Abstract
In September 2012, several cows and a calf showed decreased activity, anorexia and fever on Ishigaki Island, Okinawa Prefecture, Japan, and the cases were diagnosed as bovine ephemeral fever (BEF). We isolated BEF virus (BEFV) from one of the affected cows and then determined the complete genome sequence of the G gene, which encodes a class I transmembrane glycoprotein of BEFV. The BEFV isolate in this case, ON-3/E/12, was sorted into the same cluster as other BEFV isolates in Japan, Taiwan and China obtained in 1996−2004 and was most closely related to a 2002 Chinese isolate, JT02L, according to the phylogenetic analysis of the complete G gene. Since inactivated vaccines for BEF available in Japan are considered effective against the ON-3/E/12 isolate as well as other isolates in East Asia from 1996−2004, annual vaccination should be conducted to prevent BEF in Okinawa. Additionally, in this study, we developed an RT-PCR assay to detect the BEFV gene in Japan and neighboring countries. Our assay was able to amplify target sequences in all of the tested BEFV isolates, including 18 isolates in Japan and another isolate in Australia. The assay was found to be useful also for testing RNA samples extracted from bovine peripheral blood mononuclear cells, and the detection limit of the assay was 10 copies per tube. We believe that our assay would be an important tool for the screening of BEFV infection and the diagnosis of BEF.
Keywords: arbovirus, bovine ephemeral fever, diagnosis, molecular epidemiology, RT-PCR
Bovine ephemeral fever virus (BEFV) is classified as the type species of the genus Ephemerovirus in the family Rhabdoviridae. Virions of the BEFV consist of approximately 70 × 180-nm bullet- or cone-shaped, enveloped particles that each contains a helical nucleocapsid, comprising the negative-stranded RNA genome, a protective nucleoprotein (N), and the large (L) and small (P) subunits of an RNA-dependent RNA polymerase. The structural proteins also include a matrix (M) protein and a class I transmembrane glycoprotein (G). The G protein spans the viral envelope and is the target for the neutralizing antibody [17].
BEFV is known to cause bovine ephemeral fever (BEF) in cattle and water buffalo. The disease is characterized by the rapid onset of, and rapid recovery from, clinical signs, such as fever, anorexia, muscle stiffness, ocular and nasal discharge, salivation, depression, ruminal stasis, lameness and sternal recumbency [17]. BEFV is transmitted by both mosquitoes and Culicoides biting midges, and is widely distributed in tropical, subtropical and temperate areas of Africa, the Middle East, Australia and Asia [17]. In Japan, the first epidemic of BEF was reported in 1953 and then occurred frequently in the 1960s [6, 11], but no BEFV activity has been observed since 1992 in Japan, except for Okinawa Prefecture, in the southwestern part of Japan. Epidemics of BEF in Okinawa were reported in 1988, 1989, 2001 and 2004, but no epidemic has been reported on the main islands of Japan since 1989 [1, 7, 12].
In September 2012, several cows and a calf in Okinawa showed decreased activity, anorexia and fever, and some of the cows had reduced white blood cell count. Therefore, we conducted virological and serological investigations for these affected cows and the calf, as well as some other healthy cows on farms that had the affected animals. Additionally, we developed an RT-PCR assay to detect the BEFV gene in Japan and neighboring countries, since BEF was suspected in the cases in Okinawa yet no RT-PCR assays had been developed for screening for BEFV infection or for the molecular diagnosis of BEF in Japan.
MATERIALS AND METHODS
Sample collection and laboratory testing: Bovine blood samples were collected at three beef farms on Ishigaki Island, Okinawa Prefecture, in the middle of September 2012. The blood samples were collected from 11 animals in total, including 6 cows and a calf that showed clinical signs including fever, anorexia and nasal discharge, and 4 other cows without any clinical signs. None of the cows or the calf was vaccinated against BEF for 2 years or more. From the blood samples, peripheral blood mononuclear cells (PBMCs) were collected, and RNA was extracted from the PBMCs by using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, U.S.A.). The RNA samples were then subjected to the RT-PCR assay previously reported by Khalil et al.[8]. The PMBCs were also subjected to virus isolation with the use of BHK-21, HmLu-1 and Vero cells, respectively. In addition, paired serum samples were collected with a 12-day interval, and they were subjected to a neutralization test (NT) for the titration of antibodies against a vaccine strain of BEFV, YHL strain [7]. Titers greater than or equal to 1:2 were regarded as positive for the neutralizing antibodies.
Genetic analysis of the G gene of BEFV: RT-PCR was conducted to amplify the G gene of a BEFV isolate [7], and the PCR product was purified by using the QIAquick PCR Purification Kit (Qiagen). The product was then subjected to direct sequencing, and the nucleotide sequence of the G gene was determined. To analyze the phylogenetic relationships among the isolate in the case described here and other isolates, the sequences were aligned by the ClustalW program [14]. Then, phylogenetic trees were constructed with MEGA5 using the neighbor-joining method, and the reliability of the branching orders was evaluated by the bootstrap test (n=1,000) [13]. The nucleotide sequence data reported in this study were deposited in the DNA Data Bank of Japan (DDBJ) under the accession number AB985267.
Development of an RT-PCR assay for specific detection of BEFV gene: Primers were designed for the amplification of the G gene of BEFV, based on the nucleotide sequences of Asian and Australian BEFV isolates deposited in GenBank (Table 1). Then, RT-PCR was conducted to test the ability of the newly designed primers to amplify the target genes of Japanese and Australian BEFV isolates by using the OneStep RT-PCR Kit (Qiagen) according to the following program: 50°C for 30 min (reverse transcription), 95°C for 15 min (activation of Taq DNA polymerase, inactivation of RT enzyme and denaturation of the template cDNA), 35 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min and then 72°C for 10 min (final extension). The virus isolates tested in this study are shown in Table 2. The utility of the assay was then tested with the RNA samples extracted from bovine PBMCs as described above. We divided the samples into two groups—a group of PCR-positive samples (Group A) and another group of PCR-negative samples (Group B) —based on the results of the RT-PCR assay developed by Khalil et al.[8]. Furthermore, RNA templates containing the complete G gene sequences were synthesized by in vitro transcription to define the detection limit of the RT-PCR assay. We selected three BEFV isolates obtained since 2001 in Japan, including isolates from 2001, 2004 and 2012, and amplified the cDNA of the target sequences by RT-PCR with the OneStep RT-PCR Kit (Qiagen). The PCR products were purified and then inserted into pGEM-T Easy vector (Promega, Madison, WI, U.S.A.). The resulting plasmids were cloned into competent cells of Escherichia coli, and the E. coli was propagated in LB medium. The plasmids were then extracted from the E. coli by using the Plasmid Mini Kit (Qiagen). Partial sequences of the plasmid were amplified with T7 or SP6 promoter primers and the KOD -Ver.2- PCR Kit (Toyobo, Osaka, Japan). The PCR products were purified by using the QIAquick PCR Purification Kit (Qiagen). Each purified PCR product was used as a template for in vitro transcription with the use of the MEGAscript T7 Kit or the MEGAscript SP6 Kit (Ambion, Life Technologies, Austin, TX, U.S.A.), and the products of the in vitro transcription were purified with the RNeasy Mini Kit (Qiagen). The concentration and purity of the purified RNA were measured with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, U.S.A.). After the measurement, 10-fold dilutions were prepared from 104 to 10−1 copies/tube and were used to determine the detection limits of the RT-PCR assay.
Table 1. Oligonucleotide primers used for RT-PCR and sequencing of bovine ephemeral fever virus.
| Primer | Sequence (5’–3’) | Position | Purpose | References |
|---|---|---|---|---|
| BEFV-AO-F | GAATCATTATGGGATCGGATC | 1140–1160 | RT-PCR | This study |
| BEFV-AO-R | CCAACCTACAACAGCAGATAAAAC | 1587–1564 | ||
| EG2 | TACAACAGCAGATAAAAC | 1581–1564 | RT-PCR | [8] |
| EG3 | CATTATGGGATAGGATCC | 1144–1161 | ||
| GF1 | ATGTTCAAGGTCCTCATAATTACC | 1–24 | RT-PCR, sequencing and cloning | [7] |
| G681F | ATGGGAGGCTCCAGATATCGGG | 681–702 | Sequencing | |
| G1414F | GAGGTAATGGAGTACGATAAC | 1414–1434 | Sequencing | |
| G1481F | AAGCCAGTGAATTTAAGCCC | 1480–1499 | Sequencing | |
| GR1 | TAATGATCAAAGAACCTATCATCA | 1871–1848 | RT-PCR, sequencing and cloning | |
| G1310R | GCTTCGTTCCGGAGCTTCCT | 1329–1290 | Sequencing | |
| G589R | CTCTCACATCTGGTATCCATGTCC | 589–575 | Sequencing | |
| G410R | TTCCAAAAGCATCCAGCAGGAGGG | 410–387 | Sequencing |
Table 2. List of bovine ephemeral fever virus isolates used in this study.
| Strain | Source | Location | Year | Accession number |
|---|---|---|---|---|
| ON-3/E/12 | Bovine erythrocyte | Okinawa (Ishigaki Island), Japan | 2012 | AB985267 |
| ON-04-1 | Bovine blood | Okinawa (Ishigaki Island), Japan | 2004 | AB462044 |
| ON-BEF-01-1 | Bovine white blood cells | Okinawa (Tarama Island), Japan | 2001 | AB462041 |
| ON-BEF-01-2 | Bovine erythrocyte | Okinawa (Ishigaki Island), Japan | 2001 | AB462042 |
| ON-BEF-01-3 | Bovine erythrocyte | Okinawa (Ishigaki Island), Japan | 2001 | AB462043 |
| Onna3 | Bovine erythrocyte | Okinawa (Okinawa Island), Japan | 1989 | AB462040 |
| ON-BEF-89-1 | Bovine white blood cells | Okinawa (Ishigaki Island), Japan | 1989 | AB462037 |
| ON-BEF-89-2 | Bovine white blood cells | Okinawa (Ishigaki Island), Japan | 1989 | AB462038 |
| ON-BEF-89-3 | Bovine white blood cells | Okinawa (Ishigaki Island), Japan | 1989 | AB462039 |
| ON-BEF-88-1 | Bovine white blood cells | Okinawa (Okinawa Island), Japan | 1988 | AB462034 |
| ON-BEF-88-3 | Bovine white blood cells | Okinawa (Okinawa Island), Japan | 1988 | AB462035 |
| ON-BEF-88-4 | Bovine white blood cells | Okinawa (Okinawa Island), Japan | 1988 | AB462036 |
| Azuma | Bovine erythrocyte | Kagoshima, Japan | 1988 | AB462033 |
| Amakusa-1 | Bovine blood | Kumamoto, Japan | 1988 | AB462031 |
| Amakusa-2 | Bovine blood | Kumamoto, Japan | 1988 | AB462032 |
| Hirado-6 | Bovine plasma | Nagasaki, Japan | 1988 | AB462029 |
| Hirado-9 | Bovine plasma | Nagasaki, Japan | 1988 | AB462030 |
| YHL | Bovine blood | Yamaguchi, Japan | 1966 | AB462028 |
| BB7721 | Bovine blood | Queensland, Australia | 1968 | M94266 |
RESULTS
Findings of the laboratory testing: Among the 11 animals examined for BEFV, 6 animals (the affected calf and 5 of the affected cows) tested positive by the RT-PCR assay developed by Khalil et al. [8]. At the same time, nonspecific reactions were observed in all the tested samples (data not shown). The BEFV was isolated from a blood sample of one of the affected cows. An increase in neutralizing antibody (NtAb) titers was observed in all the affected animals by the NT using paired sera. The NtAb titers of the post sera ranged from 1:64 to ≥1:4096 in the 7 affected animals, showing 32-fold or more increases. Also, seroconversion was observed in 2 of the 4 animals without any clinical signs, and the titers of the post sera in the 2 animals were 1:4 and 1:16, respectively.
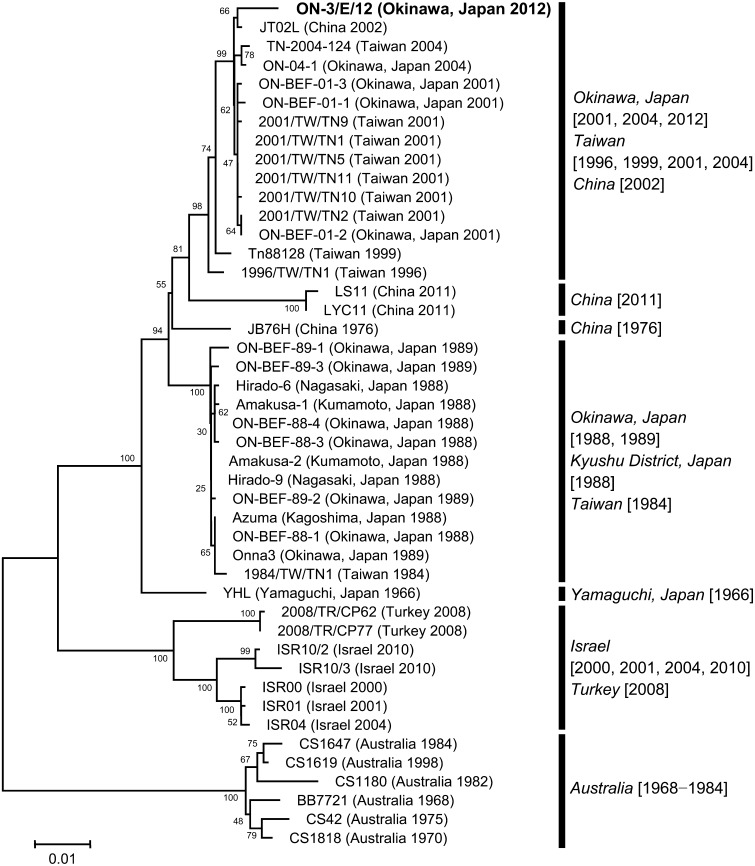
Genetic characteristics of the BEFV isolate: The BEFV isolate in the present case, ON-3/E/12, was found to be most closely related to a Chinese BEFV isolate from 2002, JT02L (Fig. 1) [20]. Also, the ON-3/E/12 isolate was closely related to several other isolates in Okinawa from 2001 and 2004, as well as several isolates in Taiwan from 1996, 1999, 2001 and 2004 (Fig. 1) [5, 7, 9, 18]. The identities of the BEFV G gene between the ON-3/E/12 isolate and another isolate in Okinawa from 2004, ON-04-1, were 98.90% and 99.34% in nucleotide and amino acid levels, respectively, and those of the G gene between the ON-3/E/12 isolate and a vaccine strain, YHL, were 96.10% and 98.51% in nucleotide and amino acid levels, respectively. The predicted amino acid sequences of the ON-3/E/12 isolate were the same as those of the ON-04-1 isolate in the neutralizing epitopes G1 (at residues 487−503), G2 (at residues 168−189) and G3 (at residues 49−63 and 215−231) [7], except for an amino acid substitution (E to D) at position 223.
Fig. 1.
Phylogenetic analysis of bovine ephemeral fever virus isolates using the complete G gene sequences performed with MEGA5. The percentage bootstrap values calculated from 1,000 replications are indicated around the internal nodes. The scale represents 1% sequence divergence.
A newly developed RT-PCR assay: The ON-3/E/12 isolate, 17 other BEFV isolates in Japan and another isolate in Australia all tested positive in our RT-PCR assay (Fig. 2). On the other hand, all the other arthropod-borne viruses of ruminants tested negative in this assay, including Akabane, Aino, Peaton, Chuzan, Ibaraki and bluetongue viruses (Fig. 2). Specific reactions were also observed with the use of RNA samples extracted from bovine PBMCs; all 6 of the samples in Group A tested positive, and one of the 5 samples in Group B, which was the sample derived from one of the affected cows, tested positive, but nonspecific bands were not observed (Fig. 3). The detection limits of the assay were 10 copies per tube, which was determined with the RNA synthesized from three BEFV isolates: ON-BEF-01-1, ON-1/B/04 and ON-3/E/12 (Fig. 4).
Fig. 2.
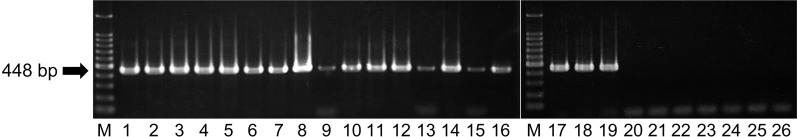
Detection of bovine ephemeral fever virus gene by RT-PCR assay. The specificity of our RT-PCR assay was checked with RNA templates extracted from bovine ephemeral fever virus and other viruses. Products of the RT-PCR assay separated on agarose gels and stained with ethidium bromide are shown. Lanes 1 to 19, bovine ephemeral fever virus (lane 1, ON-3/E/12; lane 2, ON-1/B/04; lane 3, ON-BEF-01-1; lane 4, ON-BEF-01-2; lane 5, ON-BEF-01-3; lane 6, ON-BEF-89-1; lane 7, ON-BEF-89-2; lane 8, ON-BEF-89-3; lane 9, Onna 3; lane 10, ON-BEF-88-1; lane 11, ON-BEF-88-3; lane 12, ON-BEF-88-4; lane 13, Hirado-6; lane 14, Hirado-9; lane 15, Amakusa-1; lane 16, Amakusa-2; lane 17, Azuma; lane 18, YHL; lane 19, BB7721); lane 20 Akabane virus KM-1/Br/06; lane 21, Aino virus KS-1/E/02; lane 22, Peaton virus ON-10/E/01; lane 23, Chuzan (Kasba) virus 31; lane 24, Ibaraki virus No.2; lane 25, bluetongue virus TO2-1; lane 26, negative control (RNase-free water). M, molecular mass ladder (100 bp).
Fig. 3.
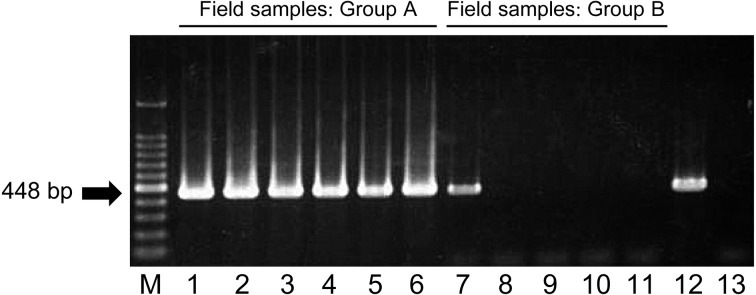
Detection of bovine ephemeral fever virus gene in RNA extracted from field-collected bovine PBMCs. Two groups of RNA samples, Group A (lanes 1 to 6) and Group B (lanes 7 to 11), were used as templates for the RT-PCR assay. Products of the RT-PCR assay separated on agarose gels and stained with ethidium bromide are shown. M, molecular mass ladder (100 bp); lane 12, positive control (BEFV YHL); lane 13, negative control (RNase-free water).
Fig. 4.
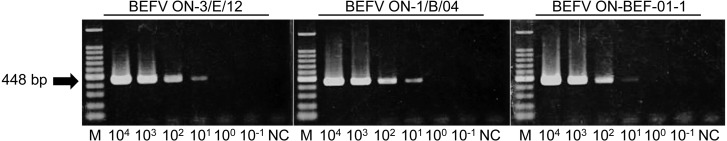
Detection limits of the RT-PCR assay for three bovine ephemeral fever virus isolates: ON-3/P/12, ON-1/B/04 and ON-BEF-01-1. Synthetic RNA of the complete G gene was prepared, and 10-fold dilutions (104 to 10−1 copies per tube) were used as templates to determine the detection limit for each isolate. Products of the RT-PCR assay separated on agarose gels and stained with ethidium bromide are shown. M, molecular mass ladder (100 bp); NC, negative control (RNase-free water).
DISCUSSION
The 7 affected animals were diagnosed as BEF according to their clinical features and both virological and serological findings. The BEFV isolate in this case, ON-3/E/12, sorted into the same cluster with other BEFV isolates in Okinawa, Taiwan and China from 1996−2004 by the phylogenetic analysis of the complete G gene [7], suggesting that all of these BEFV isolates have the same origin. The authors consider that the BEFV ON-3/E/12 isolate was introduced from tropical and subtropical zones in Asia, where the climatic conditions are more suitable for the activity of hematophagous vectors [7]. On the other hand, two Chinese BEFV isolates from 2011, LS11 and LYC11, slightly diverged from the BEFV ON-3/E/12 isolate. Origins of the two Chinese isolates may be different from that of the BEFV ON-3/E/12 isolate [20], or the two Chinese isolates might have evolved independently in mainland of China.
Since our group previously reported that the inactivated vaccine in Japan, which had been developed with the use of the YHL isolate, was still effective against the BEFV isolates from 1996−2004 [7], the vaccine is also considered to be effective against the ON-3/E/12 isolate. Very recently, Ting et al. reported that an epizootic of BEF had occurred in Taiwan in 2012−2013 [15] and that BEFV isolates in the epizootic were found to be most closely related to a Chinese isolate from 2002, JT02L, the same as the BEFV ON-3/E/12 isolate in Okinawa. Their observations indicated that the BEFV isolates in Taiwan were antigenically stable [15], suggesting in turn that vaccines are still effective in Taiwan. Based on the idea that BEFV in Japan and Taiwan shares the same gene pool with that in other East Asian countries, appropriate use of the vaccine could have prevented the field cases in Okinawa and Taiwan in 2012. Although the occurrence of BEF has been less frequent in Okinawa, there still has been some risk of BEFV introduction from other countries. Annual vaccination in Okinawa should be conducted to prevent BEF, since epidemics or outbreaks of BEF have been reported in several countries in recent years, including China, Australia and Turkey [3, 4, 10, 16, 20].
In the present study, a new RT-PCR assay was developed for the specific detection of BEFV in Japan. Primers were designed for the amplification of the G gene, because sequence data of the G gene of various BEFV isolates were available at GenBank. Since the sequences of the primer binding sites are highly conserved among the isolates, the assay would also be useful for the detection of Australian isolates other than BB7721, as well as isolates in China, Turkey and Israel. Although the bands in the PCR products generated from three Japanese BEFV isolates in the present study (Fig. 2: Lanes 9, 13, and 15) were weaker compared with those of other Japanese isolates, the sequences of the three isolates were the same as those of other Japanese isolates, which generated strong bands, in their primer binding sites. Therefore, we consider that the strength differences of PCR products resulted simply from differences in viral titers of virus solutions used for RNA extraction. Several other RT-PCR assays have been reported to date, but our assay seems to be more sensitive than those assays [2, 8, 19]; furthermore, our assay would produce fewer nonspecific reactions, since its annealing temperature is relatively high, 55°C. We believe that our assay would be useful for the screening of BEFV infection and for the molecular diagnosis of BEF in Japan and several other countries.
REFERENCES
- 1.Aizawa M., Takayoshi K., Kokuba T., Kato T., Yanase T., Yamakawa M., Tsuda T.2008. Molecular epidemiological analysis of bovine ephemeral fever virus isolated in Okinawa Prefecture. J. Jpn. Vet. Med. Assoc. 61: 363–366. doi: 10.12935/jvma1951.61.363 [DOI] [Google Scholar]
- 2.Blasdell K. R., Adams M. M., Davis S. S., Walsh S. J., Aziz-Boaron O., Klement E., Tesh R. B., Walker P. J.2013. A reverse-transcription PCR method for detecting all known ephemeroviruses in clinical samples. J. Virol. Methods 191: 128–135. doi: 10.1016/j.jviromet.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 3.Finlaison D. S., Read A. J., Kirkland P. D.2010. An epizootic of bovine ephemeral fever in New South Wales in 2008 associated with long-distance dispersal of vectors. Aust. Vet. J. 88: 301–306. doi: 10.1111/j.1751-0813.2010.00596.x [DOI] [PubMed] [Google Scholar]
- 4.Finlaison D. S., Read A. J., Zhang J., Paskin R., Kirkland P. D.2014. Application of a real-time polymerase chain reaction assay to the diagnosis of bovine ephemeral fever during an outbreak in New South Wales and northern Victoria in 2009–10. Aust. Vet. J. 92: 24–27. doi: 10.1111/avj.12139 [DOI] [PubMed] [Google Scholar]
- 5.Hsieh Y. C., Chen S. H., Chou C. C., Ting L. J., Itakura C., Wang F. I.2005. Bovine ephemeral fever in Taiwan (2001–2002). J. Vet. Med. Sci. 67: 411–416. doi: 10.1292/jvms.67.411 [DOI] [PubMed] [Google Scholar]
- 6.Inaba Y.1971. Bovine ephemeral fever. Bull. Nat. Inst. Anim. Hlth. 62: 1–15. [Google Scholar]
- 7.Kato T., Aizawa M., Takayoshi K., Kokuba T., Yanase T., Shirafuji H., Tsuda T., Yamakawa M.2009. Phylogenetic relationships of the G gene sequence of bovine ephemeral fever virus isolated in Japan, Taiwan and Australia. Vet. Microbiol. 137: 217–223. doi: 10.1016/j.vetmic.2009.01.021 [DOI] [PubMed] [Google Scholar]
- 8.Khalil S. A., Khadr A. M., Zaghawa A., Akela M. A.2001. Application of PCR and immunoperoxidase for diagnosis of bovine ephemeral fever virus in Egypt during summer 2000. pp. 135–140. In: Proceedings of the 6th Scientific Congress of Egyptian Society for Cattle Diseases, Assiut, Egypt.
- 9.Liao Y. K., Inaba Y., Li N. J., Chain C. Y., Lee S. L., Liou P. P.1998. Epidemiology of bovine ephemeral fever virus infection in Taiwan. Microbiol. Res. 153: 289–295. doi: 10.1016/S0944-5013(98)80014-1 [DOI] [PubMed] [Google Scholar]
- 10.Oğuzoğlu T. C., Ertürk A., Cizmeci S. G., Koç B. T., Akça Y.2013. A report on bovine ephemeral fever virus in Turkey: Antigenic variations of different strains of BEFV in the 1985 and 2012 outbreaks using partial glycoprotein gene sequences. Transbound. Emerg. Dis. doi: 10.1111/tbed.12187 [DOI] [PubMed] [Google Scholar]
- 11.Shirakawa H., Ishibashi K., Ogawa T.1994. A comparison of the epidemiology of bovine ephemeral fever in South Korea and south-western Japan. Aust. Vet. J. 71: 502–52. doi: 10.1111/j.1751-0813.1994.tb06153 [DOI] [PubMed] [Google Scholar]
- 12.Takayoshi K., Kokuba T., Higa H., Matukawa S., Oshiro K., Takara I, Hirata K.1991. An outbreak of bovine ephemeral fever in Okinawa Prefecture in 1989. J. Jpn. Vet. Med. Assoc. 44: 591–594. doi: 10.12935/jvma1951.44.591 [DOI] [Google Scholar]
- 13.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J. D., Higgins D. G., Gibson T. J.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ting L. J., Lee M. S., Lee S. H., Tsai H. J., Lee F.2014. Relationships of bovine ephemeral fever epizootics to population immunity and virus variation. Vet. Microbiol. 173: 241–248. doi: 10.1016/j.vetmic.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 16.Tonbak S., Berber E., Yoruk M. D., Azkur A. K., Pestil Z., Bulut H.2013. A large-scale outbreak of bovine ephemeral fever in Turkey, 2012. J. Vet. Med. Sci. 75: 1511–1514. doi: 10.1292/jvms.13-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker P. J.2005. Bovine ephemeral fever in Australia and the world. Curr. Top. Microbiol. Immunol. 292: 57–80. [DOI] [PubMed] [Google Scholar]
- 18.Wang F. I., Hsu A. M., Huang K. J.2001. Bovine ephemeral fever in Taiwan. J. Vet. Diagn. Invest. 13: 462–467. doi: 10.1177/104063870101300602 [DOI] [PubMed] [Google Scholar]
- 19.Zheng F., Lin G., Zhou J., Wang G., Cao X., Gong X., Qiu C.2011. A reverse-transcription, loop-mediated isothermal amplification assay for detection of bovine ephemeral fever virus in the blood of infected cattle. J. Virol. Methods 171: 306–309. doi: 10.1016/j.jviromet.2010.10.028 [DOI] [PubMed] [Google Scholar]
- 20.Zheng F., Qiu C.2012. Phylogenetic relationships of the glycoprotein gene of bovine ephemeral fever virus isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia. Virol. J. 9: 268. doi: 10.1186/1743-422X-9-268 [DOI] [PMC free article] [PubMed] [Google Scholar]