The Nek5 protein kinase contributes not only to uncoupling of the centrosome linker but also to integrity of the pericentriolar material and centrosomal microtubule nucleation, which together ensure the timely separation of centrosomes during early mitosis.
Abstract
Nek5 is a poorly characterized member of the NIMA-related kinase family, other members of which play roles in cell cycle progression and primary cilia function. Here, we show that Nek5, similar to Nek2, localizes to the proximal ends of centrioles. Depletion of Nek5 or overexpression of kinase-inactive Nek5 caused unscheduled separation of centrosomes in interphase, a phenotype also observed upon overexpression of active Nek2. However, separated centrosomes that resulted from Nek5 depletion remained relatively close together, exhibited excess recruitment of the centrosome linker protein rootletin, and had reduced levels of Nek2. In addition, Nek5 depletion led to loss of PCM components, including γ-tubulin, pericentrin, and Cdk5Rap2, with centrosomes exhibiting reduced microtubule nucleation. Upon mitotic entry, Nek5-depleted cells inappropriately retained centrosome linker components and exhibited delayed centrosome separation and defective chromosome segregation. Hence, Nek5 is required for the loss of centrosome linker proteins and enhanced microtubule nucleation that lead to timely centrosome separation and bipolar spindle formation in mitosis.
Introduction
The human genome encodes a family of 11 NIMA-related, or Nek, protein kinases (Moniz et al., 2011; Fry et al., 2012). Functional studies indicate that the majority have roles either in cell cycle progression or at the primary cilium (O’Connell et al., 2003; Quarmby and Mahjoub, 2005; O’Regan et al., 2007; Moniz et al., 2011; Fry et al., 2012). For example, Nek2, Nek6, Nek7, and Nek9 contribute to centrosome separation and mitotic spindle assembly; Nek1, Nek8, Nek10, and Nek11 contribute to the DNA damage response; and Nek1, Nek4, and Nek8 contribute to ciliary function.
Nek5 remains the least well characterized of this family, with the only studies to date pointing to a role in myogenic differentiation (Tadokoro et al., 2010; Shimizu and Sawasaki, 2013). Here, we demonstrate that Nek5, in common with several other family members, localizes to centrosomes. Moreover, its loss leads to premature centrosome separation, a phenotype that also results from overexpression of Nek2 (Fry et al., 1998b; Faragher and Fry, 2003). During interphase, centrosomes are held together by a proteinaceous linker that is composed of coiled-coil proteins, including C-Nap1, rootletin, Cep68, centlein, and LRRC45 (Fry et al., 1998a; Mayor et al., 2000; Bahe et al., 2005; Yang et al., 2006; Graser et al., 2007; He et al., 2013; Fang et al., 2014). This linker, which extends between and connects the proximal ends of the mother and daughter centrioles, is disassembled in late G2 as a result of phosphorylation of linker proteins by Nek2 (Hardy et al., 2014). This allows centrosomes to separate in prophase and generate a bipolar spindle capable of accurate chromosome segregation (Mardin and Schiebel, 2012).
Here, we have explored the functions of Nek5 in regulating centrosome organization. We show that it contributes not only to uncoupling of the centrosome linker but also to the integrity of the pericentriolar material (PCM) and centrosomal microtubule (MT) nucleation. Together, these processes ensure the timely separation of centrosomes in early mitosis.
Results and discussion
Nek5 is a novel component of the centrosome
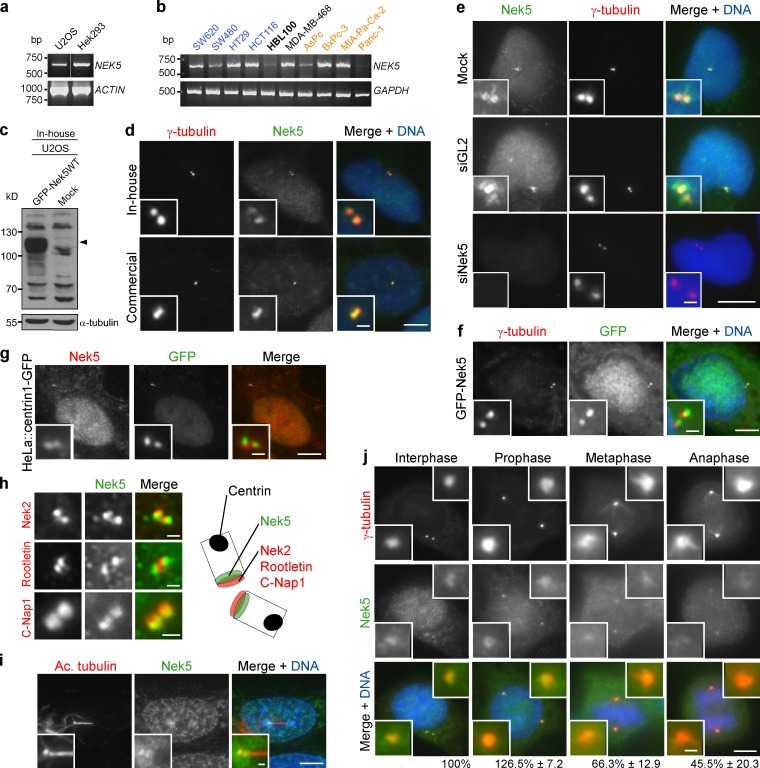
To investigate the cellular role of Nek5, we first confirmed its expression using semiquantitative RT-PCR in HEK293 and U2OS cells, as well as a panel of cancer cell lines (Fig. 1, a and b). We then generated a rabbit polyclonal antibody that detected recombinant Nek5 with a Western blot (Fig. 1 c). This antibody, as well as a commercial Nek5 antibody, stained the nucleus and centrosomes under immunofluorescence microscopy (IF; Fig. 1 d). Although these antibodies did not obviously detect endogenous Nek5 when using a Western blot, the IF signals were lost upon RNAi-mediated depletion of Nek5 (Fig. 1 e; and Fig. S1, a and b). Furthermore, recombinant GFP-tagged Nek5 localized to nuclei and centrosomes (Fig. 1 f). We then compared the centrosomal localization pattern of endogenous Nek5 with that of centrin1-GFP, a marker of centriole distal ends, and C-Nap1, rootletin, and Nek2, markers of centriole proximal ends (Fig. 1, g and h). Nek5 staining was clearly distinct from centrin but overlapped with the proximal end markers. Consistent with it being a centriolar protein, Nek5 colocalized with basal bodies, but not axonemal microtubules, in ciliated hTERT-RPE1 cells (Fig. 1 i). During mitotic progression, Nek5 was detectable at spindle poles, although staining was diminished in metaphase and anaphase (Fig. 1 j). Hence, these data provide good evidence that Nek5 is a novel component of the centrosome that localizes toward the proximal end of centrioles.
Figure 1.
Nek5 localizes to the centrosome. (a) RT-PCR of Nek5 and actin with RNA from U2OS or Hek293 cells. (b) RT-PCR of Nek5 and GAPDH with RNA from cancer cell lines (blue, colorectal; black, breast; orange, pancreatic) and immortalized breast epithelial cells (HBL100; bold). (c) Western blot analysis of U2OS cells that were mock-transfected or transfected with GFP-Nek5 for 24 h. Blots were probed with the in-house Nek5 antibody. α-Tubulin was used as loading control. (d) Staining of U2OS cells with either an in-house or commercial Nek5 antibody and γ-tubulin. (e) Staining of Nek5 (with in-house antibody) and γ-tubulin in U2OS cells transfected with Nek5- or GL2-specific siRNAs for 48 h. (f) GFP-Nek5–transfected U2OS cell stained for γ-tubulin. (g) HeLa::centrin1-GFP cell stained for Nek5. (h) U2OS cells stained for Nek2, rootletin, or C-Nap1 and Nek5. The schematic illustrates relative localization of proteins within centrioles. (i) Staining of a ciliated hTERT-RPE-1 cell for Nek5 and acetylated tubulin. (j) Staining of γ-tubulin and Nek5 in U2OS cells at the stages indicated. The intensity of Nek5 at the centrosome relative to interphase is indicated. Data are mean ± SD of at least 20 centrosomes (n = 2). Bars: (d–g and i–j, main images) 10 µm; (d–f, g, and i–j, insets, and h) 1 µm.
Nek5 is required for normal centrosome linker organization
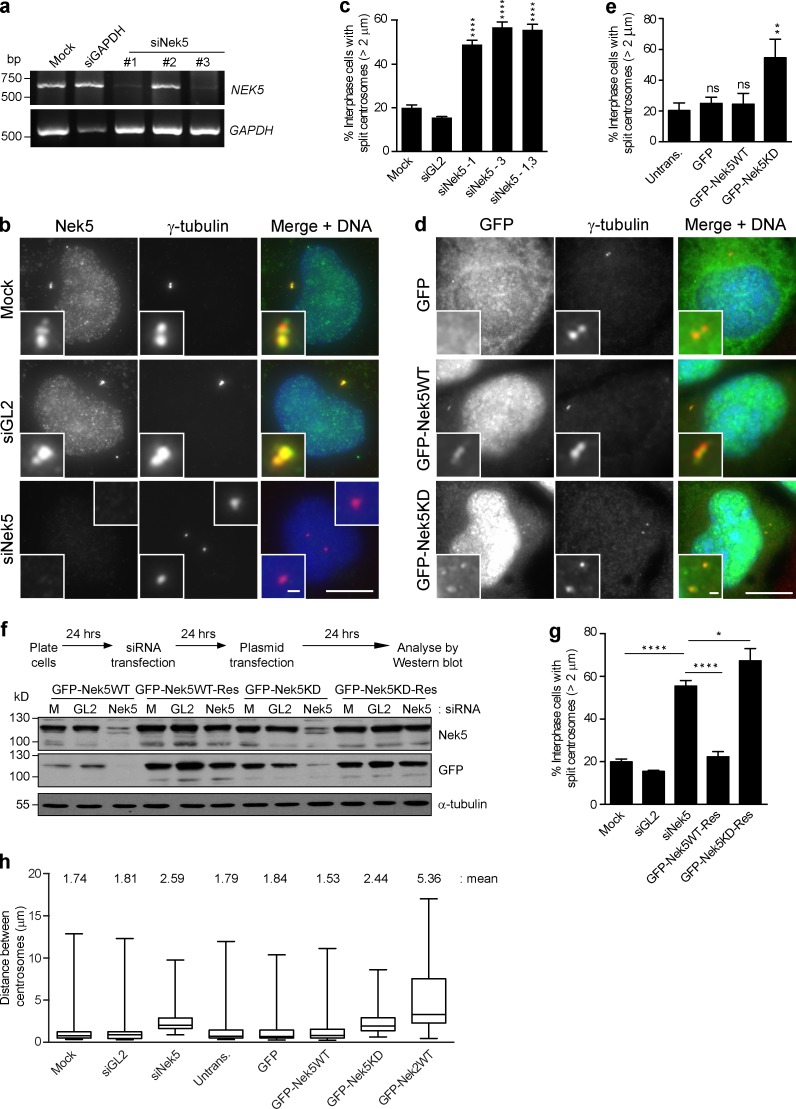
Depletion of Nek5 with two independent siRNAs led to premature centrosome separation in U2OS cells. The percentage of cells in which centrosomes were separated by >2 µm increased from 20% in controls to >55% upon Nek5 depletion (Fig. 2, a–c; and Fig. S1 b). A recombinant kinase-dead Nek5 construct (Nek5KD) was then generated with mutations in two residues that are expected to be essential for catalytic activity: K33 and D128. Although we have not been able to establish kinase assays to confirm inactivation of Nek5 by these mutations, a significant increase in the proportion of interphase cells (54.7 ± 8.2% compared with 24.3 ± 3.6% in controls) in which centrosomes were separated by >2 µm was observed upon overexpression of GFP-Nek5KD. In contrast, overexpression of wild-type Nek5 did not alter the extent of centrosome separation (Fig. 2, d and e). Moreover, RNAi-resistant wild-type, but not kinase-dead, Nek5 was able to rescue the centrosome separation defect induced by Nek5 depletion (Fig. 2, f and g). Interestingly, although the frequency of centrosome separation was increased upon Nek5 depletion or overexpression of kinase-dead Nek5, the mean distance of separation was relatively modest, being increased from 1.74 to 2.59 (Nek5 depletion) and 2.44 µm (overexpression of kinase-dead; Fig. 2 h). In contrast, overexpression of wild-type Nek2 led to a mean separation of 5.36 µm, which suggests that the mechanisms of centrosome separation that result from loss of Nek5 and overexpression of Nek2 are distinct (Fig. 2 h).
Figure 2.
Nek5 loss leads to premature centrosome separation in interphase. (a) RT-PCR of Nek5 and GAPDH with RNA collected after 72 h of siRNA treatment of U2OS cells with mock, GAPDH, or Nek5 siRNAs. Three individual Nek5 siRNAs were tested. (b) Staining of Nek5 and γ-tubulin in U2OS cells transfected with Nek5- or GL2-specific siRNAs for 48 h. (c) Quantification of cells treated as in b with centrosomes split by >2 µm. (d) Staining of γ-tubulin and DNA in U2OS cells transfected for 48 h with GFP, GFP-Nek5WT, or GFP-Nek5KD. (e) Quantification of cells with centrosomes separated by >2 µm in cells treated as in d. (f) U2OS cells were transfected with either siRNAs against Nek5 or GL2, or mock treated for 24 h before GFP-Nek5WT and GFP-Nek5KD RNAi sensitive and resistant constructs were transfected into the cells. Cells were harvested 24 h after plasmid transfection and analyzed by Western blotting with Nek5, GFP, and α-tubulin antibodies. (g) Quantification of cells with centrosomes separated by >2 µm in U2OS cells treated as in f. (h) Box and whisker plot of the distance between centrosomes in mock-, GL2-, or Nek5-depleted cells, and either untransfected cells or those expressing GFP, GFP-Nek5WT, GFP-Nek5KD, or GFP-Nek2WT. Bars: (main images) 10 µm; (insets) 1 µm. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Histograms show mean + SD (error bars); n = 3, 200 cells per experiment. Boxes represent the 25th and 75th percentile, and whiskers the 10th and 90th percentile of centrosome distances measured in 50 cells.
Nek5 is required for PCM recruitment and MT nucleation
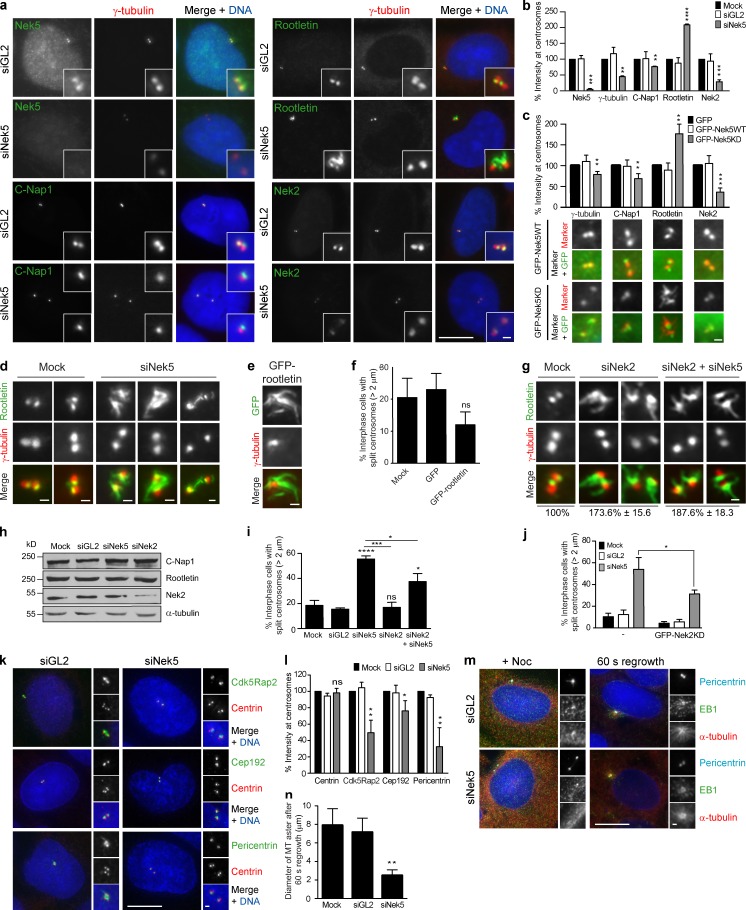
To investigate how Nek5 contributes to centrosome cohesion, the localization of centrosome linker proteins was assessed after Nek5 depletion (Fig. 3, a and b). C-Nap1 and Nek2 were both reduced in cells depleted of Nek5. In contrast, rootletin was increased twofold. Similar changes were observed in cells expressing kinase-inactive Nek5 (Fig. 3 c). The extra rootletin present at centrosomes in Nek5-depleted cells formed extended fibers (Fig. 3 d), similar to those formed upon overexpression of rootletin (Fig. 3 e; Bahe et al., 2005). However, centrosome separation was inhibited, rather than promoted, in the presence of overexpressed rootletin, possibly through cytoplasmic sequestration of Nek2 (Fig. 3 f).
Figure 3.
Nek5 loss leads to increased rootletin but reduced PCM at interphase centrosomes. (a) Staining of γ-tubulin together with Nek5, C-Nap1, rootletin, or Nek2 in U2OS cells treated with siRNAs against GL2 or Nek5 for 48 h. (b) Histogram of relative intensities of Nek5, γ-tubulin, C-Nap1, rootletin, and Nek2 in U2OS cells mock-treated or with siRNAs against GL2 or Nek5 for 48 h. (c) Histogram of relative intensities of γ-tubulin, C-Nap1, rootletin, and Nek2 in U2OS cells transfected with GFP, GFP-Nek5WT, or GFP-Nek5KD for 48 h. Representative images from GFP-Nek5WT– and GFP-Nek5KD–transfected cells stained for each protein are shown. (d) Staining of rootletin and γ-tubulin in mock- and Nek5-depleted cells. (e) Staining of GFP and γ-tubulin in cells transfected with GFP-rootletin for 24 h. (f) Quantification of cells with centrosomes separated by >2 µm in mock-, GFP-, or GFP-rootletin–transfected cells. (g) Staining of rootletin and γ-tubulin in mock-, Nek2-, or Nek2- and Nek5-depleted cells. (h) Western blot analysis of C-Nap1, rootletin, Nek2, and α-tubulin from mock-, GL2-, Nek5- or Nek2-depleted cells. (i) Quantification of cells with centrosomes separated by >2 µm in mock-, GL2-, Nek5-, Nek2-, or Nek2- and Nek5-depleted U2OS cells. (j) Quantification of cells with centrosomes separated by >2 µm in untransfected or GFP-Nek2KD–transfected mock-, GL2-, or Nek5-depleted U2OS cells. (k) Staining of centrin, Cdk5Rap2, Cep192, and pericentrin in GL2- and Nek5-depleted cells. (l) Histogram of intensities of centrin, Cdk5Rap2, Cep192, and pericentrin in U2OS cells treated with siRNAs against GL2 or Nek5 for 48 h relative to mock-treated cells. (m) Staining of pericentrin, EB1, and α-tubulin in GL2- and Nek5-depleted cells treated with nocodazole for 5 h and 60 s washout to allow MT regrowth. (n) Histogram of the diameter of MT asters formed after 60 s regrowth in mock-, GL2-, and Nek5-depleted cells. Bars: (a, k, and m, main images) 10 µm; (a, k, and m [insets], c–e, and g) 1 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Histograms show mean + SD (error bars); n = 3, 40 centrosomes (b, c, l, and n) or 200 cells (f, i, and j) per experiment.
To explore the relationship between Nek5 and Nek2 in regulating centrosome cohesion, we analyzed cells that were either singly or co-depleted of Nek2 and Nek5. We found that each condition led to an approximately twofold increase in rootletin at the centrosome (Fig. 3 g). Western blot analysis indicated that the total protein levels of C-Nap1 and rootletin were unchanged (Fig. 3 h). Nek2 depletion did not lead to an increase in centrosome separation, whereas codepletion with Nek5 led to a reduced frequency of separation relative to Nek5 depletion alone (Fig. 3 i). Consistent with this, overexpression of kinase-dead Nek2 (GFP-Nek2KD) reduced the frequency of centrosome separation in Nek5-depleted cells from 54.6 ± 10.5% upon Nek5 depletion alone to 31.3 ± 3.5% in cells also expressing GFP-Nek2KD (Fig. 3 j). Hence, we conclude that Nek2 and Nek5 have distinct but cooperative functions in regulating organization of the centrosome linker.
We had noted that γ-tubulin levels were also reduced in Nek5-depleted cells (Fig. 3, a and b). We therefore assessed the abundance of other centrosomal components to determine whether there was a general loss of centrosome integrity. The PCM components pericentrin, Cep192, and Cdk5Rap2 (Cep215) were also significantly reduced, whereas the core centriole component, centrin, was unaffected (Fig. 3, k and l; and Fig. S1 c). This suggests that loss of Nek5 primarily disrupts PCM integrity as opposed to centriole structure. Consistent with this, MT regrowth was slowed after nocodazole-induced depolymerization, with the asters formed in Nek5-depleted cells being only half the diameter of those formed in control cells after 60 s (Fig. 3, m and n).
Nek5 promotes centrosome linker disassembly in mitosis
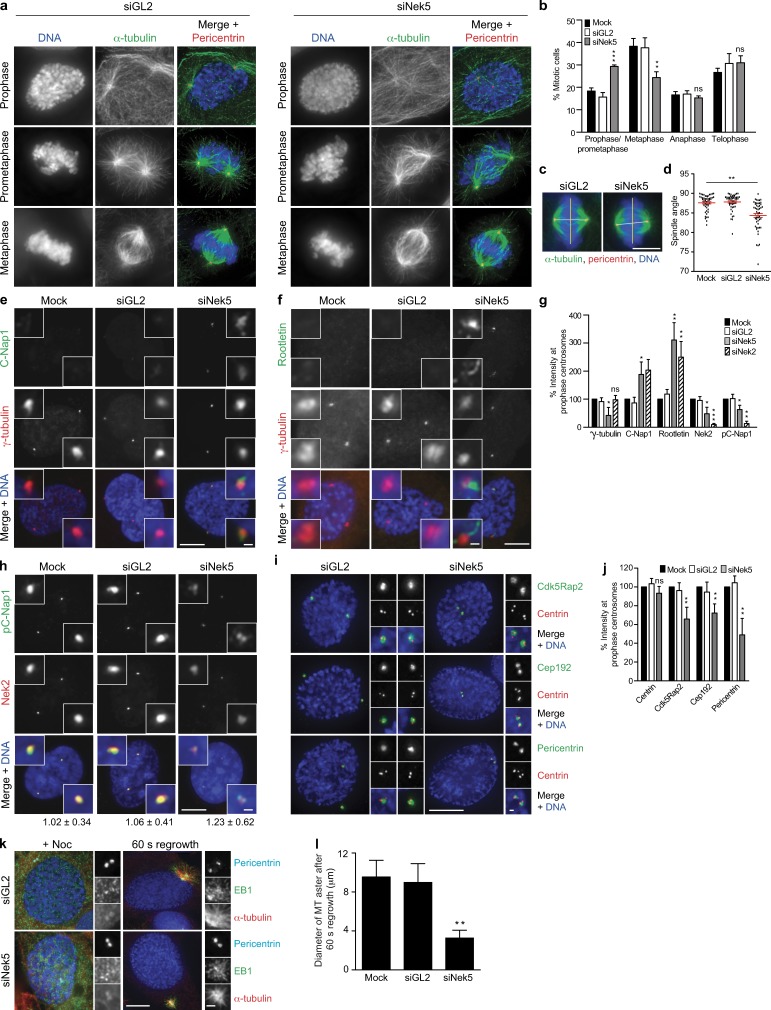
We next tested the importance of Nek5 for mitotic progression. Flow cytometry showed no major change in cell cycle profile in Nek5-depleted cells or cells overexpressing wild-type or kinase-dead Nek5 (Fig. S1 d). However, this does not rule out a more subtle cell cycle defect, and microscopy analysis revealed an increase in the relative fraction of mitotic cells in prophase/prometaphase in Nek5-depleted cells, with a concomitant decrease in metaphase cells (Fig. 4, a and b). Although no defect was observed in the spindle assembly checkpoint response to taxol (Fig. S1 e), metaphase spindles assembled in Nek5-depleted cells exhibited altered geometry, with poles often out of line with normal spindle orientation (Fig. 4, c and d). Additionally, C-Nap1 and rootletin still remained on separated centrosomes in prophase (Fig. 4, e–g). Conversely, Nek2 abundance and C-Nap1 phosphorylation were reduced at centrosomes in Nek5-depleted prophase cells. Similar increases in C-Nap1 and rootletin abundance and a decrease in phosphorylated C-Nap1 were observed at separated centrosomes in prophase after Nek2 depletion (Fig. 4 h). Thus, we propose that the absence of Nek5 hinders recruitment of Nek2 to centrosomes, leading to reduced C-Nap1 phosphorylation and inappropriate retention of centrosome linker proteins in prophase. However, although there was a decrease in phospho-C-Nap1 and Nek2 in response to Nek5 depletion, the increase in total C-Nap1 leaves it possible that there is also a change in localized activity of Nek2 at the centrosome (Fig. 4, g and h).
Figure 4.
Nek5 loss leads to retention of centrosome linker proteins and reduced MT nucleation in mitosis. (a) Staining of DNA, α-tubulin, and pericentrin in GL2- and Nek5-depleted cells at the mitotic stages indicated. (b) Histogram of cells in different stages of mitosis, as indicated, after 48 h of mock, GL2, or Nek5 depletion in U2OS cells. (c) Metaphase GL2- and Nek5-depleted cells stained for DNA, α-tubulin, and pericentrin. The yellow line indicates the plane of the metaphase plate; the white line indicates the pole-to-pole spindle axis. (d) Dot plot of the angle between the spindle axis and metaphase plate for mock-, GL2-, and Nek5-depleted cells. Each dot represents a single cell with data collected over two independent experiments, bars represents mean ± SD. (e and f) Staining of C-Nap1 or rootletin and γ-tubulin in prophase U2OS cells after mock, GL2, or Nek5 depletion. (g) Histogram of relative intensities of γ-tubulin, C-Nap1, rootletin, Nek2, and phospho-C-Nap1 (pC-Nap1) in mock-, GL2-, Nek5-, and Nek2-depleted prophase U2OS cells. Each protein was normalized to its intensity in mock-treated cells. (h) Staining of pC-Nap1 and Nek2 in prophase U2OS cells that were mock, GL2, or Nek5 depleted. The ratios of pC-Nap1/Nek2 intensities are indicated. (i) Staining of centrin, Cdk5Rap2, Cep192, and pericentrin in GL2- and Nek5-depleted prophase cells. (j) Histogram of relative intensities of centrin, Cdk5Rap2, Cep192, and pericentrin in prophase U2OS cells that were mock-, GL2-, or Nek5-depleted for 48 h. Each protein was normalized to its intensity in mock-treated cells. (k) Staining of pericentrin, EB1, and α-tubulin in GL2- and Nek5-depleted prophase cells treated with nocodazole for 5 h and 60 s washout to allow MT regrowth. (l) Histogram of the diameter of MT asters formed after 60 s of regrowth in mock-, GL2-, Nek5-, and Cep192-depleted cells. Bars: (a, c, e, f, h, and i, main images) 10 µm; (c) 5 µm; (e, f, h, i, and k, insets) 1 µm. Histograms show mean + SD (error bars); n = 3, 40 centrosomes (l) or 100 cells (b, g, and j) per experiment. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Nek5-depleted prophase cells also had reduced levels of γ-tubulin, Cdk5Rap2, Cep192, and pericentrin, and a reduced rate of MT regrowth (Fig. 4, e–g, and i–l). Thus, the spindle geometry defect may be attributed to both retention of linker proteins and reduced MT-nucleating capacity of prophase centrosomes. Indeed, Cdk5Rap2 has dual roles in both centrosome cohesion and PCM integrity, which suggests that its loss may contribute to both phenotypes observed upon Nek5 loss (Pagan et al., 2015).
Nek5 is required for timely centrosome separation and genome integrity
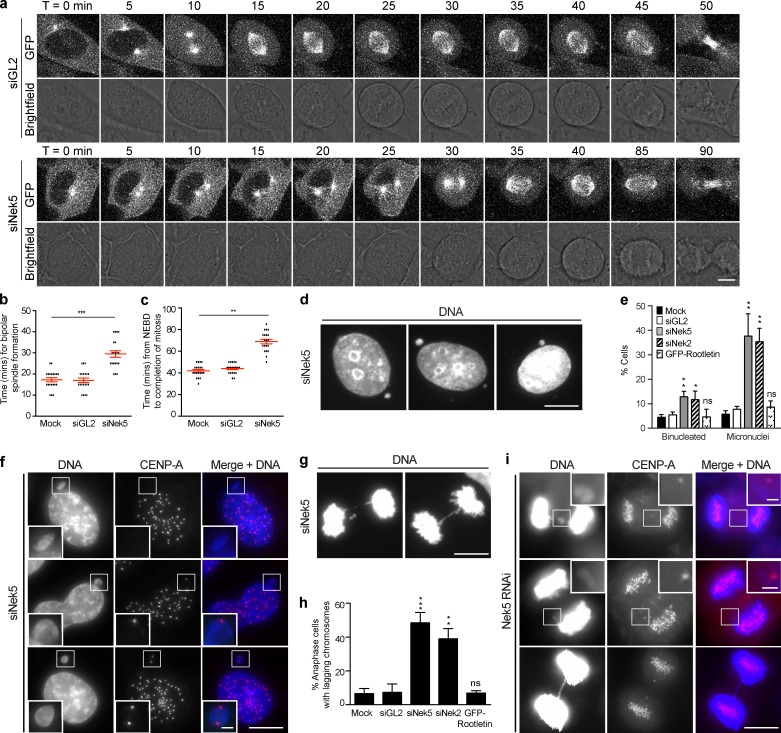
To test the impact of Nek5 depletion on the timing of centrosome separation, we performed time-lapse imaging of HeLa cells stably expressing GFP–α-tubulin. Upon mitotic entry, a bright GFP dot becomes visible as the MT organizing center (MTOC) increases nucleation activity. This MTOC then separates to opposite sides of the cell to establish a bipolar spindle. Designating the time at which a distinct MTOC is first seen as T = 0, we observed that Nek5-depleted cells take ∼15 min longer to establish a bipolar spindle with opposing poles compared with control cells (Fig. 5, a and b; and Videos 1–6). Indeed, only 48% of Nek5-depleted cells enter prometaphase with centrosomes at opposite poles, as compared with 76% of control cells (n = 25). Additionally, the time taken from nuclear envelope breakdown to anaphase onset was increased by ∼25 min in Nek5-depleted cells (Fig. 5 c). Together, these data suggest that spindle assembly is less efficient in Nek5-depleted cells, which is consistent with the increase in prophase/prometaphase duration.
Figure 5.
Nek5 loss leads to delayed centrosome separation and chromosome segregation errors in mitosis. (a) Brightfield and GFP stills from time-lapse imaging of GL2- and Nek5-depleted HeLa cells expressing GFP–α-tubulin. Time is in minutes and t = 0 was defined as the frame in which a distinct MTOC becomes visible. (b) Dot plot of time from when an MTOC becomes visible to bipolar spindle formation in mock-, GL2-, and Nek5-depleted HeLa::GFP–α-tubulin cells. (c) Dot plot of time from nuclear envelope breakdown (NEBD) to anaphase onset in mock-, GL2-, and Nek5-depleted HeLa::GFP–α-tubulin cells. (d) DNA stain in U2OS cells depleted of Nek5 for 48 h. (e) Histogram of interphase U2OS cells that were binucleated or contained micronuclei after mock, GL2, Nek5, or Nek2 depletion or GFP-rootletin overexpression. (f) Interphase Nek5-depleted U2OS cells stained for DNA and CENP-A. (g) Anaphase Nek5-depleted U2OS cells stained for DNA. (h) Histogram of anaphase cells with lagging chromosomes after mock, GL2, Nek5, or Nek2 depletion or GFP-rootletin overexpression. (i) Anaphase Nek5-depleted U2OS cells stained for DNA and CENP-A. Bars: (a, d, and f, main images) 10 µm; (g and i, main images) 5 µm; (f and i, insets) 1 µm. Histograms show mean + SD (error bars); n = 3, 100 cells per experiment. For dot plots, each dot represents a single cell with data collected over two independent experiments; bars represent mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
A block to prophase centrosome separation leads to spindle assembly via a prometaphase pathway that is dependent on motor protein–driven pole separation (Toso et al., 2009; Kaseda et al., 2012; Mchedlishvili et al., 2012). However, spindles assembled via the prometaphase pathway exhibit increased frequencies of merotelic attachments and chromosome segregation defects (Silkworth et al., 2012). We noted that upon Nek5 depletion there was a fourfold increase in interphase cells that had micronuclei, as well as a significant increase in binucleated cells (Fig. 5, d and e). These observations are consistent with chromosome segregation errors and cytokinesis failure, both of which occur in response to merotelic attachments. Micronuclei in Nek5-depleted cells contained 0, 1, or 2 centromeres, which suggests that they arise from both acentric chromosome fragments and lagging whole chromosomes (Fig. 5 f). There was also an increase in the frequency of anaphase cells with lagging chromosomes, rising from 6.6% in control cells to 48.4% upon Nek5 depletion (Fig. 5, g–i). We also observed unresolved sister chromatids that gave rise to chromosome bridges (Fig. 5, g and i). Similar results were seen upon depletion of Nek2, but not after overexpression of rootletin (Fig. 5, e and h), which supports the hypothesis that both Nek2 and Nek5 are required for timely separation of centrosomes in early mitosis.
In summary, we provide the first demonstration of a cell cycle role for Nek5. We propose that Nek5 is important for both maintenance of PCM integrity and the mitotic loss of centrosome cohesion. Together, these processes are essential for the timely separation of centrosomes at the G2/M transition. Disassembly of the centrosome linker may be achieved through cooperation with Nek2, although how this happens remains to be determined. Indeed, there are many potential substrates for these kinases, given the growing list of centrosome linker components. Nevertheless, in the absence of either Nek2 or Nek5, centrosome separation is delayed, leading to chromosome segregation errors and highlighting the importance of these kinases for genome stability.
Materials and methods
Plasmid construction and site-directed mutagenesis
A full-length Nek5 cDNA clone (GenBank accession no. BC063885) encoding a 708-residue protein was obtained from GeneCopoeia. For antibody production, a Nek5 DNA fragment corresponding to amino acids 505–708 was amplified by PCR and cloned into the pETM-11 (T7 promoter; EMBL) vector. To generate tagged mammalian expression constructs, full-length Nek5 cDNA was PCR amplified and cloned into the pEGFP-T7 (pEGFP-C1 [CMV promoter; Takara Bio Inc.] with an additional T7 promoter [Fry laboratory]) or pFLAG-CMV-2 (CMV promoter; Sigma-Aldrich) vectors. Point mutations to generate the GFP-Nek5-K33R/D128A mutant were introduced by sequential site-directed mutagenesis of the GFP-Nek5WT construct using the GeneTailor Site-Directed Mutagenesis System (Invitrogen). RNAi-resistant constructs were generated using primers designed to change the third position of each codon recognized by oligos #1 and #3 (#1 forward, CCTTCTTCAATTCATTTCAAGAAAACGGGAGACTCTTCATCGTAATGGAAT, and reverse, TTGAAATGAATTGAAGAAGGCTACAATGTT; #3 forward, TCTTTCAAGTATCTCCTCGAGATCGCCCGTCGATCAACTCGATTTTGAAAA, and reverse, TCGAGGAGATACTTGAAAGAGCTGAGATAT). GFP-rootletin (full-length human rootletin), Myc-Nek2WT (full-length wild-type human Nek2A) and GFP-Nek2KD (amino acid change, K37R, in full-length Nek2A to give inactive Nek2A) have been described previously (Fry et al., 1998b; Faragher and Fry, 2003; Bahe et al., 2005). All plasmid constructs were verified by DNA sequencing (The Protein Nucleic Acid Chemistry Laboratory, University of Leicester).
Antibody generation
For production of Nek5 antibodies, rabbits were immunized by Cambridge Research Biochemicals with bacterially expressed His-tagged protein corresponding to amino acids 505–708 of Nek5. For affinity purification, His-Nek5-507-708 was coupled to CNBr-activated Sepharose according to the manufacturer’s instructions (GE Healthcare). Antiserum was passed over the column, and after extensive washing with 10 mM Tris-HCl, pH 7.5, followed by 10 mM Tris-HCl, pH 7.5, 500 mM NaCl, specific antibodies were eluted with 100 mM glycine, pH 2.5, or 100 mM Triethylamine, pH 11.5, into tubes containing neutralizing quantities of 1 M Tris-HCl, pH 8.0.
Cell culture, drug treatments and transfection
All cell lines were maintained in a humidified 5% CO2 atmosphere at 37°C. U2OS, tetracycline-inducible U2OS::GFP-Nek2WT (USOS cells with stably incorporated GFP-Nek2WT full length under a tetracycline-inducible promoter; Faragher and Fry, 2003), Hek293, HeLa::GFP–α-tubulin (stably expressing GFP-tagged full length α-tubulin), HeLa::centrin1-GFP (stably expressing GFP-tagged full-length centrin1), as well as colorectal (SW620, SW480, HT29, HCT116), breast (HBL100), and pancreatic (MIA-Pa-Ca-2, Panc-1) cancer cell lines were cultured in DMEM (Invitrogen). Other breast (MDA-MB-468) and pancreatic (AsPc, BxPc-3) cancer cell lines were cultured in RPMI 1640 media (Invitrogen). hTERT-RPE1 cells were cultured in DMEM/Ham’s F12 supplemented with 0.4% wt/vol sodium bicarbonate solution. All cell lines were supplemented with 10% vol/vol FBS and penicillin-streptomycin (100 U/ml and 100 µg/ml, respectively). Primary cilia formation was induced in hTERT-RPE1 cells by culturing cells in serum-free medium for 48 h. To induce expression of Nek2 in the U2OS::GFP-Nek2WT cell line, doxycycline was added at 1 µg/ml for 24 h. To arrest cells in mitosis, cells were treated with 10 µM taxol (Sigma-Aldrich). For MT regrowth experiments, cells were treated with 20 µM nocodazole (Sigma-Aldrich) for 5 h, then cells were washed and incubated in culture medium without nocodazole at 37°C for the indicated time. For transient transfections, 1 µg of plasmid DNA was complexed with Lipofectamine 2000 (Invitrogen) in serum-free OptiMEM (Gibco) according to the manufacturer’s instructions and added to cells at 60% confluency. Cells were fixed and analyzed 24–48 h after transfection.
RNAi
siRNA oligonucleotides specific to Nek5 (#1, GAGAAUGGCAGGCUGUUUAUU; #2, GAAUAAACCCUACAACAAUUU; #3, GACCGACCAUCCAUAAAUUUU; GE Healthcare or QIAGEN), Nek2 (GAAAGGCAAUACUUAGAUG and AAACAUCGUUCGUUACUAU; Thermo Fisher Scientific), GAPDH (AM4631; Ambion), or luciferase GL2 (D-001100-01-20; Thermo Fisher Scientific) were transfected at 50 nmol into cells using Oligofectamine or Lipofectamine 2000 (Invitrogen) in serum-free OptiMEM (Gibco) according to the manufacturer’s instructions. Cells were analyzed 48–72 h after transfection. Unless otherwise indicated, Nek5 depletions were undertaken with siRNA oligonucleotides #1 and #3 combined.
Immunofluorescence microscopy
Cells were extracted with 0.2% NP-40 in 80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA for 30 s before fixation with methanol at −20°C for 10 min. After rehydration with PBS for 20 min, cells were blocked in 1% wt/vol BSA-PBS and incubated with primary antibodies for 1 h at room temperature followed by a 45-min incubation with secondary antibodies. Secondary antibodies used were Alexa Fluor 488, 594, and 647, FITC or Rhodamine-red goat or donkey anti–rabbit, goat or donkey anti–mouse, and donkey anti–rat IgGs (1 µg/ml; Invitrogen or Jackson ImmunoResearch Laboratories, Inc.). DNA was stained with Hoechst 33258 or DAPI (Sigma-Aldrich). Coverslips were mounted with 3% wt/vol N-propyl-gallate, 80% vol/vol glycerol in PBS, or ProLong Gold (Invitrogen). Images were captured as single focal planes under oil at room temperature on a microscope (TE300; Nikon) equipped with a digital camera (ORCA-R2; Hamamatsu Photonics) and a 100× oil objective lens, NA 1.4, or a microscope (IX71; Olympus) equipped with a camera (Orca AG; Hamamatsu Photonics) and a 100× oil objective lens, NA 1.35, with 1 × 1 binning, using Volocity software (Improvision). Post-imaging merges and individual channel images were exported as TIFs for publication and cropped using Photoshop CS6 (Adobe). All intensity and distance measurements were performed using ImageJ (v1.48). Alternatively, images were captured under oil at room temperature as three-dimensional datasets acquired on an imaging system (DeltaVision RT; Applied Precision) equipped with a microscope (IX70; Olympus), a CCD camera (CoolSNAP 1,024 × 1,024; Roper Scientific), and a 60×, NA 1.42, Plan-Apochromat oil objective lens with 1 × 1 binning. Z stacks (25 sections, 0.2 µm) were collected and computationally deconvolved (Constrained Iterative Deconvolution algorithm, enhanced ratio method) with the softWoRx software package (v3.5; Applied Precision) and exported as TIFs as two-dimensional maximum intensity projections. Images were converted from 16- to 8-bit using ImageJ (v1.48) and cropped using Photoshop CS6 (Adobe). Mouse monoclonal antibodies used were as follows: Nek5 (ab77451, 1:250; Abcam), γ-tubulin (T6557, 1:1,000; Sigma-Aldrich), Nek2 (610594, 1:500; BD), acetylated tubulin (T6793, 1:2,000; Sigma-Aldrich), GFP (11814460001, 1:2,000; Roche), CENP-A (ab13939, 1:400; Abcam), centrin (04-1624, 1:1,000; EMD Millipore), and α-tubulin (T6199, 1:1,000; Sigma-Aldrich). Rat monoclonal antibody was used against EB1 (ab53358, 1:250; Abcam). Rabbit polyclonal antibodies were used against Nek5 (this study, 1:200), C-Nap1 (raised against his-tagged amino acids 1978–2442 of human C-Nap1, 1:750; Fry et al., 1998a), pC-Nap1 (raised against a phospho-peptide corresponding to residues LLEKpSLAQRVQC of human C-Nap1, 1:1,000; Hardy et al., 2014), rootletin (raised against MBP-tagged amino acids 1651–2018 of human rootletin, 1:1,000; Hardy et al., 2014), GFP (ab6556, 1:1,000; Abcam), phospho-Histone H3 (06-570, 1:500; EMD Millipore), Cep192 (sc-84784, 1:250; Santa Cruz Biotechnology, Inc.), pericentrin (ab4448, 1:1,000; Abcam), and Cdk5Rap2 (IHC-00063, 1:500; Bethyl Laboratories, Inc.).
Live cell microscopy
Time-lapse imaging was performed at 37°C under oil on a confocal microscope (TCS SP5; Leica) equipped with an inverted microscope (DMI 6000B; Leica) using a 63× oil objective lens, NA 1.4. Cells were maintained on the stage at 37°C in a 5% CO2 atmosphere using a microscope temperature control system (Life Imaging Services). HeLa cells stably expressing GFP–α-tubulin (HeLa::GFP–α-tubulin) were cultured in 35-mm glass-bottomed dishes (CellView) consisting of four compartments (Greiner Bio-One) such that each treatment condition could be imaged in the same session. Cells were treated with siRNAs for 40 h, then imaged every 5 min for 18 h. Z stacks composed of twenty 1-µm steps were acquired at each time point, and images were captured from two positions per compartment of the 35-mm dish. Stacks were processed into maximum-intensity projections using LAS-AF software (Leica) and images were processed using ImageJ (v1.48).
Flow cytometry
For flow cytometry, cells were fixed with ice-cold 70% ethanol at −20°C overnight. After fixation, cells were washed twice with PBS then resuspended in PBS containing 200 µg/ml RNase A and 40 µg/ml propidium iodide. Cytometry was performed on a FACSCanto (BD).
Western blotting
Whole-cell extracts were prepared with radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, and 1 mM EDTA, with protease and phosphatase inhibitor cocktails). Extracts were boiled in loading buffer for 5 min. Proteins were separated on 8 or 12% SDS PAGE gels and transferred to nitrocellulose membranes (GE Healthcare) for analysis. Blots were detected by ECL (GE Healthcare). Primary antibodies used were: rabbit polyclonals against Nek5 (this study; 1:500), C-Nap1 (raised against his-tagged amino acids 1978–2442 of human C-Nap1; 1:1,000; Fry et al., 1998a), and rootletin (raised against MBP-tagged amino acids 1651–2018 of human rootletin, 1:1,000; Hardy et al., 2014); mouse monoclonals against Nek5 (ab77451, 1:500; Abcam), α-tubulin (T5168, 1:10,000; Sigma-Aldrich), Nek2 (610594, 1:1,000; BD), and GFP (11814460001, 1:5,000; Roche). HRP-conjugated goat anti–mouse or anti–rabbit secondary antibodies were used at 1:5,000 (Jackson ImmunoResearch Laboratories, Inc.).
RT-PCR
To prepare RNA for RT-PCR, cells were homogenized with TRIzol reagent (Sigma-Aldrich). 1 (GAPDH, actin) or 5 µg (Nek5) of total RNA was then used for first strand cDNA synthesis with Superscript III reverse transcription (Invitrogen) according to the manufacturer’s instructions. 25% of the first strand reaction was then used in the PCR reaction mixture, with 25 µl total volume, containing 1.5 mM MgCl2, 0.2 mM dNTPs, 0.8 µM of each forward and reverse primer, and 1 U Taq DNA polymerase (Invitrogen). 30 cycles of PCR were used after denaturation at 95°C for 2 min. Cycles consisted of 95°C for 1.5 min, annealing at 50°C for 2 min, and elongation at 72°C for 1 min, followed by one final extension at 72°C for 10 min. The primer pairs used were as follows: Nek5 forward, CTTAAACATCCTTTTGAGGGTAAC; and reverse, TACTCAGCAGGCCATTGACTTGGG; GAPDH forward, CGGAGTCAACGGATTTGGTCGTAT; and reverse, ATGGACTGTGGTCATGAGTCCTTC; and actin forward, TGATGGTGGGCATGGGTCAG; and reverse, CATTTGCGGTGGACGATGGAG. PCR products were analyzed by agarose gel electrophoresis.
Statistical analysis
All quantitative data represent mean and SD of at least three independent experiments in which at least 100–200 cells were quantified. For protein intensity measurements, the intensity of each protein was normalized to its intensity in control-treated cells. 40 centrosomes were quantified for each protein for each condition in three independent experiments. Statistical analyses were performed using a one-tailed unpaired Student’s t test assuming unequal variance. P-values are defined as: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For box and whisker plots, boxes represent the 25th and 75th percentile, and whiskers the 10th and 90th percentile of centrosome distances measured in 50 cells. For dot plots, each dot represents a single cell with data collected over two independent experiments, and the bar represents mean ± SD.
Online supplemental material
Fig. S1 shows Western blot, IF, RT-PCR, and flow cytometry data that support conclusions drawn from the figures in the manuscript. Videos 1–6 show time-lapse imaging movies of GL2 and Nek5-depleted HeLa cells expressing GFP–α-tubulin as they progress into mitosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412099/DC1.
Supplementary Material
Acknowledgments
We acknowledge support of the University of Leicester Core Biotechnology Services for access to imaging facilities, plasmid construction, and DNA sequencing.
This work was supported by the Wellcome Trust (082828; to A.M. Fry), the Biotechnology and Biological Sciences Research Council (A.M. Fry), a Science Foundation Ireland Principal Investigator award 10/IN.1/B2972 (to C.G. Morrison), Canadian Institutes of Health Research (MOP-123468; to L. Pelletier), and the Krembil Foundation (to L. Pelletier).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- IF
- immunofluorescence microscopy
- MT
- microtubule
- MTOC
- MT organizing center
References
- Bahe S., Stierhof Y.D., Wilkinson C.J., Leiss F., and Nigg E.A.. 2005. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 171:27–33 10.1083/jcb.200504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Zhang D., Yin H., Zheng L., Bi X., and Yuan L.. 2014. Centlein mediates an interaction between C-Nap1 and Cep68 to maintain centrosome cohesion. J. Cell Sci. 127:1631–1639 10.1242/jcs.139451 [DOI] [PubMed] [Google Scholar]
- Faragher A.J., and Fry A.M.. 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell. 14:2876–2889 10.1091/mbc.E03-02-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., and Nigg E.A.. 1998a. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Meraldi P., and Nigg E.A.. 1998b. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17:470–481 10.1093/emboj/17.2.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., O’Regan L., Sabir S.R., and Bayliss R.. 2012. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125:4423–4433 10.1242/jcs.111195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., and Nigg E.A.. 2007. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120:4321–4331 10.1242/jcs.020248 [DOI] [PubMed] [Google Scholar]
- Hardy T., Lee M., Hames R.S., Prosser S.L., Cheary D.M., Samant M.D., Schultz F., Baxter J.E., Rhee K., and Fry A.M.. 2014. Multisite phosphorylation of C-Nap1 releases it from Cep135 to trigger centrosome disjunction. J. Cell Sci. 127:2493–2506 10.1242/jcs.142331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Huang N., Bao Y., Zhou H., Teng J., and Chen J.. 2013. LRRC45 is a centrosome linker component required for centrosome cohesion. Cell Reports. 4:1100–1107 10.1016/j.celrep.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Kaseda K., McAinsh A.D., and Cross R.A.. 2012. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol. Open. 1:12–18 10.1242/bio.2011012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin B.R., and Schiebel E.. 2012. Breaking the ties that bind: new advances in centrosome biology. J. Cell Biol. 197:11–18 10.1083/jcb.201108006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T., Stierhof Y.D., Tanaka K., Fry A.M., and Nigg E.A.. 2000. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 151:837–846 10.1083/jcb.151.4.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mchedlishvili N., Wieser S., Holtackers R., Mouysset J., Belwal M., Amaro A.C., and Meraldi P.. 2012. Kinetochores accelerate centrosome separation to ensure faithful chromosome segregation. J. Cell Sci. 125:906–918 10.1242/jcs.091967 [DOI] [PubMed] [Google Scholar]
- Moniz L., Dutt P., Haider N., and Stambolic V.. 2011. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 6:18 10.1186/1747-1028-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.J., Krien M.J., and Hunter T.. 2003. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 13:221–228 10.1016/S0962-8924(03)00056-4 [DOI] [PubMed] [Google Scholar]
- O’Regan L., Blot J., and Fry A.M.. 2007. Mitotic regulation by NIMA-related kinases. Cell Div. 2:25 10.1186/1747-1028-2-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan J.K., Marzio A., Jones M.J., Saraf A., Jallepalli P.V., Florens L., Washburn M.P., and Pagano M.. 2015. Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat. Cell Biol. 17:31–43 10.1038/ncb3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby L.M., and Mahjoub M.R.. 2005. Caught Nek-ing: cilia and centrioles. J. Cell Sci. 118:5161–5169 10.1242/jcs.02681 [DOI] [PubMed] [Google Scholar]
- Shimizu K., and Sawasaki T.. 2013. Nek5, a novel substrate for caspase-3, promotes skeletal muscle differentiation by up-regulating caspase activity. FEBS Lett. 587:2219–2225 10.1016/j.febslet.2013.05.049 [DOI] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Paul R., Mogilner A., and Cimini D.. 2012. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell. 23:401–411 10.1091/mbc.E11-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro D., Takahama S., Shimizu K., Hayashi S., Endo Y., and Sawasaki T.. 2010. Characterization of a caspase-3-substrate kinome using an N- and C-terminally tagged protein kinase library produced by a cell-free system. Cell Death Dis. 1:e89 10.1038/cddis.2010.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A., Winter J.R., Garrod A.J., Amaro A.C., Meraldi P., and McAinsh A.D.. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184:365–372 10.1083/jcb.200809055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Adamian M., and Li T.. 2006. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol. Biol. Cell. 17:1033–1040 10.1091/mbc.E05-10-0943 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.