Abstract
The original pink-eyed dilution (p) on chromosome 7 is a very old spontaneous mutation in mice. The oculocutaneous albinism II (Oca2) gene has previously been identified as the p gene. Oca2 transcripts have been shown to be absent in the skin of SJL/J mice with the original p mutant allele (Oca2p); however, the molecular genetic lesion underlying the original Oca2p allele has never been reported. The NCT mouse (commonly known as Nakano cataract mouse) has a pink-eyed dilution phenotype, which prompted us to undertake a molecular genetic analysis of the Oca2 gene of this strain. Our genetic linkage analysis suggests that the locus for the pink-eyed dilution phenotype of NCT is tightly linked to the Oca2 locus. PCR cloning and nucleotide sequence analysis indicates that the NCT mouse has a nonsense nucleotide substitution at exon 7 of the Oca2 gene. Examination of three mouse strains (NZW/NSlc, SJL/J, and 129X1/SvJJmsSlc) with the original Oca2p allele revealed the presence of a nonsense nucleotide substitution identical to that in the NCT strain. RT-PCR analysis revealed that the Oca2 transcripts were absent in the skin of NCT mice, suggesting intervention of the nonsense-mediated mRNA decay pathway. Collectively, the data in this study indicate that the nonsense nucleotide substitution in the Oca2 gene underlies the Oca2p allele. Our data also indicate that the NCT mouse can be used not only as a cataract model, but also as a model for human type II oculocutaneous albinism.
Keywords: mouse, nonsense mutation, oculocutaneous albinism II, pink-eyed dilution
Introduction
The original pink-eyed dilution (p) on chromosome 7 is a very old spontaneous mutation present in a various mouse strains (e.g., SJL/J, 129/J, P/J, and FS/Ei) [2]. In addition to the original mutant allele, more than 80 spontaneous, chemically induced, and radiation-induced mutant alleles have been identified and generated to date (http://www.informatics.jax.org/allele/summary?markerId=MGI:97454). Homozygotes of these mutant alleles generally have coat and eye pigment dilution due to reduced melanin content in the melanocytes and retinal epithelium, even though the degree of dilution varies depending on the allele. Humans with type II oculocutaneous albinism (OCA, type II) exhibit a similar phenotype to the p mutant mice (http://omim.org/entry/203200). The locus for OCA, type II has furthermore been mapped to a region of chromosome 15q that is orthologous to the region of mouse chromosome 7 containing the p locus. On the basis of these observations, the p mutants have been proposed to provide as mouse models for type II oculocutaneous albinism.
The mouse p (Oca2) [5] and human OCA2 [4] genes have been identified and characterized. Oca2 is approximately 300 kb in size and contains 24 exons that code for an integral melanosomal membrane protein [14, 16]. Despite extensive studies in the past 2 decades, the function of the OCA2 protein and the mechanisms by which defects in the Oca2 gene lead to the pink-eyed dilution phenotype remain unknown. It was reported that the OCA2 is an anion transporter that controls the acidification of melanosomes [1, 13]. Other studies suggest that the OCA2 is involved in regulating the maturation of melanosomes and stabilizing or trafficking of melanosomal membrane proteins including tyrosinase that plays a key role in melanin synthesis [3, 15, 18]. In addition to the functions in the melanosome, OCA2 regulates the proliferative activities of epidermal melanoblasts [8].
Cloning of the Oca2 gene led to the elucidation of mutations underlying p mutant alleles, including missense nucleotide substitutions (e.g. Oca2p-18J), deletions (e.g. Oca2p-J), inversions (e.g. Oca2p-25H), and insertions (e.g. Oca2p-un). Unlike in the cases of these mutant alleles, the molecular genetic basis of the original p allele (Oca2p) has not yet been elucidated. Southern blot analysis of the genomic DNA of the mouse strains with the original p allele using an Oca2 probe demonstrated a hybridization pattern distinct from that of mouse strains with the wild-type allele [2, 5]. However, approximately the same number of hybridizing fragments was observed in the original p mutant strains. It thus seems that the original p mutation is not a result of a deletion, duplication, inversion, insertion, or translocation in the Oca2 gene. Intriguingly, northern blot analysis demonstrated that Oca2 transcripts were missing in the skin of the SJL/J mouse strain [5]. The molecular genetic basis of the absence of Oca2 transcripts in the mouse strains with the original p allele, however, has not been elucidated.
The NCT inbred mouse strain (commonly known as Nakano cataract mouse) is a mutant mouse model for hereditary cataract [6, 12, 17]. In addition to cataracts, NCT mice exhibit a pink-eyed dilution phenotype. In a previous study, we revealed that cataracts in the NCT mouse are caused by a hypomorphic mutation of the coproporphyrinogen oxidase (Cpox) gene [11]; however, the genetic features of the pink-eyed dilution phenotype of the NCT mouse, including the chromosomal position and identity of the mutant gene, remained unknown. In order to characterize these genetic features, genetic linkage analysis of the pink-eyed dilution phenotype in NCT mice was conducted in this study. Based on the finding of this analysis, PCR cloning and mutation screening of the Oca2 gene was carried out. The Oca2 gene was furthermore examined in other mouse strains with the original Oca2p allele.
Materials and Methods
Mouse strains
NCT mice were obtained from Laboratory Animal Resource Bank, National Institute of Biomedical Innovation, Japan. These mice were derived from a colony maintained at Kitasato University [6]. A congenic BALB.NCT-Cpoxnct mouse strain, in which the mutant Cpoxnct locus for cataract was introduced into the background of a BALB/c strain [10], was obtained from RIKEN BRC through the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan (RBRC00422). These mice have an albino coat (Tyrc, Tyrp1b, nonagouti (a)). SJL/J mice were purchased from Charles River Laboratories Japan (Yokohama, Japan). C57BL/6J, BALB/c, NZW/NSlc, and 129X1/SvJJmsSlc mice were purchased from Japan SLC (Hamamatsu, Japan). All experimental procedures involving mice were carried out in accordance with the Regulations for Animal Experimentation of Shinshu University.
Immunohistochemistry of the skin
Dorsal dermal specimens were obtained from 12 week-old C57BL/6J and NCT mice. Formalin-fixed paraffin-embedded tissues were sliced into 3 µm thick sections. Hematoxylin and eosin (HE) staining and immunohistochemical staining of tyrosinase-related protein 1 (TRP1) were performed as described [19]. For heat-mediated antigen retrieval, slides were incubated at 95 – 100°C in Target Retrieval Solution, pH 9 (S2367, DAKO, Glostrup, Denmark) for 40 min. Slides were incubated with polyclonal anti-TRP1 antibody (sc25543, Santa Cruz Biotechnology, Dallas, TX, USA) overnight at 4°C. Anti-rabbit IgG antibody (VECTASTAIN ABC kit, Vector Laboratories, Burlingame, CA, USA) was used as secondary antibody.
Measurement of hair melanin content
Dorsal hairs were collected form 12 week-old C57BL/6J, NCT, and BALB/c mice. Total melanin content in the hair was measured as described [9]. Hair sample (1 mg) was suspended in 100 µl of distilled water in a 15-ml screw-capped conical test tube, to which 900 µl Soluene-350 (Perkin-Elmer, Waltham, MA, USA) was added. The tube was vortex-mixed and heated at 100°C (boiling water bath) for 15 min. The tube was vortex-mixed again and was heated at 100°C (boiling water bath) for an additional 15 min to solubilize melanin. The mixture was centrifuged at 4,000 g for 3 min, and the supernatant was analyzed for optical density at 500 nm. Analysis of variance was used to examine any significant differences in melanin content among the strains. Subsequent post-hoc tests to determine significant differences in the pair-wise comparisons were performed using the Tukey-Kramer test.
Genetic linkage analysis of pink-eyed dilution in NCT mice
A female BALB.NCT-Cpoxnct mouse (homozygous for the normal Oca2 allele) was crossed with a male NCT mouse to give (BALB.NCT-Cpoxnct × NCT)F1 mice. A female F1 mouse was crossed to a male NCT to give (BALB.NCT-Cpoxnct × NCT)F1 × NCT backcross mice. Backcross mice were diagnosed for pink-eyed dilution phenotype at 4 weeks of age by visual inspection, and then sacrificed. Liver genomic DNA was extracted, and mice were genotyped for the D7Mit346 microsatellite marker locus by PCR amplification and agarose gel electrophoresis.
PCR cloning and nucleotide sequence analysis of the Oca2 gene
Primer pairs for PCR-amplification and sequencing of the 24 exons and putative promoter of the mouse Oca2 gene are listed in Table 1. These primers were designed based on the mouse genome sequence (GRCm38/mm10). Genomic DNA was isolated from the liver of mice using standard methods. PCR amplification was performed using a TaKaRa LA Taq DNA polymerase (TAKARA BIO INC., Otsu, Japan) using the methods recommended by the manufacturer. The resulting PCR products were purified using an UltraClean PCR Clean Up Kit (Mo Bio Laboratories, Carlsbad, CA, USA) and sequenced using a BigDye Cycle Sequencing FS Ready Reaction Kit (Life Technologies, Grand Island, NE, USA) and an ABI 310 automated sequencer.
Table 1. Nucleotide sequence of oligonucleotide primers used for analysis of the mouse Oca2 gene.
| Target | Forwad primer | Reverse primer |
|---|---|---|
| Exon 1 | 5′-GGAAAGCAGCTCCCTTGGCT-3′ | 5′-TTACCCTACTAGCTTCCCAG-3′ |
| Exon 2 | 5′-GGAGACACCCACATACTGTC-3′ | 5′-GCCCATCATGAAGGGATTAGTACCCA-3′ |
| Exon 3 | 5′-AGTGCACTCCTGCAAGCATC-3′ | 5′-GCCTGTCAAAGATCTGACAG-3′ |
| Exon 4 | 5′-CTGTCTTGTAGACATGCTCAG-3′ | 5′-GCTAACAGAAGCTATGTGCAGCC-3′ |
| Exon 5 | 5′-GCCATGTCATGCATACTCAGTG-3′ | 5′-GTCCCAACATAGGCTCTCTTG-3′ |
| Exon 6 | 5′-CAATCAAGGCTCATCGCAGC-3′ | 5′-GACCTTTGAGAAGCCTGAACCT-3′ |
| Exon 7 | 5′-CAGTGGTAAGCAGGCCAAGT-3′ | 5′-CCGTTGGCTTCCATCAAATCC-3′ |
| Exon 8 | 5′-CCCACAGGTTCTACATTCAAGG-3′ | 5′-GTGCTATCAGACTTCTCCAG-3′ |
| Exon 9 | 5′-CAGGTCTGTACTCAGGGCTTTTC-3′ | 5′-CCTAGAATCCGTAGGAATTGCTCC-3′ |
| Exon 10 | 5′-GAGCATCCTTTCACAGTCAC-3′ | 5′-ATGCCTCTATGAATTGTGGGCC-3′ |
| Exon 11 | 5′-CAATGTCTGCGCTTGCAGTAAG-3′ | 5′-GATAGGATCAGCAACTGTGGG-3′ |
| Exon 12 | 5′-GCTTCAAGACTGATCAAGCC-3′ | 5′-GGTCTTGATGTGTAGCCCTAAC-3′ |
| Exon 13 | 5′-GATAAGGAGACCTTGGCACAC-3′ | 5′-CATTTGCTCCAAACTTGGCC-3′ |
| Exon 14 | 5′-CGATGTGCCTTCCTGTACAAG-3′ | 5′-GCAAGAACCTTGGTTCACAG-3′ |
| Exon 15 | 5′-GTCATGTAAGCACTGGCTTTTC-3′ | 5′-CTAAGAATGCAGCATCCCAGG-3′ |
| Exon 16 | 5′-GCTTCCACGATGACTTTCAGAG-3′ | 5′-C-CTTCTGCTAAGTTGCTACACTG3′ |
| Exon 17 | 5′-CTTACAAGGCTGTGCCAGGATCCAC-3′ | 5′-GCCATGATTCAGAATGCCTTG-3′ |
| Exon 18 | 5′-CCTCTGAGGGACTCATAGGA-3′ | 5′-CACAGCAGCCTTTGGTACAG-3′ |
| Exon 19 | 5′-CTATCGAGACAAGAGAAGGCTG-3′ | 5′-GCCTTAGGCTTCACTCTGCAAAC-3′ |
| Exon 20 | 5′-GAAAGGATAACCTGTGCCTACC-3′ | 5′-CATCTCTAGGTTACCCAGGG-3′ |
| Exon 21 | 5′-CCAAGGTATAGGTATGGCCC-3′ | 5′-CAACAGAGGAGCTGGTTTCATGCC-3′ |
| Exon 22 | 5′-GCCAGTGCATTTAACGGCTG-3′ | 5′-GGAAGTCTCCAGGATTCTCC-3′ |
| Exon 23 | 5′-GGGACCTTCTCCTTAGCATC-3′ | 5′-GTGCCTTGATCTGAGCTCATC-3′ |
| Exon 24 | 5′-CCTTACGTAAAGATTCTGGGGC-3′ | 5′-GCCATGTATATCATGGTGAGC-3′ |
| cDNA (Set 1) | 5′-CTGGAAAACTACTCTGTGAACCTC-3′ | 5′-AGCCTCCCTGCTCACTATCT-3′ |
| cDNA (Set 2) | 5′-CCAGAAGATCAGCGCTTTGC-3′ | 5′-GGAGATAGCACATCCCAATG-3′ |
| Tyr cDNA | 5′-TGAACAATGGCTGCGAAGGC-3′ | 5′-GCAGCAATAACAGCTCCCAC-3′ |
Reverse transcription-PCR analysis of Oca2 in the skin of mice
Messenger RNA (mRNA) was extracted from the dorsal skin of mice at 12 weeks of age using the QuickPrep Micro mRNA Purification Kit (Amersham Biosciences, Buckinghamshire, England). First-strand complementary DNA (cDNA) was synthesized using a First-Strand cDNA Synthesis Kit (Amersham Biosciences). A cDNA fragment for the Oca2 gene was then amplified by PCR using 2 sets of primers (Table 1). These primers were designed based on the reported sequence for the mouse Oca2 (NM_021879). The primer set 1 could amplify a region that contains the nonsense substitution in NCT. The primer set 2 was designed to amplify a downstream region of the nonsense substitution. PCR amplification was performed using TaKaRa LA Taq DNA polymerase in 25 µl reactions according to the manufacturer’s instructions. PCR reactions were carried out using the following conditions: initial denaturation at 94°C for 1 min, followed by 40 cycles at 94°C for 20 s, 55°C for 15 s, and 72°C for 1 min. A 10 µl aliquot of each PCR product was subjected to agarose gel electrophoresis. The PCR products were purified and directly sequenced with the primers.
Results
NCT mice have pink-eyed dilution phenotype
NCT mice have a pink-eyed dilution appearance (Fig. 1). Melanin granules were observed in hair bulbs of both wild type C57BL/6J and NCT mice (Fig. 2A). Immunohistochemical staining of the dorsal dermis revealed no significant difference between wild type C57BL/6J and NCT mice in expression of melanocyte-specific protein TRP1 in hair bulbs, where follicular melanocytes localize (Fig. 2B). These results suggest that decent amount of melanocytes are present in the NCT skin, consistent with the observation in pink-eyed dilution mutant mice [8]. Hair melanin level in NCT mice was reduced to 11% of that in C57BL/6J mice (Fig. 2C), consistent with the result in pink-eyed dilution mutant mice [7]. Overall, these results suggest that NCT is a pink-eyed dilution mutant.
Fig. 1.
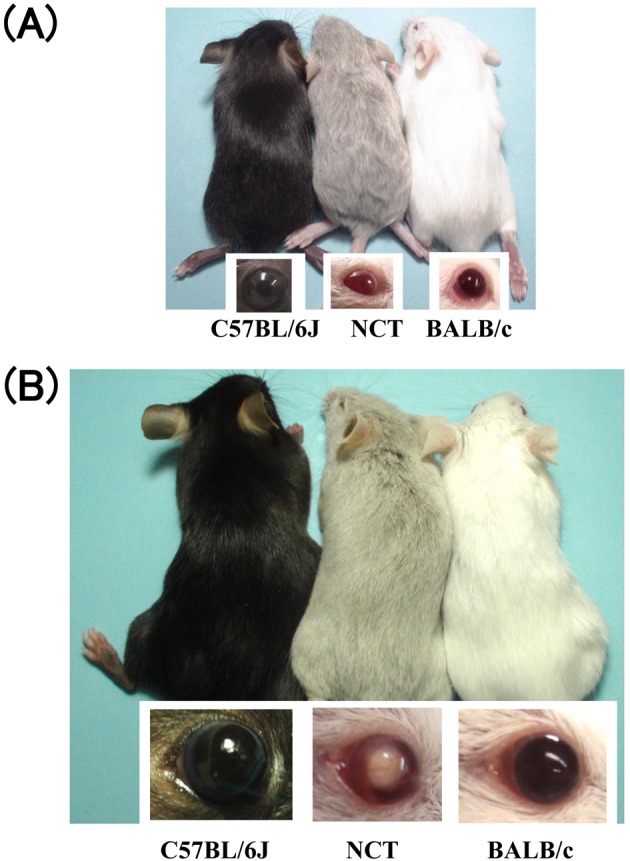
Image of C57BL/6J, NCT, and BALB/c mice at 2 weeks (A) and 12 weeks (B) of age. Note that the NCT mice exhibit pink-eyed dilute coat color phenotype. The 12-week-old NCT has also cataracts.
Fig. 2.
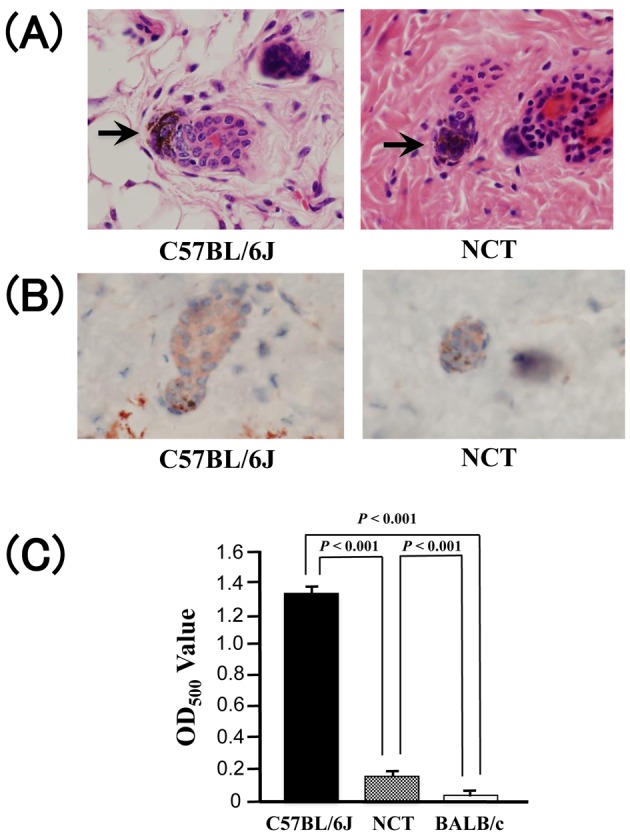
(A) Hematoxylin and eosin staining of the dorsal dermis of 12-week-old C57BL/6J and NCT mice (original magnification ×400). Melanin granules in hair bulb are indicated by arrows. (B) Immunohistochemical staining of TRP1 in the dorsal dermis of 12-week-old C57BL/6J and NCT mice (original magnification ×400). (C) Comparison of hair melanin levels between 12-week-old C57BL/6J, NCT, and BALB/c (mean ± SD; n = 3).
The locus for the pink-eyed dilution phenotype of NCT mice is tightly linked to the Oca2 gene
All (BALB.NCT-Cpoxnct × NCT)F1 mice had black nonagouti coats and eyes at 4 weeks of age, suggesting that the pink-eyed dilution of NCT is genetically recessive. (BALB.NCT-Cpoxnct × NCT)F1 × NCT backcross mice (N = 42) exhibited either normal (N = 17) or pink-eyed dilution (N = 25) phenotypes. This segregation ratio was not statistically different from the expected ratio of 1:1 for the Mendelian recessive trait (χ2 = 1.52; P>0.10). Because the mouse Oca2 (p) gene, mutations of which are responsible for the pink-eyed dilution phenotype, is located at 33.44 cM on chromosome 7, genetic linkage of the pink-eyed dilution phenotype and a D7Mit346 marker locus (33.88 cM) was examined in the backcross mice. A 100% concordance was observed between the pink-eyed dilution phenotype and homozygosity for the NCT allele of D7Mit346 (Fig. 3). These findings suggest that the pink-eyed dilution phenotype of NCT mice is caused by a mutation in the Oca2 (p) gene.
Fig. 3.
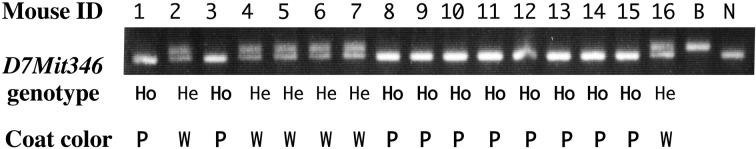
Genetic linkage of pink-eyed dilution and a D7Mit346 marker locus. Only the data for 16 (BALB.NCT-Cpoxnct × NCT)F1 × NCT backcross mice are shown. B and N stand for BALB.NCT-Cpoxnct and NCT mice, respectively. Ho and He stand for the homozygous and heterozygous states at the D7Mit346 marker locus, respectively. P and W stand for pink-eyed dilution and wild-type phenotypes, respectively. Note that there is 100% concordance between the pink-eyed dilution phenotype and homozygosity for the NCT allele of D7Mit346.
NCT mice have a nonsense nucleotide substitution in the Oca2 gene
PCR cloning of the exons of the Oca2 gene from the NCT mouse genome was carried out. When compared to the mouse reference genome sequence (GRCm38/mm10), eighteen nucleotide substitutions were identified in the upstream region of NCT mouse Oca2 (Table 2). The effects of these nucleotide substitutions on the promoter activity of the mouse Oca2 gene are unclear. Elucidation of these effects will ultimately require further experimental analysis.
Table 2. List of nucleotide alteration found in the Oca2 gene of mouse strains with the original Oca2p allele.
| RGCm38 coodinate |
Position in the gene |
Reference | NCT | NZW/ NSlc |
SJL/J | 129X1/ SvJJmsSlc |
Effect | SNP reference ID |
|---|---|---|---|---|---|---|---|---|
| 56,239,117 | Upstream | G | A | A | A | A | rs32975435 | |
| 56,239,155 | Upstream | C | T | T | T | T | rs32975437 | |
| 56,239,216 | Upstream | T | G | G | G | G | rs32975438 | |
| 56,239,238 | Upstream | T | C | C | C | C | rs46887862 | |
| 56,239,239 | Upstream | T | A | A | A | A | rs50866175 | |
| 56,239,254 | Upstream | G | T | T | T | T | rs50711255 | |
| 56,239,277 | Upstream | T | C | C | C | C | rs32975439 | |
| 56,239,299 | Upstream | G | A | A | A | A | rs108518501 | |
| 56,239,318 | Upstream | A | G | G | G | G | rs32975440 | |
| 56,239,329 | Upstream | A | G | G | G | G | rs32975441 | |
| 56,239,404 | Upstream | G | C | C | C | C | rs32975443 | |
| 56,239,413 | Upstream | T | C | C | C | C | rs49155013 | |
| 56,239,448 | Upstream | A | C | C | C | C | rs32976114 | |
| 56,239,488 | Upstream | G | A | A | A | A | rs32976116 | |
| 56,239,538 | Upstream | C | T | T | T | T | rs32976117 | |
| 56,239,604 | Upstream | T | A | A | A | A | rs50836782 | |
| 56,239,607 | Upstream | A | G | G | G | G | rs50507765 | |
| 56,239,626 | Upstream | T | C | C | C | C | rs32976118 | |
| 56,239,800 | exon 1 | G | A | A | A | A | non-coding | rs32976120 |
| 56,239,830 | exon 1 | T | G | G | G | G | non-coding | rs32976121 |
| 56,239,850 | exon 1 | T | C | C | C | C | non-coding | rs32976122 |
| 56,254,868 | exon 2 | C | T | T | T | T | silent | rs32975605 |
| 56,254,910 | exon 2 | T | C | C | C | C | silent | rs32065217 |
| 56,280,859 | exon 4 | T | A | A | A | A | silent | rs51221903 |
| 56,293,785 | exon 7 | A | T | T | T | T | silent | rs32980939 |
| 56,293,847 | exon 7 | C | T | T | T | T | missense (p.A256V) | rs32980940 |
| 56,293,864 | exon 7 | C | T | T | T | T | nonsense (p.R262X) | rs32980941 |
| 56,295,441 | exon 8 | C | G | G | G | G | missense (p.L266V) | rs31110637 |
| 56,295,446 | exon 8 | C | T | T | T | T | silent | rs31471555 |
| 56,295,506 | exon 8 | T | C | C | C | C | silent | rs32976441 |
| 56,297,217 | exon 9 | A | C | C | C | C | silent | rs31936191 |
| 56,324,663 | exon 14 | A | G | G | G | G | silent | rs50144561 |
| 56,327,313 | exon 15 | G | C | C | C | C | silent | rs50712907 |
| 56,328,831 | exon 16 | T | C | C | C | C | silent | rs32977966 |
| 56,413,733 | exon 20 | A | T | T | T | T | silent | rs32979629 |
| 56,429,368 | exon 23 | A | C | C | C | C | missense (p.K794Q) | rs31090430 |
| 56,535,958 | exon 24 | G | A | A | A | A | missense (p.V811M) | rs32985345 |
In the coding exons (exons 2-24), 16 nucleotide substitutions were identified, including 11 silent substitutions, 4 missense substitutions, and most importantly one nonsense substitution in exon 7 (Table 2). Analysis of the effects of the p.A256V, p.L266V, p.K794Q, and p.V811M amino acid substitutions caused by the missense substitutions with the SIFT algorithm (http://siftdna.org/www/SIFT_BLink_submit.html) predicted them to be tolerated with scores of 0.22, 1.00, 1.00, and 1.00, respectively, suggesting that they are not pathogenic. The nonsense substitution was a C to T substitution in the codon for the 262nd amino acid of the OCA2 protein (p.R262X) (Fig. 4).
Fig. 4.
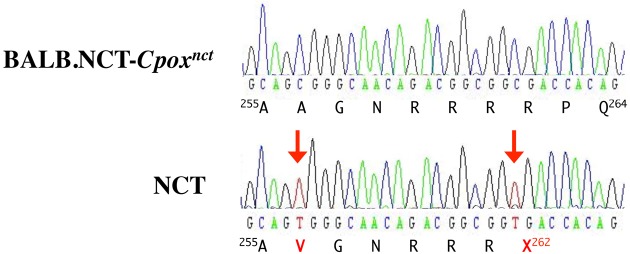
Sequence chromatograms of the PCR products corresponding to a portion of exon 7 of the Oca2 gene from BALB.NCT-Cpoxnct and NCT mice. Amino acid sequences encoded by the nucleotide sequences are also shown. Nucleotide substitutions in NCT are indicated by arrows.
Other mouse strains with the original Oca2p allele have a nonsense nucleotide substitution identical to that found in NCT mice
All nucleotide substitutions identified in NCT mice were accounted in the mouse SNP database (http://www.ncbi.nlm.nih.gov/snp/; Table 2), and the ID number of the identified nonsense substitution is rs32980941. The database revealed that only the NZW/LacJ mouse strain has the same thymine nucleotide substitution as the NCT strain at rs32980941, while 14 other strains (BALB/cByJ, A/J, AKR/J, C3H/HeJ, DBA/2J, FVB/NJ, 129S1/SvImJ, KK/HlJ, NOD/LtJ, PWD/PhJ, WSB/EiJ, MOLF/EiJ, BTBR T+ Itpr3tf/J, and CAST/EiJ) have a cytosine nucleotide at that position (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi?ss=ss48239412). Intriguingly, all 14 above-mentioned strains have the wild-type allele at the Oca2 locus, while the NZW/LacJ strain has the original Oca2p allele (http://www.informatics.jax.org/external/festing/mouse/docs/NZW.shtml). This strongly suggests that the nonsense nucleotide substitution is the primary cause of the original Oca2p mutant allele. Genotypes of other mouse strains with the original Oca2p allele for rs32980941 were, however, not present in the database. Next, NZW/NSlc and two representative mouse strains with the original Oca2p allele, SJL/J and 129X1/SvJJmsSlc, were assessed, and it was found that the three strains possessed the nonsense nucleotide substitution in exon 7 of the Oca2 gene (Table 2). Moreover, these 3 strains shared the identical nucleotide substitutions at all 36 other positions with NCT.
Only traces of the Oca2 transcripts are expressed in the NCT skin
A previous report demonstrated that Oca2 transcripts were absent in the skin of SJL/J mice [5]. To corroborate whether this is the case in the skin of NCT, RT-PCR analysis was conducted. Consistent with the previous report, no RT-PCR product was evident from the skin of NCT (Fig. 5A). Roughly equal amount of RT-PCR products for a melanocyte-specific marker tyrosinase (Tyr) were obtained from the skin of C57BL/6J and NCT, suggesting that the absence of Oca2 transcripts in the skin of NCT was not due to extreme reduction in melanocyte quantity in the skin.
Fig. 5.
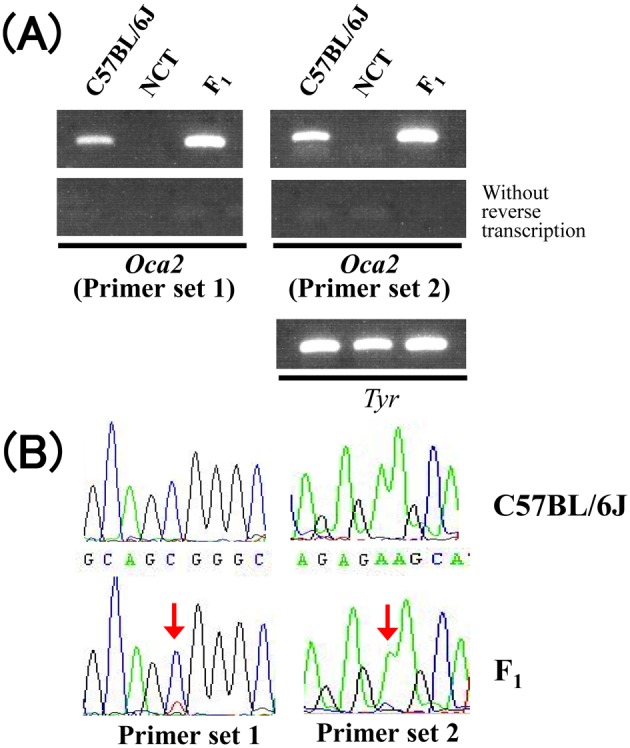
(A) Agarose gel image of the RT-PCR products for Oca2 and Tyr (internal control) mRNA derived from the skin of C57BL/6J, NCT, and (C57BL/6J × NCT)F1 (F1) mice. (B) Sequence chromatograms of the RT-PCR products for Oca2 from C57BL/6J and (C57BL/6J × NCT)F1 (F1) mice. Overlaps of 2 different nucleotides are indicated by arrows.
Abundant Oca2 RT-PCR products were obtained also from the skin of a (C57BL/6J × NCT)F1 mouse that was heterozygous for the wild and mutant Oca2 alleles (Fig. 5A). Nucleotide polymorphisms between the 2 alleles allowed us to distinguish transcripts derived from the 2 alleles. Indeed, nucleotide sequence analysis of the PCR products revealed overlaps of 2 peaks at the polymorphic nucleotides, indicating that the PCR products were a mixture of wild and mutant types (Fig. 5B). However, the peak heights of chromatograms for the mutant products were greatly lower than that for the wild products. These results indicated that down-regulation occurs intrinsically and exclusively to the mutant Oca2 transcripts in melanocytes.
Discussion
The data obtained in this study indicate that the NCT mouse is an authentic pink-eyed dilution mutant. Most importantly, the data in this study also indicate that the nonsense nucleotide substitution in exon 7 of the Oca2 gene underlies the mouse original pink-eyed dilution allele (Oca2p). The absence of Oca2 transcripts in the skin of mice with the Oca2p allele [5] can be accounted for by the action of the nonsense-mediated mRNA decay pathway, which eliminates mRNA transcripts that contain premature stop codons. There remains a possibility that a trace amount of full-length Oca2 transcripts and truncated OCA2 protein encoded by the transcripts are expressed in the NCT skin; however, it is highly unlikely that the truncated OCA2 protein retains normal function. Our data corroborate that the mouse strains with the original Oca2p allele represent bona fide models for human type II oculocutaneous albinism. One might wonder why the molecular genetic basis of the original Oca2p allele has not been elucidated for more than 20 years since identification of the Oca2 as a causative gene of the mouse pink-eyed dilution. Obviously, absence of Oca2 transcripts in the skin of mice with the original Oca2p allele [5] should have impeded molecular cloning and nucleotide sequencing of the transcripts. Molecular cloning of the Oca2 gene from the mouse genome should have been a laborious task, because the size of the gene is as large as 300 kb. To our knowledge, there had been no report of molecular cloning of the mouse Oca2 gene until the mouse genome project was completed. Strangely enough, the genotype of SJL/J and 129X1/SvJ for the SNP rs32980941 was not present in the current mouse SNPs database. The reason for the absence of information is not clear; however, it might suggest that the database is still incomplete.
The present data further indicate that the NCT mouse has the original Oca2p mutant allele, although it is not clear how the NCT mouse acquired the Oca2p allele. The first report of the NCT mouse dates as far back as 1960 [12]. This study reported a cataractous mouse in a colony that was obtained from the National Institute of Genetics, Japan. However, details of the colony, including the strain name, are not described. The fact that NCT, NZW/NSlc, SJL/J, and 129X1/SvJ share the identical nucleotide substitutions in the Oca2 negates the possibility that the nonsense nucleotide substitution for the pink-eyed dilution phenotype occurred independently in these strains. Rather, it is highly likely that the nonsense substitution has derived from a single mutation event. It has been suggested that the original Oca2p mutation occurred in Japanese wild mice, Mus musculus molossinus, long before the laboratory mouse strains were established [2]. It is further assumed that the mutant allele was subsequently inherited to ancestors of several modern mouse strains including SJL/J, 129/J, P/J, and FS/Ei. As a result, these mouse strains retain different extents of the chromosome 7 segment centered around the Oca2p locus derived from Mus musculus molossinus [2]. It is likely that NCT has the original Oca2p allele derived from Mus musculus molossinus, because NCT shares the identical nucleotide substitutions with the NZW/NSlc, SJL/J, and 129X1/SvJJmsSlc strains. Regardless of the history of the Oca2p allele of NCT, our data indicate that the NCT mouse can be used not only as a cataract model, but also as a model for human type II oculocutaneous albinism.
Acknowledgments
We thank Dr. Furutachi and the Laboratory Animal Resource Bank, National Institute of Biomedical Innovation, Japan, for providing the NCT mice. We also thank RIKEN BRC and the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan, for providing the BALB.NCT-Cpoxnct mice (RBRC00422).
References
- 1.Brilliant M.H.2001. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment. Cell Res. 14: 86–93. doi: 10.1034/j.1600-0749.2001.140203.x [DOI] [PubMed] [Google Scholar]
- 2.Brilliant M.H., Ching A., Nakatsu Y., Eicher E.M.1994. The original pink-eyed dilution mutation (p) arose in Asiatic mice: implications for the H4 minor histocompatibility antigen, Myod1 regulation and the origin of inbred strains. Genetics 138: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen K., Manga P., Orlow S.J.2002. Pink-eyed dilution protein controls the processing of tyrosinase. Mol. Biol. Cell 13: 1953–1964. doi: 10.1091/mbc.02-02-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durham-Pierre D., Gardner J.M., Nakatsu Y., King R.A., Francke U., Ching A., Aquaron R., del Marmol V., Brilliant M.H.1994. African origin of an intragenic deletion of the human P gene in tyrosinase positive oculocutaneous albinism. Nat. Genet. 7: 176–179. doi: 10.1038/ng0694-176 [DOI] [PubMed] [Google Scholar]
- 5.Gardner J.M., Nakatsu Y., Gondo Y., Lee S., Lyon M.F., King R.A., Brilliant M.H.1992. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science 257: 1121–1124. doi: 10.1126/science.257.5073.1121 [DOI] [PubMed] [Google Scholar]
- 6.Hara A., Matsumoto M., Uga S.1999. Morphological study on cataractogenesis of the Nakano mouse lens. Graefes Arch. Clin. Exp. Ophthalmol. 237: 249–255. doi: 10.1007/s004170050226 [DOI] [PubMed] [Google Scholar]
- 7.Hirobe T., Ito S., Wakamatsu K.2011. The mouse pink-eyed dilution allele of the P-gene greatly inhibits eumelanin but not pheomelanin synthesis. Pigment. Cell Melanoma. Res. 24: 241–246. doi: 10.1111/j.1755-148X.2010.00783.x [DOI] [PubMed] [Google Scholar]
- 8.Hirobe T., Kawa Y., Mizoguchi M., Ito S., Wakamatsu K.2002. Effects of genic substitution at the pink-eyed dilution locus on the proliferation and differentiation of mouse epidermal melanocytes in vivo and in vitro. J. Exp. Zool. 292: 351–366. doi: 10.1002/jez.10051 [DOI] [PubMed] [Google Scholar]
- 9.Ito S., Nakanishi Y., Valenzuela R.K., Brilliant M.H., Kolbe L., Wakamatsu K.2011. Usefulness of alkaline hydrogen peroxide oxidation to analyze eumelanin and pheomelanin in various tissue samples: application to chemical analysis of human hair melanins. Pigment. Cell Melanoma. Res. 24: 605–613. doi: 10.1111/j.1755-148X.2011.00864.x [DOI] [PubMed] [Google Scholar]
- 10.Matsuzawa A., Wada E.1988. Retarded and distinct progress of lens opacification in congenic hereditary cataract mice, Balb/c-nct/nct. Exp. Eye Res. 47: 705–711. doi: 10.1016/0014-4835(88)90038-3 [DOI] [PubMed] [Google Scholar]
- 11.Mori M., Gotoh S., Taketani S., Hiai H., Higuchi K.2013. Hereditary cataract of the Nakano mouse: involvement of a hypomorphic mutation in the coproporphyrinogen oxidase gene. Exp. Eye Res. 112: 45–50. doi: 10.1016/j.exer.2013.04.005 [DOI] [PubMed] [Google Scholar]
- 12.Nakano K., Yamamoto S., Kutsukake G., Ogawa H., Nakajima A., Takano E.1960. Hereditary cataract in mice. Jpn. J. Clin. Ophthalmol 14: 1772–1776. [Google Scholar]
- 13.Puri N., Gardner J.M., Brilliant M.H.2000. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J. Invest. Dermatol. 115: 607–613. doi: 10.1046/j.1523-1747.2000.00108.x [DOI] [PubMed] [Google Scholar]
- 14.Rosemblat S., Durham-Pierre D., Gardner J.M., Nakatsu Y., Brilliant M.H., Orlow S.J.1994. Identification of a melanosomal membrane protein encoded by the pink-eyed dilution (type II oculocutaneous albinism) gene. Proc. Natl. Acad. Sci. USA 91: 12071–12075. doi: 10.1073/pnas.91.25.12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosemblat S., Sviderskaya E.V., Easty D.J., Wilson A., Kwon B.S., Bennett D.C., Orlow S.J.1998. Melanosomal defects in melanocytes from mice lacking expression of the pink-eyed dilution gene: correction by culture in the presence of excess tyrosine. Exp. Cell Res. 239: 344–352. doi: 10.1006/excr.1997.3901 [DOI] [PubMed] [Google Scholar]
- 16.Sitaram A., Piccirillo R., Palmisano I., Harper D.C., Dell’Angelica E.C., Schiaffino M.V., Marks M.S.2009. Localization to mature melanosomes by virtue of cytoplasmic dileucine motifs is required for human OCA2 function. Mol. Biol. Cell 20: 1464–1477. doi: 10.1091/mbc.E08-07-0710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takehana M.1990. Hereditary cataract of the Nakano mouse. Exp. Eye Res. 50: 671–676. doi: 10.1016/0014-4835(90)90112-8 [DOI] [PubMed] [Google Scholar]
- 18.Toyofuku K., Valencia J.C., Kushimoto T., Costin G.E., Virador V.M., Vieira W.D., Ferrans V.J., Hearing V.J.2002. The etiology of oculocutaneous albinism (OCA) type II: the pink protein modulates the processing and transport of tyrosinase. Pigment. Cell Res. 15: 217–224. doi: 10.1034/j.1600-0749.2002.02007.x [DOI] [PubMed] [Google Scholar]
- 19.Uchiyama R., Uhara H., Uchiyama A., Ogawa E., Takazawa Y., Ashida A., Koga H., Hayashi K., Kiniwa Y., Okuyama R.2014. 5-Hydroxymethylcytosine as a useful marker to differentiate between malignant melanomas and benign melanocytic nevi. J. Dermatol. Sci. 73: 161–163. [DOI] [PubMed]


