During anaphase B, Imp1-mediated transport of the AAA-ATPase Cdc48 protein at the membrane domain surrounding the mitotic spindle midzone promotes spindle midzone dissolution in fission yeast.
Abstract
During each cell cycle, the mitotic spindle is efficiently assembled to achieve chromosome segregation and then rapidly disassembled as cells enter cytokinesis. Although much has been learned about assembly, how spindles disassemble at the end of mitosis remains unclear. Here we demonstrate that nucleocytoplasmic transport at the membrane domain surrounding the mitotic spindle midzone, here named the midzone membrane domain (MMD), is essential for spindle disassembly in Schizosaccharomyces pombe cells. We show that, during anaphase B, Imp1-mediated transport of the AAA-ATPase Cdc48 protein at the MMD allows this disassembly factor to localize at the spindle midzone, thereby promoting spindle midzone dissolution. Our findings illustrate how a separate membrane compartment supports spindle disassembly in the closed mitosis of fission yeast.
Introduction
Like most fungi, the fission yeast Schizosaccharomyces pombe segregates chromosomes within the nucleus in the process known as closed mitosis (Zheng et al., 2007; Zhang and Oliferenko, 2013). As fission yeast cells enter mitosis, the cytoplasmic microtubules rearrange to form the intranuclear mitotic spindle, which is responsible for chromosome segregation. The mitotic spindle is assembled from the spindle pole bodies (SPBs), specialized microtubule-organizing organelles functionally analogous to metazoan centrosomes. Upon mitotic entry, the SPBs are inserted into the nuclear envelope to form a bipolar mitotic spindle (Ding et al., 1997; Tallada et al., 2009). SPBs eventually separate and move to opposite sides of the nucleus. During anaphase A, each set of sister chromatids segregates toward opposite spindle poles, followed by separation of the poles and karyokinesis during anaphase B (Hagan, 1998).
By the end of mitosis, the mitotic spindle is disassembled and cytokinesis produces two independent cells (Krapp et al., 2004). Rapid spindle disassembly at the end of mitosis is crucial for cell proliferation (Woodruff et al., 2012). However, mechanisms that contribute to spindle microtubule depolymerization at the end of mitosis are poorly understood (Sagolla et al., 2003).
During anaphase B, the central internuclear region is coincident with the central spindle domain, a region where the antiparallel interpolar microtubules (ipMTs) interdigitate at their plus ends to form the mitotic spindle midzone (Maddox et al., 2000). The size and integrity of the mitotic spindle are maintained by a vast array of microtubule-binding proteins that regulate microtubule dynamics (e.g., XMAP215, CLASP, and EB1) or cross-link the antiparallel ipMTs at the midzone (e.g., Ase1; Glotzer, 2009). The analysis of mutants defective in spindle disassembly in budding yeast indicates that this process is achieved by functionally overlapping subprocesses such as degradation of cross-linking proteins leading to disengagement of the spindle halves, arrest of spindle elongation, and initiation of ipMT depolymerization. These subprocesses are largely driven by the anaphase-promoting complex/cyclosome (APC/C), Aurora B kinase, and kinesin-8 (Woodruff et al., 2010). Here, we show that nucleocytoplasmic transport in a restricted nuclear membrane domain, the midzone membrane domain (MMD), is required for proper spindle disassembly in the fission yeast, revealing yet another layer of regulation.
Results
Imp1 depletion leads to hyperextended mitotic spindles
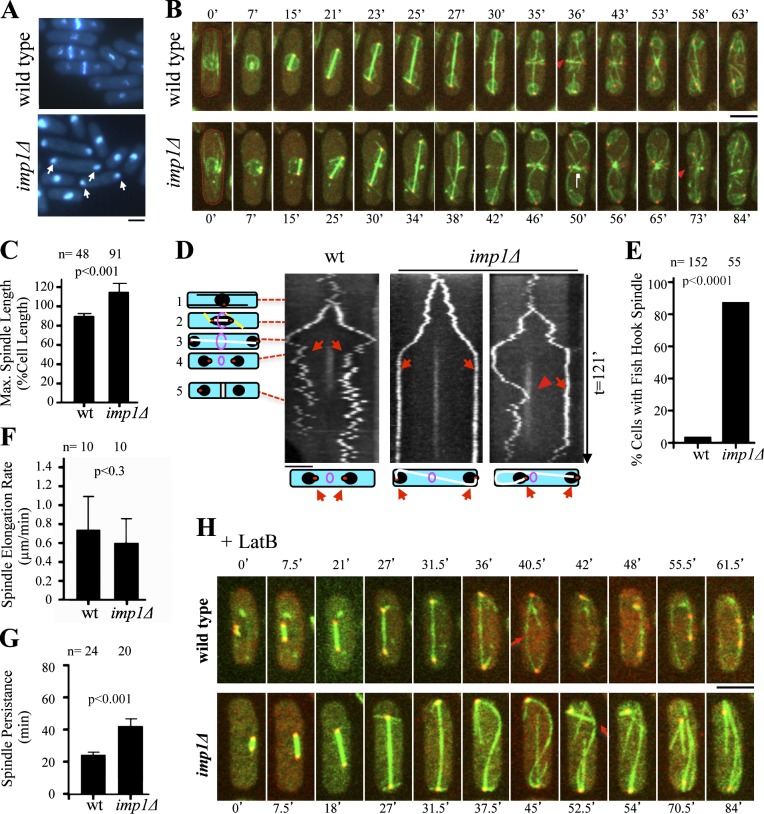
The fission yeast importin-α Imp1 was identified in a genome-wide screen for cell cycle regulators (Tallada et al., 2002). Importin-α plays important roles in nucleocytoplasmic transport, nuclear envelope structure, and mitotic spindle assembly (Clarke and Zhang, 2001; Dasso, 2001; Geles et al., 2002; Schatz et al., 2003; Wozniak et al., 2010). To further analyze the role of Imp1 in cell cycle regulation, we first characterized phenotypic defects of Imp1-depleted cells. Cells lacking Imp1 were viable, but we observed that postmitotic nuclei were often mispositioned in the daughter cells, remaining at the cell tips (Fig. 1 A). Similar results were previously described (Umeda et al., 2005). This phenotype is diagnostic of possible defects in microtubule dynamics (Daga and Chang, 2005). To check this possibility, we expressed GFP-Atb2 (a microtubule marker), Sid2-Tom (a protein kinase located at the SPB and the division septum), and Cut11-GFP (a trans-membrane protein of the nuclear envelope) in wild-type and imp1Δ cells and followed the dynamics of microtubule, SPBs, and nuclear membrane by in vivo time-lapse microscopy as cells enter mitosis. As shown in Fig. 1 B, in a normal cell cycle, the mitotic spindle assembled early in mitosis and elongated during anaphase B until a maximum length. The spindle then rapidly disassembled before cytokinesis. Interestingly, we found that late anaphase B spindles in Imp1-depleted cells were consistently longer relative to those of wild-type cells (see Fig.1 B). The mean maximum spindle length in wild-type cells was 90% of the total cell length. In the imp1Δ strain, the mitotic spindle reached 115% of the cell length (Fig. 1 C). This may explain the abnormal localization of the nuclei at the cell tips in Imp1-depleted cells. Time course representation of the Sid2-Tom marker (kymograph) showed that in the wild-type control, the two SPBs (and the attached nuclei) moved toward the tips of the cell during mitosis and then relocalized to the middle of each daughter cell during cytokinesis. In contrast, the SPBs and nuclei remained at the cell tips in imp1Δ cells with hyperextended spindles (Fig. 1 D). Hyperextended mitotic spindles often appeared fishhook shaped (Fig. 1, B and E), a structure which eventually located one of the SPBs (and the corresponding nucleus) at the division site (Fig. 1, B and D). Therefore, we conclude that the loss of the importin-α Imp1 results in hyperextended mitotic spindles.
Figure 1.
Imp1 is required for mitotic spindle disassembly. (A) DAPI and Calcofluor white staining of wild-type and imp1Δ S. pombe cells. In asynchronous cultures, 21% of imp1Δ cells show nuclei at the cell tips (arrows), whereas only 9% of these cells are found in the wild-type strain. (B) Time-lapse fluorescence images of GFP-Atb2 (spindle), Cut11-GFP (nuclear envelope), and Sid2-Tom (SPBs and medial division ring) in wild-type and imp1Δ cells. Red arrowheads indicate initiation of spindle disassembly as cytokinesis is initiated in wild-type cells (t = 36 min) and spindle breakdown by the constricting actomyosin ring in imp1Δ cells (t = 73 min). (C) Mean maximal spindle length (percentage of the cell length) in wild-type and imp1Δ cells. (D) Kymographs of wild-type (left) and imp1Δ cells (middle and right) expressing Sid2-Tom. Schematized cells on the left represent cell cycle stages: 1, interphase; 2, early anaphase B; 3, late anaphase B; 4, ring contraction and daughter nuclei reposition; 5, end of cytokinesis. Although nuclei are repositioned in wild-type cells (left, arrows), in imp1Δ cells, hyperextended spindles persisted during cytokinesis (middle, arrows) and eventually, a fishhook-shaped spindle localizes one SPB and its associated nucleus close to the division site (right, arrowhead). Schematized cells (bottom) represent the structure of the corresponding mitotic spindles. (E) Frequency (%) of fishhook-shaped mitotic spindles in wild-type and imp1Δ cells. (F) Spindle elongation rate in wild-type and imp1Δ cells. (G) Average time of mitotic spindle persistence in wild-type and imp1Δ cells. (H) Time-lapse fluorescence images of GFP-Atb2 and Sid2-Tom in wild-type and imp1Δ cells in the presence of latrunculin B. Arrows indicate initiation of spindle disassembly as cytokinesis is initiated in wild-type cells, and spindle breakdown is caused by bending forces in imp1Δ cells. Bars, 5 µm. Graphs represent mean and standard deviation. n is the total number of cells scored from at least three independent experiments.
Abnormal spindle length may result from unbalanced assembly/disassembly activities. Either an increase in the spindle elongation rate or a defect or decrease in the rate of disassembly would yield hyperextended spindles. A detailed analysis of the mitotic spindle dynamics in the imp1Δ strain indicated that neither spindle initiation nor elongation (slightly reduced in imp1Δ cells) were significantly affected (Fig. 1, B and F). However, mitotic spindles in imp1Δ cells persisted significantly longer into mitosis than those of wild-type cells (Fig. 1, B, C, and G), suggesting that spindle disassembly was severely affected in these cells. Consistently, we observed that the mitotic spindle remained assembled during cytokinesis until it was finally cut by the actomyosin contractile ring in 76% (n = 29) of the imp1Δ cells, leading to an aberrant disassembly (Fig. 1 B). When we blocked actomyosin ring formation, using latrunculin B (Gachet et al., 2004), wild-type cells disassembled the spindle at the end of mitosis. In imp1Δ cells, the spindle was still present after mitosis until finally breaking, possibly because of bending forces (Fig. 1 H). Therefore, the mitotic spindle disassembly fails at the end of mitosis in Imp1-depleted cells, suggesting that importin-α Imp1 is required for this step in fission yeast.
Imp1 is essential for spindle midzone dissolution
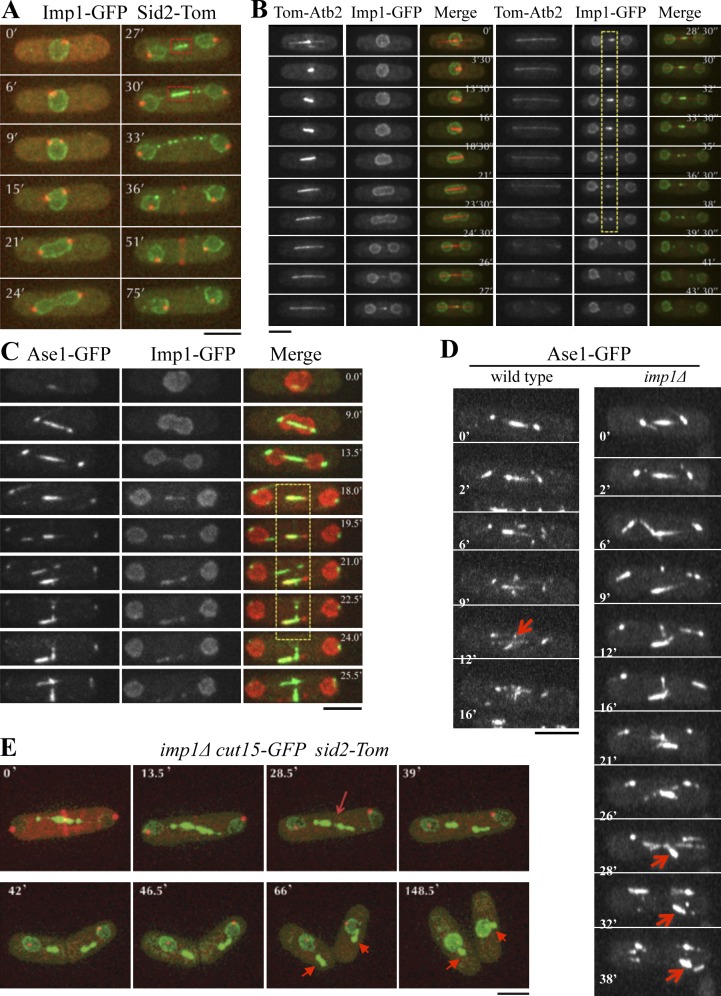
To further understand how Imp1 influences mitotic spindle disassembly, we analyzed its localization in S. pombe cells using the Imp1-GFP–tagged construct (coexpressed with Sid2-Tom to visualize SPBs and medial ring in the same cells). As shown in Fig. 2 A, Imp1 was found in the cytoplasm and the nucleoplasm, but more intensely in the nuclear envelope. As cells proceed to anaphase B, the nucleus elongates and eventually assumes a dumbbell shape, with the two segregating nuclei linked by an internuclear bridge late in mitosis. Intriguingly, Imp1 was transiently found at the central region of this mitotic internuclear bridge (Fig. 2 A). A similar localization to Imp1 has been previously described for the other S. pombe importin-α, Cut15 (Matsusaka et al., 1998). To analyze this internuclear localization in greater depth, we visualized Imp1 and mitotic spindles in cells simultaneously expressing Imp1-GFP and the microtubule marker Tom-Atb2. Interestingly, localization of Imp1 at the central internuclear region was coincident with the central spindle domain (Fig. 2 B), a region where the antiparallel ipMTs interdigitate at their plus ends to form the mitotic spindle midzone (Maddox et al., 2000). Ase1, a key protein involved in cross-linking antiparallel ipMTs at this region, was used as a midzone marker (Loïodice et al., 2005; Yamashita et al., 2005; Fu et al., 2009). Live-imaging analysis of Imp1-Tom– and Ase1-GFP–expressing cells confirmed that Imp1 specifically localized at the spindle midzone domain (Fig. 2 C).
Figure 2.
Imp1 regulates spindle disassembly at the midzone. (A) Time-lapse fluorescence images of Imp1-GFP– and Sid2-Tom–expressing cells. During anaphase B, Imp1-GFP localizes at the central internuclear bridge (dashed squares at 27 and 30 min). (B) Time-lapse fluorescence images of a representative cell expressing Imp1-GFP and the microtubule marker Tom-Atb2. Imp1-GFP colocalizes with the spindle midzone, where antiparallel microtubules interdigitate (dashed area). (C) Colocalization of Imp1-Tom and the spindle midzone marker Ase1-GFP by time-lapse fluorescence microscopy (merge, dashed area). (D) Localization of Ase1-GFP by time-lapse fluorescence in wild-type and imp1Δ cells. Arrows indicate Ase1-GFP disappearance from spindle midzone during normal spindle disassembly in wild-type cells and Ase1-GFP persistence at the midzone domain in imp1Δ cells. (E) Time-lapse fluorescence images of imp1Δ cells coexpressing Cut15-GFP and Sid2-Tom, showing assembled midzone fragments after mitosis (arrows). Bars, 5 µm.
In budding yeast, the mitotic spindle disassembles by depolymerization of the ipMTs from their plus ends at the midzone, leading to spindle midzone dissolution and separation of the spindle halves at the end of mitosis (Maddox et al., 2000). Thus, the location of Imp1 at the midzone region is consistent with the proposed role for importin-α in spindle disassembly in fission yeast. To further study a possible role of Imp1 in spindle midzone disassembly, we followed fluorescence of the Ase1-GFP marker in wild-type and imp1Δ living cells. As shown in Fig. 2 D, wild-type cells efficiently dissolved the spindle midzone late in anaphase B. However, in Imp1-depleted cells, the midzone Ase1-GFP protein continued to decorate the central spindle domain and remained assembled even at the end of cytokinesis, after spindle breakage. By using importin-α Cut15-GFP as a central domain marker (and the Sid2-Tom SPBs and medial ring marker), we confirmed that Imp1-depleted cells retained assembled midzone fragments after cytokinesis (Fig. 2 E). Therefore, these results indicate that Imp1 is specifically required for midzone spindle disassembly late in mitosis.
Nucleocytoplasmic transport activity controls spindle midzone disassembly
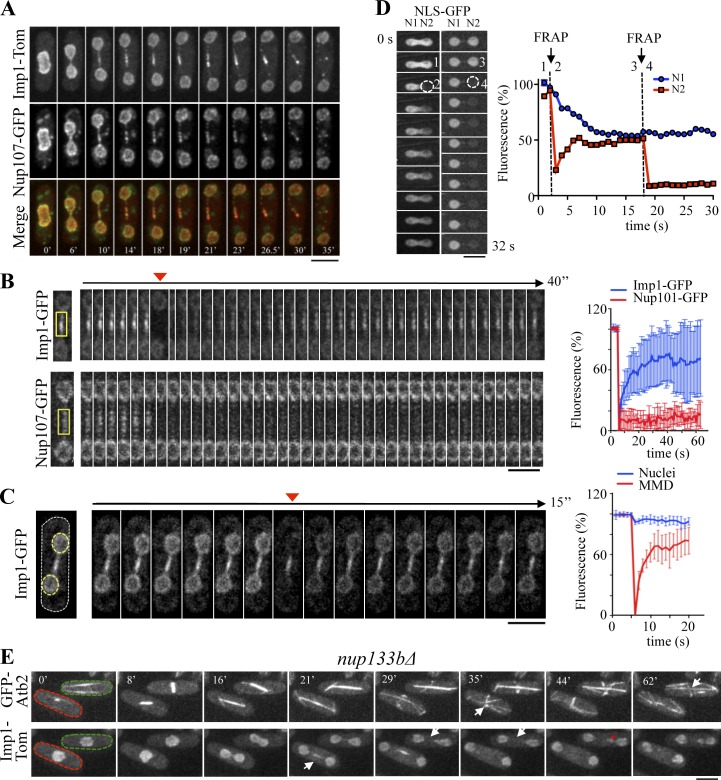
Imp1 is a member of the importin-α family, which functions in transport through the nuclear pore (Umeda et al., 2005). We wondered whether nuclear pores were also found at this internuclei region of the central spindle domain. The simultaneous expression of Imp1-Tom and the nucleoporin Nup107-GFP or Nup61-GFP, canonical nuclear pore components (Baï et al., 2004; Chen et al., 2004; Wozniak et al., 2010), showed that Imp1 colocalized with nuclear pores at the nuclear envelope along the cell cycle and also at the central spindle midzone region during anaphase B (Fig. 3 A and Fig. S1 A). During the closed mitosis of S. pombe cells, the nuclear membrane envelops the elongating mitotic spindle as the daughter nuclei segregate (Zhang and Oliferenko, 2013). The colocalization of nuclear pore complexes and Imp1 at the specific membrane domain surrounding the spindle midzone suggests that functional nucleocytoplasmic transport activity takes place at this specific domain.
Figure 3.
The MMD represents a differentiated nuclear envelope region with nucleocytoplasmic transport activity. (A) Colocalization of Imp1-Tom and the nuclear pore marker Nup107-GFP by time-lapse fluorescence microscopy. (B) FRAP time-lapse images of Imp1-GFP– and Nup107-GFP–expressing cells. Arrowhead indicates image acquired immediately after photobleaching the indicated regions (yellow squares). Time course fluorescence recovery (percentage related to initial fluorescence intensity) is represented (right, n = 8). (C) FRAP time-lapse images of a Imp1-GFP–expressing cell. Arrowhead indicates image acquired immediately after photobleaching both daughter nuclei (yellow-dashed circles). Fluorescence recovery of imp1-GFP signal measured either at spindle midzone (MMD) or at nuclei is shown as a percentage of initial fluorescence (right, n = 5). (D) Photobleaching of nucleoplasmic NLS-GFP (dashed circle) during early (1, prebleach; 2, bleach) and late (3, prebleach; 4, bleach) anaphase B in a representative cell from four independent experiments. Graph represents fluorescence intensity levels (percentage related to initial values) over time in bleached (red) and nonbleached (blue) regions. (E) Time-lapse fluorescence images of nup133bΔ cells coexpressing GFP-Atb2 and Imp1-Tom. The red-dashed cell forms a normal MMD region during anaphase B (arrow at 21 min), which is followed by mitotic spindle disassembly (arrow at 35 min). The green-dashed cell, however, undergoes mitosis without a detectable MMD region (arrow at 29 and 35 min), which correlates with persistence of mitotic spindle at late cytokinesis (arrow at 62 min). In this cell, nuclear missegregation is also observed (asterisk at 44 min), a feature that may explain the abnormal chromosome segregation previously described in this strain (Baï et al., 2004). Bars, 5 µm.
To study whether this midzone membrane region represents a functional domain of the nuclear envelope, we followed the dynamics of nuclear pores and Imp1 at this domain by FRAP experiments. Nucleoporins remain integrated at the nuclear pore for a long time, but nuclear pore complexes are mobile through the nuclear envelope in yeast (Bucci and Wente, 1997). Accordingly, we determined that fluorescence did not recover (in our experimental time scale) upon photobleaching Nup107-GFP in the entire nuclear envelope (Fig. S1 B), but it rapidly recovered when bleaching only a nuclear envelope region flanking the internuclei membrane bridge (Fig. S1 C). Thus, as expected, nuclear pores can freely diffuse at the nuclear envelope. However, Nup107-GFP fluorescence was not recovered after bleaching the medial region (Fig. 3 B), suggesting that diffusional mobility of nuclear pores from the adjacent nuclear membranes to this region is highly restricted during anaphase B.
In contrast to the nucleoporins, importins are rapidly exchanged at the nuclear pores as a result of their transport activity (Cardarelli et al., 2009). Accordingly, fluorescence rapidly recovered after photobleaching the internuclear membrane region in Imp1-GFP expressing–cells, with a half-time equilibration of 10 s (Fig. 3 B). Recovery dynamics in the internuclear region were similar to those observed in the flanking nuclear envelops in these cells (Fig. 3 C), indicating that Imp1 undergoes high nucleocytoplasmic transport activity in both the nuclear envelope and the internuclear region. Importantly, fluorescence in the central membrane domain was not altered by bleaching Imp1-GFP fluorescence at the adjacent nuclear regions (Fig. 3 C), suggesting that Imp1-GFP diffusion from the internuclear to the flanking nuclear region is restricted. Therefore, the membrane region surrounding the spindle midzone represents a compartmentalized domain, here named the MMD. By using a nuclear localization signal (NLS)–GFP fusion protein (Shibuya et al., 1999), photobleaching experiments indicated that this nucleoplasmic protein freely diffuses between the segregating nuclei until anaphase B, when exchange of nucleoplasmic NLS-GFP was extremely reduced by the dumbbell morphology of the dividing nucleus (Fig. 3 D). Thus, midzone localization of proteins regulating spindle disassembly in the S. pombe closed mitosis likely requires active transport, either through microtubule-mediated plus-end motors from the nucleoplasm of the flanking nuclei and/or by nucleocytoplasmic transport through nuclear pores at the MMD.
To determine whether nucleocytoplasmic transport activity at the MMD controls spindle disassembly, we studied microtubule dynamics and Imp1-GFP localization at the MMD in cells lacking the Nup133b nucleoporin. In nup133bΔ mutants, proper nuclear pore distribution is impaired (Baï et al., 2004) and, occasionally, cells lacking nuclear pores at the MMD were produced. By coexpressing Imp1-Tom and GFP-Atb2 in the nup133bΔ strain, we observed that cells with a normal MMD region (based on Imp1-GFP localization) followed normal mitotic spindle assembly and disassembly dynamics (reported by the GFP-Atb2 marker). However, cells without nuclear pores at the MMD failed to disassemble the spindle at the end of mitosis (spindles 115% of the cell length, n = 5; Fig. 3 E). Hence, nuclear pore–mediated nucleocytoplasmic transport activity at the MMD region likely regulates spindle disassembly.
Importin-α/β–NLS substrate complexes mediate conventional nucleocytoplasmic transport activity at the MMD
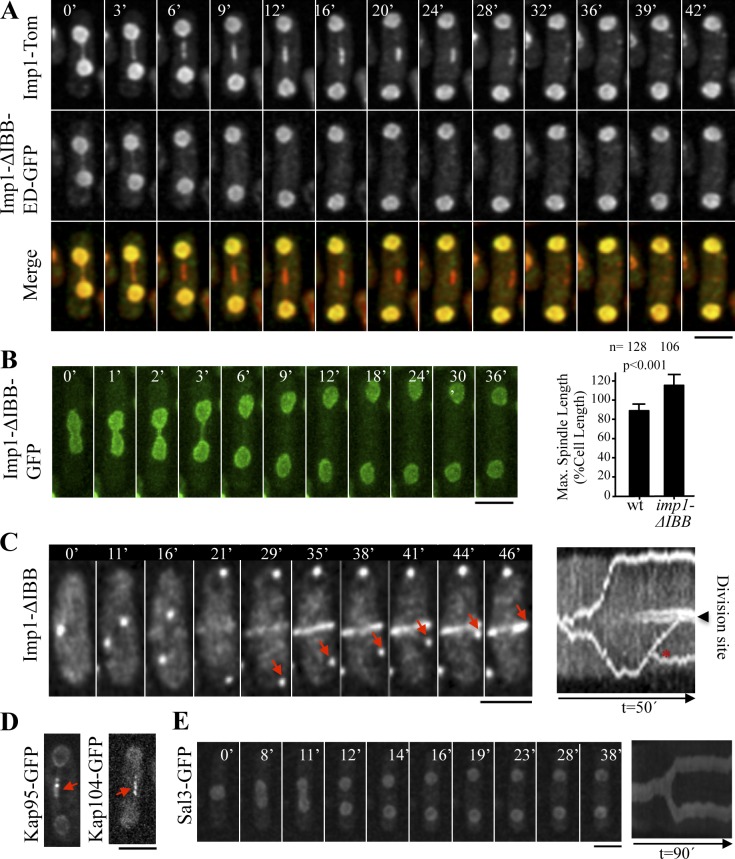
Two nuclear import pathways are well characterized in eukaryotic cells: (1) the classical pathway, which involves the conventional NLSs and the receptors importin-α/karyopherin-α and importin-β/karyopherin-β1 and (2) the karyopherin-β2 pathway (Chook and Süel, 2011). In the classical nucleocytoplasmic transport mechanism, importin-α interacts in the cytoplasm with proteins targeted by an NLS, acting as an adaptor to link the cargo to importin-β. The resulting importin-α/β–NLS substrate ternary complex migrates into the nucleoplasm through the nuclear pore in an importin-β–dependent manner. Once the importin-α/β–NLS substrate complex reaches the nucleoplasm, importin-β interacts directly with Ran-GTP, resulting in the release of importin-α and the NLS-bearing cargo (Lange et al., 2007). Interestingly, it has also been shown that importin-α can migrate into the nucleus in an importin-β– and Ran-independent manner (Miyamoto et al., 2002). According to this importin-β–independent transport mechanism, a budding yeast importin-α mutant that lacks the importin-β binding domain (IBB) and that cannot bind NLS cargoes (ED domain) localizes at the nucleus as does wild-type importin-α (Miyamoto et al., 2002). To examine the possibility that Imp1 could also function in an importin-β–independent pathway in fission yeast, we examined the subcellular localization of an Imp1 mutant (GFP tagged) that lacks the importin-β binding domain and that cannot bind NLS-tagged proteins (the Imp1-Δ-BB-ED mutant; Gruss et al., 2001; Sato and Toda, 2007). Results shown in Fig. 4 A reveal that Imp1-ΔIBB-ED-GFP is efficiently localized at the nucleus, indicating that in S. pombe cells there is also an importin-β–independent mechanism for nuclear import of importin-α (Sato and Toda, 2007). However, to our surprise, Imp1-ΔIBB-ED-GFP was not localized at the MMD region. Identical results were obtained when expressing the Imp1-ΔIBB-GFP mutant version, lacking only the importin-β binding domain (Fig. 4 B), showing that importin-β drives MMD localization of Imp1 and its associated cargoes. Remarkably, we found that in this strain, cells exhibited hyperextended mitotic spindles (Fig. 4 B). Kymograph analysis showed that mitotic SPB dynamics in Imp1-ΔIBB cells (Fig. 4 C) are identical to those found in imp1Δ cells (Fig. 1 D), indicating that importin-β association is essential for Imp1 to undergo nucleocytoplasmic transport at the MMD region for proper spindle disassembly.
Figure 4.
Imp1-mediated transport in the MMD region is importin-β–NLS cargo dependent. (A) Time-lapse fluorescence images of a cell coexpressing Imp1-Tom and Imp1-ΔIBB-ED-GFP. Only Imp1-Tom is found at the MMD. (B) Time-lapse fluorescence images of a cell expressing Imp1-ΔIBB-GFP. (graph) Mean maximal spindle length (percentage of the cell length) in wild-type and Imp1-ΔIBB cells (graph represents mean and standard deviation; n is the total number of cells scored from two independent experiments). (C) Time-lapse fluorescence images of Sid2-Tom in Imp1-ΔIBB cells. Arrows indicate the position of the SPB after cytokinesis is initiated. Kymograph of this cell is shown. Asterisk denotes a signal of a nearby cell. (D) MMD localization (arrows) of Kap95-GFP and Kap104-GFP importin-β proteins during anaphase B. (E) Time-lapse fluorescence images of a cell expressing Sal3-GFP importin-β protein. Kymograph of this cell is shown. Bars, 5 µm.
The S. pombe genome contains 12 genes coding for predicted importin-β proteins. We analyzed the cellular location of some of these proteins (Kap95, Kap104, Kap109, Kap113, Kap114, and Sal3) and found that Kap95 and Kap104 localized at the MMD during anaphase B (Fig. 4 D), whereas the remaining importin-β proteins analyzed did not (see Fig. 4 E for Sal3). Therefore, Imp1 transport of NLS proteins through nuclear pores at the MMD region might be dependent on a specific importin-β subunit, likely Kap95 or Kap104.
Ran-dependent nucleocytoplasmic transport activity at the MMD
The small GTPase Ran is required for all forms of active import and export activities. In the classical import pathway, NLS-tagged proteins form stable importin-α/β complexes in the cytoplasm, where the concentration of Ran-GTP is low, but dissociate from their carriers in the nucleus, where the concentration of Ran-GTP is high. Thereafter, the importin-β–Ran-GTP complex and importin-α bound to its exportin Cse1 and Ran-GTP are reexported to the cytoplasm for another round of import (Lange et al., 2007).
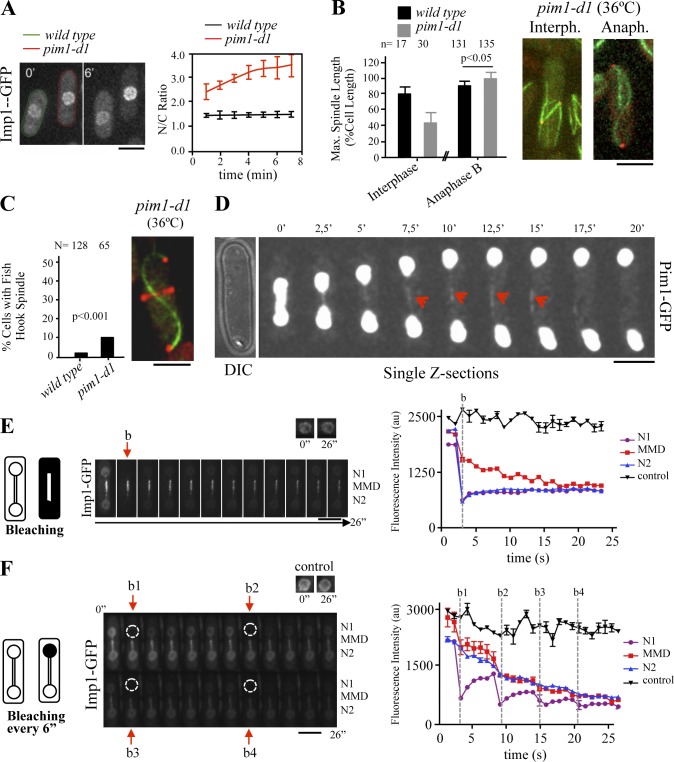
In fission yeast, the nucleotide-bound state of the Ran GTPase (Spi1) is regulated by a nuclear guanine nucleotide exchange factor, Pim1 (Ran-GEF), and a cytoplasmic GTPase activating protein, Rna1 (Ran-GAP; Sazer and Dasso, 2000; Salus et al., 2002). To determine whether spindle disassembly driven by nucleocytoplasmic transport activity at the MMD is Ran dependent, we followed the mitotic spindle dynamics in a compromised Ran GTPase system by using the pim1-d1 temperature-sensitive mutant.
In the pim1-d1 mutant strain, the nuclear Ran-GEF activity is defective and, consequently, the nucleocytoplasmic transport is severely impaired (Salus et al., 2002). By using an Imp1-GFP construct, we first confirmed that inactivation of Pim1 in pim1-d1 cells compromised Imp1-dependent nucleocytoplasmic transport. Accordingly, Imp1 rapidly accumulated at the nucleus upon shift to restrictive temperature in these cells (Fig. 5 A). Interestingly, when Pim1 was inactivated during interphase, we observed that GFP-Atb2–expressing cells assembled an abnormally short mitotic spindle when they are in mitosis (Fig. 5 B). In contrast, in cells that were at anaphase B, Pim1 inactivation caused formation of hyperextended, frequently fishhook-shaped mitotic spindles, similar to those observed in Imp1-depleted cells (Fig. 5, B and C). Short spindles in pim1-d1 cells that entered mitosis during Pim1 inactivation is in agreement with previous results, where Ran and importin-α and -β were implicated in spindle assembly (Fleig et al., 2000; Dasso, 2001; Schatz et al., 2003; Sato and Toda, 2007). In the same way, the existence of hyperextended mitotic spindles in Pim1-deficient anaphase B cells is consistent with the requirement of Ran-dependent nucleocytoplasmic transport activity for spindle disassembly. Accordingly, when using single z-section analysis, fluorescent signal from Pim1-GFP at the spindle midzone was detected (Fig. 5 D).
Figure 5.
Ran-dependent nucleocytoplasmic transport activity at the MMD. (A) Imp1-GFP localization in wild-type (lectin labeled) and pim1-d1 cells (green- and red-dashed cells, respectively) at 0 and 6 min after temperature shift form 25°C (permissive temperature for pim1-d1) to 36°C (restrictive temperature for pim1-d1). Time course accumulation of Imp1-GFP at the nucleus (ratio of nuclear to cytoplasmic fluorescence intensity) in wild-type (lectin labeled) and pim1-d1 cells after temperature shift from 25°C to 36°C (graph represents mean and standard deviation of at least six cells scored from three independent experiments). (B) Maximal spindle length (percentage of the cell length) in wild-type and pim1-d1 cells after temperature shifting during either interphase or anaphase B (graph represents mean and standard deviation; n is the total number of cells scored from at least three independent experiments). Fluorescence images of representative interphase and anaphase B pim1-d1 cells (at 36°C) expressing GFP-Atb2 and Sid2-Tom are shown. (C) Frequency (%) of fishhook-shaped mitotic spindles in wild-type and pim1-d1 (at 36°C) cells (graph represents mean and standard deviation; n is the total number of cells scored from three independent experiments). Representative fishhook-shaped mitotic spindles in a pim1-d1 GFP-atb2 sid2-Tom cell (at 36°C) is shown. (D) Time-lapse fluorescence images of a representative Pim1-GFP cell. Single z-sections in which Pim1-GFP signal can be observed at the MMD (arrowhead) are shown. (E) Time-lapse fluorescence images of Imp1-GFP–expressing cells. Arrow (b) indicates image acquired immediately after photobleaching Imp1-GFP in the entire cell, except the MMD compartment (see schematized cells). Graph shows time course fluorescence intensity (FLIP assay, arbitrary units) in both nuclei and the MMD region (N1, N2, and MMD, respectively) in a representative cell of at least four cells scored in three independent experiments. Nonbleached nucleus is used as a control. (F) Time-lapse fluorescence images of Imp1-GFP–expressing cells. Arrows (b1–4) indicate image acquired immediately after photobleaching Imp1-GFP in one of the nucleus (dashed circles; see schematized cells). Graph shows time course fluorescence intensity (FLIP assay, arbitrary units) in the MMD compartment and the flanking nuclei (MMD, N1, and N2, respectively) in a representative cell of at least four cells scored in three independent experiments. Nonbleached nucleus is used as a control. Bars, 5 µm.
To study Imp1 export dynamics, fluorescence loss in photobleaching (FLIP) assays were performed in the Imp1-GFP strain. These assays indicated that the imported Imp1-GFP is not retained in the MMD compartment (Fig. 5 E and Fig. S2). Instead, it exits the midzone region with kinetics similar to the kinetics of Imp1-GFP exit from the nucleus (Fig. 5 F). Therefore, this importin-α is subjected to import/export cycles at the MMD region, as expected in a conventional Ran-GTPase–dependent manner. However, the mechanism by which the small Ran GTPase operates at the MMD nucleoplasm will be an important question to tackle in future studies.
Identification of Imp1-dependent NLS cargoes
The role of Imp1 in spindle disassembly can be understood by considering the function of Imp1 itself in nucleocytoplasmic transport activity. In a simple model, during anaphase B, importin-α Imp1 binds spindle disassembly factors in the cytoplasm. These factors (in complexes with specific importin-β subunits) are transported through the nuclear pore in the MMD, and then unloaded in the spindle midzone in a Ran-GTP–dependent manner, becoming competent to promote spindle disassembly. Proteins and mechanisms that regulate disassembly at the midzone in fission yeast are poorly understood. However, a vast array of factors has been described to regulate this process in budding yeasts and other eukaryotes (Fridman et al., 2009; Woodruff et al., 2010). To identify possible Imp1-dependent disassembly factors, we first analyzed localization and dynamics of several these proteins (GFP tagged) in wild-type and imp1Δ fission yeast cells, such as Klp5 and Klp6 proteins of the kinesin-8 family (West et al., 2001; Unsworth et al., 2008), the Ark1 (Aurora B) kinase (Buvelot et al., 2003; Woodruff et al., 2010), and the Clp1 phosphatase (Cueille et al., 2001; Trautmann et al., 2001; Fu et al., 2009), which are known regulators of spindle disassembly in budding yeast. All these proteins localized at the spindle midzone during anaphase B in S. pombe cells, supporting a role in microtubule midzone dynamics also. However, none of them depend on Imp1 transport for midzone localization (Fig. S3). This suggests that there are other, unidentified Imp1 cargoes transported at the MMD region to trigger spindle midzone disassembly.
The BIOGrid database lists 368 different proteins that have been found to physically interact with Srp1, the Imp1/Cut15 homologue in budding yeast (Yano et al., 1992; Küssel and Frasch, 1995; Loeb et al., 1995). In S. pombe, only a single protein, the transcription factor Pap1, which also interacts with the Cut15 importin, has been shown to interact with Imp1 (Umeda et al., 2005). To identify Imp1-dependent factors involved in spindle disassembly in S. pombe cells, we purified Imp1–3xHA complexes and identified Imp1–3xHA binding proteins by mass spectrometry. Interestingly, importin-β Kap95 and Kap104, which are highly enriched at the MMD region (Fig. 4 D); nucleoporins Nup60, Nup61, and Nup104; Imp1 itself; and the other importin-α Cut15 were among the most abundant Imp1-associated proteins (Fig. S4 and Table S1). This result agrees with a conventional role for Imp1 in fission yeast in which importin-α/β–NLS substrate complexes cross the nuclear membrane through the nuclear pore in an importin-β–dependent manner (Lange et al., 2007).
The efficient identification of different components of the nucleocytoplasm transport machinery indicates that the assay may be useful to identify putative Imp1-binding NLS cargoes. Importantly, the AAA-ATPase Cdc48, a protein that is involved in spindle disassembly during mitotic exit in both budding yeast and Xenopus laevis egg extracts (Cao et al., 2003; Cao and Zheng, 2004; Cheeseman and Desai, 2004), and regulatory subunits Rpn501, Rpn7, Rpn8, Rpt5, and Rpt4 of the 26S of the proteasome complex (Wilkinson et al., 1998) were also identified in Imp1–3xHA purifications (Table S1).
Imp1-dependent transport of the spindle disassembly factor Cdc48 in the MMD
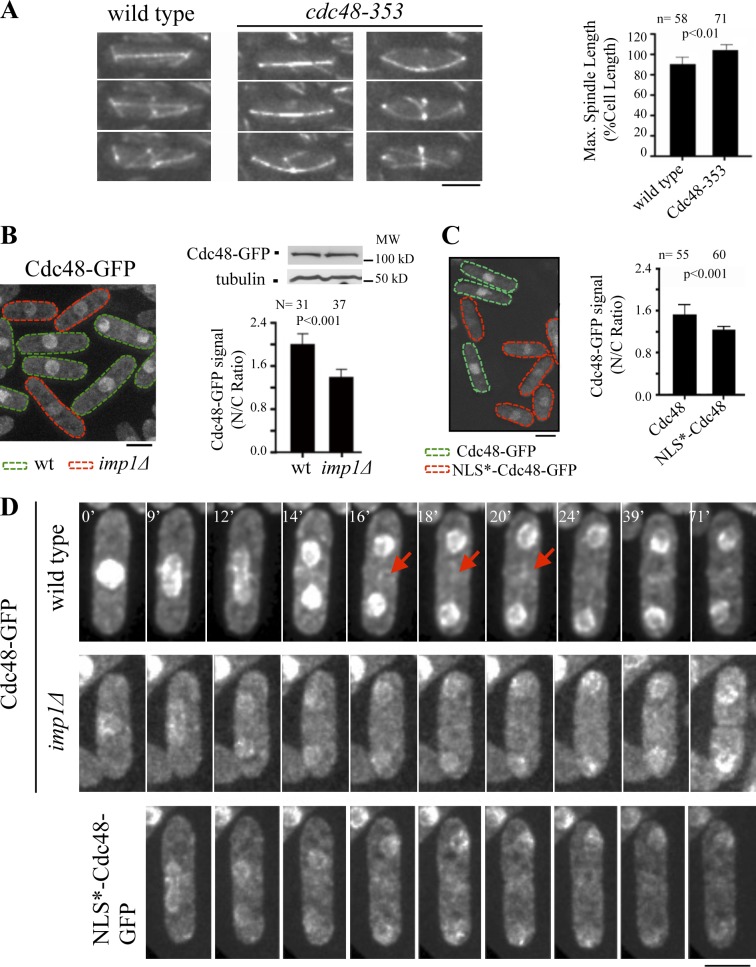
To determine a possible role for Cdc48 in mitotic spindle disassembly in fission yeast, we analyzed spindle dynamics in the S. pombe cdc48-353 temperature-sensitive strain (Ikai and Yanagida, 2006), expressing GFP-Atb2 as a microtubule marker. At the semi-permissive temperature of 32°C, cdc48-353 cells showed hyperextended mitotic spindles that elongated up to 104% of the cell length (Fig. 6 A), in contrast to 90% in a wild-type background (Fig. 1 C). This result indicates that, like in Saccharomyces cerevisiae and X. laevis (Cao et al., 2003), Cdc48 is also required for spindle disassembly in S. pombe cells.
Figure 6.
Imp1-dependent transport of the AAA-ATPase Cdc48 protein. (A) Time-lapse fluorescence images of wild-type and cdc48-353 cells expressing GFP-Atb2 at 32°C (semi-restrictive temperature for the cdc48-353 strain) as cells enter cytokinesis. (graph) Mean maximal spindle length (percentage of the cell length) in wild-type and cdc48-353 cells at 32°C. (B) Cdc48-GFP localization in asynchronous wild-type (lectin labeled) and imp1Δ cells (green- and red-dashed cells, respectively) incubated at 25°C. (graph) Quantification of Cdc48-GFP signal at the nucleus (ratio of nuclear to cytoplasmic fluorescence intensity) in asynchronous wild-type (lectin labeled) and imp1Δ cells. Western blot analysis of Cdc48-GFP (tubulin is used as a control) in cell extract of both strains is shown. (C) Cdc48-GFP localization in asynchronous Cdc48-GFP (lectin labeled) and NLS*-Cdc48-GFP (inactivated NLS) cells (green- and red-dashed cells, respectively) incubated at 25°C. (graph) Quantification of Cdc48-GFP signal at the nucleus (ratio of nuclear to cytoplasmic fluorescence intensity) in asynchronous Cdc48+ (lectin labeled) and NLS*-Cdc48 mutant cells incubated at 25°C. (D) Localization of Cdc48-GFP by time-lapse fluorescence in wild-type (top) and imp1Δ cells (middle). Arrows indicate Cdc48 midzone localization in wild-type cells. Localization of NLS*-Cdc48-GFP by time-lapse fluorescence in wild-type background (bottom). Bars, 5 µm. Graphs represent mean and standard deviation. n is the total number of cells scored from at least three independent experiments.
In budding yeast, Cdc48 is mostly associated with the nuclear envelope and enters the nucleus during late G1 (Madeo et al., 1998). Using a GFP-tagged Cdc48 construct, we determined that this protein also localized at the nucleus in S. pombe cells. In agreement with being an Imp1 cargo, nuclear Cdc48-GFP signal decreased 30% in the imp1Δ strain in comparison to wild type (Fig. 6 B). Sequence analysis determined that fission yeast Cdc48 has a canonical NLS, ranging from residues 36 to 41 at its N-terminal domain. Mutational inactivation of this NLS domain (NLS*-Cdc48-GFP) significantly reduced its nuclear localization (Fig. 6 C), indicating that, independently of other nuclear localization domains, this NLS signal confers to Cdc48 conventional nucleocytoplasmic transport activity.
To follow Cdc48 localization dynamics, we used time-lapse microscopy. In wild-type cells, GFP-tagged Cdc48 localized to the nucleus throughout the cell cycle. However, we also observed that it transiently localized to the MMD region during mitosis (Fig. 6 D). Time course analysis in Imp1-depleted cells showed that this AAA-ATPase can also be detected at the nucleoplasm and the nuclear membrane, but failed to localize at the spindle midzone region (Fig. 6 D). Analysis of the NLS-deficient Cdc48-GFP construct recapitulated this result (Fig. 6 D). We therefore conclude that in S. pombe cells, Cdc48 is a spindle midzone disassembly factor whose transport at the MMD is Imp1 dependent.
A molecular mechanism for the MMD in the control of the mitotic spindle disassembly
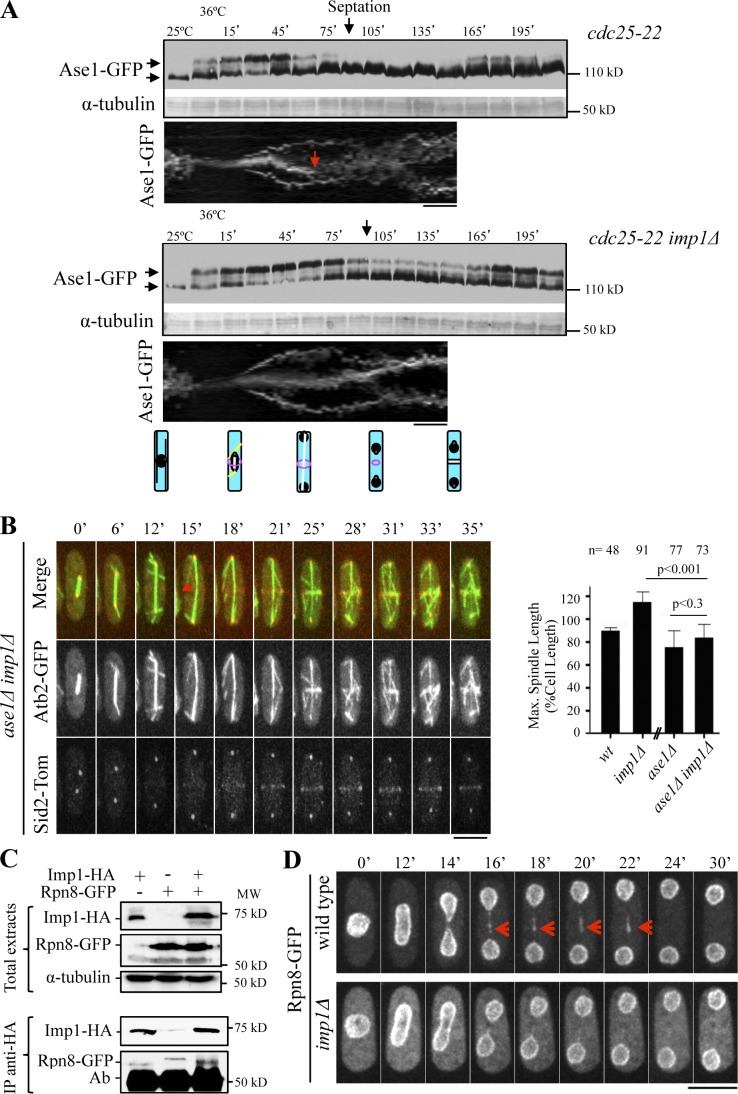
It is established that in S. cerevisiae cells, Cdc48 targets Ase1 for degradation at the end of mitosis (Juang et al., 1997; Cao and Zheng, 2004). Because Ase1 bundles microtubules at the spindle midzone, its degradation is essential for spindle disassembly in this organism (Woodruff et al., 2010). To test whether Cdc48 is required for Ase1 regulation at the spindle midzone in S. pombe cells, we analyzed Ase1-GFP levels in total cell extracts from cdc25-22 and cdc25-22 imp1Δ cells (which lack Cdc48 protein at the spindle midzone) synchronized by G2 block and release. Kymographs were used to show Ase1-GFP localization throughout the cell cycle. In cdc25-22 cells, as previously reported (Loïodice et al., 2005), a high molecular mass form of Ase1 (likely phosphorylated Ase1) appears as cells enter into mitosis and disappears at the time that the spindle midzone is dissolved during mitotic exit before septation (Fig. 7 A). However, in cdc25-22 imp1Δ cells, the high molecular mass Ase1 fraction and midzone-localized Ase1 remained at high levels during septation, until the end of cytokinesis (Fig. 7 A). From these results, we hypothesize that Cdc48 import activity by Imp1 at the MMD is required to remove phosphorylated Ase1 from the spindle midzone, either by promoting Ase1 dephosphorylation or its degradation. In agreement with this hypothesis, we determined that depletion of Ase1 rescued hyperextension of mitotic spindles in the imp1Δ strain, yielding similar maximal spindle length in ase1Δ and ase1Δ imp1Δ cells (Fig. 7 B).
Figure 7.
Imp1-dependent levels of the spindle-stabilizing protein Ase1. (A) Western blot analysis of Ase1-GFP protein levels through the cell cycle in synchronized cdc25-22 and cdc25-22 imp1Δ cells. α-Tubulin is used as a control. Maximal septation index values are indicated. Kymographs of cdc25-22 and cdc25-22 imp1Δ cells expressing Ase1-GFP. Red arrow indicates midzone dissolution in cdc25-22 cells. Schematized cells (bottom) representing interphase, early anaphase, late anaphase B, ring contraction and daughter nuclei repositioning, and the end of cytokinesis, respectively, are shown. (B) Time-lapse fluorescence images of ase1Δ imp1Δ cells expressing Atb2-GFP and Sid2-Tom. Arrowhead indicates the initiation of spindle disassembly. (graph) Maximal spindle length in ase1Δ and ase1Δ imp1Δ cells, compared with wild-type and imp1Δ cells (from Fig. 1C). (C) Levels of Imp1-HA and Rpn8-GFP (Western blot analysis) in cells expressing Imp1-HA, Rpn8-GFP, or both tagged proteins in total extracts and in Imp1-HA immunopurified complexes. (D) Time-lapse fluorescence images of Rpn8-GFP in wild-type (top) and imp1Δ (bottom) cells. Arrows indicate Rpn8-GFP midzone localization in wild-type cells. Bars, 5 µm. Graphs represent mean and standard deviation. n is the total number of cells scored from three independent experiments.
Significantly, regulatory subunits of the 26S proteasome complex were found among the proteins associated with Imp1 (Table S1). Further analysis of Rpn8-GFP indicated that this proteasome regulatory factor is imported into the nucleus by Imp1 (Fig. S5). We also determined that Rpn8 physically interacts with Imp1 (Fig. 7 C) and, furthermore, that its midzone localization depends on this importin-α (Fig. 7 D). Hence, we suggest that Imp1 transport activity at the MMD serves to assemble the protein degradation machinery at the midzone region, allowing APC/C-targeted proteins to be rapidly degraded during spindle dissolution.
Discussion
Nucleocytoplasmic transport at the MMD
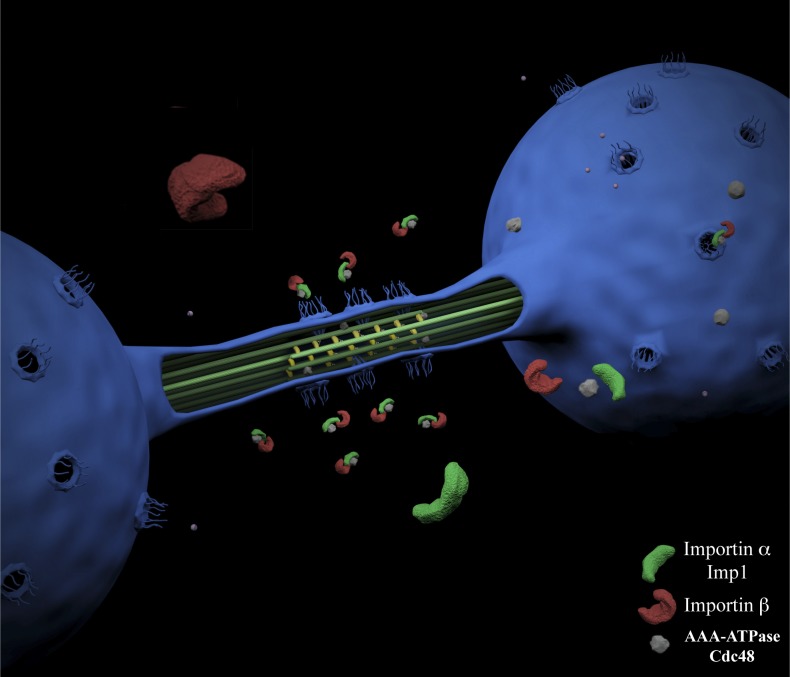
In this paper, we investigate how nucleocytoplasmic transport is required for mitotic spindle disassembly in the closed mitosis of fission yeast. In S. pombe cells, spindle disassembly occurs late in mitosis, during the dumbbell shape of the anaphase B nucleus (Sagolla et al., 2003). Using photobleaching, we show that at this stage the nuclear envelope surrounding the mitotic spindle midzone constrains nucleoplasmic diffusion at the midzone compartment (Fig. 3 D). A similar compartmentalization has been shown during anaphase in budding yeast, where this morphological constraint functions as a diffusion barrier that allows the asymmetric segregation of nucleoplasmic components between the mother and the daughter cell (Boettcher et al., 2012). In fission yeast, this compartmentalization also has clear functional consequences for the cell. By limiting the nucleoplasmic diffusion at the midzone region, access of disassembly factors required for midzone dissolution necessarily depends on active transport mechanisms. Accordingly, spindle disassembly defects observed in Imp1-depleted cells (Figs. 1 and 2), nuclear pore localization studies (Fig. 3 and Fig. S1), import/export dynamics of importin-α (Figs. 3 and 5), specific importin β subunit association (Fig. 4 and Table S1), and Pim1 localization/function analysis (Fig. 5), support the requirement of nucleocytoplasmic transport activity at the MMD for the spatiotemporal control of the spindle disassembly in fission yeast. In particular, we found that the spindle disassembly factor Cdc48 is transported by Imp1 through nuclear pores at the MMD and unloaded in the mitotic spindle midzone during mitosis exit (Fig. 6), supporting the key role that nucleocytoplasmic transport activity plays at the MMD in spindle midzone dissolution. A model representing the proposed nucleocytoplasmic transport activity at the MMD is shown in Fig. 8.
Figure 8.
A model of the nucleocytoplasmic transport activity at the MMD. Picture representing the anaphase B nucleus of an S. pombe cell with several nuclear pore complexes localized at the MMD. Association of NLS cargoes to Imp1 allows this importin-α adaptor, in complexes with specific impotin β subunits, the transport of disassembly factors such as Cdc48 through the nuclear pore. Disassembly factors are then unloaded at the midzone nucleoplasmic compartment to promote spindle midzone dissolution.
Triggering midzone dissolution by the NLS-Cdc48 disassembly factor
In budding yeast, the APC/C is essential for disassembly of the mitotic spindle late in mitosis (Juang et al., 1997; Woodruff et al., 2010). Cdc48 is required for the degradation of at least two key APC/C substrates involved in mitotic exit in X. laevis and budding yeast cells: the polo kinase Cdc5 and the microtubule binding protein Ase1 (Cao et al., 2003; Cheeseman and Desai, 2004). Our results also establish a role for Cdc48 as a disassembly factor in fission yeast (Fig. 6). Furthermore, we determined that, as previously reported (Loïodice et al., 2005), a slow migrating Ase1 band (likely Ase1-P) disappears as the midzone is dissolved, before septation. This Ase1 variant persists in Imp1-depleted cells (lacking Cdc48 at the midzone) until the end of cytokinesis (Fig. 7 A). Thus, we hypothesize that in S. pombe cells, Imp1-dependent transport of Cdc48 at the MMD likely promotes removal of this Ase1-P protein from the spindle midzone late in anaphase B (Fig. 7 B and Fig. 8).
Whether Cdc48 targets Ase1 for proteasome degradation in S. pombe cells is not yet known. Regulation of Ase1 is also achieved by phosphorylation involving some major cell-cycle regulatory kinases (CDK1 and Aurora B; Duellberg et al., 2013). Because Cdc48 regulates activity and stability of Aurora B (Heallen et al., 2008; Dobrynin et al., 2011), it could promote Ase1 disappearance either by directing this Ase1 variant to the 26S proteasome for degradation and/or by indirectly controlling its phosphorylation state. Further studies will be required to investigate the mechanisms controlling Ase1 degradation/dephosphorylation to provide important information about spindle disassembly in S. pombe cells.
During mitosis, a variety of nuclear proteins are recognized and ubiquitinated by the APC/C E3 ubiquitin ligase, which targets them for degradation by the 26S proteasome (Juang et al., 1997; Woodruff et al., 2010). Accordingly, Srp1-mediated transport of proteasome subunits in the nucleus suggests that protein degradation could occur within this organelle (Chen et al., 2011). In S. pombe, the proteasome localizes at the nucleus, but, intriguingly, it localizes at the mitotic spindle midzone too (Wilkinson et al., 1998). By using the Rpn8 subunit, we demonstrated that proteasome localization at the midzone depends on Imp1 activity (Figs. 7 and S5). Hence, independently of mechanisms regulating Ase1, we propose that Imp1-dependent transport of 26S proteasome components could serve to assemble the protein degradation machinery at the midzone, allowing APC/C-targeted proteins to be rapidly degraded at this compartment for efficient mitotic spindle disassembly (Woodruff et al., 2010).
The MMD, a necessary domain in closed mitosis
Eukaryotic cell division uses different forms of mitosis for accurate chromosome segregation (Sazer, 2010). In closed mitosis, the nuclear membrane that envelops the mitotic spindle during nuclei segregation generates a physical barrier that, as proposed in this study, requires specialization at the MMD to promote spindle disassembly at the end of mitosis. In open and semi-open mitosis, a membranous spindle matrix surrounds the mitotic spindles (Tsai et al., 2006; Ma et al., 2009; Zheng, 2010). It will be interesting to elucidate whether the MMD and this membranous matrix have any functional/evolutionary equivalence.
Nucleocytoplasmic transport specialization has been reported in higher eukaryotes (Adams and Wente, 2013). During evolution, the importin-α gene has undergone multiple rounds of duplications and lineage-specific expansions. In animals, most importin-α proteins belong to one of three conserved clades, referred as α1, α2, and α3 (Mason et al., 2009), expressed in distinct temporal and spatial patterns undergoing specialized functions (Hogarth et al., 2006; Pumroy and Cingolani, 2015). In Arabidopsis thaliana, only one of the ten members of the importin-α family mediates Agrobacterium tumefaciens transformation (Bhattacharjee et al., 2008), indicating that also in plants importin-α evolved to carry out specialized roles. In budding yeast there is only one gene coding for importin-α, named SRP1 (Yano et al., 1992; Kuüssel and Frasch, 1995; Loeb et al., 1995). However, fission yeast has two importin-α genes, cut15 and imp1 (Matsusaka et al., 1998; Umeda et al., 2005). The specialized role of Imp1 in spindle disassembly described here likely represents an early step of the evolutionary diversion of this protein in eukaryotic cells. Altogether our data support that, in S. pombe, a separate membrane domain (MMD) surrounding the spindle midzone supports spindle disassembly. The role of Imp1 in the transport of specific NLS cargoes at this domain may provide new insights into mechanisms that lead to specialized nucleocytoplasmic transport events.
Materials and methods
Yeast strains, genetic procedures, and media
The S. pombe strains used in this study are listed in Table S2. ase1-deleted and ase1-GFP strains were provided by P.T. Tran (Centre de Recherche, Institut Curie, Paris, France). The pim1-d1 strain was provided by S. Sazer (Baylor College of Medicine, Houston, TX). Ark1-GFP, Klp5-GFP, and Klp6-GFP constructs were provided by Y. Watanabe (University of Tokyo, Tokyo, Japan). Plasmids with Imp1 variants carrying IBB and ED mutations were provided by T. Toda (Cancer Research UK, London Research Institute, London, UK). Sal3-GFP and Cdc48 alleles were provided by the Yeast National Bioresearch Project (Japan). Strains were generated by genetic crosses and tested by segregation of markers or PCR. Experiments in liquid culture were performed in minimal medium (EMM), supplemented as required. Experiments used a starting cell density of 2–4 × 106 cells/ml, corresponding to the mid-exponential growth phase. The nmt promoters were repressed in EMM medium with 5 µg/ml thiamine and expressed in EMM without thiamine (Maundrell, 1993). Deletion of endogenous imp1+ was performed by the PCR-based gene-targeting method with uracil as selection marker (Bähler et al., 1998). Imp1-Tom, Cdc48-GFP, Rpn8-GFP, and Pim1-GFP C terminus tagging were performed at their native loci by homologous recombination using plasmids pFA6a-tdT-NatMX6 and pFA6A-GFP(S65T)-KanMX6 (Bähler et al., 1998). Oligonucleotides NLS1-5 (5′-CTTTTTTTTAGAATTCGATGCTAACAAGCTCAGTACATATAGTGCAGAAGACACAGCAACAGCTATTCTTCGTGCAGCAGCAGCACCAAA-3′) and NLS1-3 (5′-CATGGTATTGGACGACAATGTGATTACTGAGTTATCATCGTTGGTAGCGTCATCGACCACTAGAGAATTTGGTGCTGCTGCTGCACGAAG-3′) were used for the mutational inactivation of the Cdc48 NLS (LRKKRKP to LRAAAAP) located between residues 36 and 42 in pREP41-Cdc48-GFP multicopy plasmid following described procedures (Edelheit et al., 2009). DNA sequencing was used to confirm the mutation (NLS*-Cdc48-GFP). Multicopy plasmid pREP41-GFP-Imp1-ΔIBB-ED carrying the putative cargo-binding sites of Imp1 (D192R E393R) was used for expression of the Imp1-ΔIBB-ED variant (Sato and Toda, 2007). The plasmid was repressed in EMM with 5 µg/ml thiamine and expressed in EMM without thiamine. To make the GFP-Imp1-ΔIBB construct, an ApaI-BspEI 0.9-kb fragment containing the ED mutations was replaced with a wild-type sequence in the pREP41-GFP-Imp1-ΔIBB-ED plasmid. Double mutants were constructed by tetrad dissection or random spore analysis. Transformation was achieved by the lithium acetate protocol (Norbury and Moreno, 1997). Synchronous cultures were generated by a block and release strategy using the cdc25-22 mutation (Moreno et al., 1991). Cells were arrested in a water bath at 36°C for 3 h and then shifted back to 25°C. Aliquots were taken every 20 min to analyze the percentage of septated cells.
Microscopy and image analysis
Live-cell imaging was performed with a spinning disk confocal microscope (IX-81; Olympus; CoolSnap HQ2 camera, Plan Apochromat 100×, 1.4 NA objective [Roper Scientific]) or a Delta Vision microscope (IX-71; Olympus; CoolSnap HQ camera, Plan Apochromat 60×, 1.42 NA objective [Applied Precision]) in EMM supplemented as required. Images were acquired and analyzed with Metamorph software (Molecular Devices), ImageJ (National Institutes of Health), and Softworx (Applied Precision). Time-lapse experiments at 25°C were imaged in 35-mm glass-bottom culture dishes (P35-1.5-10-C; MatTek) coated with soybean lectin (Sigma-Aldrich) and immersed in 3 ml of medium. cdc48-353 live-cell imaging was performed at 32°C mounted on a FCS2 chamber (Bioptechs) coated with soybean lectin and using an objective heater (Bioptechs) for temperature control during imaging. pim1-d1 cells were grown at 25°C and mounted on a FCS2 chamber as described for cdc48-353. This led to a temperature shift of the cells from 25 to 36°C 2 min after mounting.
For spinning disk time-lapse experiments, cells were imaged at 30–60-s intervals in 20 z-series with a step size of 0.3 µm. For Delta Vision time-lapse experiments, cells were imaged in 12 z-series with a step size of 0.5 µm.
Cell wall and chromatin regions were visualized by Calcofluor white (Sigma-Aldrich) and DAPI (Sigma-Aldrich), respectively. Latrunculin B (Toronto Chemicals) was added at a final concentration of 10 µM.
Kymographs were constructed using ImageJ software. Spindle lengths were measured using GFP-Atb2 as a marker. Graphs and statistical analyses were generated using GraphPad Prism version 5 (GraphPad Software).
FRAP and FLIP analysis
FRAP and FLIP techniques were performed on the spinning disk confocal microscope. Images were taken at one optical section, exposed for 100 ms, and captured at 1-s intervals for 100 s total. In FRAP and FLIP experiments, at least five images were captured before bleaching. Laser power ranged from 10 to 20% in all experiments.
Western blotting
Protein extracts from Cdc48-GFP and Cdc48-GFP Imp1Δ cells, or from Rpn8-GFP and Rpn8-GFP Imp1Δ cells, were prepared at sequential time points, collected by centrifugation, washed with STOP buffer (0.9% NaCl, 1 mM NaN3, 10 mM EDTA, and 50 mM NaF), and frozen in liquid nitrogen. Total protein extracts were prepared by Fast-prep vortexing with glass beads (Sigma-Aldrich) as described previously (Moreno et al., 1991). Protein extracts from Ase1-GFP Cdc25-22 and Ase1-GFP Imp1Δ Cdc25-22 were prepared as described previously (Zapata et al., 2014). In brief, 1.6 ml of sample was collected at every time point and centrifuged at 13,000 rpm for 15 s. The supernatant was removed and 200 µl of glass beads were added before freezing in liquid nitrogen. 120 µl of sample buffer (65 mM Tris-HCl, pH 6.8, 3% SDS, 10% glycerol, 50 mM NaF, 100 mM β-glycerophosphate, 5% 2-mercaptoethanol, 2 mM PMSF, and Bromophenol blue) were added to lyse the cells in a Multibeater-8 at top speed for 2 min. The samples were placed in boiling water for 5 min and immediately centrifuged for 5 min at 13,000 rpm. Extracts were loaded on 9% SDS-PAGE and blotted on nitrocellulose. Blots were probed with mouse anti-GFP (Roche) at 1:2,000 dilution and mouse anti–α-tubulin (Sigma-Aldrich) at 1:10,000 dilution followed by anti-mouse IgG conjugates (Sigma-Aldrich) at 1:5,000 dilution. The ECL system (GE Healthcare) was used to detect anti–α-tubulin and Ase1-GFP, whereas Supersignal (Thermo Fisher Scientific) was used to detect Cdc48-GFP and Rpn8-GFP.
Coimmunoprecipitation assays
Cells expressing Imp1-HA, Rpn8-GFP, or both were grown at 25°C. 1 liter of each cell culture was pelleted, washed once in stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, and 1 mM NaN3, pH 8), and frozen on liquid nitrogen. Total protein extracts of these strains were prepared from 3–5 × 109 cells using HB buffer (50 mM Hepes-KOH, pH 7.6, 75 mM β-glycerol phosphate, 50 mM NaF, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 0.25% Tween-20, and 1 mM PMSF; Moreno et al., 1991). Immunoaffinity beads were made by binding mouse anti-HA monoclonal antibodies (Santa Cruz Biotechnology, Inc.) to protein A beads (Bio-Rad Laboratories) overnight at 4°C. Cell extracts (2 mg of total protein) were incubated with the immunoaffinity beads for 4 h at 4°C. The beads were washed four times with 1 ml of HB buffer and resuspended in sample buffer. Immunoprecipitates were resolved on SDS-PAGE (12%), blotted, and probed with anti-HA (1:1,000; F-7; Santa Cruz Biotechnology, Inc.) or anti-GFP (1:1,000) antibody (Roche). Coimmunoprecipitation assays were also performed in cells expressing Imp1–3xHA and Cdc48-GFP, but significant Cdc48-GFP signal was not obtained in the different Western blot methods used.
Purification of endogenous Imp1–3xHA complexes
To prepare anti-HA control beads, 450 µg of affinity-purified anti-HA antibody was incubated with 500 µl protein A–agarose at room temperature on a rotator for 2 h in a 1.5-ml tube containing antibody-binding buffer. After binding, the beads were washed into extract buffer (50 mM Hepes, pH 7.6, 100 mM β-glycerolphosphate, 100 mM KCl, 5% glycerol, 1 mM MgCl2, 1 mM EGTA, and 1% Tween-20) and transferred to 15-ml conical tubes. To purify endogenous Imp1–3xHA–containing complexes, 4 liters of the Imp1–3xHA strain were grown to OD595 = 0.5 and cells were harvested by centrifugation. The cells were resuspended in 50 ml of 50 mM Hepes-KOH, pH 7.6, and combined into two 50-ml conical tubes, pelleted again, and frozen in liquid nitrogen. Cell lysis was achieved by grinding the cells in liquid nitrogen using a mortar and pestle. To prepare the extract, 15 g of the ground cell powder was placed in a prechilled beaker and allowed to warm until the powder on the sides of the beaker was just beginning to thaw. 20 ml of extract buffer containing 1 mM PMSF was added and the powder was resuspended by stirring rapidly into solution using a spatula. The remaining procedures were performed at 4°C except where noted. The extract was stirred using a magnetic stir bar for 10 min and then centrifuged for 5 min at 10,000 rpm in a JA 20 rotor and 75 min at 45,000 rpm in a 60 Ti rotor (Beckman Coulter). 15 ml of clarified extract was added to each tube containing the antibody-coated beads and incubated at 4°C on a rotator for 2–3 h. The beads were then washed three times with 15 ml of extract buffer without PMSF, transferred to a 1-ml Dispo column (Bio-Rad Laboratories), and washed with five 1-ml aliquots of extract buffer without Tween-20. The Imp1–3xHA–containing complexes were then eluted from the beads at room temperature by the addition of five 250-µl aliquots of elution buffer (50 mM Hepes, 5% glycerol, 1 mM MgCl2, 1 mM EGTA, and 350 µg/ml HA dipeptide) with a 15-min incubation between addition of each aliquot. Fractions 2–5 were pooled, TCA precipitated, and resuspended in 50 µl of sample buffer. For Coomassie blue staining, 15 µl was run in 10% SDS-PAGE. For Western blotting, 1/10 of the total elution was loaded.
Mass spectrometry and sample preparation
To identify subunits of the Imp1–3xHA complex in the total elution from the large-scale immunoaffinity chromatography, 190 µl of eluate was mixed with 760 µl methanol in a 1.6-ml microcentrifuge tube and vortexed; 190 µl of chloroform was added and the tube was vortexed again. 570 µl of water was then added. The solution was vortexed and spun for 5 min at top speed in a microfuge. The aqueous layer was then removed and discarded, with care taken not to disturb the interface. 570 µl methanol was then added to precipitate the proteins and mixed by gentle vortexing, and the proteins were collected by centrifugation for 5 min at top speed in a microfuge. The supernatant was removed by aspiration and the pellet was dried. Identification of proteins present in the fraction, recovery, and trypsin digestion of proteins from Coomassie blue–stained polyacrylamide gels were performed as described previously (Shevchenko et al., 1996). In-gel digestions were performed with sequencing grade-modified trypsin (Promega) overnight in 50 mM ammonium bicarbonate at 37°C. Peptides were extracted twice with 50% acetonitrile and 1% formic acid, dried in a speedvac (Thermo Fisher Scientific), and desalted by solid phase extraction on handmade C18 columns. Desalted peptides were dried in a speedvac, resuspended in 5% formic acid, and analyzed by nanospray liquid chromatography tandem mass spectrometry on a LTQ Orbitrap Discovery mass spectrometer (Thermo Fisher Scientific). Peptides were separated by reverse-phase HPLC on a hand-packed column (packed with 5 µm, 200 Å pores, Magic C18AQ resin; Michrom BioResources) using a 65-min gradient of 5–27% buffer B (97% ACN and 0.125% FA) with an in-column flow rate of 0.5–1.0 µl/min. Peptides were detected using a Top 10 method. For each cycle, one full mass spectrometry scan of m/z = 300–1,500 was acquired in the Orbitrap at a resolution of 30,000 at m/z = 400 with AGC target = 106. Each full scan was followed by the selection of up to 10 of the most intense ions for CID and tandem mass spectrometry analysis in the linear ion trap. Selected ions were excluded from further analysis for 60 s. Ions with charge 1+ or unassigned were also rejected. Maximum ion accumulation times were 1,000 ms for each full mass spectrometry scan and 150 ms for tandem mass spectrometry scans. Lockmass, using atmospheric polydimethylsiloxane (m/z = 371.1012) as an internal standard, was used in all runs to normalize mass spectrometry precursor masses.
MS2 spectra were searched using SEQUEST against a composite database containing the translated sequences of all predicted open reading frames of S. pombe and its reversed complement, using the following parameters: a precursor mass tolerance of ±20 ppm, 1.0 D product ion mass tolerance, tryptic digestion, up to two missed cleavages, static modifications of carbamidomethylation on cysteine (+57.0214), and a dynamic modification of methionine oxidation (+15.9949). Peptide spectral matches were filtered to 1% FDR using the target–decoy strategy (Elias and Gygi, 2007) combined with linear discriminant analysis using SEQUEST scoring parameters including Xcorr, ΔCn′, precursor mass error, and charge state (Huttlin et al., 2010).
Online supplemental material
Fig. S1 shows the colocalization of Imp1-Tom and Nup61-GFP and time-lapse images of a representative Nup107-GFP–expressing cell after photobleaching one entire nucleus or only a nuclear region. Fig. S2 shows time-lapse fluorescence images of a representative cell after photobleaching Imp1-GFP in the two nuclei. Fig. S3 shows kymographs of Imp1-independent factors involved in spindle midzone dynamics. Fig. S4 shows Imp1–3xHA–associated proteins separated by SDS-PAGE. Fig. S5 shows the role of Imp1 in the nuclear localization of Rpn8. Table S1 shows the list of proteins identified by mass spectrometry potentially associated to Imp1–3xHA complexes. Table S2 shows the genotype of the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412144/DC1.
Supplementary Material
Acknowledgments
We are grateful to Takashi Toda, Phong T. Tran, Shelley Sazer, Rosa Aligué, Yoshinori Watanabe, the Yeast National Bioresearch Project (Japan), and Iain Hagan for providing strains, plasmids, and reagents. We thank Juan Mata, Silvia Salas-Pino, and Ana Belen Iglesias-Romero for useful comments and discussions and Laura Tomas-Gallardo, Maria del Valle Rubio, and Victor Carranco for technical help.
This work was supported by grants BFU2013-46923-P and BFU2010-21310 to J. Jimenez and R.R. Daga, respectively, from the Ministerio de Economía y Competitividad of the Spanish Government.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- FLIP
- fluorescence loss in photobleaching
- ipMT
- interpolar microtubule
- MMD
- midzone membrane domain
- NLS
- nuclear localization signal
- SPB
- spindle pole body
References
- Adams R.L., and Wente S.R.. 2013. Uncovering nuclear pore complexity with innovation. Cell. 152:1218–1221 10.1016/j.cell.2013.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. III, Steever A.B., Wach A., Philippsen P., and Pringle J.R.. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Baï S.W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., and Doye V.. 2004. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell. Biol. 24:6379–6392 10.1128/MCB.24.14.6379-6392.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Lee L.-Y., Oltmanns H., Cao H., Veena, Cuperus J., and Gelvin S.B.. 2008. IMPa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell. 20:2661–2680 10.1105/tpc.108.060467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher B., Marquez-Lago T.T., Bayer M., Weiss E.L., and Barral Y.. 2012. Nuclear envelope morphology constrains diffusion and promotes asymmetric protein segregation in closed mitosis. J. Cell Biol. 197:921–937 10.1083/jcb.201112117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M., and Wente S.R.. 1997. In vivo dynamics of nuclear pore complexes in yeast. J. Cell Biol. 136:1185–1199 10.1083/jcb.136.6.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S.Y., Vermaak D., and Biggins S.. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339 10.1083/jcb.200209018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., and Zheng Y.. 2004. The Cdc48/p97-Ufd1-Npl4 complex: its potential role in coordinating cellular morphogenesis during the M-G1 transition. Cell Cycle. 3:422–424 10.4161/cc.3.4.814 [DOI] [PubMed] [Google Scholar]
- Cao K., Nakajima R., Meyer H.H., and Zheng Y.. 2003. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 115:355–367 10.1016/S0092-8674(03)00815-8 [DOI] [PubMed] [Google Scholar]
- Cardarelli F., Bizzarri R., Serresi M., Albertazzi L., and Beltram F.. 2009. Probing nuclear localization signal-importin α binding equilibria in living cells. J. Biol. Chem. 284:36638–36646 10.1074/jbc.M109.036699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2004. Cell division: AAAtacking the mitotic spindle. Curr. Biol. 14:R70–R72 10.1016/j.cub.2003.12.048 [DOI] [PubMed] [Google Scholar]
- Chen L., Romero L., Chuang S.M., Tournier V., Joshi K.K., Lee J.A., Kovvali G., and Madura K.. 2011. Sts1 plays a key role in targeting proteasomes to the nucleus. J. Biol. Chem. 286:3104–3118 10.1074/jbc.M110.135863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.Q., Du X., Liu J., Balasubramanian M.K., and Balasundaram D.. 2004. Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast. 21:495–509 10.1002/yea.1115 [DOI] [PubMed] [Google Scholar]
- Chook Y.M., and Süel K.E.. 2011. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta. 1813:1593–1606 10.1016/j.bbamcr.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P.R., and Zhang C.. 2001. Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 11:366–371 10.1016/S0962-8924(01)02071-2 [DOI] [PubMed] [Google Scholar]
- Cueille N., Salimova E., Esteban V., Blanco M., Moreno S., Bueno A., and Simanis V.. 2001. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114:2649–2664. [DOI] [PubMed] [Google Scholar]
- Daga R.R., and Chang F.. 2005. Dynamic positioning of the fission yeast cell division plane. Proc. Natl. Acad. Sci. USA. 102:8228–8232 10.1073/pnas.0409021102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M.2001. Running on Ran: nuclear transport and the mitotic spindle. Cell. 104:321–324 10.1016/S0092-8674(01)00218-5 [DOI] [PubMed] [Google Scholar]
- Ding R., West R.R., Morphew D.M., Oakley B.R., and McIntosh J.R.. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 8:1461–1479 10.1091/mbc.8.8.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrynin G., Popp O., Romer T., Bremer S., Schmitz M.H., Gerlich D.W., and Meyer H.. 2011. Cdc48/p97-Ufd1-Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells. J. Cell Sci. 124:1571–1580 10.1242/jcs.069500 [DOI] [PubMed] [Google Scholar]
- Duellberg C., Fourniol F.J., Maurer S.P., Roostalu J., and Surrey T.. 2013. End-binding proteins and Ase1/PRC1 define local functionality of structurally distinct parts of the microtubule cytoskeleton. Trends Cell Biol. 23:54–63 10.1016/j.tcb.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Edelheit O., Hanukoglu A., and Hanukoglu I.. 2009. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 9:61 10.1186/1472-6750-9-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J.E., and Gygi S.P.. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 4:207–214 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- Fleig U., Salus S.S., Karig I., and Sazer S.. 2000. The fission yeast Ran GTPase is required for microtubule integrity. J. Cell Biol. 151:1101–1111 10.1083/jcb.151.5.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman V., Gerson-Gurwitz A., Movshovich N., Kupiec M., and Gheber L.. 2009. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 10:387–393 10.1038/embor.2009.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Ward J.J., Loiodice I., Velve-Casquillas G., Nedelec F.J., and Tran P.T.. 2009. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell. 17:257–267 10.1016/j.devcel.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y., Tournier S., Millar J.B., and Hyams J.S.. 2004. Mechanism controlling perpendicular alignment of the spindle to the axis of cell division in fission yeast. EMBO J. 23:1289–1300 10.1038/sj.emboj.7600156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geles K.G., Johnson J.J., Jong S., and Adam S.A.. 2002. A role for Caenorhabditis elegans importin IMA-2 in germ line and embryonic mitosis. Mol. Biol. Cell. 13:3138–3147 10.1091/mbc.E02-02-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M.2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., and Mattaj I.W.. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell. 104:83–93 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Hagan I.M.1998. The fission yeast microtubule cytoskeleton. J. Cell Sci. 111:1603–1612. [DOI] [PubMed] [Google Scholar]
- Heallen T.R., Adams H.P., Furuta T., Verbrugghe K.J., and Schumacher J.M.. 2008. An Afg2/Spaf-related Cdc48-like AAA ATPase regulates the stability and activity of the C. elegans Aurora B kinase AIR-2. Dev. Cell. 15:603–616 10.1016/j.devcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth C.A., Calanni S., Jans D.A., and Loveland K.L.. 2006. Importin α mRNAs have distinct expression profiles during spermatogenesis. Dev. Dyn. 235:253–262 10.1002/dvdy.20569 [DOI] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., and Gygi S.P.. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 143:1174–1189 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikai N., and Yanagida M.. 2006. Cdc48 is required for the stability of Cut1/separase in mitotic anaphase. J. Struct. Biol. 156:50–61 10.1016/j.jsb.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Juang Y.L., Huang J., Peters J.M., McLaughlin M.E., Tai C.Y., and Pellman D.. 1997. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 275:1311–1314 10.1126/science.275.5304.1311 [DOI] [PubMed] [Google Scholar]
- Krapp A., Gulli M.P., and Simanis V.. 2004. SIN and the art of splitting the fission yeast cell. Curr. Biol. 14:R722–R730 10.1016/j.cub.2004.08.049 [DOI] [PubMed] [Google Scholar]
- Küssel P., and Frasch M.. 1995. Yeast Srp1, a nuclear protein related to Drosophila and mouse pendulin, is required for normal migration, division, and integrity of nuclei during mitosis. Mol. Gen. Genet. 248:351–363 10.1007/BF02191602 [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., and Corbett A.H.. 2007. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282:5101–5105 10.1074/jbc.R600026200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb J.D., Schlenstedt G., Pellman D., Kornitzer D., Silver P.A., and Fink G.R.. 1995. The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. USA. 92:7647–7651 10.1073/pnas.92.17.7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loïodice I., Staub J., Setty T.G., Nguyen N.P., Paoletti A., and Tran P.T.. 2005. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell. 16:1756–1768 10.1091/mbc.E04-10-0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Tsai M.Y., Wang S., Lu B., Chen R., Iii J.R., Zhu X., and Zheng Y.. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 11:247–256 10.1038/ncb1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Bloom K.S., and Salmon E.D.. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41 10.1038/71357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Schlauer J., Zischka H., Mecke D., and Fröhlich K.U.. 1998. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol. Biol. Cell. 9:131–141 10.1091/mbc.9.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D.A., Stage D.E., and Goldfarb D.S.. 2009. Evolution of the metazoan-specific importin α gene family. J. Mol. Evol. 68:351–365 10.1007/s00239-009-9215-8 [DOI] [PubMed] [Google Scholar]
- Matsusaka T., Imamoto N., Yoneda Y., and Yanagida M.. 1998. Mutations in fission yeast Cut15, an importin α homolog, lead to mitotic progression without chromosome condensation. Curr. Biol. 8:1031–1034 10.1016/S0960-9822(07)00425-3 [DOI] [PubMed] [Google Scholar]
- Maundrell K.1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 123:127–130 10.1016/0378-1119(93)90551-D [DOI] [PubMed] [Google Scholar]
- Miyamoto Y., Hieda M., Harreman M.T., Fukumoto M., Saiwaki T., Hodel A.E., Corbett A.H., and Yoneda Y.. 2002. Importin α can migrate into the nucleus in an importin β- and Ran-independent manner. EMBO J. 21:5833–5842 10.1093/emboj/cdf569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., and Nurse P.. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Norbury C., and Moreno S.. 1997. Cloning cell cycle regulatory genes by transcomplementation in yeast. Methods Enzymol. 283:44–59 10.1016/S0076-6879(97)83006-6 [DOI] [PubMed] [Google Scholar]
- Pumroy R.A., and Cingolani G.. 2015. Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem. J. 466:13–28 10.1042/BJ20141186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagolla M.J., Uzawa S., and Cande W.Z.. 2003. Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 116:4891–4903 10.1242/jcs.00796 [DOI] [PubMed] [Google Scholar]
- Salus S.S., Demeter J., and Sazer S.. 2002. The Ran GTPase system in fission yeast affects microtubules and cytokinesis in cells that are competent for nucleocytoplasmic protein transport. Mol. Cell. Biol. 22:8491–8505 10.1128/MCB.22.24.8491-8505.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., and Toda T.. 2007. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 447:334–337 10.1038/nature05773 [DOI] [PubMed] [Google Scholar]
- Sazer S.2010. Nuclear membrane: nuclear envelope PORosity in fission yeast meiosis. Curr. Biol. 20:R923–R925 10.1016/j.cub.2010.10.005 [DOI] [PubMed] [Google Scholar]
- Sazer S., and Dasso M.. 2000. The ran decathlon: multiple roles of Ran. J. Cell Sci. 113:1111–1118. [DOI] [PubMed] [Google Scholar]
- Schatz C.A., Santarella R., Hoenger A., Karsenti E., Mattaj I.W., Gruss O.J., and Carazo-Salas R.E.. 2003. Importin α-regulated nucleation of microtubules by TPX2. EMBO J. 22:2060–2070 10.1093/emboj/cdg195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., and Mann M.. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Shibuya T., Tsuneyoshi S., Azad A.K., Urushiyama S., Ohshima Y., and Tani T.. 1999. Characterization of the ptr6+ gene in fission yeast: a possible involvement of a transcriptional coactivator TAF in nucleocytoplasmic transport of mRNA. Genetics. 152:869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallada V.A., Daga R.R., Palomeque C., Garzón A., and Jimenez J.. 2002. Genome-wide search of Schizosaccharomyces pombe genes causing overexpression-mediated cell cycle defects. Yeast. 19:1139–1151 10.1002/yea.902 [DOI] [PubMed] [Google Scholar]
- Tallada V.A., Tanaka K., Yanagida M., and Hagan I.M.. 2009. The S. pombe mitotic regulator Cut12 promotes spindle pole body activation and integration into the nuclear envelope. J. Cell Biol. 185:875–888 10.1083/jcb.200812108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., Wolfe B.A., Jorgensen P., Tyers M., Gould K.L., and McCollum D.. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11:931–940 10.1016/S0960-9822(01)00268-8 [DOI] [PubMed] [Google Scholar]
- Tsai M.Y., Wang S., Heidinger J.M., Shumaker D.K., Adam S.A., Goldman R.D., and Zheng Y.. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 311:1887–1893 10.1126/science.1122771 [DOI] [PubMed] [Google Scholar]
- Umeda M., Izaddoost S., Cushman I., Moore M.S., and Sazer S.. 2005. The fission yeast Schizosaccharomyces pombe has two importin-α proteins, Imp1p and Cut15p, which have common and unique functions in nucleocytoplasmic transport and cell cycle progression. Genetics. 171:7–21 10.1534/genetics.105.042598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth A., Masuda H., Dhut S., and Toda T.. 2008. Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics. Mol. Biol. Cell. 19:5104–5115 10.1091/mbc.E08-02-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom T., Troxell C.L., and McIntosh J.R.. 2001. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 12:3919–3932 10.1091/mbc.12.12.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C.R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J.P., McIntosh J.R., and Gordon C.. 1998. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 17:6465–6476 10.1093/emboj/17.22.6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., and Barnes G.. 2010. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J. Cell Biol. 191:795–808 10.1083/jcb.201006028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., and Barnes G.. 2012. Spindle assembly requires complete disassembly of spindle remnants from the previous cell cycle. Mol. Biol. Cell. 23:258–267 10.1091/mbc.E11-08-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R., Burke B., and Doye V.. 2010. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell. Mol. Life Sci. 67:2215–2230 10.1007/s00018-010-0325-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Sato M., Fujita A., Yamamoto M., and Toda T.. 2005. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol. Biol. Cell. 16:1378–1395 10.1091/mbc.E04-10-0859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Oakes M., Yamaghishi M., Dodd J.A., and Nomura M.. 1992. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5640–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata J., Dephoure N., Macdonough T., Yu Y., Parnell E.J., Mooring M., Gygi S.P., Stillman D.J., and Kellogg D.R.. 2014. PP2ARts1 is a master regulator of pathways that control cell size. J. Cell Biol. 204:359–376 10.1083/jcb.201309119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., and Oliferenko S.. 2013. Remodeling the nuclear membrane during closed mitosis. Curr. Opin. Cell Biol. 25:142–148 10.1016/j.ceb.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Zheng Y.2010. A membranous spindle matrix orchestrates cell division. Nat. Rev. Mol. Cell Biol. 11:529–535 10.1038/nrm2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Schwartz C., Magidson V., Khodjakov A., and Oliferenko S.. 2007. The spindle pole bodies facilitate nuclear envelope division during closed mitosis in fission yeast. PLoS Biol. 5:e170 10.1371/journal.pbio.0050170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.