Late endosome-loaded dynein–snapin complexes drive amphisome retrograde transport upon fusion of autophagosomes with late endosomes in distal axons.
Abstract
Efficient degradation of autophagic vacuoles (AVs) via lysosomes is an important cellular homeostatic process. This is particularly challenging for neurons because mature acidic lysosomes are relatively enriched in the soma. Although dynein-driven retrograde transport of AVs was suggested, a fundamental question remains how autophagosomes generated at distal axons acquire dynein motors for retrograde transport toward the soma. In this paper, we demonstrate that late endosome (LE)–loaded dynein–snapin complexes drive AV retrograde transport in axons upon fusion of autophagosomes with LEs into amphisomes. Blocking the fusion with syntaxin17 knockdown reduced recruitment of dynein motors to AVs, thus immobilizing them in axons. Deficiency in dynein–snapin coupling impaired AV transport, resulting in AV accumulation in neurites and synaptic terminals. Altogether, our study provides the first evidence that autophagosomes recruit dynein through fusion with LEs and reveals a new motor–adaptor sharing mechanism by which neurons may remove distal AVs engulfing aggregated proteins and dysfunctional organelles for efficient degradation in the soma.
Introduction
Autophagy is an important cellular process to eliminate defective organelles and aggregated proteins over a neuron’s lifetime. Impaired autophagy is associated with several age-related neurodegenerative disorders (Yue et al., 2009; Rubinsztein et al., 2011; Nixon, 2013; Schneider and Cuervo, 2014). Autophagy undergoes stepwise maturation: bulk cytoplasmic components and organelles are engulfed within double-membrane organelles termed autophagosomes followed by fusion with late endosomes (LEs) into hybrid organelles called amphisomes or fusion with lysosomes into autolysosomes for degradation (Klionsky and Emr, 2000; Levine and Klionsky, 2004; Shen and Mizushima, 2014; Tooze et al., 2014). Newly formed autophagosomes in distal axons are transported to the soma for degradation (Maday et al., 2012). Neurons are highly polarized cells with long axons where kinesin and dynein motors drive long-distance transport of organelles (Hirokawa et al., 2010). Neurons face special challenges to deliver autophagic vacuoles (AVs) generated at distal processes toward proximal regions and the soma, where mature acidic lysosomes are relatively enriched (Cai et al., 2010; Lee et al., 2011). Therefore, retrograde transport is critical to maintain cellular homeostasis essential for neuronal development, survival, and synaptic function.
Emerging evidence supports that dynein motors drive AV retrograde transport from axonal terminals to the soma. Mutation in dynein motors or inhibition of the motor activity reduces autophagic clearance (Ravikumar et al., 2005; Jahreiss et al., 2008; Kimura et al., 2008; Katsumata et al., 2010). However, these studies raise a mechanistic question: How do autophagosomes in distal axons acquire their retrograde motility? Specifically, it is unknown whether dynein motors are directly recruited to autophagosomes or indirectly associated with amphisomes upon fusion of autophagosomes with LEs. Our previous study reveals that snapin acts as an adaptor recruiting dynein motors to LEs and thus plays a key role in mediating LE retrograde transport (Cai et al., 2010). Here, by using live rat dorsal root ganglion (DRG) neurons as the cell model combined with interrupting the formation of amphisomes and dynein–snapin coupling, we reveal a new mechanism underlying recruitment of dynein motors to AVs. Our study demonstrates that LE-loaded dynein–snapin complexes drive amphisome retrograde transport after autophagosome fuses with LE in distal axons. This motor sharing mechanism is crucial for neurons to maintain effective autophagic clearance through lysosomal degradation. Therefore, our study highlights a new cellular pathway to reduce autophagic stress in distal axons associated with several major neurodegenerative diseases.
Results and discussion
Amphisomes are the predominant AVs moving retrogradely in axons
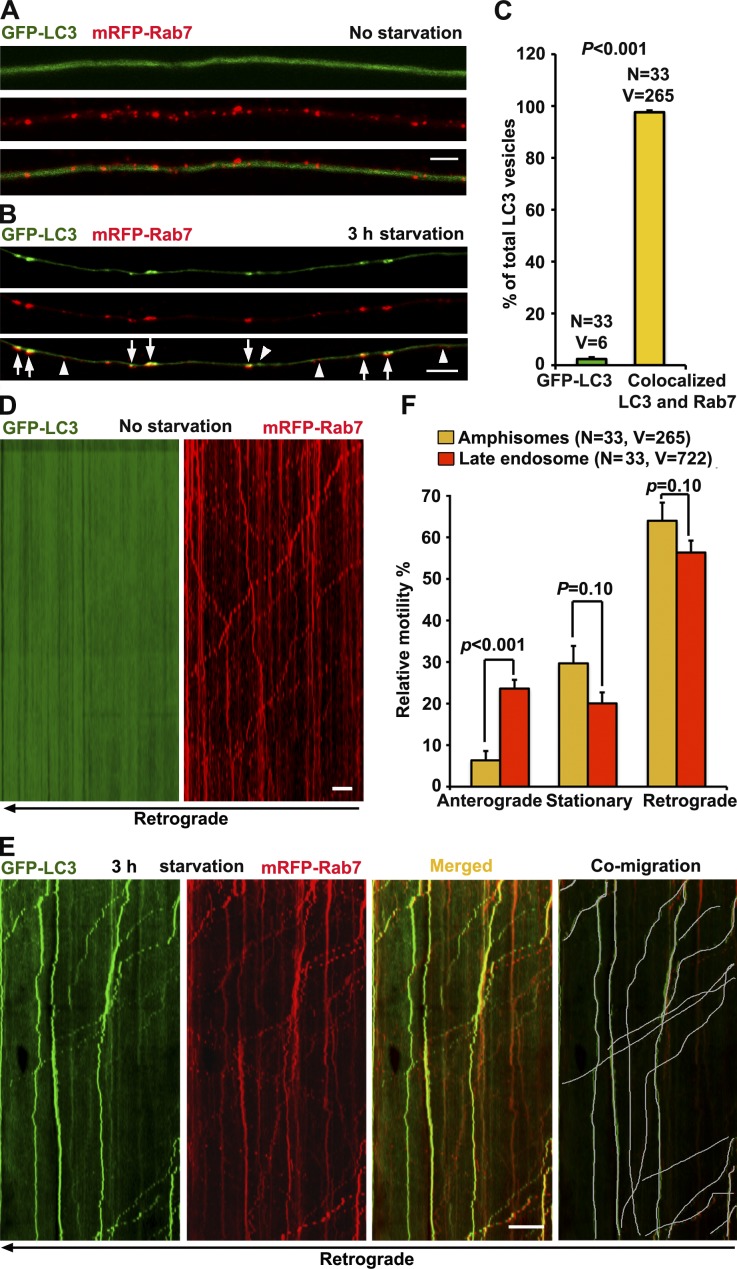
Because mature acidic lysosomes are mainly enriched in the soma and proximal regions, we assume that the majority of autophagosomes in distal axons undergo (a) fusion with LEs to form amphisomes, and then (b) retrograde transport toward the soma where they mature into autolysosomes for degradation. To test our hypothesis, we examined whether autophagosomes mature into amphisomes by fusion with LEs. We used cultured DRG neurons because almost all neurites are tau-positive axons (Perlson et al., 2009) and microtubules in the neurites are uniformly polarized with plus ends outward (Maday et al., 2012). DRG neurons isolated from postnatal day 8–10 (P8–10) rats were cotransfected with autophagy marker GFP-LC3 and LE marker mRFP-Rab7 at day in vitro 0 (DIV0) and imaged at DIV3 after autophagy induction in serum-free medium for 3 h (starvation) as previously described (Maday et al., 2012). Under control conditions, GFP-LC3 was diffuse as cytosolic LC3-I, whereas Rab7-labeled LEs appeared as vesicular structures distributed along the same axons (Fig. 1 A). Upon starvation, GFP-LC3 was efficiently recruited to autophagosomes as lipidated LC3-II (Fig. 1 B). To our surprise, almost all the autophagosomes (97.62% ± 0.65) were colocalized with Rab7-labeled LEs under starvation conditions (Fig. 1 C), suggesting effective formation of amphisomes by fusion of the two organelles. Only a small portion (2.38% ± 0.65) of LC3-II puncta was not labeled with Rab7. Thus, amphisomes are the predominant form of axonal AVs after a 3-h starvation. Our observations are consistent with previous studies that the majority of axonal autophagosomes are colocalized with Rab7 but not with early endosomal markers Rab5 and RhoB (Lee et al., 2011; Maday et al., 2012).
Figure 1.
Axonal amphisomes are the predominant AVs moving retrogradely. (A–C) Majority of autophagosomes in DRG axons target LEs after 3-h starvation. DRG neurons were cotransfected with GFP-LC3 and mRFP-Rab7 at DIV0 and imaged at DIV3 after incubation with serum (control; A) or serum-free medium (starvation; B) for 3 h. Images were taken from the middle segment of axons. Arrows indicate amphisomes colabeled with LC3 and Rab7, whereas arrowheads point out AV or LE alone. (D and E) Dual-channel kymographs showing comigration of GFP-LC3 and mRFP-Rab7 during 5-min time-lapse imaging. Vertical lines represent stationary organelles; slanted lines or curves to the right (negative slope) represent anterograde movement; to the left (positive slope), they indicate retrograde movement. An organelle is considered stationary if it remains immotile (displacement ≤ 10 µm). Under control conditions, GFP-LC3 was diffused, whereas LEs predominantly transported toward the soma (D). Under starvation, amphisomes (LC3 and Rab7) moved retrogradely (white lines in E). (F) Quantitative analysis showing that amphisomes and LEs share the similar predominant retrograde motility in DRG axons. Data were quantified from the total number of vesicles (V) in the total number of neurons (N) from greater than three experiments. Error bars: SEM. Student’s t test. Bars: (A and B) 5 µm; (D and E) 10 µm.
Next, we monitored the retrograde motility of axonal AVs in live DRG neurons. Under nonstarvation, GFP-LC3 was diffuse, whereas LEs moved toward the soma along the same axon (Fig. 1 D). Upon starvation, amphisomes colabeled with LC3 and Rab7 displayed predominant retrograde motility from distal axons to the soma (63.98 ± 0.38%, SEM), whereas anterograde ones accounted for only 6.34 ± 2.24% (Fig. 1, E and F). Strikingly, amphisomes and LEs showed similar retrograde motility in the same axons (Fig. 1 F). These observations prompted us to propose the hypothesis that autophagosomes acquire their retrograde motility by sharing LE-loaded dynein motors upon their fusion. Thus, any deficiency in LE transport may impair amphisome retrograde transport, consequently reducing autophagic clearance in neurons.
LE-loaded dynein–snapin complex drives amphisome retrograde trafficking
Dynein is the primary motor driving retrograde transport of both LEs and AVs from distal axons to the soma (Cai et al., 2010; Katsumata et al., 2010; Lee et al., 2011; Maday et al., 2012). Snapin serves as an adaptor that recruits dynein motors to LEs by binding the dynein intermediate chain (DIC; Cai et al., 2010). We immunoisolated LE organelles from adult mouse brains using anti-Rab7–coated magnetic beads. Because snapin-null mice exhibit embryonic and neonatal death, we generated snapin flox/flox conditional knockout (cKO) mice in which the snapin gene was deleted from the frontal cortex and hippocampus by Cre expression. We detected reduced recruitment of dynein DIC, but not dynactin p150Glued, onto the purified LEs in snapin cKO mouse brains (Fig. S1, A and B). As controls, similar amounts of LE membrane proteins Rab7 and LAMP-2 were attached to LEs from wild-type (WT) and snapin cKO mouse brains. In addition, an anti-snapin antibody coimmunoprecipitated dynein subunits dynein heavy chain (DHC), DIC, and dynactin p150Glued from brain homogenates (Fig. S1 C), thus confirming snapin’s role in connecting dynein motors to LEs via binding to DIC.
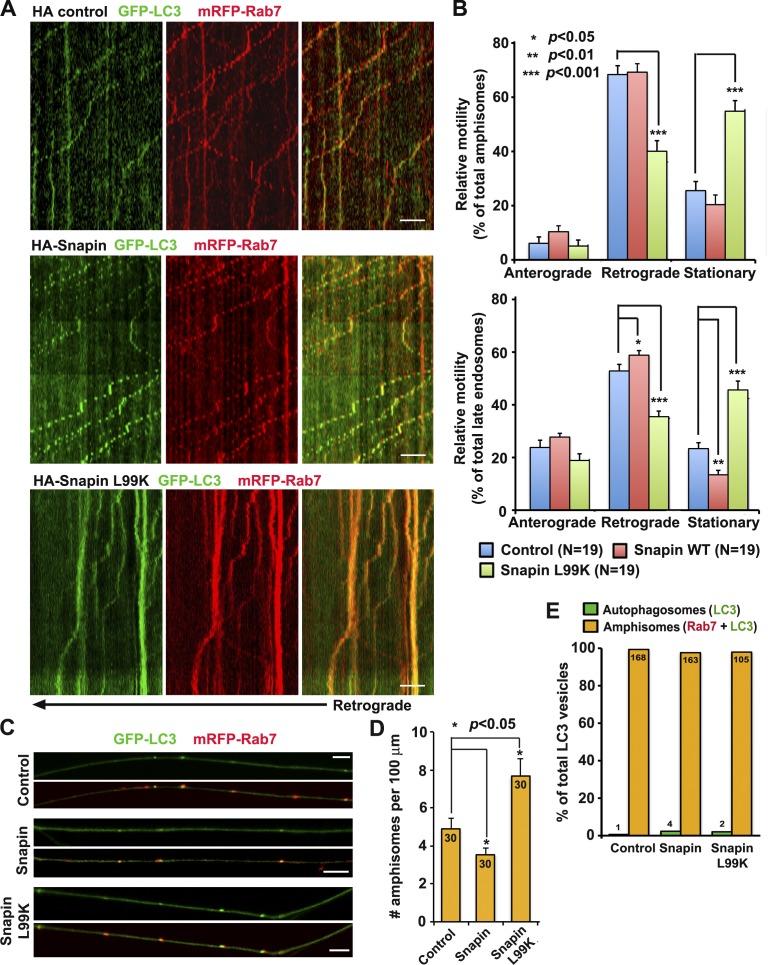
To test whether LE-loaded dynein–snapin complexes are shared by autophagosomes, we examined the retrograde transport of amphisomes after disrupting DIC–snapin coupling. Impairing DIC–snapin coupling by expressing HA-snapin-L99K, a dominant-negative mutant defective in DIC binding (Cai et al., 2010), reduced retrograde transport of both amphisomes (40.07 ± 3.89%) and LEs (35.48 ± 2.13%) along the same axons relative to control neurons expressing HA vector (68.37 ± 3.25% for amphisomes and 52.84 ± 2.5% for LEs; P < 0.001; SEM), thus resulting in increased stationary pools of both organelles (Fig. 2, A and B). Consequently, more AVs were accumulated in axons expressing HA-snapin-L99K compared with that of control neurons (Fig. 2, C and D). Disrupting DIC–snapin coupling has no detectable impact on kinesin-driven anterograde transport of both organelles (Fig. 2 B) and did not impair the fusion between autophagosomes and LEs (Fig. 2 E). Furthermore, expressing snapin-L99K did not alter mitochondrial motility in DRG neurons (Fig. S1, D and E). These results suggest that LE-loaded dynein–snapin complexes selectively drive axonal retrograde transport of LEs and amphisomes.
Figure 2.
LE-loaded dynein–snapin complexes drive amphisome retrograde trafficking. (A and B) Kymographs (A) and quantitative analysis (B) showing impaired retrograde transport of amphisomes by disrupting DIC–snapin coupling. DRG neurons were cotransfected with GFP-LC3 and mRFP-Rab7 along with HA-snapin, HA-snapin-L99K, or HA vector at DIV0 and time-lapse imaged for 3 min at DIV3. The total number of neurons (N) examined is indicated in parentheses from greater than three experiments. (C and D) Images (C) and quantitative analysis (D) showing that disrupted snapin–DIC coupling increases the density of axonal amphisomes. The total number of neurons examined for each group is 30 from more than three experiments. (E) Disrupting snapin–DIC coupling had no significant effect on fusion between autophagosomes and LEs. Data were quantified from total number of AVs denoted in or above the bars from greater than three experiments. Mann–Whitney test (B) and Student’s t test (D and E). Error bars: SEM. Bars: (A) 10 µm; (C) 5 µm.
Autophagosomes acquire retrograde motility by fusion with LEs
To test whether autophagosomes acquire retrograde motility upon fusion with LEs, we applied a molecular tool that specifically retains autophagosomes by blocking their fusion with LEs. Although bafilomycin A1 or Rab7 mutants were reported to block this fusion event, they may have a broad effect in membrane trafficking (Jäger et al., 2004; Fader et al., 2008; Klionsky et al., 2008). Recent studies in mammalian cells and Drosophila melanogaster established that syntaxin 17 (Stx17) mediates the fusion of autophagosomes with LE/lysosomal organelles by forming a SNARE fusion complex with SNAP29 and VAMP8 (CG1599/Vamp7 in flies; Itakura et al., 2012; Hamasaki et al., 2013; Takáts et al., 2013; Guo et al., 2014; Mizushima, 2014). Stx17 knockdown impairs the degradation of autophagosomes. Stx17-siRNAs were rigorously tested for (a) effective suppression of endogenous Stx17, (b) fusion blockage, and (c) no off-target effect (Itakura et al., 2012). Unlike SNAP29 and VAMP8, which associate with LEs and lysosomes, Stx17 selectively targets autophagosomes, so that manipulating its expression would have less impact on endolysosomal trafficking.
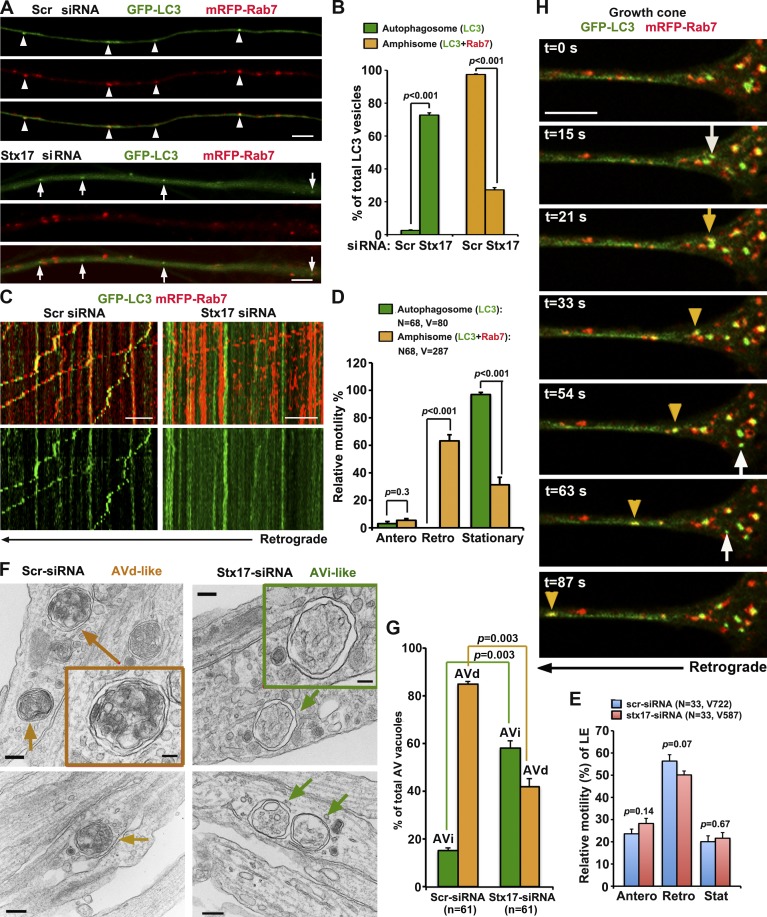
We first verified the knockdown efficiency of Stx17-siRNA#1 relative to a scrambled siRNA (Scr-siRNA) in HEK293T cells and DRG neurons (Fig. S2, A and B). Depleting Stx17 expression in DRG neurons effectively blocks formation of axonal amphisomes at DIV3 after a 3-h starvation (Fig. 3, A and B). There was a robust increase in the number of autophagosomes and a decrease in amphisomes in axons expressing Stx17-siRNA#1 relative to Scr-siRNA (P < 0.001). Among GFP-LC3–labeled AVs, only 27.25% of them are colabeled with mRFP-Rab7 with Stx17 knockdown, whereas in control axons, 97.44% of them are colabeled with Rab7. The phenotypes were further confirmed using a second independent Stx17-siRNA#2 in DRG neurons (Fig. S2, C–F). The phenotypes were also effectively rescued in neurons coexpressing a siRNA-resistant Stx17 silent mutant (Stx17*; Fig. S2, I–K). These results indicate that Stx17 knockdown effectively interferes with the formation of amphisomes in distal axons and further confirm that targeting GFP-LC3 to LEs is a SNARE-mediated fusion process, thus ruling out nonspecific clustering of GFP-LC3 and mRFP-Rab7.
Figure 3.
Autophagosomes acquire retrograde motility by fusion with LEs. (A and B) Stx17 knockdown blocks formation of amphisomes. (A) Imaging was performed on the middle to distal segments of axons. Arrowheads indicate amphisomes (yellow) colabeled with LC3 and Rab7, whereas arrows indicate autophagosomes (green) without Rab7 labeling. Note a robust increase in autophagosomes and decrease in amphisomes in axons expressing Stx17-siRNA (B). The number of autophagosomes or amphisomes was expressed as a percentage of the total LC3-labeled AVs. Data were quantified from 28 axons in each group and a total number of 293 AVs in greater than three experiments. (C–E) Relative motility of amphisomes, autophagosomes, and LEs. (C, right) Note that in neurons with Stx17 knockdown, autophagosomes (green) were largely stationary, whereas LEs (red) underwent predominant retrograde transport, some of which passed through stationary autophagosomes. (C and E) Depleting Stx17 had no effect on the motility of LEs. Quantification was performed from the total number of AVs or LEs (V, vesicles) from the total number of neurons (N) in greater than three experiments. (F and G) TEM analysis showing early stage AVi versus late-stage AVd in DRG neurons. In Stx17 knockdown neurites, the majority of AV-like structures are early stage autophagosomes (AVi; green box and arrows) and were easily found. In control neurons, the majority of AVs are late-stage autophagic vacuoles (AVd) containing electron-dense material and small vesicles (orange box and arrows), suggesting fusion with LEs. Data were collected from 61 micrographs (5 × 5 µm) for each condition. (H) Dynamic de novo autophagosomal biogenesis, fusion, and retrograde transport in a DRG neuron growth cone. Time-lapse imaging was taken once every 3 s with acquisition exposure time at 150 ms in a Nikon spinning-disk confocal. White arrows point to the appearance of new autophagosomes within growth cone at 15, 54, and 63 s; the yellow arrow denotes fusion events, and yellow arrowheads indicate a retrograde motile amphisome between 33 and 87 s (also see Video 1). Error bars: SEM. Unpaired Student’s t test. Bars: (A and H) 5 µm; (C) 10 µm; (F, main images) 200 nm; (F, boxes) 100 nm.
Next, we examined the relative motility of both autophagosomes (LC3 alone) and amphisomes (LC3 and Rab7) in DRG axons expressing Scr-siRNA or Stx17-siRNA. We pooled all LC3-labeleld AVs together and regrouped into autophagosomes and amphisomes. The majority of amphisomes (63.24 ± 4.45%, SEM) underwent retrograde transport. Surprisingly, almost all autophagosomes (96.93% ± 1.53) were stationary (Fig. 3, C and D). Notably, LEs moved passing through stationary AV clusters along the same axons where the fusion was blocked by Stx17 knockdown, thus excluding axonal roadblocks of those larger AV clusters. The altered motility was further confirmed using a second independent Stx17-siRNA#2 (Fig. S2, G and H). There is no detectable difference in the LE retrograde transport along axons expressing scrambled or Stx17-targeted siRNAs (Fig. 3 E). These results support our hypothesis that amphisome formation is a prerequisite step to acquire retrograde motility; blocking the fusion impairs retrograde transport of autophagosomes, but not LEs, from distal axons to the soma. Thus, our study provides evidence that autophagosomes and LEs share dynein motor machinery upon their fusion into amphisomes.
To test whether Stx17 knockdown impacts autophagosomal formation in DRG neurons, a phenotype reported in nonneuronal cells (Hamasaki et al., 2013), we used transmission EM (TEM) after Stx17 knockdown and starvation. Strikingly, the majority (57.10 ± 3.36%) of AVs in neurites are early stage initial AVs (AVi) with sealed double-membrane bilayers separated by an electron-lucent cleft (Fig. 3, F and G). These AVi vacuoles contain intact cytosol and/or organelles without any signs of degradation and were readily found in most micrographs examined. Nonsealed phagophore-like structures were hardly observed under Stx17 knockdown conditions. In neurons transfected with Scr-siRNA, the majority (84.84 ± 1.12%) of AVs are those engulfing electron-dense material and small vesicles, suggesting late-stage degradative AVs (AVd) after fusion with late endocytic organelles. These unique ultrastructural features of AVi versus AVd are consistent with the currently accepted ultrastructural characterization of AVs (Klionsky et al., 2012). Thus, our TEM analysis suggests that Stx17 knockdown in DRG neurons has no detectable effect on the formation of intact AVi, consistent with the observations in HeLa cells after Stx17 knockdown (Itakura et al., 2012). It is possible that siRNA knockdown suppresses but does not abolish Stx17 expression; remaining Stx17 may support AV biogenesis (Hamasaki et al., 2013) but not be sufficient for SNARE-dependent organelle fusion.
We performed time-lapse imaging to monitor dynamic de novo autophagosomal biogenesis, fusion with LE, and retrograde transport in both growth cones (Fig. 3 H and Video 1) and along distal axon shafts (Fig. S3 A) of live DRG neurons immediately after starvation. Autophagosomes were generated within growth cones and distal axonal shafts and then quickly fused with LE and moved away from growth cones or transported toward the soma. The time frame for these steps is ~1 min. However, some AVs remained stationary if they did not fuse with LEs during the recording time.
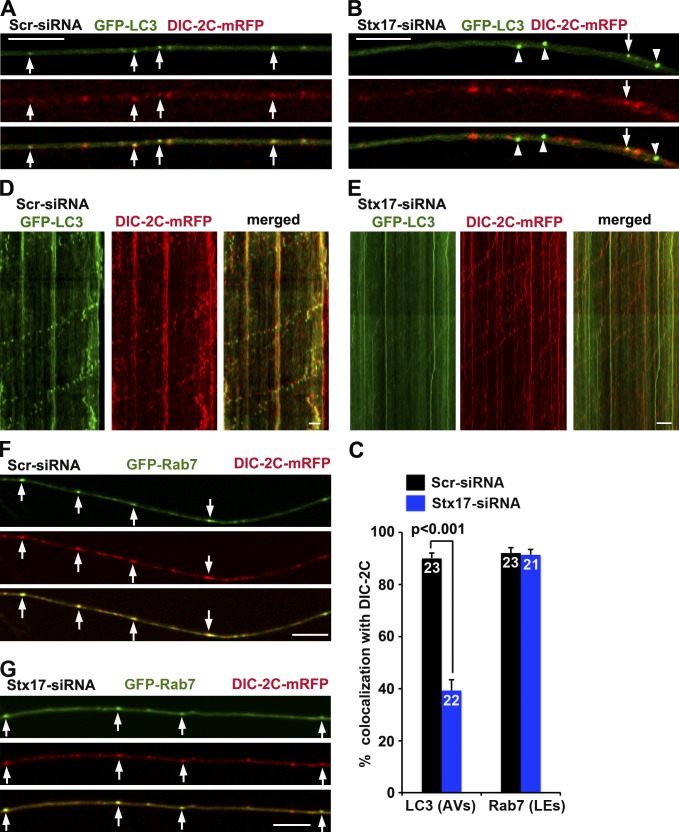
To determine whether dynein motors are recruited to amphisomes but not autophagosomes, we performed time-lapse imaging in DRG neurons by expressing DIC-2C–mRFP, a dynein subunit (Ha et al., 2008) that targets motile AVs (Maday et al., 2012). Depleting Stx17 significantly decreased the recruitment of DIC-2C to AVs (P < 0.001) relative to control neurons, in which DIC-2C was effectively recruited to AVs (Fig. 4, A–C). Inhibiting, but not abolishing, the fusion with two independent Stx17-siRNAs is consistent with some remaining amphisomes recruiting DIC-2C. In addition, although the majority of AVs coretrograde transport with DIC-2C in control neurons, most AVs in Stx17 knockdown neurons were stationary with apparent detachment of DIC-2C (Fig. 4, D and E). Quantitative analysis shows that the majority (90.11%) of retrograde motile AVs recruit DIC-2C, whereas only small portion (21.37%) of stationary AVs are labeled by DIC-2C (Fig. S3 B), thus providing a nice correlation between AV retrograde motility and DIC recruitment. As a control, blocking fusion has no observable effect on DIC recruitment to LEs (Fig. 4, C, F, and G). To further confirm that LEs recruit DIC via snapin–DIC coupling, we coexpressed DIC-2C–mRFP and GFP-Rab7 with snapin-L99K or HA vector in DRG neurons. Disrupting snapin–DIC coupling reduced the recruitment of DIC to LEs (Fig. S3, C and D), consistent with immune-isolation assays showing reduced association of endogenous DIC with the purified LE membranes in snapin cKO mouse brains (Fig. S1, A and B). Thus, our live-imaging analysis further indicates that autophagosomes acquire retrograde transport by recruiting dynein motors upon fusing with LEs.
Figure 4.
Blocking fusion reduces the recruitment of dynein motors to AVs. (A–C) Representative images (A and B) and quantitative analysis (C) showing that blocking fusion reduces DIC recruitment to AVs. Arrows indicate AVs containing DIC-2C (A), whereas arrowheads indicate AVs not labeled with DIC-2C (B). (D and E) AVs (green) comigrate with DIC-2C in retrograde direction in control neurons (D), whereas depleting Stx17 immobilizes AVs by reducing DIC recruitment but has no effect on DIC retrograde motility along the same axons (E). The retrograde direction is toward the left. (F and G) Blocking fusion has no observable effect on DIC recruitment to LEs by quantitative analysis (C). Arrows indicate LEs containing DIC-2C. The total neuron numbers for quantification were indicated inside the bars. Error bars: SEM. Mann–Whitney test. Bars, 10 µm.
Aberrant accumulation of AVs in snapin-deficient neuronal processes
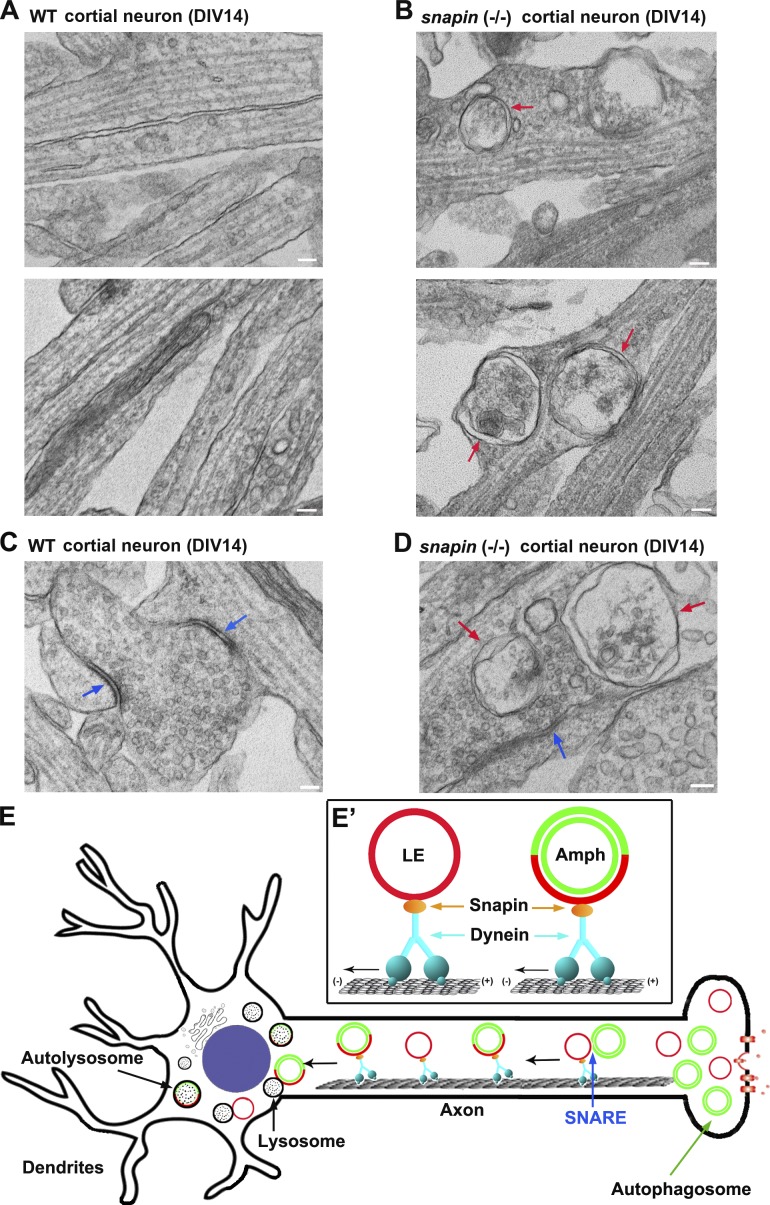
Our previous study showed that disrupting DIC–snapin coupling impairs LE retrograde transport (Cai et al., 2010), thus raising the question of whether impaired dynein–snapin coupling retains AVs at distal axons. To address this issue, we examined cortical neuron ultrastructure at DIV14 from WT and snapin-null mice (Fig. 5, A–D). We observed that a striking number of double-membrane AVs accumulated within snapin−/− neurites (4.13 ± 0.47 vs. WT, 0.15 ± 0.05 per 10-µm length; P < 0.001) and presynaptic terminals (1.43 ± 0.13 relative to WT, 0.06 ± 0.03 per terminal; P < 0.001). These AV-like structures were rarely observed in WT neurons. The aberrant AV accumulation in distal neurites reflects defective retrograde transport, thus impairing autophagic clearance through the lysosomal system. Interestingly, immobilized AVs have a tendency to be clustered in axons (Fig. 5 B), consistent with the observation showing clustered AVs in Stx17 knockdown neurons (Fig. 3 F).
Figure 5.
Aberrant accumulation of AVs in snapin-deficient neuronal processes. (A–D) TEM showing aberrant accumulation of double-membrane AVs along neurites (B) and at presynaptic terminals (D) of snapin−/− cortical neurons at DIV14. Red arrows indicate AV-like structures, which are not readily observed in WT neurons. Blue arrows point to synaptic active zones. Images were representative from 35–50 electron micrographs from three pairs of mice. Bars, 100 nm. (E) A model of LE-loaded dynein–snapin complex driving AV for retrograde trafficking along axons. (E′) Autophagosomes (green) at distal axons acquire the dynein motor complex from LEs (red) upon their fusion into amphisomes. Therefore, dynein–snapin complexes mediate amphisomes, but not autophagosomes, for long-distance retrograde trafficking to the soma, where mature acidic lysosomes are mainly located.
In DRG neurons, sequential recruitment of MT plus end–binding proteins and the dynactin complex facilitate loading of dynein onto MT plus ends, thus initiating retrograde transport for multiple cargos including LEs (Moughamian et al., 2013; Maday et al., 2014). In the current study, we propose a motor–adapter sharing model in which LE-loaded dynein–snapin complex is shared by amphisomes (Fig. 5 E). Distally generated autophagosomes acquire the dynein motor complex from LEs upon their fusion into amphisomes. Therefore, dynein–snapin (motor–adaptor) complexes mediate amphisomes for retrograde transport to the soma, where mature acidic lysosomes are mainly located. In addition to snapin, other dynein adaptors or scaffolding proteins may also be shared by LEs and AVs in different types of neurons through a similar mechanism. For example, a motor scaffolding protein, JIP1, interacts with LC3 and maintains AV retrograde transport through inactivating anterograde motor KIF5 (Fu et al., 2014); JIP1 knockdown also disrupts LE retrograde transport. Huntingtin, a DIC-binding protein (Caviston et al., 2007), mediates transport of both endosomes and AVs along axons (Caviston et al., 2011; Wong and Holzbaur, 2014). RILP, a Rab7-interacting lysosomal protein, recruits dynactin to LEs (Johansson et al., 2007) and was also implicated in autophagic clearance in Purkinje neurons (Bains et al., 2011). Our study reveals a new mechanism underlying recruitment of dynein motors to amphisomes, thus driving AV transport from distal axons toward the soma. This trafficking route is crucial for neurons to maintain effective autophagic clearance through lysosomal degradation in the soma. Mechanistic insight into this fundamental process will advance our understanding of several major neurodegenerative diseases associated with autophagic stress in distal axons and at synapses.
Materials and methods
Antibodies and constructs
Source of the antibodies are as follows: control mouse IgG, monoclonal anti-Rab7, rabbit polyclonal anti-LAMP2, and rabbit polyclonal anti-Stx17 (Sigma-Aldrich), monoclonal anti-dynein IC74 (EMD Millipore), monoclonal anti-dynactin p150Glued (BD), rabbit polyclonal anti-DHC (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-EEA1 (Cell Signaling Technology), and rabbit polyclonal anti-snapin (Synaptic Systems). mRFP-Rab7 (promoter: cytomegalovirus [CMV]; backbone: pmRFP-C3) was purchased from Addgene. Full-length snapin was cloned into the Eco RI–Kpn I sites of the pCMV-HA vector (Takara Bio Inc.) using standard PCR techniques. Site-directed mutagenesis to generate HA-snapin-L99K was performed using standard PCR techniques. GFP-LC3 was subcloned into pEGFP-C3 with CMV promoter. DIC-2C–mRFP (promoter: CMV; backbone: mRFP-N1) is a gift from K. Pfister (University of Virginia, Charlottesville, VA). These constructs were verified by DNA sequencing.
siRNA
Stealth RNAi oligonucleotides were used for siRNA knockdown experiments (Invitrogen). The sequences were described in Itakura et al. (2012) as follows: rat Stx17-siRNA #1 antisense, 5′-AAUUAAGUCCGCUUCUAAGGUUUCC-3′, and sense, 5′-GGAAACCUUAGAAGCGGACUUAAUU-3′; rat Stx17-siRNA #2 antisense, 5′-UCGCAGGCUUCAAAGUGGCAGGAAU-3′, and sense: 5′-AUUCCUGCCACUUUGAAGCCUGCGA-3′; and Scr-siRNA antisense, 5′-UCCACUAAUGAUCGGACUUACUUG-3′, and sense, 5′-ACAAGUAAGUCCGAUCAUUAGUGGA-3′. An siRNA-resistant Stx17 silent mutant (Stx17*) was created by substituting three nucleotides in the Stx17-siRNA#1–targeting region (A528G, G531A, and C534T).
Cell culture and transfection
HEK293T cells were grown in high-glucose DMEM containing sodium pyruvate and l-glutamine supplemented with 10% fetal bovine serum and penicillin–streptomycin (Invitrogen). Subconfluent HEK293T cells were transfected with Stealth RNAi oligonucleotides (10 nM) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Cells were transfected and maintained for additional 3 d followed by biochemical analysis.
DRG neurons were isolated from postnatal 8–10-d Sprague–Dawley rat spinal cords in Hank’s buffered salt solution and digested in 2.5 U/ml dispase II (Roche) and 200 U/ml collagenase (Worthington Biochemical Corporation) at 37°C for 30 min followed by a 35-min shaking at room temperature. Neurons were then collected with 70-µm nylon cell strainer (Falcon). Before plating, DRG neurons were transfected with various plasmid DNA and/or Stealth RNAi oligonucleotides. For GFP-LC3 and mRFP-Rab7 cotransfection, the plasmid concentration ratio is 1:1; for cotransfection of GFP-LC3 and mRFP-Rab7 with HA, HA-snapin, or HA-snapin-L99K (Fig. 2) or Stx17* (Fig. S2), the ratio is 1:1:3; for cotransfection of DIC2C-mRFP with GFP-LC3 or GFP-Rab7 (Fig. 4), the ratio is 1:2; for cotransfection of GFP-Rab7 and DIC2C-mRFP with HA or HA-snapin-L99K, the ratio is 1:2:2 (Fig. S3); for cotransfection of DsRed-Mito and HA or HA-snapin-L99K, the ratio is 1:3 (Fig. S1). The concentration of siRNA for transfection in DRG neurons is 30 nM. We used a Nucleofector (Lonza) for transfection according to the manufacturer’s specifications and plated on coverslips/two-chambered coverglass coated with poly-ornithine (Sigma-Aldrich; diluted 1:3 in ddH20) and laminin (Roche; diluted 1:50 in PBS). Neurons were then maintained in Neurobasal medium supplemented with 10% fetal bovine serum, 2 mM GlutaMAX, and B27 (Invitrogen) for 3 d at 37°C in a 5% CO2 incubator.
Live-cell imaging and image analysis
Before imaging, DRG neurons were transferred to Neurobasal medium with 2 mM GlutaMAX and B27 (without 10% fetal bovine serum) for a 3-h starvation at 37°C in a 5% CO2 incubator. Imaging was performed in FluoroBrite DMEM media (Life Technologies) with 2 mM GlutaMAX and B27. Neurons were time-lapse imaged under an environmental chamber in which temperature was maintained at 37°C and CO2 concentration at 5%. DRG neurons were visualized with a microscope (LSM 510; Carl Zeiss) equipped with a Plan Neofluar 40× NA 1.3 oil immersion objective. Single-color time-lapse images of 1,024 × 1,024 pixels (8 bit) were taken consecutively every 5 s with red channel. Dual-color time-lapse images of 1,024 × 1,024 pixels (8 bit) were taken consecutively every 2 s with green channel followed by red channel. Imaging duration varies from 1.5 to 10 min. Kymographs were generated by ImageJ (National Institutes of Health) and converted to QuickTime (Apple) videos. Background subtraction and adjustment of brightness/contrast were applied to the whole image. A vesicle was considered stationary if it moved a net distance ≤10 µm for the duration of the entire time-lapse imaging. A motile one at either direction was counted only if the net displacement was ≥10 µm.
For the video, DRG neurons plated on a two-chambered cover glass (Thermo Fisher Scientific) were subjected to imaging after medium was changed into prewarmed FluoroBrite DMEM media (Life Technologies) with 2 mM GlutaMAX and B27. Dual-color time-lapse images were taken once every 3 s with acquisition exposure time at 150 ms (green channel followed by red channel) by a spinning-disk confocal microscope (A1R; Nikon) with the Perfect Focus System (PFS; Apochromat 100×, 1.49 NA oil immersion objective; Nikon) in an environmental chamber at 37°C. QuickTime video was generated by ImageJ, and adjustments of brightness/contrast were applied to all the frames.
EM
Cultured mouse cortical neurons at DIV14 were fixed at room temperature with EM fixative (2% glutaraldehyde and 2% paraformaldehyde in 0.1 N Na+ cacodylate buffer); cultured rat DRG neurons at DIV3 were starved in serum-free medium for 3 h and fixed at room temperature with 4% glutaraldehyde in 0.1 N Na+ cacodylate buffer for 30 min. Samples were then stored at 4°C overnight and then treated with osmium tetroxide, en bloc mordanted with uranyl acetate, dehydrated through a series of graded ethanol washes, and embedded in epoxy resins. Thin sections were stained with uranyl acetate and lead citrate (Electron Microscopy Facility, National Institute of Neurological Disorders and Stroke [NINDS], National Institutes of Health). Images were acquired on an electron microscope (200CX; JEOL) and analyzed using ImageJ.
Immunoisolation of LEs
Animal care and use were carried out in accordance with National Institutes of Health guidelines and approved by the National Institutes of Health, NINDS/National Institute on Deafness and Other Communication Disorders, Animal Care and Use Committee. Whole brains from adult WT and conditional snapin-deficient mice were collected for immunoisolation of LEs. The tissues were gently homogenized with ice-cold homogenization buffer (0.25 M sucrose, 10 mM Tris, pH 7.4, 1 mM EGTA, and protease inhibitor) using a glass rod with three to four gentle strokes of the pestle of the 30-ml Dounce homogenizer and then centrifuged at 700 g for 10 min, after which the supernatant was collected. This step was repeated, and the first and second supernatants were combined. The supernatant was centrifuged at 20,000 g for 10 min. The pellet was resuspended in homogenization buffer using a glass rod and recentrifuged at 20,000 g for 10 min. The resultant crude light membrane pellets were then suspended in homogenization buffer and subjected to immunoisolation with superparamagnetic beads (Dynabeads sheep anti–mouse IgG; Invitrogen) coated with an anti-Rab7 monoclonal antibody or control mouse IgG (Sigma-Aldrich), as previously described (Cai et al., 2010; Ye and Cai, 2014). In brief, after washing the superparamagnetic beads twice for 5 min with isolation buffer (Ca2+- and Mg2+-free PBS with 0.1% BSA and 2 mM EDTA), the beads were incubated with 2 µg anti-Rab7 monoclonal antibody or control mouse IgG at 4°C for 2 h on a rotator. After incubation, wash the beads with the isolation buffer twice and once with incubation buffer. Approximately 400-µg light membrane fractions from brain homogenates were mixed with incubation buffer (PBS, pH 7.4, with 5% fetal bovine serum and 2 mM EDTA) containing magnetic beads (final reaction volume of 1 ml) and incubated for 4 h at 4°C on a rotator. After incubation, beads were collected with a magnetic device (Magnetic Particle Concentrator; Invitrogen), washed five times with incubation buffer and three times with PBS for 10 min each at 4°C, and then resolved by 4–12% Bis-Tris NuPAGE for sequential Western blots on the same membranes after stripping between each antibody application.
Online supplemental material
Fig. S1 shows impaired recruitment of dynein DIC to LE membranes in snapin cKO mouse brains. Fig. S2 demonstrates that depleting Stx17 blocks formation of amphisomes and immobilizes AVs along axons. Fig. S3 shows dynamic autophagosomal biogenesis, fusion, and retrograde transport along the distal axon shaft of a DRG neuron. Video 1 shows time-lapse images of dynamic autophagosomal biogenesis, fusion, and retrograde transport in the growth cone of a DRG neuron. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412046/DC1.
Supplementary Material
Acknowledgments
We thank J. di Giovanni and members of the Sheng laboratory for technical assistance and discussions, S. Cheng and V. Crocker at the National Institute of Neurological Disorders and Stroke (NINDS) Electron Microscopy Facility for TEM analysis, V. Schram at the National Institute of Child Health and Human Development Light Imaging Facility for the Nikon spinning-disk confocal microscope, K. Pfister (University of Virginia, Charlottesville, VA) for sharing DIC-2C–mRFP, and D. Schoenberg for proof editing.
X.-T. Cheng is a PhD student working at the National Institutes of Health (NIH) through the joint NIH PhD program with Shanghai Jiao Tong University. The work was supported by the Intramural Research Program of NINDS, NIH ZIA HS003029 and ZIA NS002946 (Z.-H. Sheng), and NIH grant R01NS089737 (Q. Cai).
The authors declare no competing financial interests.
Author contributions: X.-T. Cheng conducted most of the imaging experiments and the data analysis; B. Zhou conducted time-lapse images of de novo autophagosomal biogenesis, fusion, and retrograde transport; M.-Y. Lin and Q. Cai designed and performed snapin biochemical analysis. Z.-H. Sheng is the senior author who designed the project; X.-T. Cheng and Z.-H. Sheng wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- AV
- autophagic vacuole
- AVd
- degradative AVs
- AVi
- initial AVs
- cKO
- conditional knockout
- CMV
- cytomegalovirus
- DHC
- dynein heavy chain
- DIC
- dynein intermediate chain
- DIV
- day in vitro
- DRG
- dorsal root ganglion
- LE
- late endosome
- Scr-siRNA
- scrambled siRNA
- Stx17
- syntaxin 17
- TEM
- transmission EM
- WT
- wild type
References
- Bains M., Zaegel V., Mize-Berge J., and Heidenreich K.A.. 2011. IGF-I stimulates Rab7-RILP interaction during neuronal autophagy. Neurosci. Lett. 488:112–117 10.1016/j.neulet.2010.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Lu L., Tian J.-H., Zhu Y.-B., Qiao H., and Sheng Z.-H.. 2010. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 68:73–86 10.1016/j.neuron.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J.P., Ross J.L., Antony S.M., Tokito M., and Holzbaur E.L.F.. 2007. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. USA. 104:10045–10050 10.1073/pnas.0610628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J.P., Zajac A.L., Tokito M., and Holzbaur E.L.F.. 2011. Huntingtin coordinates the dynein-mediated dynamic positioning of endosomes and lysosomes. Mol. Biol. Cell. 22:478–492 10.1091/mbc.E10-03-0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., and Colombo M.I.. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 9:230–250 10.1111/j.1600-0854.2007.00677.x [DOI] [PubMed] [Google Scholar]
- Fu M.-M., Nirschl J.J., and Holzbaur E.L.F.. 2014. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev. Cell. 29:577–590 10.1016/j.devcel.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A.L., Zhang Z., et al. 2014. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16:1215–1226 10.1038/ncb3066 [DOI] [PubMed] [Google Scholar]
- Ha J., Lo K.W.-H., Myers K.R., Carr T.M., Humsi M.K., Rasoul B.A., Segal R.A., and Pfister K.K.. 2008. A neuron-specific cytoplasmic dynein isoform preferentially transports TrkB signaling endosomes. J. Cell Biol. 181:1027–1039 10.1083/jcb.200803150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Niwa S., and Tanaka Y.. 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68:610–638 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., and Eskelinen E.-L.. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837–4848 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- Jahreiss L., Menzies F.M., and Rubinsztein D.C.. 2008. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 9:574–587 10.1111/j.1600-0854.2008.00701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., and Neefjes J.. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 176:459–471 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K., Nishiyama J., Inoue T., Mizushima N., Takeda J., and Yuzaki M.. 2010. Dynein- and activity-dependent retrograde transport of autophagosomes in neuronal axons. Autophagy. 6:378–385 10.4161/auto.6.3.11262 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., and Yoshimori T.. 2008. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33:109–122 10.1247/csf.08005 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., and Emr S.D.. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Elazar Z., Seglen P.O., and Rubinsztein D.C.. 2008. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 4:849–850 10.4161/auto.6845 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 8:445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Sato Y., and Nixon R.A.. 2011. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J. Neurosci. 31:7817–7830 10.1523/JNEUROSCI.6412-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., and Klionsky D.J.. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 6:463–477 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Maday S., Wallace K.E., and Holzbaur E.L.F.. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Twelvetrees A.E., Moughamian A.J., and Holzbaur E.L.. 2014. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 84:292–309 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N.2014. Sugar modification inhibits autophagosome-lysosome fusion. Nat. Cell Biol. 16:1132–1133 10.1038/ncb3078 [DOI] [PubMed] [Google Scholar]
- Moughamian A.J., Osborn G.E., Lazarus J.E., Maday S., and Holzbaur E.L.. 2013. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J. Neurosci. 33:13190–13203 10.1523/JNEUROSCI.0935-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R.A.2013. The role of autophagy in neurodegenerative disease. Nat. Med. 19:983–997 10.1038/nm.3232 [DOI] [PubMed] [Google Scholar]
- Perlson E., Jeong G.B., Ross J.L., Dixit R., Wallace K.E., Kalb R.G., and Holzbaur E.L.. 2009. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J. Neurosci. 29:9903–9917 10.1523/JNEUROSCI.0813-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Acevedo-Arozena A., Imarisio S., Berger Z., Vacher C., O’Kane C.J., Brown S.D.M., and Rubinsztein D.C.. 2005. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 37:771–776 10.1038/ng1591 [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Mariño G., and Kroemer G.. 2011. Autophagy and aging. Cell. 146:682–695 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Schneider J.L., and Cuervo A.M.. 2014. Autophagy and human disease: emerging themes. Curr. Opin. Genet. Dev. 26:16–23 10.1016/j.gde.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.-M., and Mizushima N.. 2014. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem. Sci. 39:61–71 10.1016/j.tibs.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze S.A., Abada A., and Elazar Z.. 2014. Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb. Perspect. Biol. 6:a018358 10.1101/cshperspect.a018358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.C., and Holzbaur E.L.F.. 2014. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 34:1293–1305 10.1523/JNEUROSCI.1870-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., and Cai Q.. 2014. Snapin-mediated BACE1 retrograde transport is essential for its degradation in lysosomes and regulation of APP processing in neurons. Cell Reports. 6:24–31 10.1016/j.celrep.2013.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Friedman L., Komatsu M., and Tanaka K.. 2009. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim. Biophys. Acta. 1793:1496–1507 10.1016/j.bbamcr.2009.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.