Abstract
Oca2p-cas (oculocutaneous albinism II; pink-eyed dilution castaneus) is a coat color mutant gene on mouse chromosome 7 that arose spontaneously in wild Mus musculus castaneus mice. Mice homozygous for Oca2p-cas usually exhibit pink eyes and gray coat hair on the non-agouti genetic background, and this ordinary phenotype remains unchanged throughout life. During breeding of a mixed strain carrying this gene on the C57BL/6J background, we discovered a novel spontaneous mutation that causes darkening of the eyes and coat hair with aging. In this study, we developed a novel mouse model showing this unique phenotype. Gross observations revealed that the pink eyes and gray coat hair of the novel mutant young mice became progressively darker in color by approximately 3 months after birth. Light and transmission-electron microscopic observations revealed a marked increase in melanin pigmentation of coat hair shafts and choroid of the eye in the novel mice compared to that in the ordinary mice. Sequence analysis of Oca2p-cas revealed a 4.1-kb deletion involving exons 15 and 16 of its wild-type gene. However, there was no sequence difference between the two types of mutant mice. Mating experiments suggested that the novel mutant phenotype was not inherited in a simple fashion, due to incomplete penetrance. The novel spontaneous mutant mouse is the first example of progressive hair darkening animals and is an essential animal model for understanding of the regulation mechanisms of melanin biosynthesis with aging.
Keywords: animal model, coat color, hair darkening, mouse, Oca2
Introduction
Many genetic loci affecting coat color in mice have so far been reported [6, 26]. Oca2(oculocutaneous albinism II) on mouse chromosome 7 is one of the oldest coat color loci. At this locus, 88 alleles have so far been reported, 20 spontaneous, 63 chemically and radiation induced, one gene-trapped, one transgenic and three targeted alleles [6]. Of these, Oca2p (oculocutaneous albinism II; pink-eyed dilution, old symbol p) is the first original spontaneous allele that arose in Japanese wild mice (Mus musculus molossinus), and it was introduced into common laboratory strains as a result of fancy mouse trade in the 19th century [4]. Mice homozygous for respective Oca2 alleles are generally characterized by hypopigmentation in both eyes and coat hair, the degree of which varies among alleles. Chemical analysis of melanin in Oca2p dorsal hair revealed that eumelanin synthesis is greatly inhibited but that pheomelanin synthesis is not inhibited [10]. Some specific alleles produce cleft palate, reproductive, endocrine or neurological disorders, and/or lethality [6].
In humans, OCA2 is one of several types of albinism and it is caused by mutations in the human orthologous OCA2 of the mouse Oca2 gene [18, 23]. The functions of the human OCA2 protein and also the mouse OCA2 protein are currently not clear. It has been suggested, however, that OCA2 may be involved either in the supply of substrates to the tyrosinase enzyme in melanin biosynthesis [2] or in the intracellular trafficking of this enzyme during melanosome maturation [27]. The inactivation of contiguous genes on human chromosome 15q11–q13, a region syntenic with the mouse Oca2 chromosomal region, is associated with human Prader-Willi syndrome (PWS) and Angelman syndrome (AS) [7]. The Oca2p−30PUb mouse carrying a large deletion between Oca2 and Ipw loci, in which some candidate genes for PWS are involved, is thought to be an excellent animal model for PWS and related syndromes [22].
Oca2p-cas (oculocutaneous albinism II; pink-eyed dilution castaneus) in mice is a spontaneous mutation that arose in second-generation descendants produced from brother-sister matings between wild M. m. castaneus mice captured in Bandar Lampung City (Sumatra), Indonesia [28]. Similar to the phenotype of the original Oca2p mice, mice homozygous for Oca2p-cas usually exhibit pink eyes and gray coat hair on the non-agouti genetic background, and this ordinary phenotype remains unchanged throughout life. The homozygotes are fully fertile and viable, and they display neither abnormal behavior nor abnormal external appearance [28]. Surprisingly, during breeding of a mixed strain (named B6;Cg-Oca2p-cas) carrying Oca2p-cas as a homozygous state on the genetic background of the C57BL/6J inbred strain, one of the authors (A. Ishikawa) discovered a novel spontaneous mutation that causes progressive darkening of the eye and coat colors with aging (Fig. 1).
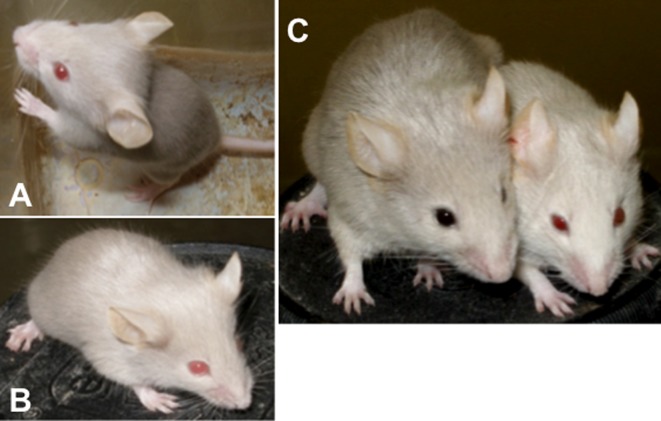
Fig. 1.
External appearances of novel and ordinary Oca2p-cas/Oca2p-cas mutant mice on the non-agouti background. (A) Young novel mutant mouse at one month after birth before becoming darker in color. (B) Young ordinary mutant mouse at one month of age. (C) Adult novel mouse at 5 months of age (left side), which grew from the young mouse shown in (A). The eyes and coat hair became darker than those of the unchanged ordinary littermate (right side).
Several mutants displaying progressive hair graying phenotypes with aging have been reported in animals [25, 26]. For example, gray horses are born colored, gradually lose hair pigmentation, and finally become almost completely white by the age of 6–8 years. This graying phenotype is inherited in an autosomal dominant fashion and is caused by a 4.6-kb duplication in intron 6 of the STX17(syntaxin 17) gene [21]. Similarly, progressive graying in dogs is characterized by a progressive dilution of eumelanin from black to gray with age. This phenotype is controlled by an autosomal dominant gene, but its causative gene has not yet been identified [12]. The gray horses have a high incidence of melanomas and vitiligo-like skin depigmentation [21], whereas the gray dogs do not have such additional skin abnormalities [12]. In mice, graying with aging is caused by melanocyte dysfunction resulting from maternal transmission of murine leukemia viruses at pre- or early-postnatal stages [19]. Bcl2(B cell leukemia/lymphoma 2) null mice show hair graying in the second hair follicle cycle, due to incomplete maintenance of melanocyte stem cells [20]. On the other hand, no mutants becoming progressively darker with age have so far been reported in animals. Hence, our novel spontaneous Oca2p-cas/Oca2p-cas mouse must be the only animal with a progressive darkening phenotype and it could serve as an excellent animal model for further understanding of the regulation mechanisms of melanin biosynthesis with aging.
In this study, we developed the novel Oca2p-cas/Oca2p-cas mouse model and described morphological and genetic characteristics of the novel mutant phenotype. We also characterized the Oca2p-cas gene at the molecular level because there has been no report on the nucleotide sequence of this gene.
Materials and Methods
Animals
All animal work was carried out in accordance with the guidelines for the care and use of laboratory animals of the Graduate School of Bioagricultural Sciences, Nagoya University, Japan. C57BL/6JJcl (called B6 hereafter) mice were purchased from Clea Japan (Tokyo). Novel and ordinary Oca2p-cas/Oca2p-cas mutant mice were maintained at the Laboratory of Animal Genetics, Graduate School of Bioagricultural Sciences, Nagoya University. For mating experiments, a novel mutant male was crossed to B6 females and ordinary mutant females were crossed to B6 males. Obtained F1 mice were crossed to each other to produce subsequent F2, F3 and other generations. To phenotype the progenies obtained, their eye and coat colors were checked twice a week by the naked eye.
All mice were reared in an environment with a temperature of 23 ± 2°C and a light/dark cycle of 12:12. Commercial chow pellets (CA-1, Clea Japan) and tap water were provided ad libitum. Young mice were weaned at about 3 weeks after birth.
Light microscopic analysis of coat hair
Coat hair samples were plucked out of the dorsal region of nine novel and five ordinary mutants and three B6 mice at 16 days and about 4 months of age, and the samples were wrapped in filter paper and stored at room temperature. The hair samples were dehydrated in 90%, 95% and 99% ethanol for 30 min each time and then in 100% ethanol for 60 min. After that, they were cleared in ethanol-xylene for 60 min and in xylene for 60 min and then embedded in Canada balsam.
Light microscopic analysis of the eyes and skin
Five novel and four ordinary mutants and five B6 mice at 4–6 months of age were anesthetized with Avertin and euthanized by exsanguination. Fresh eyes and skin of these mice were excised and immediately fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. The fixed tissue samples were embedded in paraffin. Sample blocks were cut into 4-µm-thick sections and deparaffinized through the use of xylene and an ethanol series. Some sections were dyed with hematoxylin-eosin for structural examination of the eyes and skin, and others were used for Fontana-Masson staining to identify melanin in these samples. Fontana-Masson staining was performed according to the method of Masson [16] with slight modifications of the concentrations of ammonial silver nitrate and sodium thiosulfate, being 0.5% and 0.12%, respectively.
Transmission electron microscopic analysis of the eyes and skin
Under Avertin anaesthesia, three novel and three ordinary mutants and two B6 mice at about 5 months of age were perfused with 0.9% saline followed by 5% glutaraldehyde in 0.05M cacodylate buffer, pH 7.4. The eyes and skin were excised, trimmed to about 1 mm3, and immersed in the same glutaraldehyde solution. Tissue samples were post-fixed with 1% osmium tetroxide/1.5% potassium ferrocyanide [24], dehydrated, and embedded in epoxy resin. Ultrathin sections were cut using an Ultracut N (Reichert-Nissei, Wein, Austria), stained using uranyl acetate followed by lead citrate, and examined using a Hitachi H-7650 transmission electron microscope (Hitachi Ltd., Tokyo).
cDNA sequencing
After sacrificing novel mutant, ordinary mutant and B6 control mice (two individuals each) by decapitation, both eyes were immediately dissected and stored in RNAlater (Life Technologies, Carlsbad, CA, USA). Total RNA was extracted from the eyes using TRIzol reagent (Life Technologies), and 1 µg of total RNA was used for first-strand cDNA synthesis with the Superscript III First-Strand Synthesis System for RT-PCR (Life Technologies) according to the manufacturer’s instructions.
Mouse Oca2 cDNA was amplified with eight pairs of primers listed in Supplementary data 1, which were designed on the basis of the cDNA sequence for B6 Oca2(NCBI Reference Sequence NM_021879.2). Polymerase chain reaction (PCR) was performed in a 25-µl reaction volume containing 15 ng cDNA and 0.5 units of KOD-Plus-Neo polymerase (TOYOBO, Osaka) under the conditions of step-down cycles: initial denaturation of 2 min at 94°C; 5 cycles of 98°C for 10 s and 74°C for 30 s/kb; 5 cycles of 98°C for 10 s and 72°C for 30 s/kb; 5 cycles of 98°C for 10 s and 70°C for 30 s/kb; 25 cycles of 98°C for 10 s and 68°C for 30 s/kb; and a final extension at 68°C for 7 min. The PCR products were purified from agarose gels using a Gel-M gel extraction Kit (Viogene, Umeå, Sweden). Cycle sequencing was performed with the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Tokyo) according to the manufacturer’s instructions. Nucleotide sequences were determined using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Genomic DNA sequencing
Genomic DNA was isolated from ear clips of the novel and ordinary mutants and B6 (two individuals each) by the standard phenol-chloroform method. Genomic fragments around the promoter region of Oca2 and around the deletion region revealed by the above-described cDNA sequencing were amplified with primers (Supplementary data 1) designed on the basis of the genomic sequence of B6 chromosome 7 (Reference Sequence NC_000073.5). PCR and sequence analyses were performed as described in the above section. The sequence data obtained were assembled with ATGC ver.5 sequence assembly software (Genetyx Corporation, Tokyo).
Prediction of the effects of nonsynonymous substitutions on protein function
Effects of nonsynonymous substitutions detected in the above-described cDNA sequence analysis on function of the OCA2 protein were investigated with two web-based computational software programs, SIFT [15] and PolyPhen-2 [1]. SIFT predicts tolerated and deleterious substitutions for nonsynonymous substitutions based on the evolutionary conservation of amino acids within protein families [15]. PolyPhen-2 predicts possible impact of an amino acid substitution on the structure and function of a protein using straightforward physical and comparative considerations [1]. Since PolyPhen-2 was developed for human proteins, this software was implemented after converting the positions of amino acid substitutions in our mouse study to the corresponding positions of the human protein.
RT-qPCR analysis
Total RNA was extracted from the eyes of novel and ordinary mutant males (three individuals each) and one B6 control male using Trizol as described earlier. Genomic DNA contamination was removed by the gDNA Eraser enzyme and then first-strand cDNA was synthesized from 1 µg of total RNA by reverse transcription using a PrimeScript® RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Otsu) according to the manufacturer’s instructions. Quantitative real-time PCR (RT-qPCR) was performed in a 10.0-µl reaction volume on the StepOnePlusTM Real-Time PCR system (Applied Biosystems) with SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara Bio). Primer sequences for Oca2 and two endogenous control genes, Actb (actin, beta) and B2m (beta-2 microglobulin), were designed with Primer Express® version 3.0 (Applied Biosystems) and are listed in Supplementary data 1. Cycle conditions were 95°C for 30 s and 40 cycles of 95°C for 5 s and 60°C for 30 s. All samples were analyzed in two duplicates. Quantitative relative standard curves for Oca2 and endogenous genes, with four serial dilution points of the B6 control cDNA (20 ng, 4 ng, 0.8 ng and 0.16 ng), were used to determine the Oca2 expression levels normalized to the endogenous controls. Dissociation curves, PCR amplification efficiencies and R2 values were examined to determine the precision of RT-qPCR.
Results
Origin
The Oca2p-cas/Oca2p-cas mutant mice had been maintained on the genetic background of the wild M. m. castaneus mouse by inbreeding at the Laboratory of Animal Genetics, Nagoya University [28]. As the mutant mice faced extinction at around the F15 generation of inbreeding due to the poor breeding, a B6;Cg-Oca2p-cas mixed strain was created using a cross-intercross system in which the mutant mice were crossed to B6 and then wild-type mice obtained were intercrossed to each other to produce the recessive mutant phenotype. This cross-intercross system was repeated at least 4 times, and thereafter inbreeding was continued to fix the mutant phenotype. During inbreeding of B6;Cg-Oca2p-cas, the mutant mice were again crossed to B6 because of the poor breeding. The novel mutant mice becoming darker in color with aging were first identified at the F15+N4+F11+F7 generation from the discovery of the original Oca2p-cas/Oca2p-cas mice.
Two new sublines, named B6;Cg-Oca2p-cas/1Nga and B6;Cg-Oca2p-cas/2Nga, were created by inbreeding of the progeny obtained from mating experiments (see below). The latter subline was mostly fixed for the novel darkening mutant phenotype and the former was mostly fixed for the ordinary unchanged mutant phenotype. Genotyping of microsatellite markers revealed that the minimum introgressed interval derived from the wild M. m. castaneus mouse is 6.71 cM between Saa1 (at 30.54 cM from the centromere of chromosome 7) and D7Mit277 (37.25 cM) and that the maximum introgressed interval is 19.78 cM between D7Mit247(23.90 cM) and D7Mit122 (43.68 cM) in which both sides of recombination regions are included. There were no differences in lengths of maximum and minimum introgressed intervals between the two sublines.
External appearances
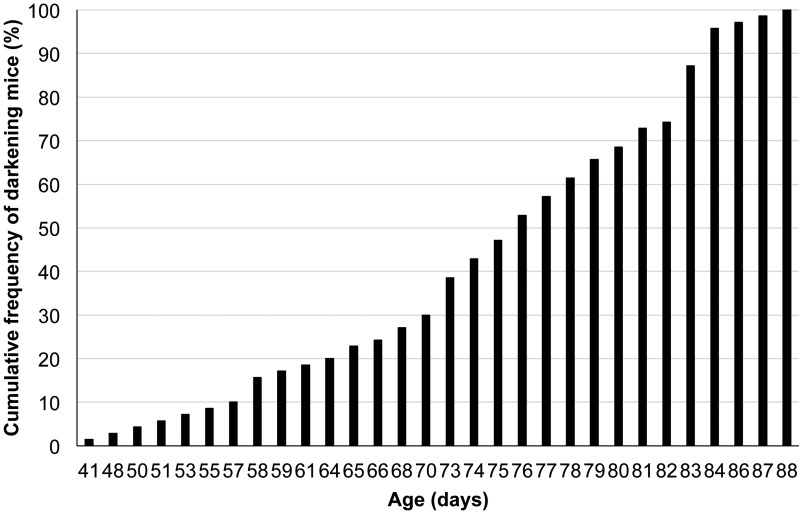
As shown in Fig. 1A, novel Oca2p-cas/Oca2p-cas young mice had pink eyes and gray coat hair on the non-agouti genetic background. These phenotypic characteristics were not distinguishable by the naked eye from those of ordinary Oca2p-cas/Oca2p-cas young mice (Fig. 1B). During growth to adulthood, the eyes and coat hair of the novel mutant mice became progressively darker in color, making a sharp contrast with the external appearance of the ordinary mutants that remained unchanged throughout life (Fig. 1C). Gross observations using the eye color change as an indicator revealed that the novel mutant mice gradually became dark by 41–88 days of age (Fig. 2). No additional visible abnormalities such as wavy hair and small body size were observed in the novel mutant mice, although microphthalmia and cataracts of one or both eyes occasionally appeared in the novel mutants as well as the ordinary mutants. There were no clear sex differences in the above external appearances and also in histological appearances that are described below.
Fig. 2.
Cumulative frequency distribution of novel mutant mice that displayed darkening of the eyes. A total of 70 mice was observed.
Histological appearances
In mice, there are four types of coat hair: monotrich, awl, auchene and zigzag [26]. We light-microscopically examined melanin pigmentation of coat hair with a focus on the awl and zigzag hairs because these hairs were easily detectable in microscopic specimens by greater numbers of those hairs than those of monotrich and auchene hairs. The pigmentation of awl hairs in both novel and ordinary mutant mice was markedly decreased compared to the fully pigmented awl hair of B6 mice (Fig. 3A). At 16 days of age, the awl hair of the novel mutant mice (Fig. 3C) was similar in degree of pigmentation to that of the ordinary mutant mice (data not shown). At about 4 months of age, the degree of pigmentation in the ordinary mice remained unchanged (Fig. 3B). In contrast, the novel mice at 4 months of age showed markedly increased pigmentation (Fig. 3D) but not as much as that of B6 mice (Fig. 3A). The same tendency was observed for zigzag and other hair types (data not shown). However, not all hairs in each hair type showed increased pigmentation with aging. That is, the pigmentation of some hairs remained unchanged, indicating the incomplete penetrance of the causative gene (s) responsible for the novel mutant phenotype.
Fig. 3.
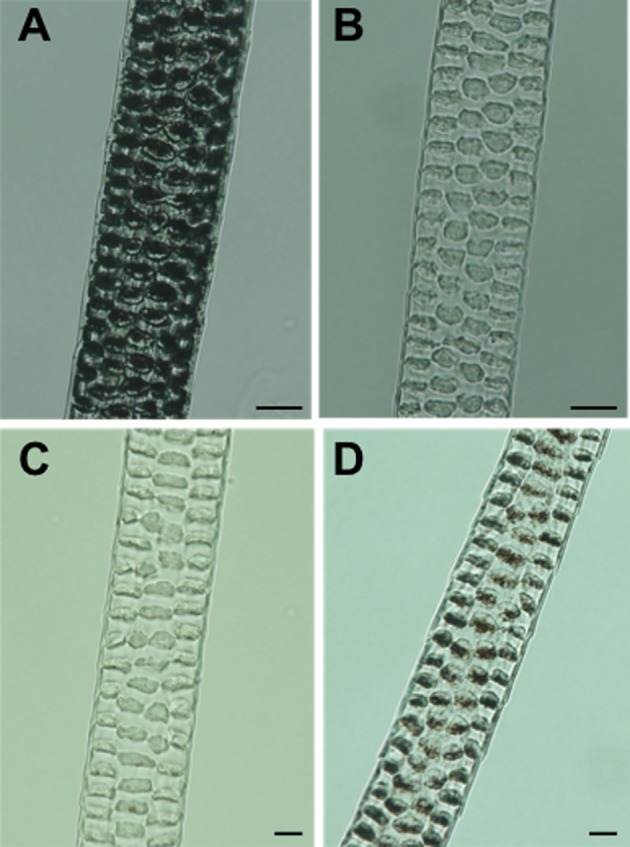
Light micrographs of middle regions of dorsal awl hairs. (A) B6 non-agouti control mouse at about 4 months of age. (B) Ordinary Oca2p-cas/Oca2p-cas mutant mouse at about 4 months of age. (C) Novel Oca2p-cas/Oca2p-cas mutant mouse at 16 days of age. (D) The same novel mouse at about 4 months of age. Melanin pigmentation of the awl hair was obviously increased with aging. Scale bar=10 µm.
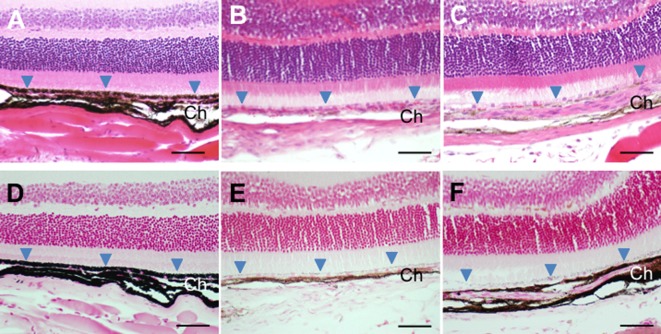
Light-microscopic observations of the hematoxylin-eosin-stained eyes obtained from adult mice at 4–6 months of age revealed that there was little pigmentation in the regions composed of choroid and retinal pigment epithelium in the ordinary mutant mice, whose eyes were red in color (Fig. 4B), compared to those regions well pigmented in B6 control mice with black eyes (Fig. 4A). In contrast, the choroid of the novel mutant adults whose eyes had already turned black in color appeared to be slightly pigmented (Fig. 4C). Fontana-Masson staining revealed that melanin staining was very strong in both regions of choroid and retinal pigment epithelium in B6 mice (Fig. 4D) whereas it was weaker in those of both types of mutant mice (Figs. 4E and F). The choroid of the novel mutant mice appeared to be more heavily stained than that of the ordinary mutant mice. However, there seemed to be no pigmentation difference in retinal pigment epithelium between the two types of mutant mice (Figs. 4E and F). Light-microscopically, it remained ambiguous whether pigmentation was truly increased in one or both regions of choroid and retinal pigment epithelium in the novel mice compared to those of the ordinary mice.
Fig. 4.
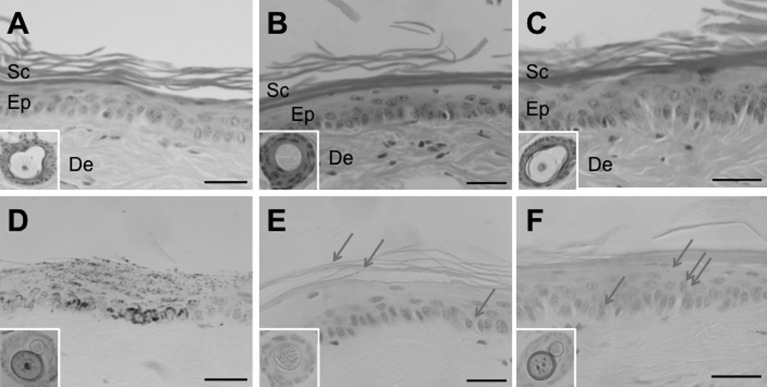
Hematoxylin-eosin-strained (A-C) and Fontana-Masson-stained light micrographs (D-F) of eyes from adult mice. (A, D) B6 control. (B, E) Ordinary mutant. (C, F) Novel mutant. Arrowheads show retinal pigment epithelium. Ch: choroid; Scale bar=50 µm.
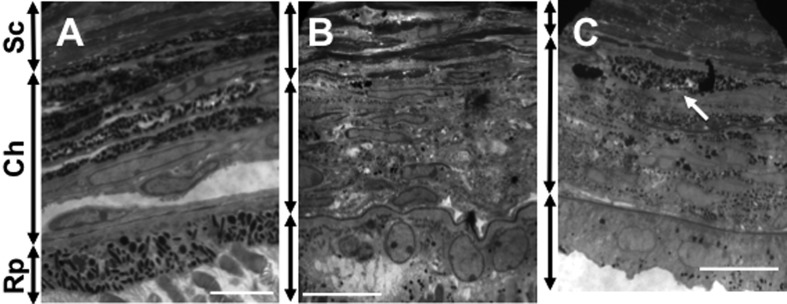
To clarify this issue, we performed transmission electron microscopic analysis. In the B6 control mice, many well-pigmented melanosomes were observed in both the choroid and retinal pigment epithelium (Fig. 5A). In the ordinary mutants, fewer melanosomes were observed in both regions (Fig. 5B). In contrast, little pigmentation was seen in the retinal pigment epithelium, whereas accumulation of pigmentation occurred in the choroid of the novel mutants. The number of the choroidal melanosomes appeared to be intermediate between those of the B6 and ordinary mice (Fig. 5C).
Fig. 5.
Transmission electron micrographs of eyes from adult mice. (A) B6 control. (B) Ordinary mutant. (C) Novel mutant. Pigment accumulation (arrow) is visible in the choroid of the novel mutant mouse in (C). Sc: sclera; Ch: choroid; Rp: retinal pigment epithelium; Scale bar=10 µm.
In adult mice, it is known that melanocytes, which synthesize melanin pigments, are mostly confined to hair follicles in the trunk skin sprouting coat hair, although a few melanocytes remain in the epidermis of the hairy trunk skin; in contrast, it is known that many melanocytes are present in the nonhairy skin of the tail, ears and other regions [9]. We hence focused on histological analysis of the tail skin.
Light-microscopically, no clear histological abnormality was observed in the hematoxylin-eosin-stained tail skin of either type of mutant mice compared to that of B6 control mice at 4–6 months of age (Figs. 6A, B and C). On the other hand, Fontana-Masson staining revealed that B6 mice had abundant epidermal melanin throughout the epidermis (Fig. 6D). In contrast, melanin staining was very weak in both types of mutant mice. There was no obvious difference in degree of epidermal melanin deposition between the two types of mutant mice (Figs. 6E and F). However, the hair follicle of the novel mutant (Fig. 6F inset) was more heavily stained than that of the ordinary mutant (Fig. 6E inset), coinciding with the pigmentation difference in hair shafts between the two types of mutant mice (Figs. 3B and D).
Fig. 6.
Light micrographs of tail skin from adult mice stained with hematoxylin and eosin (A–C) and stained with Fontana and Masson (D–F). (A, D) B6 control. (B, E) Ordinary mutant. (C, F) Novel mutant. Insets indicate hair follicles that were used as positive controls for Fontana-Masson staining. Arrows indicate melanin pigmentation in the two types of mutant mice. De: dermis; Ep: epidermis; Sc: stratum corneum; Scale bar=20 µm.
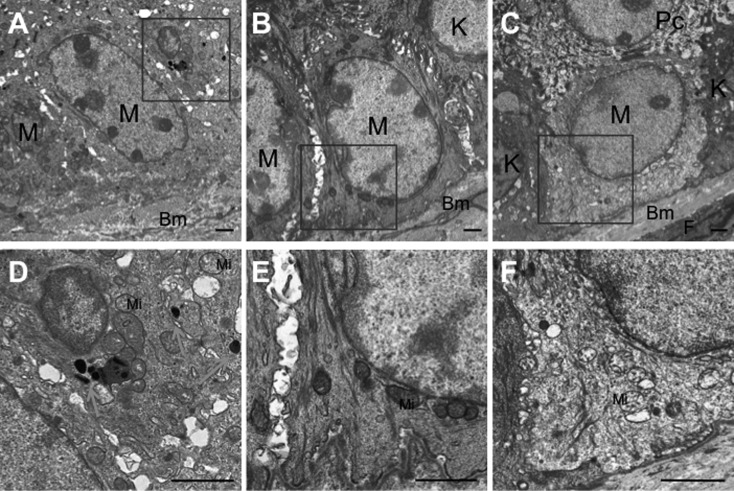
Transmission electron microscopic analysis of the B6 tail epidermis revealed well-pigmented melanosomes in the melanocyte (Figs. 7A and D). However, as expected from the above-described results of Fontana-Masson staining, it was not easy to identify melanosomes in the epidermis of both types of mutant mice (Figs. 7B, C and E). A few melanosome-like granules were observed in the novel mutant mice (Fig. 7F), although it was unclear whether they were true melanosomes or not in the present study.
Fig. 7.
Transmission electron micrographs of tail epidermis from adult mice. (A, D) B6 control. (B, E) Ordinary mutant. (C, F) Novel mutant. (D), (E) and (F) are magnified views of squared areas in (A), (B) and (C), respectively. Arrows show melanosomes. M: melanocyte; K: keratinocyte; F: fibroblast; Pc: prickle cell; Bm: basal membrane; Mi: mitochondria; Scale bar=1 µm.
Sequence analysis
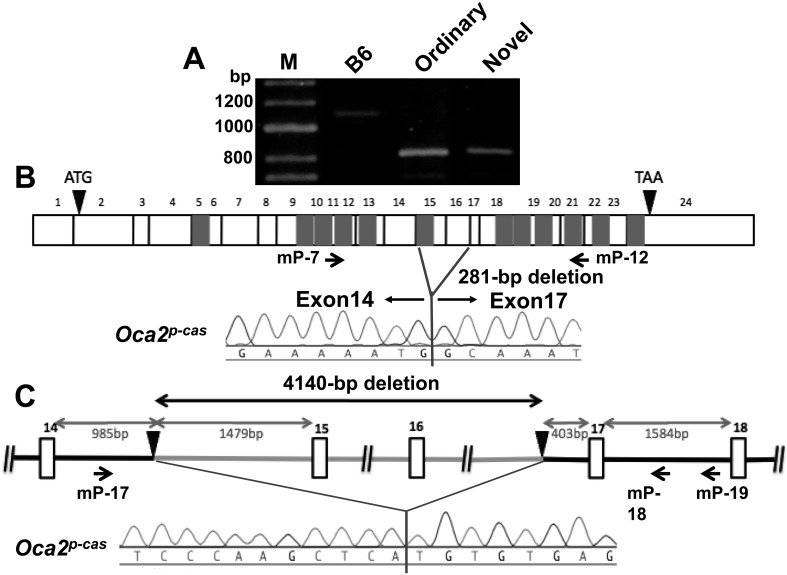
To find SNPs or other nucleotide changes in the Oca2p-cas mutant gene, we first performed RT-PCR analysis using eye cDNAs from the B6 wild-type control mice and the two types of mutant mice as templates, with eight pairs of primers (Supplementary data 1) covering the entire coding sequence of the wild-type Oca2+ gene. When a primer set of mP-7 and mP-12 was used, RT-PCR amplified a fragment in both types of mutant mice that was approximately 300-bp smaller in size than that of B6 mice (Fig. 8A), indicating that both types of mutant mice had a deletion. Direct sequencing of the RT-PCR products revealed a 281-bp deletion of exons 15 and 16 in the Oca2p-cas mutant gene (Fig. 8B). This deletion resulted in a reading frameshift and then creation of a new stop codon on exon 18. Hence, a total number of amino acids deduced from the Oca2p-cas cDNA sequence was 549, contrasting to 833 in the wild-type Oca2+ cDNA (Supplementary data 2). To identify the breakpoints of the deletion at the genomic DNA level, we carried out PCR with primers that amplified the genomic DNA region including the deletion region seen in cDNA. Direct sequencing of the PCR products of Oca2p-cas revealed a 4140-bp deletion including the entire exons 15 and 16 (Fig. 8C).
Fig. 8.
Consequences of the Oca2p-cas mutation. (A) Agarose gel electrophoretic patterns of RT-PCR products that were amplified eye cDNAs from B6 control mice and ordinary and novel mutant mice with primers mP-7 and mP-12 (Supplementary data 1) indicated by horizontal arrows. M: HyperLadder II (Bioline, London) molecular marker. (B) Schematic representation of the Oca2p-cas deletion mutation shown on the cDNA sequence. Positions of the translation initiation codon (ATG) and stop codon (TAA) are shown by black arrowheads. White boxes denote each exon. Twelve gray boxes denote putative transmembrane domains. Direct sequencing of the RT-PCR products revealed a 281-bp deletion of exons 15 and 16. (C) Schematic representation of the Oca2p-cas deletion mutation shown on the genomic DNA sequence. Black arrowheads represent 5′ and 3′ breakpoints of the deletion. Positions and directions of three primers, mP-17, mP-18 and mP-19 (Supplementary data 1), used for PCR amplification are indicated by horizontal arrows. Direct sequencing of the PCR products revealed a 4,140-bp deletion including exons 15 and 16.
As summarized in Table 1, RT-PCR analysis revealed five synonymous and three nonsynonymous SNPs in the coding region of the Oca2p-cas gene. Several additional SNPs and a 24-bp deletion were found in the 5′- and 3′-untranslated regions. At eight of the SNP loci found, the wild-type and mutant alleles were segregating within the Oca2p-cas gene of both types of mutant mice (Table 1). Potential effects of the amino acid substitutions caused by the three nonsynonymous SNPs (Leu266Val, Val283Met and Ser296Phe) on function of the OCA2 protein were investigated with two web-based software programs, SIFT and PolyPhen-2. Both programs predicted that none of the amino acid substitutions could inflict possible damage on OCA2 protein function.
Table 1. Summary of nucleotide changes and deduced amino acid changes found in Oca2p-cas cDNA.
| Exon | Position in | Nucleotide change | Deduced amino acid change b) |
|
|---|---|---|---|---|
| cDNA a) | Amino acid | |||
| 1 | 60 | 5’-UTR | T>G | – |
| 1 | 80 | 5’-UTR | T>C | – |
| 5 | 691 | 187 | T>T or C | Ser |
| 7 | 835 | 235 | A>A or T | Gly |
| 8 | 926 | 266 | C>C or G | Leu>Leu or Val |
| 8 | 931 | 267 | C>C or T | Thr |
| 8 | 977 | 283 | G>G or A | Val>Val or Met |
| 9 | 1017 | 296 | C>C or T | Ser>Ser or Phe |
| 9 | 1144 | 338 | T>T or A | Thr |
| 14 | 1489 | 453 | A>G | Glu |
| 15–16 | 1,618–1,899 | 497–590 | 4,140-bp deletion | Frameshift |
| 18 | 2,059 | 643–644 | Termination | Stop codon |
| 20 | 2,197 | 689 | A>T | 3’-UTR |
| 23 | 2,510 | 794 | A>C | 3’-UTR |
| 24 | 2,561 | 811 | G>G or A | 3’-UTR |
| 24 | 2,675–2,698 | 3’-UTR | 24-bp deletion | – |
| 24 | 2,744 | 3’-UTR | C>T | – |
a)Position in the mouse Oca2+ cDNA (NM_021879.2). The coding DNA sequence is located at positions from 131 to 2632. b)See Supplementary data 2 for detailed amino acid alignments of the wild-type Oca2+ and mutant Oca2p-cas alleles. UTR: untranslated region; –: not applied.
The mouse Oca2 promoter region has not yet been characterized. Referring to the human OCA2 promoter region positioned −435 to +80 upstream from its transcriptional start site [5], we PCR-amplified an approximately 1-kb upstream region of the mouse Oca2 transcriptional start site using genomic DNAs from novel, ordinary and B6 mice. Direct sequencing of the PCR products revealed 28 SNPs (Supplementary data 3). In the promoter region, four putative DNA binding motifs, OSE2 (osteoblast-specific cis-acting element 2), RUNX1 (runt-related transcription factor 1), zinc finger and homeodomain, were identified. Among the 28 SNPs revealed, one was found within the RUNX1 motif. In addition, five-times CA repeats were inserted in the Oca2p-cas mutant allele. The total number of CA repeats was 28 times in the mutant allele, whereas it was 23 times in the wild-type allele.
Taken together, a large number of nucleotide substitution mutations and structural mutations, consisting of SNPs, deletions and insertions, were observed within the Oca2p-cas gene. It was noteworthy, however, that there were no sequence differences in both the cDNA and promoter region of Oca2p-cas between novel and ordinary mutant mice. The cDNA and promoter sequences of Oca2p-cas were deposited in the DDBJ/EMBL/GenBank databases with accession numbers AB716353 and AB716354, respectively.
RT-qPCR analysis
As expected from the no sequence differences described above, mRNA expression level of the Oca2p-cas gene was not significantly different between novel and ordinary mutant mice at P=0.404 (Supplementary data 4). However, it was approximately 0.4-times higher that of the wild-type Oca2+ gene of B6 control mice (data not shown). Percent PCR efficiency and R2 values were 110.9% and 0.995 for Oca2, 94.4% and 0.999 for Actb, and 98.9% and 0.999 for B2m, respectively.
Mating experiments
To investigate the mode of inheritance of the novel mutant phenotype, we first crossed novel mutant mice to B6 wild-type mice, producing 38 F1 mice with the wild-type phenotype only (Table 2). Crosses between these wild-type F1 mice produced 298 F2 progeny in which 231 wild-type, 51 ordinary and 16 novel mice were segregated. This observed segregation ratio statistically met with the expected ratio of 12:3:1 based on assumptions that the novel mutant phenotype is controlled by two autosomal recessive genes, Oca2p-cas and an unknown modifier gene, and that this phenotype is recessive to the ordinary mutant phenotype. Subsequent crosses between novel and ordinary mice and crosses between ordinary mice at F2 and F3 generations unexpectedly produced some progeny displaying the novel-like phenotype, in disagreement with the above assumptions. The emergence of these novel-like progeny might be explained by the supposition that the ordinary mice used for the crosses were carriers for the modifier gene, though we did not carry out a progeny test to confirm the genotype of the ordinary mice crossed. However, this supposition was not able to explain why a notable number of progeny displaying the ordinary-like phenotype still appeared in progenies from crosses between novel mice at F7 and F8 generations.
Table 2. Segregation of wild-type, ordinary-mutant and novel-mutant phenotypes in progenies at F1, F2 and subsequent generations obtained from crosses of ordinary and novel Oca2p-cas/Oca2p-cas mutant mice with B6 control mice.
| CrossFemale × Male | No. of mice | Phenotype of progeny a) | Expected segregation ratio b) | χ2 | P | |||
|---|---|---|---|---|---|---|---|---|
| Wild type | Ordinary | Novel | Wild-type : Ordinary : Novel | |||||
| C57BL/6J × Novel | 38 | 38 | 0 | 0 | 1:0:0 | – | – | |
| F1 × F1 | 298 | 231 | 51 | 16 | 12:3:1 | 1.047 | 0.50<P<0.70 | |
| Ordinary F2 × Ordinary F2 | 65 | 0 | 54 | 11 | – | – | – | |
| Ordinary F2 × Novel F2 | 46 | 0 | 35 | 11 | – | – | – | |
| Novel F2 × Ordinary F2 | 7 | 0 | 5 | 2 | – | – | – | |
| Novel F2 × Novel F2 | 9 | 0 | 0 | 9 | 0:0:1 | – | – | |
| Ordinary F3 × Ordinary F3 | 50 | 0 | 49 | 1 | – | – | – | |
| Ordinary F3 × Novel F3 | 8 | 0 | 8 | 0 | – | – | – | |
| Novel F3 × Ordinary F3 | 11 | 0 | 8 | 3 | – | – | – | |
| Novel F3 × Novel F3 | 5 | 0 | 0 | 5 | 0:0:1 | – | – | |
| Novel F7 × Novel F7 | 44 | 0 | 14 | 30 | 0:0:1 | – | – | |
| Novel F8 × Novel F8 | 18 | 0 | 8 | 10 | 0:0:1 | – | – | |
| Ordinary × C57BL/6J | 33 | 33 | 0 | 0 | 1:0:0 | – | – | |
| F1 × F1 | 163 | 111 | 52 | 0 | 3:1:0 | 4.141 | 0.02<P<0.05 | |
| Ordinary F2 × Ordinary F2 | 94 | 0 | 88 | 6 | 0:1:0 | – | – | |
| Ordinary F3 × Ordinary F3 | 46 | 0 | 46 | 0 | 0:1:0 | – | – | |
| Novel F3 × Ordinary F3 | 8 | 0 | 6 | 2 | 0:1:0 | – | – | |
a)The phenotype was classified at 4 months after birth by eye color. b)The expected segregation ratio is based on the hypothesis that the novel mutant phenotype is controlled by two independent autosomal recessive genes, Oca2p-cas and a modifier; some cases highlighted by bold letters, however, did not meet the expected segregation ratio.
On the other hand, crosses between wild-type F1 mice obtained from crosses of ordinary mutants and B6 produced progeny in which the segregation ratio of wild-type to ordinary phenotypes was slightly skewed from the expected ratio of 3:1 for an unknown reason (Table 2). Crosses between ordinary F2 mice unexpectedly produced six F3 mice exhibiting the novel-like phenotype, again in disagreement with the above assumptions for the simple genetic control of the novel mutant phenotype.
As described earlier, both B6;Cg-Oca2p-cas/2Nga and B6;Cg-Oca2p-cas/1Nga sublines have been established from the descendants of the above mating experiments. In B6;Cg-Oca2p-cas/2Nga, only mice with the novel mutant phenotype were selected and inbred with each other at each generation. Likewise, only mice displaying the ordinary mutant phenotype were selected and inbred in B6;Cg-Oca2p-cas/1Nga. However, mice displaying the opposite-like phenotype occasionally emerged in both sublines (data not shown). Taken together, these results suggested that the novel mutant phenotype is under genetic control but that its mode of inheritance may be complex in nature.
Discussion
We discovered a novel spontaneous mouse mutant, the eyes and coat hair of which become progressively darker in color by approximately 3 months of age, from the B6;Cg-Oca2p-cas mixed strain carrying the Oca2p-cas allele derived from wild M. m. castaneus mice. Our histological analyses revealed that melanin pigmentation of coat hair shafts was markedly increased in the novel darkening mutant adults compared to that in ordinary unchanged mutant adults. Interestingly, ocular pigmentation in the novel adults was increased in the choroid, whereas no change in pigmentation was observed in the retinal pigment epithelium. Developmentally, it is well known that cutaneous melanocytes of the coat hair are derived from neural crest cells and that choroidal melanocytes are also of neural crest origin [3]. Melanoblasts that differentiate into these neural crest-derived melanocytes migrate from the neural tube to corresponding target tissue sites. In contrast, retinal pigment epithelial cells are derived from neuroectodermal cells of the developing embryo forebrain, and their precursor cells do not migrate [3]. Thus, a great divergence in the developmental process of melanocytes exists between the cutaneum/choroid and the retinal pigment epithelium. This developmental divergence clearly explains our histological results that an increase in melanin pigmentation was restricted to the cutaneous/choroidal melanocytes.
Our nucleotide sequence analyses revealed a variety of base substitution mutations, consisting of synonymous and nonsynonymous SNPs, insertions and deletions, in the promoter and coding regions of the Oca2p-cas gene. Of these, the most serious mutation that would definitely affect OCA2 protein function is a large deletion of the genomic region involving the entire exons 15 and 16. The OCA2 protein is predicted to have 12 transmembrane domains [7, 23]. This large deletion led to the deficiency of the 6th transmembrane domain and furthermore caused a reading frameshift. This frameshift created a new stop codon on the 7th transmembrane domain, and hence it produced many amino acid substitutions between the 7th and 12th transmembrane domains. The OCA2p-cas protein would surely have no normal biological function.
In humans, it has been reported that the noncoding region around the SNP rs12913832, which is located in intron 86 of the nonpigment gene HERC2(hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 2), acts as a melanocyte-specific enhancer and that a long-range chromatin loop between this enhancer and the OCA2 promoter regulates OCA2 expression [29]. This scenario would be true in the case of the mouse Herc2 gene. Herc2 is contained in the wild-derived, introgressed interval of both B6;Cg-Oca2p-cas/1Nga and B6;Cg-Oca2p-cas/2Nga sublines that we developed in this study. Our RT-qRCR analysis revealed no significant difference in Oca2p-cas expression between novel and ordinary mutant mice, suggesting that the sequence difference in Herc2 enhancer is unlikely to be found between the two types of mutant mice.
We found an SNP within the RUNX1 binding site on the Oca2p-cas promoter. Very recently, genome-wide studies using chromatin immunoprecipitation followed by sequencing (ChIP-seq) in both human lymphoblastoid cell lines [13, 14, 17] and mouse macrophages [8] have shown that SNPs within transcription factor binding sites can cause histone modifications and that these histone modifications influence gene expression, finally resulting in phenotypic differences between individuals, although not all regulatory variants will lead to differences in traits and/or gene expression levels. It is thus likely that the RUNX1 SNP causes differential expression between the wild-type Oca2+ allele and the mutant Oca2p-cas allele observed in this study. Alternatively, since the Herc2 gene of B6 is of M. m. domesticus origin (examined by Mouse Phylogeny Viewer [30]) and that of the mutant mice is derived from M. m. castaneus, a sequence difference in Herc2 enhancer between the two subspecies may cause the differential expression between the two alleles by reason of the above-mentioned chromatin-loop formation.
The segregation pattern of the novel darkening phenotype at the F2 generations in our mating experiments, at first, suggested that this phenotype was controlled by two unlinked autosomal recessive genes: one is Oca2p-cas and the other is an unknown gene. However, this simple recessive mode of inheritance cannot explain why unexpected novel- or ordinary-like mutant phenotypes emerged at F3 and subsequent generations and also why mutant phenotypic variation still appeared within each of the B6;Cg-Oca2p-cas/1Nga and B6;Cg-Oca2p-cas/2Nga sublines. Since histone modifications influencing gene expression have been reported to be heritable across generations [8, 13, 14, 17], we speculate that the emergence of the unexpected phenotypes was caused by some epigenetic factors such as histone modification and DNA methylation. We believe that the expression of an unknown recessive gene, not Oca2p-cas, controlling the novel mutant phenotype was subjected to epigenetic modification because results of the present study and a previous mating experiment [28] demonstrated that Oca2p-cas has no phenotypic modification caused by epigenetic or other environmental factors. Genome-wide epigenetic analyses of mice in both sublines may provide some evidence for this expectation.
As the B6;Cg-Oca2p-cas strain, in which the novel mutant mice were found, and subsequent sublines (B6;Cg-Oca2p-cas/1Nga and B6;Cg-Oca2p-cas/2Nga) are mixed strains, we cannot rule out the possibility that, in addition to the target regions, these strains may have some genomic regions derived from wild M. m. castaneus. It is unclear whether the unknown recessive gene is originated from wild M. m. castaneus or B6 in the present study. However, it is likely that this unknown recessive gene is originated from wild M. m. castaneus because it has been demonstrated that many novel natural variant genes still remain undiscovered in a gene pool of M. m. castaneus [11]. In addition, the unknown recessive gene must have incomplete penetrance because not all hairs of the novel mice appeared to show the darkening behavior. We are now planning to map the unknown gene by a combined approach of next-generation sequencing analysis of mice in both sublines and mating experiments between them.
In conclusion, our novel mouse mutant that spontaneously occurred in the B6;Cg-Oca2p-cas mixed strain carrying the Oca2p-cas gene derived from wild M. m. castaneus is the first example of progressive hair darkening animals. Melanin pigmentation of the cutaneous and choroidal melanocytes was increased by approximately three months after birth. Oca2p-cas had a deletion of the 4.1-kb genomic DNA region involving entire exons 15 and 16 of the wild-type Oca2 gene, resulting in a reading frameshift and then creation of a new stop codon on exon 18. The mating experiments suggested that the novel mutant phenotype was not inherited in a simple way because either complex epigenetic modification and/or incomplete penetrance have caused the unexpected phenotypic variation in mice obtained from some crosses. As the OCA2 protein function remains elusive in mice and humans, the novel mutant mouse is an essential animal model to study complex genetic or epigenetic factors that switch on melanin biosynthesis in early adult life.
Supplementary Material
Acknowledgments
We thank H. Miyake for guidance and technical support in transmission electron microscopic analysis. This work was supported by Grants-in-Aids for Scientific Research from the Japan Society for the Promotion of Science to A. Ishikawa.
References
- 1.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R.2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ancans J., Tobin D.J., Hoogduijn M.J., Smit N.P., Wakamatsu K., Thody A.J.2001. Melanosomal pH controls rate of melanogenesis, eumelanin/phaeomelanin ratio and melanosome maturation in melanocytes and melanoma cells. Exp. Cell Res. 268: 26–35. doi: 10.1006/excr.2001.5251 [DOI] [PubMed] [Google Scholar]
- 3.Boissy R.E., Hornyak T.J.2006. Extracutaneous melanocytes. pp. 91–107. In: The Pigmentary System: Physiology and Pathophysiology, 2nd ed. (Nordlund, J.J., Boissy, R.E., Hearing, V.J., King, R.A., Oetting, W.S., and Ortonne, J.-P. eds.), Blackwell Publishing, Massachusetts. [Google Scholar]
- 4.Brilliant M.H., Ching A., Nakatsu Y., Eicher E.M.1994. The original pink-eyed dilution mutation (p) arose in Asiatic mice: implications for the H4 minor histocompatibility antigen, Myod1 regulation and the origin of inbred strains. Genetics 138: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiberg H., Troelsen J., Nielsen M., Mikkelsen A., Mengel-From J., Kjaer K.W., Hansen L.2008. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 123: 177–187. doi: 10.1007/s00439-007-0460-x [DOI] [PubMed] [Google Scholar]
- 6.Eppig J.T., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E., Mouse Genome Database Group2012. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic. Acids Res. 40: D881–D886. doi: 10.1093/nar/gkr974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner J.M., Nakatsu Y., Gondo Y., Lee S., Lyon M.F., King R.A., Brilliant M.H.1992. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science 257: 1121–1124. doi: 10.1126/science.257.5073.1121 [DOI] [PubMed] [Google Scholar]
- 8.Heinz S., Romanoski C.E., Benner C., Allison K.A., Kaikkonen M.U., Orozco L.D., Glass C.K.2013. Effect of natural genetic variation on enhancer selection and function. Nature 503: 487–492. doi: 10.1038/nature12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirobe T.1984. Histochemical survey of the distribution of the epidermal melanoblasts and melanocytes in the mouse during fetal and postnatal periods. Anat. Rec. 208: 589–594. doi: 10.1002/ar.1092080414 [DOI] [PubMed] [Google Scholar]
- 10.Hirobe T., Ito S., Wakamatsu K.2011. The mouse pink-eyed dilution allele of the P-gene greatly inhibits eumelanin but not pheomelanin synthesis. Pigment. Cell Melanoma Res. 24: 241–246. doi: 10.1111/j.1755-148X.2010.00783.x [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa A.2013. Wild mice as bountiful resources of novel genetic variants for quantitative traits. Curr. Genomics 14: 225–229. doi: 10.2174/1389202911314040001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaelin C.B., Barsh G.S.2013. Genetics of pigmentation in dogs and cats. Annu. Rev. Anim. Biosci. 1: 125–156. doi: 10.1146/annurev-animal-031412-103659 [DOI] [PubMed] [Google Scholar]
- 13.Kasowski M., Kyriazopoulou-Panagiotopoulou S., Grubert F., Zaugg J.B., Kundaje A., Liu Y., Boyle A.P., Zhang Q.C., Zakharia F., Spacek D.V., Li J., Xie D., Olarerin-George A., Steinmetz L.M., Hogenesch J.B., Kellis M., Batzoglou S., Snyder M.2013. Extensive variation in chromatin states across humans. Science 342: 750–752. doi: 10.1126/science.1242510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilpinen H., Waszak S.M., Gschwind A.R., Raghav S.K., Witwicki R.M., Orioli A., Migliavacca E., Wiederkehr M., Gutierrez-Arcelus M., Panousis N.I., Yurovsky A., Lappalainen T., Romano-Palumbo L., Planchon A., Bielser D., Bryois J., Padioleau I., Udin G., Thurnheer S., Hacker D., Core L.J., Lis J.T., Hernandez N., Reymond A., Deplancke B., Dermitzakis E.T.2013. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science 342: 744–747. doi: 10.1126/science.1242463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P., Henikoff S., Ng P.C.2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 16.Masson P.1970. Human Tumors. Histology, Diafnosis and Technique, 2nd ed. Wayne-State University Press, Detroit. [Google Scholar]
- 17.McVicker G., van de Geijn B., Degner J.F., Cain C.E., Banovich N.E., Raj A., Lewellen N., Myrthil M., Gilad Y., Pritchard J.K.2013. Identification of genetic variants that affect histone modifications in human cells. Science 342: 747–749. doi: 10.1126/science.1242429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montoliu L., Grønskov K., Wei A.H., Martínez-García M., Fernández A., Arveiler B., Morice-Picard F., Riazuddin S., Suzuki T., Ahmed Z.M., Rosenberg T., Li W.2014. Increasing the complexity: new genes and new types of albinism. Pigment. Cell Melanoma Res. 27: 11–18. doi: 10.1111/pcmr.12167 [DOI] [PubMed] [Google Scholar]
- 19.Morse H.C., 3rd, Yetter R.A., Stimpfling J.H., Pitts O.M., Fredrickson T.N., Hartley J.W.1985. Greying with age in mice: relation to expression of murine leukemia viruses. Cell 41: 439–448. doi: 10.1016/S0092-8674(85)80017-9 [DOI] [PubMed] [Google Scholar]
- 20.Nishimura E.K., Granter S.R., Fisher D.E.2005. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307: 720–724. doi: 10.1126/science.1099593 [DOI] [PubMed] [Google Scholar]
- 21.Pielberg G.R., Golovko A., Sundström E., Curik I., Lennartsson J., Seltenhammer M.H., Druml T., Binns M., Fitzsimmons C., Lindgren G., Sandberg K., Baumung R., Vetterlein M., Strömberg S., Grabherr M., Wade C., Lindblad-Toh K., Pontén F., Heldin C.H., Sölkner J., Andersson L.2008. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nat. Genet. 40: 1004–1009. doi: 10.1038/ng.185 [DOI] [PubMed] [Google Scholar]
- 22.Resnick J.L., Nicholls R.D., Wevrick R., Prader-Willi Syndrome Animal Models Working Group2013. Recommendations for the investigation of animal models of Prader-Willi syndrome. Mamm. Genome 24: 165–178. doi: 10.1007/s00335-013-9454-2 [DOI] [PubMed] [Google Scholar]
- 23.Rinchik E.M., Bultman S.J., Horsthemke B., Lee S.T., Strunk K.M., Spritz R.A., Avidano K.M., Jong M.T.C., Nicholls R.D.1993. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361: 72–76. doi: 10.1038/361072a0 [DOI] [PubMed] [Google Scholar]
- 24.Russel L., Burguet S.1977. Ultrastructure of leydig cells as revealed by secondary tissue treatment with a ferrocyanide-osmium mixture. Tissue Cell 9: 751–766. doi: 10.1016/0040-8166(77)90040-4 [DOI] [PubMed] [Google Scholar]
- 25.Searle A.G.1968. Comparative Genetics of Coat Color in Mammals. Academic Press, New York. [Google Scholar]
- 26.Silvers W.K.1979. The Coat Colors of Mice. Springer-Verlag, New York. [Google Scholar]
- 27.Toyofuku K., Valencia J.C., Kushimoto T., Costin G.E., Virador V.M., Vieira W.D., Ferrans V.J., Hearing V.J.2002. The etiology of oculocutaneous albinism (OCA) type II: the pink protein modulates the processing and transport of tyrosinase. Pigment. Cell Res. 15: 217–224. doi: 10.1034/j.1600-0749.2002.02007.x [DOI] [PubMed] [Google Scholar]
- 28.Tsuji A., Wakayama T., Ishikawa A.1995. A new spontaneous allele at the pink-eyed dilution (p) locus discovered in Mus musculus castaneus. Exp. Anim. 44: 347–351. doi: 10.1538/expanim.44.347 [DOI] [PubMed] [Google Scholar]
- 29.Visser M., Kayser M., Palstra R.J.2012. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 22: 446–455. doi: 10.1101/gr.128652.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J.R., de Villena F.P.M., McMillan L.2012. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinformatics 13:(Suppl 3): S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.