely studied anti-tumor agents. β-lapachone was known to affect cell metabolism by redox cycling and consuming NAD(P)H (Pink et al., 2000), while shikonin was reported to inhibit pyruvate kinase M2 (Chen et al., 2011); however, as far as we are aware, the other compounds have never been directly linked to energy metabolism. Of particular note is the Akt inhibitor, also known as KP372-1 (Figure 3F). This compound is not only the most potent agent in increasing the NAD+/NADH ratio, but also the most potent cancer cell cytotoxic compound, which decreased the viability of a wide range of cancer cells of different origin at concentrations as low as 100 nM. Importantly, and by contrast, the compound showed low toxicity toward various primary cells (Figures 3B, 3C and 3G).
Figure 3. SoNar-based high-throughput screen for compounds affecting cancer cell metabolism.
(A) Schematic of the drug screen. H1299 cells expressing SoNar were incubated with compounds in 384 well plates for 4 min and fluorescence measured. (B) Effects of 5,501 compounds on the index ratio, (ZCompound-ZControl)/(ZOxamate-ZPyruvate). The mean value and s.d. of the experimental population screened is expressed as ZScores. Orange, low concentration; Green, high concentration. (C) 12 lowest indexed hits that increase intracellular NAD+/NADH ratio and their effects on cell viability. (D and E) Fluorescence images of H1299 cells expressing SoNar (D) or cpYFP (E) before and 4 min after treatment with different hit compounds (10 μM). Images were pseudocolored with the ratio of fluorescence excited at 425 nm and 482 nm. Scale bar, 10 μm. (F) Chemical structure of KP372-1. (G) Effects of different concentrations of KP372-1 on the viability of 5 primary cell cultures and 21 cancer cells. All experiments were performed in H1299 cells unless otherwise indicated. See also Figure S3 and Table S2.
KP372-1 affects tumor metabolism and suppresses tumor growth in vivo
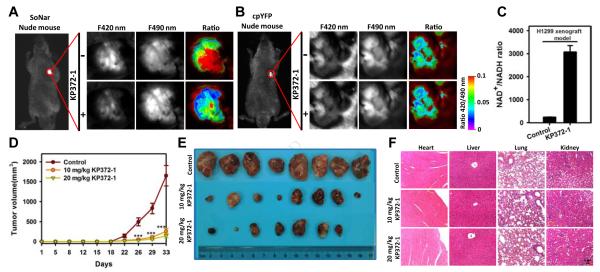
We then proceeded to study the effect of KP372-1 on tumor metabolism in vivo. Pharmacokinetic studies after a single intravenous or intragastric dose (Figure S4A) demonstrated that KP372-1 has good bioavailability with relatively low clearance, a long half-life, and good volume of distribution (Table S3). In tumor-bearing mice in which H1299 cells stably expressing SoNar were subcutaneously implanted, administration of KP372-1 by tail vein injection significantly decreased the ratio of SoNar fluorescence with excitation at 420 and 490 nm in the tumor (Figures 4A and S4B), an effect that did not recover during the one hour time course of observation. As a control, minimal changes of fluorescence were observed in cpYFP-expressing tumor tissues with KP372-1 treatment (Figures 4B and S4B). These results suggest that KP372-1 readily enters tumor tissue and induces persistent oxidation of NADH in cancer cells (Figure 4C).
Figure 4. KP372-1 induces NADH oxidation and impairs tumor growth in vivo without apparent toxicity.
(A and B) In vivo ratiometric fluorescence imaging of H1299 xenografts expressing SoNar (A) or cpYFP (B) in response to KP372-1. Images were pseudocolored with the ratio of fluorescence excited at 420 nm and 490 nm. Scale bar, 2 mm. (C) Quantification of the NAD+/NADH ratio in H1299 xenografts according to SoNar fluorescence. Error bars represent SD. (D) H1299 tumor-growth curves of untreated or KP372-1-treated mice. Error bars denote SEM (n=8 in each group). (E) Photographs of tumors after dissection at day 33. (F) H&E staining for the indicated tissues in animals treated with vehicle, 10 mg/kg KP372-1, or 20 mg/kg KP372-1 daily for 33 days. See also Figure S4 and Table S3.
We next observed that KP372-1 profoundly suppressed the growth of the H1299 xenografts implanted subcutaneously into nude mice versus vehicle (Figures 4D, 4E and S4C). Staining sections of harvested xenografts revealed that KP372-1 reduced proliferation (Ki67 staining), increased caspase-3 activity, and induced apoptosis (TUNEL staining) (Figure S4D), consistent with in vitro studies that KP372-1 increases apoptosis and caspase-3 activation in H1299 cells (Figures S4E-S4G). KP372-1 treated H1299 cells showed significant mitochondrial membrane permeabilization and enhanced Bax localization in mitochondria, suggesting apoptosis occurred through a mitochondrial pathway (Figures S4H and S4I). These characteristics of mitochondrial apoptosis also occurred in KP372-1 treated A549 cells, another non-small-cell lung cancer (NSCLC) cell line (Figures S4J-S4M).
No toxic effects or other indicators of illness, including significant weight loss (Figure S4N) or tissue damage (macroscopic or microscopic), were observed in these mice. Pathological review of sections and an apoptosis assay of heart, liver, lung, and kidney also showed no signs of cytotoxicity (Figures 4F and S4P). The results of blood, liver, and kidney function tests were also normal (Figure S4O). These studies suggest the safety of KP372-1 for use as anti-cancer therapy.
KP372-1 induces significant oxidative stress and activates the pentose phosphate pathway
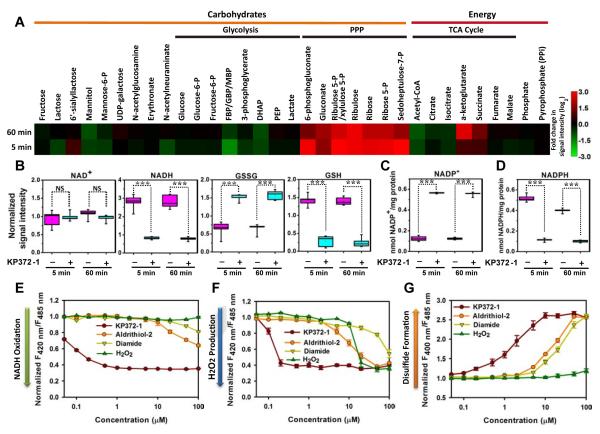
We performed metabolomic analysis to gain insight into how KP372-1 affected cancer cell metabolism, and identified 269 metabolites of known identity (Figures 5A and S5A; Table S4). Most prominently, many intermediates in the pentose phosphate pathway (PPP) increased 7-8 fold at 5 min and sustained these elevations at 60 min in response to KP372-1 (Figure 5A; Table S4). There were minimal changes in the levels of glycolytic metabolites, including lactate, in response to KP372-1, with the exception of a precipitous decrease of fructose-1,6-diphosphate (Figure 5A; Table S4). Biochemical analysis of cell culture medium and cell lysate indicated that both intracellular and extracellular lactate production remained constant (Figures S5C and S5F), again suggesting that direct perturbation of glycolysis was not significant. Biochemical analysis showed that both intracellular and extracellular pyruvate increased significantly in response to KP372-1 treatment (Figures S5B and S5E), consistent with the increased cytosolic NAD+/NADH ratio (Figure S5D). For metabolites involved in the TCA cycle, α-ketoglutarate and succinate increased at 5 min and further increased by 60 min (Figure 5A), suggesting cumulative effects over time. Metabolomic studies validated the decrease of NADH level and increase of NAD+ level. KP372-1 also markedly lowered glutathione (GSH) levels and elevated NADP+ and oxidized GSH (GSSG) levels (Figures 5B and S5G). Although NADPH was not identified in the metabolomic analysis, conventional biochemical assay showed that cellular NADPH levels decreased 4.5-fold , whereas NADP+ levels increased 4.5-fold after 5 min of KP372-1 treatment (Figures 5C and 5D).
Figure 5. KP372-1 induces significant oxidative stress and activates the pentose phosphate pathway.
(A) Metabolite profiles of KP372-1-treated cells shown as a heat map. Increased metabolite concentrations are shown in red, whereas decreased metabolite concentrations are shown in green. H1299 cells were treated with 5 μM KP372-1 for 5 min and 60 min, respectively. (B) KP372-1-induced oxidation of intracellular NADH and GSH. Data obtained by metabolomic studies as panel A (n=5 in each group). ***p < 0.001. NS, not significant. (C and D) Intracellular NADP+ (C) and NADPH (D) levels with or without 5 μM KP372-1 in H1299 cells. Data obtained by a single extraction method and in vitro biochemical assay (n=5 in each group). ***p < 0.001. NS, not significant. (E-G) Fluorescence responses of cells expressing SoNar (E), Hyper (F), and roGFP1 (G) to different concentrations of KP372-1 and other oxidants. Fluorescence was measured immediately after chemical addition and normalized to the control condition. All experiments were performed in H1299 cells; error bars represent SEM. See also Figure S5 and Table S4.
The dose-dependent effects of KP372-1 on intracellular NADH, H2O2, and disulfide redox state were further explored by genetically encoded sensors, including SoNar, Hyper (Belousov et al., 2006), and roGFP1 (Dooley et al., 2004). KP372-1 induced significant oxidative stress via oxidizing NADH, generating H2O2, and oxidizing thiols to disulfides, respectively, 2 to 3 orders of magnitude more efficiently than H2O2 or thiol oxidants, such as aldrithiol-2 and diamide (Figures 5E-5G, S5H-S5J, S5L, and S5M). In controls, none of these compounds affected the fluorescence of cpYFP-expressing cells (Figure S5K). Kinetic studies showed that the decrease of NADH occurred immediately after KP372-1 addition, followed by H2O2 production and then thiol oxidation (Figure S5N). Overall, these results suggest that KP372-1 has a prominent impact on cellular redox state with NAD(P)H oxidation being the vanguard event. Oxidative stress is necessary for KP372-1 induced cell death, which was largely blocked by overexpressing catalase in the cell, or by treating the cell with N-acetylcysteine, an general antioxidant, or Tiron, a cell permeable superoxide scavenger (Figures S5O and S5P).
NQO1 catalyzes NAD(P)H-dependent KP372-1 redox cycling and promotes cancer cell death
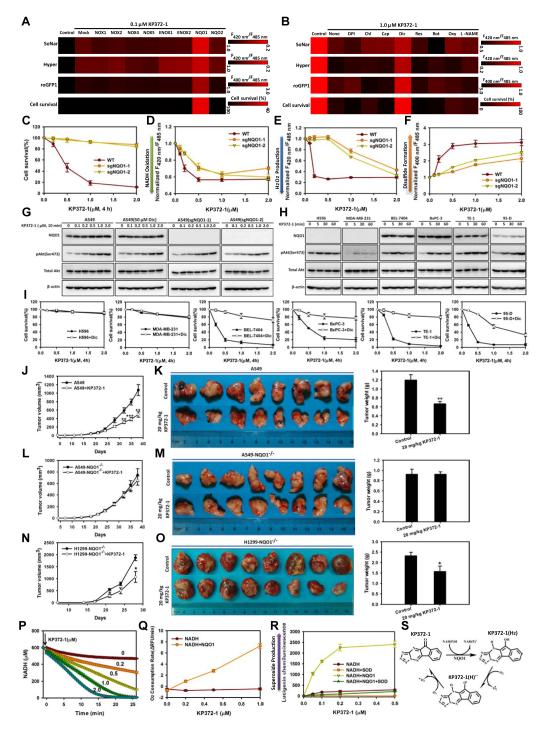
We considered that KP372-1 may activate NAD(P)H-dependent reactive oxygen species (ROS)-generating enzymes in view of its potency in inducing oxidative stress, and explored the functions of such oxidases by overexpressing them in H1299 cells (Figures 6A, S6A and S6F). We found that only overexpression of NQO1 boosted KP372-1’s effects on intracellular NADH oxidation, H2O2 production, thiol oxidation, and cell death (Figures 6A, S6A and S6C). NQO1 is a promising therapeutic target in cancer therapy and highly expressed in many cancer cell lines (Bey et al., 2007; Cullen et al., 2003; Pink et al., 2000), including the A549 NSCLC cell line (Bey et al., 2007) (Figure S6H). In A549 cells, KP372-1-induced oxidative stress and cell death were largely inhibited by dicoumarol, a known inhibitor of NQO1 (Pink et al., 2000) (Figures 6B, S6B and S6D). In contrast, pharmacologic inhibitors of NAD(P)H oxidase, ENOX1, ENOX2, NQO2, mitochondrial respiratory chain complex I, xanthine oxidase, and nitric oxide synthase showed minimal effects on KP372-1-induced oxidative stress and cell death (Figures 6B and S6B). Consistent with the SoNar measurement, data calculated from the measured concentrations of the pyruvate and lactate, and the equilibrium constants of lactate dehydrogenase (Williamson et al., 1967) also showed that cytosolic NAD+/NADH ratio was dose dependently increased in the cells treated with KP372-1, which is promoted by NQO1 expression (Figure S6E), or repressed by the NQO1 inhibitor dicoumarol (Figure S6G). We next generated isogenic, NQO1 knockout A549 cell lines using the CRISPR-Cas9 System (Cong et al., 2013; Ran et al., 2013). These NQO1 knockout lines, confirmed by sequencing and western blotting, were much more resistant to oxidative stress and cell death induced by KP372-1 (Figures 6C-6G, S6I). Similarly, these NQO1 knockout lines were also much more resistant to oxidative stress and cell death induced by β-lapachone (Figure S6J), another hit compound in our screen (Figures 3B and 3C) and an antitumor agent known to target to NQO1 (Bey et al., 2007; Pink et al., 2000). Interestingly, the only two cancer cell lines resistant to KP372-1 toxicity in this study, MDA-MB-231 cells and H596 cells, were also NQO1-negative (Figures 6H and 6I), while the killing effects of KP372-1 on other cancer cell lines expressing NQO1 were effectively attenuated by dicoumarol (Figures 6H and 6I). β-lapachone showed a similar cytoxicity profile as KP372-1 and was similarly antagonized by dicoumarol (Figure S6K).
Figure 6. NQO1 catalyzes NAD(P)H-dependent KP372-1 redox cycling and promotes cancer cell death.
(A) Overexpression of NQO1, but not other NAD(P)H oxidases, increases KP372-1-induced NADH oxidation, H2O2 production, thiol oxidation, and cell death in H1299 cells. (B) The NQO1 inhibitor dicumarol, but not other pharmacologic inhibitors of ROS-generating enzymes, decrease KP372-1-induced NADH oxidation, H2O2 production, thiol oxidation, and cell death in A549 cells. For A and B, after cells were treated with KP372-1, SoNar fluorescence was measured immediately, while Hyper and roGFP1 fluorescence were measured after 10 min. (C-F) Knockout of NQO1 decreased KP372-1-induced cell death (C), NADH oxidation measured by SoNar fluorescence (D), H2O2 production measured by Hyper fluorescence (E), and thiol oxidation measured by roGFP1 fluorescence (F) in A549 cells. (G) The effect of NQO1 inhibitor dicumarol or NQO1 knockout on KP372-1-induced phosphorylation of Akt in A549 cells. Cells were treated with different concentrations of KP372-1 for 10 min. (H) Phosphorylation of Akt in different cells treated with 0.5 μM KP372-1 at the indicated times. (I) The effect of NQO1 inhibitor dicumarol on KP372-1-induced cell death. Cells were exposed to a 4-h pulse of KP372-1 either with or without 50 μM dicumarol and then grown for 24 h. (J-O) The effects of KP372-1 on tumor growth of WT A549 (J and K), NQO1 knockout A549 (L and M) and NQO1 knockout H1299 (N and O) xenografts. Tumor-growth curves (J, L and N) were measured in untreated or KP372-1-treated mice, error bars denote SEM (n=8 in each group). Xenografts were dissected at day 38 (K and M) or day 28 (O), photographed (left) and weighted (right). Error bars indicate SEM. (P) NQO1-catalyzed NADH oxidation in the presence of different concentrations of KP372-1. NADH was measured by its endogenous fluorescence. The concentrations of KP372-1 are indicated adjacent to each curve. (Q) Oxygen consumption of NADH solution (600 μM) in the presence or absence of 1 μg/ml NQO1 and different concentrations of KP372-1. Oxygen levels were measured with a BD Oxygen Biosensor System. (R) Superoxide production by a lucigenin chemiluminescence assay in an assay solution containing 600 μM NADH and KP372-1, in the presence or absence of 1 μg/ml NQO1 and/or 30 U/ml superoxide dismutase. (S) Hypothetical model for KP372-1-mediated redox cycling. Error bars represent SEM. See also Figure S6.
It is known that a subset of NSCLC cell lines and tumors exhibit KEAP1 mutations which activate NRF2 (Dai et al., 2013; Singh et al., 2006). In these cells NRF2 then induces expression of NQO1, as well as a palette of antioxidant genes (Gorrini et al., 2013; Sporn and Liby, 2012; Thimmulappa et al., 2002). In KEAP wild type cells, there is a good correlation between the cytotoxicity of KP372-1 and NQO1 expression (Figures S6L and S6M). In KEAP1 mutant cells, however, there is no clear correlation between the cytotoxicity of KP372-1 and the KEAP mutation (Figures S6L and S6M), suggesting the effects of overexpressed NQO1 may be counterbalanced by the enhanced antioxidant mechanism (Figures S6N and S6O).
The role of NQO1 in KP372-1 toxicity is further supported by in vivo data, which showed that the growth of the A549 xenografts was significantly suppressed by KP372-1 compared with vehicle (Figures 6J and 6K). In contrast, the growth of NQO1 knockout A549 xenografts was not suppressed by KP372-1 compared with vehicle (Figures 6L and 6M). Similarly, growth of NQO1 knockout H1299 xenografts was much less suppressed by KP372-1 (Figures 6N, 6O and S6P), compared to wild type H1299 xenografts (Figures 4D and 4E, S4C).
KP372-1 is known to inhibit Akt kinase (Koul et al., 2006; Mandal et al., 2005; Mandal et al., 2006; Zeng et al., 2006). Interestingly, our data showed that the effects of KP372-1 on the fluorescence of SoNar or roGFP1 were absent when A549 and H1299 cells were treated with different Akt inhibitors (Figures S6Q-S6T). Surprisingly and in contradistinction to previous reports that KP372-1 inhibited Akt phosphorylation in cancer cells (Koul et al., 2006; Mandal et al., 2005; Mandal et al., 2006; Zeng et al., 2006), we found that KP372-1 treatment dramatically, transiently and dose-dependently increased phosphorylation of Akt in A549 and H1299 cells (Figures 6G and S6U), which was not significantly affected by NQO1 inhibition or NQO1 knockout(Figure 6G). Such enhanced Akt phosphorylation induced by KP372-1 also occurred to varying extents in other cancer cell lines irrespective of their NQO1 expression level (Figure 6H). These data showed that KP372-1’s action on NAD+/NADH redox, oxidative stress, and viability of cancer cells was not due to the inhibition of this kinase.
NQO1 is a NAD(P)H-dependent oxidoreductase that catalyzes the 2-electron reduction of a broad range of quinones. β-lapachone was previously reported to exert it anti-tumor function by “bioactivation” by NQO1 to an unstable hydroquinone that rapidly undergoes a two-step oxidation back to the parent compound, perpetuating a futile redox cycle and generating superoxide in the process (Pink et al., 2000; Reinicke et al., 2005). We found that NAD(P)H was rapidly oxidized in the presence of both KP372-1 and trace amounts of NQO1 in vitro, monitored by either NAD(P)H endogenous fluorescence (Figures 6P and S6V) or SoNar (Figure S6W). The oxidation of NADH was accompanied by oxygen consumption (Figure 6Q) and generation of superoxide and hydrogen peroxide (Figures 6R and S6X). In these studies, greater than 1000 molar equivalents of NADH were oxidized per molar equivalent of KP372-1 in 30 min (Figure 6J), suggesting that KP372-1 underwent NQO1-dependent redox cycling. In H1299 cells, oxygen consumption also significantly increased in the presence of KP372-1, which further increased with overexpression of NQO1 (Figure S6Y). Taken together, our findings provide evidence that NQO1-mediated futile redox cycling of KP372-1 (Figure 6S) induced oxidative stress and cell death in cancer cells.
DISCUSSION
The SoNar sensor described herein represents a substantial improvement for live cell NAD+/NADH measurement over existing reporters. First, SoNar fluorescence is intrinsically ratiometric with two excitation wavelengths having opposing responses to NAD+ and NADH, allowing quantitative determination of NAD+ or NADH separately as well as ratiometrically. To our knowledge, such a dual response property is unprecedented among genetically encoded sensors developed to date. In addition to its role in energy metabolism, NAD+ also participates in a wide range of cellular processes, acting as precursor of signaling molecules such as cADPR and ADPR, and as a co-substrate of regulatory enzymes of significant importance, such as mono- or poly-ADP-ribose polymerases and Sirt family deacylases. Many of these enzymes are also important targets for drug discovery. SoNar is capable of reporting NAD+ levels directly in vitro independent of NADH, and can be utilized in simple, continuous enzymatic assays, which may be amenable to high-throughput screening. Second, SoNar is an authentic reporter of the NAD+/NADH ratio under physiological conditions. SoNar had an apparent Kd of ≈5.0 μM for NAD+, much lower than intracellular NAD+ concentration, allowing the sensor to report the NAD+/NADH ratio under physiological conditions accurately unaffected by the total NAD(H) pool. By contrast, the Frex sensor reports only NADH levels, while Peredox reports the cytosolic NADH level, partly compensated by the NAD + level rather than the precise NAD+/NADH ratio (Hung et al., 2011). Third, SoNar displays a superior rapid response and large dynamic range, and is very useful for the real-time tracking of subtle differences in cellular metabolic states. SoNar displays a 1500% fluorescence change under different NAD+/NADH ratios in vitro, almost 10-fold greater than that of Peredox and 2-fold greater than that of Frex, making it one of the most responsive genetic encoded sensors currently available. SoNar responded well to conventional metabolic perturbations such as extraneous glucose, pyruvate, and lactate supplementation, or inhibition of energy metabolism pathways such as mitochondrial respiration, the mitochondrial NADH shuttle, glycolysis, and LDH. In live cell studies, the sensor displayed an 8-fold dynamic range, and was able to detect the response to added pyruvate as low as 5 μM, substantially outperforming either Frex or Peredox sensors. SoNar also performed nicely in in vivo imaging studies, exhibiting intense fluorescence and the ability to detect increases or decreases of NAD+/NADH levels within normal physiological conditions. Our study showed that Peredox was not able to detect the decrease of the NAD+/NADH ratio when cells were treated with lactate or an LDH inhibitor, as it was almost fully saturated by NADH in cells in the presence of 1 mM glucose (Hung et al., 2011). This deficiency complicates Peredox’s usage for in vivo imaging, considering the normal range of blood glucose concentration.
Various studies suggested that the intracellular NAD+/NADH pool is greatly reduced in many cancer cells compared to non-cancer cells, inferred by the lower lifetime of NADH endogenous fluorescence and increased mitochondrial free NADH, or lower ratio of the fluorescence intensity of FAD-to-NADH (Ostrander et al., 2010; Pradhan et al., 1995; Skala et al., 2007; Yu and Heikal, 2009); however, to our knowledge, the intracellular NAD+/NADH ratio had not been rigorously quantified due to lack of appropriate quantitative non-invasive methodology. By monitoring SoNar’s fluorescence, we provide direct evidence that the cytosolic NAD+/NADH ratio is significantly reduced in certain cancer cell lines, such as H1299, U87, and MDA-MB-468. Further dissection of the metabolic pathways governing this ratio using enzyme inhibitors showed that these cancer cells were able to utilize low levels of glucose and exhibit high glycolytic activity, consistent with a previous report that H1299 cells overexpress pyruvate kinase M2 and display high rates of glycolysis (Christofk et al., 2008). For reference, we attach a diagram that explains the central role NAD+/NADH plays in relevant metabolic pathways in this study (Figure S2K).
The intense fluorescence and large dynamic range of SoNar enabled its usage in robust high-throughput and real-time screening assays of small-molecule libraries that may affect cellular metabolism, which may be useful as probes for metabolic studies and as lead compounds for drug discovery. In contrast to many other high-throughput assays in drug discovery that target a single protein or enzyme, SoNar is multi-targeted as it is capable of reporting subtle perturbations in many pathways affecting energy metabolism, including glycolysis and mitochondrial respiration. Furthermore, the rapid response and high sensitivity greatly reduce the costs of and time needed for screening. Most of the compounds that we screened did not affect intracellular NAD+ redox states; however, protein kinase inhibitor and phosphokinase inhibitor libraries had a significant percentage of hits, highlighting the regulatory roles of protein phosphorylation in cell metabolism. It is interesting that quite a few compounds among FDA approved drugs and the NIH clinical collection also affect cellular metabolism. Further analysis may help to identify whether the effects of these drugs on metabolism are related to their anti-tumor mechanism and efficacy.
It is also interesting that only a small fraction of compounds that decrease the NAD+/NADH ratio exhibited obvious cytotoxicity, whereas the majority of the compounds that increase the NAD+/NADH ratio exhibited obvious cancer cell toxicity, among which β-lapachone, shikonin, and fascaplysin were potent, widely studied anti-tumor agents. Consistent with their effects on their NAD+/NADH ratio as reported by the SoNar sensor, β-lapachone is known to be reduced by NQO1, which then enters into futile cycling, consumes NAD(P)H, and generates superoxide, while shikonin inhibits cancer cell glycolysis by targeting tumor pyruvate kinase M2 (Chen et al., 2011). The other compounds identified were not previously linked to cellular metabolism. The most potent compound that increases the NAD+/NADH ratio and the most potent selective cancer killing compound, KP372-1, was previously reported to exert inhibitory effects on PDK1/Akt signaling pathways, suppressing cell proliferation and inducing/sensitizing cells to apoptosis (Koul et al., 2006; Mandal et al., 2005; Mandal et al., 2006; Zeng et al., 2006). Intriguingly, we found that the compound actually enhanced Akt phosphorylation, contradictory to previous reports, and that various specific Akt inhibitors had no significant impact on the NAD+/NADH redox level. KP372-1 induced significant oxidative stress as shown by NAD(P)H depletion, H2O2 generation, and thiol oxidation. The PPP was activated to replenish NADPH for antioxidant enzyme catalysis. Further genetic and chemical screening showed that KP372-1 was reduced by NQO1 in the presence of NAD(P)H, which enters into a futile redox cycle and generates superoxide; its antitumor activity was blocked by a specific NQO1 inhibitor or in isogenic NQO1 knock out cells. These data show that KP372-1’s action on NAD+/NADH redox, oxidative stress and viability of cancer cells was due to NQO1-mediated futile redox cycling, and not the inhibition of Akt signaling. Kinase inhibitors have been very popular for dissecting signal transduction pathways and drug discovery. Although investigators are aware that inhibition of an off-target kinase may contribute to the observed biological effects of these inhibitors, the results reported here provide an interesting example that a distinct redox enzyme and mechanism are responsible for the selective cancer cell killing effects of a well-known kinase inhibitor rather than kinase inhibition itself. NQO1 was highly expressed in numerous cancer cells (Bey et al., 2007; Cullen et al., 2003; Pink et al., 2000). β-lapachone, an antitumor agent containing the quinone pharmacophore, , can kill cancer cells overexpressing NQO1 regardless of cell cycle phase or p53 status. The quinone pharmacophore is particularly common in antitumor drugs, such as mitomycin C, β-lapachone, and 17-N-allylamino-17-demethoxygeldanamycin 17-AAG (Siegel et al., 2012). Unlike these compounds, KP372-1 does not contain a quinone pharmacophore. In view of the NQO1-catalyzed redox cycling of β-lapachone (Pink et al., 2000; Reinicke et al., 2005), we speculate the possible reaction mechanism of KP372-1, NAD(P)H, and NQO1 as follows (Figure 6S): KP372-1 contains a quinone mimic and can be reduced to the “hydro” form by NAD(P)H via a 2-electron reduction catalyzed by NQO1. The “hydro” form is presumably unstable and spontaneously auto-oxidizes to its original parent form, probably through an intermediate (KP372-1(H).-), which consumes oxygen and generates superoxide. This redox cycling causes rapid and profound oxidation of NAD(P)H.
Most current NQO1 substrates developed for cancer therapy have problems in pharmacokinetics, tolerability, delivery, and bioavailability. For example, β-lapachone suffers from poor aqueous solubility and a short half-life in vivo (~20 minutes), , which necessitates the use hydroxypropyl-β-cyclodextrin (HPβCD) as a solubility aid at the doses poorly tolerated (Bair, 2012; Blanco et al., 2010; Huang et al., 2012). We reasoned that KP372-1 could be a better candidate in future NQO1-dependent chemotherapies, as it is one magnitude more potent than β-lapachone; is capable of being administered orally; has a long half-life, large volume of distribution, and excellent bioavailability; and is very well tolerated by mice at the doses required to detect a significant anticancer effect. By in vivo imaging of tumor metabolism using the SoNar sensor, we also demonstrated that KP372-1 enters the tumor rapidly. Furthermore, identification of a non-quinone redox cycling agent like KP372-1 expands our scope and understanding of NQO1 and redox cycling agents. These compound and their analogues can be further modified and investigated to develop better anti-tumor drugs.
In summary, we report that SoNar is a superior tool for tracking the intracellular NAD+/NADH redox state and, therefore, subtle changes in cell metabolic status, as it is intensely fluorescent, is intrinsically ratiometric, and has a large dynamic range. These properties make it an ideal reagent for metabolic studies and drug discovery in cells or animals. Interestingly, we identified a potent NQO1-activated redox cycling and anti-tumor agent, KP372-1, which was previously used as (believed to be) an Akt inhibitor. Further studies on KP372-1 and its analogues might lead to therapeutic options for drug-resistant cancers. With the help of the SoNar sensor, we also identified many other compounds, including clinically used drugs that affect cellular metabolism. Although not investigated in this study owing to practical considerations of time, effort, and compound availability, it is possible that the clinical benefits of these drugs are less attributable to their reported targets or mechanism of action, and, perhaps, more likely a consequence of effects on cellular metabolism.
EXPERIMENTAL PROCEDURES
Please see the Supplemental Experimental Procedures for additional details
Gene Construction and Protein Characterization
Gene construction and characterization of SoNar in vitro are described in Supplemental Experimental Procedures.
Live-Cell Fluorescence Measurement Using Microplate Reader
Cells were harvested by trypsinization and counted by hemocytometer. Cells were washed and suspended in PBS (HyClone), and aliquots of cells were incubated at 37 oC with different compounds during the measurement. Dual-excitation ratios were obtained by a Synergy 2 Multi-Mode Microplate Reader (BioTek). Detailed procedures can be provided in the Supplemental Experimental Procedures.
Fluorescence Microscopy
For fluorescence microscopy, H1299-SoNar-, H1299-cpYFP-, H1299-Hyper-, and H1299-roGFP1-expressing cells were plated on a 35 mm glass-bottom dish with phenol red-free growth medium. Images were acquired using a high-performance fluorescent microscopy system equipped with Nikon Eclipse Ti-E automatic microscope, monochrome cooled digital camera head DS-Qi1 Mc-U2, and the highly stable Sutter Lambda XL light source. Raw data were exported to ImageJ software as 12 bit TIF for analysis. Detailed procedures can be provided in the Supplemental Experimental Procedures.
Imaging NADH Metabolism in Living Animals
H1299-SoNar- and H1299-cpYFP-expressing cells (10×106) suspended in 0.1 ml serum-free RPMI1640 with an equal volume of BD Matrigel Matrix (BD Biosciences-356237) were inoculated s.c. in the right axilla of each mouse. When tumor size reached 0.6-1.2 cm in diameter, mice were anaesthetized intraperitoneally with sodium pentobarbital (100 mg/kg body weight), placed in a holding chamber, and given an intravenous tail vein injection with 100 mM sodium pyruvate (Invitrogen) or 600 μM KP372-1 (Echelon Biosciences Inc.) suspended in 0.9% NaCl in a total volume of 0.3 ml. The dual-excitation ratio imaging in tumor tissues expressing SoNar or cpYFP was obtained by a Kodak Multispectral FX imaging system (Carestream Molecular Imaging) with excitation filters 420 BP 20 nm and 490 BP 20 nm, and an emission filter 535 BP 50 nm (for both excitation wavelengths).
Chemical Screen and Analysis
All screened compounds were purchased from the National Compounds Resource center (China) as stock solutions in DMSO. The majority of the compounds were arrayed in 96-well plates and diluted to 3 μM or 30 μM with PBS containing 25 mM glucose. We then transferred 25 μl chemical solutions from 96-well plates into black 384-well flat bottom plate (Greiner Bio-one, Germany) using an 8-channel pipette and then immediately added 50 μl H1299-SoNar-expressing cell suspensions into each well using am 8-channel electronic pipette (50-1250 μl), resulting in 1 μM and 10 μM final concentrations for most compounds. Each plate contained the following control wells: 4-8 wells DMSO-only control, 2-4 wells controls with 1 mM pyruvate, and 2-4 wells controls with 5 mM oxamate. Fluorescence intensity was measured immediately by a Synergy 2 Multi-Mode Microplate Reader (Bio Tek) with 420 nm or 485 nm excitation, and 528 nm emission wavelengths. Analysis of screening data is described in Supplemental Experimental Procedures. The I value for every compound performed in our screen can be found in Table S2.
Metabolomics Analysis
The metabolomic analysis was performed by Metabolon, Inc. (Durham, NC, USA). Detailed procedures can be found in the Supplemental Experimental Procedures.
Xenograft Studies
To test the antitumor activity of KP372-1 in vivo, nude mice (nu/nu, male 6- to 8-week-old, Shanghai SLAC Laboratory Animal Co. Ltd.) were injected subcutaneously with 5×106 cells. Animals were divided randomly into control (vehicle), 10 mg/kg prevention groups, or 20 mg/kg prevention groups of eight each. Treatment was performed once daily by intragastric administration of KP372-1. Tumor formation was assessed every 2–3 days with calipers in all the groups. After 4-6 weeks, the tumors were dissected and weighed.
Statistical Analysis
Data are presented either as a representative example of a single experiment repeated at least in triplicate or as three or more experiments. Data obtained are represented as mean values SD or mean values ± SEM. All P values were obtained using unpaired two-tailed Student’s t-test. Values of p<0.05 were considered statistically significant (*0.01 < p < 0.05; **0.001 < p < 0.01; ***p < 0.001).
Supplementary Material
Highlights.
SoNar sensor is capable of tracking subtle changes of NAD+/NADH in vitro and in vivo
SoNar enables high-throughput screening for new agents targeting tumor metabolism
KP372-1 is identified as a potent NQO1-dependent redox cycling anti-tumor agent
ACKNOWLEDGMENTS
We thank Gary Yellen for the Peredox vector; Jim Remington for the roGFP vector; Jiahuai Han for the cDNA of Enox and Nox families; Hongbin Ji, Jianwen Liu, Jing Zheng, Ping Shi, Xuni Cao, Zhaoyang Ye, and Jia Yu for different cell lines; Guiping Li, Qiongyu Yan, Chunyuan Zhou, Qiuning Lin, Lei Xu, Rongkun Tao, Jianhua Wang, Zhengda Chen and Wei Wang for technical assistance; and Stephanie Tribuna for secretarial assistance. This work was supported by the 973 Program (2013CB531200), NSFC (31225008, 91313301, 31071260, 31170815, 31470833 and 91013012), Specialized Research Fund for the Doctoral Program of Higher Education (20100074110010), Shanghai Science and Technology Commission (12JC1402900, 11DZ2260600, 15YF1402600), Dawn Program of the Shanghai Education Commission (11SG31), State Key Laboratory of Bioreactor Engineering, the 111 Project (B07023), and the Fundamental Research Funds for the Central Universities; and NIH grants (to JL) HL061795, HL084734, and HL108630.
Footnotes
AUTHOR CONTRIBUTIONS
Y. Y. and Q.H. designed the sensor, Y.Y. and Y.Z. designed the in vitro, in vivo imaging, metabolic screening, and target identification experiments. Y.Z., Q.H., N.S., A.W and Y.Z performed experiments. F.C, H.H., X.C., H.Z, X.H, K.Y., Q.Z., X.W., J.Y., L.Z, X.Q., L.C., Y.T gave technical support and conceptual advice. Y.Y., Y.Z., and J.L. analyzed the data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bair J. Dissertation. 2012. The development of deoxynyboquinone as a personalized anticancer compound. [Google Scholar]
- Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- Bey EA, Bentle MS, Reinicke KE, Dong Y, Yang CR, Girard L, Minna JD, Bornmann WG, Gao J, Boothman DA. An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc Natl Acad Sci U S A. 2007;104:11832–11837. doi: 10.1073/pnas.0702176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E, Bey EA, Khemtong C, Yang SG, Setti-Guthi J, Chen H, Kessinger CW, Carnevale KA, Bornmann WG, Boothman DA, et al. Beta-lapachone micellar nanotherapeutics for non-small cell lung cancer therapy. Cancer Res. 2010;70:3896–3904. doi: 10.1158/0008-5472.CAN-09-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Chen J, Xie J, Jiang Z, Wang B, Wang Y, Hu X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene. 2011;30:4297–4306. doi: 10.1038/onc.2011.137. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen JJ, Hinkhouse MM, Grady M, Gaut AW, Liu J, Zhang YP, Weydert CJ, Domann FE, Oberley LW. Dicumarol inhibition of NADPH:quinone oxidoreductase induces growth inhibition of pancreatic cancer via a superoxide-mediated mechanism. Cancer Res. 2003;63:5513–5520. [PubMed] [Google Scholar]
- Dai B, Yoo SY, Bartholomeusz G, Graham RA, Majidi M, Yan S, Meng J, Ji L, Coombes K, Minna JD, et al. KEAP1-dependent synthetic lethality induced by AKT and TXNRD1 inhibitors in lung cancer. Cancer Res. 2013;73:5532–5543. doi: 10.1158/0008-5472.CAN-13-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- Huang X, Dong Y, Bey EA, Kilgore JA, Bair JS, Li LS, Patel M, Parkinson EI, Wang Y, Williams NS, et al. An NQO1 substrate with potent antitumor activity that selectively kills by PARP1-induced programmed necrosis. Cancer Res. 2012;72:3038–3047. doi: 10.1158/0008-5472.CAN-11-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul D, Shen R, Bergh S, Sheng X, Shishodia S, Lafortune TA, Lu Y, de Groot JF, Mills GB, Yung WK. Inhibition of Akt survival pathway by a small-molecule inhibitor in human glioblastoma. Mol Cancer Ther. 2006;5:637–644. doi: 10.1158/1535-7163.MCT-05-0453. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Kim S, Younes MN, Jasser SA, El-Naggar AK, Mills GB, Myers JN. The Akt inhibitor KP372-1 suppresses Akt activity and cell proliferation and induces apoptosis in thyroid cancer cells. Br J Cancer. 2005;92:1899–1905. doi: 10.1038/sj.bjc.6602595. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mandal M, Younes M, Swan EA, Jasser SA, Doan D, Yigitbasi O, McMurphey A, Ludwick J, El-Naggar AK, Bucana C, et al. The Akt inhibitor KP372-1 inhibits proliferation and induces apoptosis and anoikis in squamous cell carcinoma of the head and neck. Oral Oncol. 2006;42:430–439. doi: 10.1016/j.oraloncology.2005.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander JH, McMahon CM, Lem S, Millon SR, Brown JQ, Seewaldt VL, Ramanujam N. Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status. Cancer Res. 2010;70:4759–4766. doi: 10.1158/0008-5472.CAN-09-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink JJ, Planchon SM, Tagliarino C, Varnes ME, Siegel D, Boothman DA. NAD(P)H:Quinone oxidoreductase activity is the principal determinant of beta-lapachone cytotoxicity. J Biol Chem. 2000;275:5416–5424. doi: 10.1074/jbc.275.8.5416. [DOI] [PubMed] [Google Scholar]
- Pradhan A, Pal P, Durocher G, Villeneuve L, Balassy A, Babai F, Gaboury L, Blanchard L. Steady state and time-resolved fluorescence properties of metastatic and non-metastatic malignant cells from different species. J Photochem Photobiol B. 1995;31:101–112. doi: 10.1016/1011-1344(95)07178-4. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinicke KE, Bey EA, Bentle MS, Pink JJ, Ingalls ST, Hoppel CL, Misico RI, Arzac GM, Burton G, Bornmann WG, et al. Development of beta-lapachone prodrugs for therapy against human cancer cells with elevated NAD(P)H:quinone oxidoreductase 1 levels. Clin Cancer Res. 2005;11:3055–3064. doi: 10.1158/1078-0432.CCR-04-2185. [DOI] [PubMed] [Google Scholar]
- Siegel D, Yan C, Ross D. NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem Pharmacol. 2012;83:1033–1040. doi: 10.1016/j.bcp.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS medicine. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature reviews. Drug discovery. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Biochem. 2006;352:282–285. doi: 10.1016/j.ab.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Heikal AA. Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B. 2009;95:46–57. doi: 10.1016/j.jphotobiol.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Samudio IJ, Zhang W, Estrov Z, Pelicano H, Harris D, Frolova O, Hail N, Jr., Chen W, Kornblau SM, et al. Simultaneous inhibition of PDK1/AKT and Fms-like tyrosine kinase 3 signaling by a small-molecule KP372-1 induces mitochondrial dysfunction and apoptosis in acute myelogenous leukemia. Cancer Res. 2006;66:3737–3746. doi: 10.1158/0008-5472.CAN-05-1278. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, Yu Z, Xu L, Wang X, Yang Y, Loscalzo J. Genetically Encoded Fluorescent Sensors for Intracellular NADH Detection. Cell Metab. 2011;14:555–566. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang Y. Profiling metabolic states with genetically encoded fluorescent biosensors for NADH. Curr Opin Biotechnol. 2015;31C:86–92. doi: 10.1016/j.copbio.2014.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.