Abstract
OBJECTIVE
A subarachnoid hemorrhage is neurologically devastating, with 50% of patients becoming disabled or deceased. Advent of Guglielmi detachable coils in 1995 permitted endovascular treatment of cerebral aneurysms. Coiling is efficacious and safe, but durability needs improvement, as nearly 20% of patients require further invasive intervention secondary to aneurysm recurrence. The aim of this study is to develop an in vitro model of endothelial cell (EC) proliferation and differentiation on four types of platinum-based coils, using gene expression profiling to understand EC biology as they colonize and differentiate on coils.
METHODS
Human umbilical vein ECs were grown in vitro on platinum coil segments. Growth patterns were assessed as a function of coil type. Gene expression profiles for coil attached versus coil unattached ECs were determined using immunohistochemistry and gene array analysis.
RESULTS
ECs showed rapid, robust attachment to all coil types. Some detachment occurred within 24–48 hours. Significant growth of remaining attached cells occurred during the next week, creating a confluence on coils and within coil grooves. Similar growth curve results were obtained with human brain ECs on platinum-based coil surfaces. Differentiation markers in attached cells (α1, α2, β1 integrins) were expressed on immunostaining, whereas microarray gene expression revealed 48 up-regulated and 68 down-regulated genes after 24-hour growth on coils. Major pathways affected as a function of time of colonization on coils and coil type included those involved in regulation of cell cycle and cell signaling.
CONCLUSIONS
We developed an in vitro model for evaluating endothelialization of platinum coils to optimize coil design to support robust EC colonization and differentiation.
Keywords: Cerebral aneurysms, Coils, Endothelial cells, Subarachnoid hemorrhage, Tissue engineering
INTRODUCTION
Thirty-five thousand patients suffer from aneurysmal subarachnoid hemorrhage yearly within the United States. The International Subarachnoid Aneurysm Trial (ISAT) reported a lower probability of death and disability for patients undergoing coiling of cerebral aneurysms as opposed to micro-surgery (15). Although ISAT built the platform for launching endovascular treatment of cerebral aneurysms as a mainstay therapy, the durability of this treatment method remains inferior to that of microsurgery. Based on follow-up cerebral angiography, 10%–30% of all coiled aneurysms will show recurrence (1, 18). Recurrence of coiled aneurysms leads to increased morbidity secondary to retreatments, angiographic follow-up, and the real risk of rerupture.
The goal of endovascular occlusion of aneurysms is to introduce platinum-based coils into the cerebral aneurysm until the entire aneurysm volume is filled with thrombus and coils. Volumetric analysis shows that most aneurysms are only filled 20%–40% with coils, and the rest represents acute thrombus (1). Histopathologic analyses based on autopsy of patients who harbored coiled aneurysms show that within the first 4 weeks, the aneurysm houses organized thrombus, which then develops into minimal fibrous tissue. Endothelial proliferation seems to occur at 3 months after embolization (3, 4, 21). At 12 months after the procedure, coils seem to be embedded in fibrous tissue with endothelialization occurring over the neck of the aneurysm (3, 4, 21). Recurrence takes place when the endothelialization process does not occur across the neck of the aneurysm and thus, the pulsatility is transmitted to the coil/thrombus mass leading to coil compaction as the thrombus dissolves. Our goal is to understand the interaction of platinum coils with endothelial cells (ECs), with the future aim of creating an intra-aneurysmal environment for EC proliferation across the neck of the aneurysm.
MATERIALS AND METHODS
Endovascular Coils
To facilitate growth curves, we purchased the following coils: Guglielmi detachable coils (GDCs) and Matrix coils from Boston Scientific, Natick, Massachusetts, USA; Cerecyte coils from Micrus Endovascular, San Jose, California, USA; and HydroCoils from MicroVention, Tustin, California, USA. Coil fragments measured 1 cm (each was weighed using an analytical balance), and were used for cellular attachment, growth analysis, immunohistochemistry, and gene array analysis.
Human Umbilical Vein EC Isolation
Umbilical cords were obtained from the Labor and Delivery Department, Thomas Jefferson University Hospital, Philadelphia, Pennsylvania, USA. Human umbilical vein ECs (HUVEC) were isolated from the cords using a collagenase digestion protocol, as described previously (9).
Cell Culture
HUVEC were cultured on tissue culture flasks coated with 0.2% gelatin. Cells were fed a complete media consisting of Media 199 (GIBCO, Carlsbad, California, USA), 10% fetal bovine serum (FBS; HyClone, Logan, Utah, USA), 50 μg/mL EC growth supplement, 50 μg/mL heparin sodium salt from porcine intestinal mucosa (Grade I-A; Sigma-Aldrich, St. Louis, Missouri, USA), 1% penicillin-streptomycin (GIBCO), and 0.1% Fungizone (GIBCO). EC growth supplement was isolated from bovine hypothalami, as described previously (12). HUVECs were used through passage 6. Similar growth curve experiments were also performed with human cerebral microvascular ECs (hCMEC/D3) at less than 10 passages (27). These cells were cultured for 2 weeks to reach confluency, at which time they were trypsinized and diluted to 1,000,000 cells/mL for experimentation. Similar cellular media, warming tray, and techniques were used for the growth curve assessment.
EC Seeding of Endovascular Coils
EC were trypsinized from confluent cultures and 1.0 mL of a 1 × 106 cells/mL suspension was placed in a sterile Eppendorf tube. One coil segment (~1 cm) was added to each tube and the weight measured. Samples were then placed in a tissue culture incubator on a rocking platform for 4–6 hours, and were gently rotated to ensure cell contact with the coil. Coils were then removed from the cell suspension and rinsed by dipping them once into a well of a 12-well tissue culture plate containing Hanks balanced salt solution at 2 mL/well. Coils were then placed into complete media in a 12-well tissue culture plate and returned to the tissue culture incubator. Culture medium was replaced every 2 days thereafter.
Measurement of EC Growth on Coils
At various times during culture (typically at days 1, 3, 7, 14, and 28), cells were removed from the coils by trypsinization and counted. Coils were placed in 0.5 mL trypsin/coil for 5–10 minutes; the resultant cell suspension was mixed 1:1 with complete media to neutralize the trypsin. Cells released from the coils were counted using a hemacytometer, and expressed as cells bound/milligram of coil. Micrographs of cell-populated coils were also taken before and after trypsinization using inverted microscopy.
Immunohistochemical Staining of EC-populated Coils
After ECs proliferated on GDCs for 7 days, coil segments and unattached cells that had become confluent within the wells were fixed using 10% buffered formalin. These two populations (attached and unattached cells) were incubated with primary anti-bodies to α1 integrin, α2 integrin, β1 integrin, or platelet EC adhesion molecule overnight at 4°C. Samples were labeled with fluorescein isothiocyanate-conjugated secondary antibodies and imaged using fluorescent microscopy.
Examination of EC Gene Expression by Gene Array Analysis
Total RNA Isolation
DNA-free total RNA of cultured cells was isolated using the RNeasy Micro Kit (Qiagen, Valencia, California, USA) according to manufacturer’s instructions. In brief, 1 × 105 cells from duplicate cultures (control and experimental) were pelleted, lysed in RNA lysis tissue buffer containing 1% (vol/vol) β-mercaptoethanol. DNase-treated RNA was ethanol precipitated and quantified on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA), followed by RNA quality assessment by analysis on an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, California, USA).
Microarray Methods
Ribo-single-primer-isothermal-amplification-based RNA amplifications and target preparations were performed according to the manufacturer’s instructions (Ovation Biotin System; NuGEN Technologies, San Carlos, California, USA). Briefly, first-strand complementary DNA (cDNA) was synthesized from 50 ng of total RNA using reverse transcription with a unique oligo (dT)/RNA chimeric primer. In the second step, DNA/RNA heteroduplex double-strand cDNA was generated with DNA polymerase. In the third step, SPIA linear isothermal DNA amplification process was performed using DNA/RNA chimeric primer, DNA polymerase, and RNase H in a homogenous isothermal assay that provides efficient amplification of DNA sequence.
Fragmentation and Biotin Labeling
In the first step, DNA amplification products were fragmented by chemical and enzymatic fragmentation that yields single-stranded cDNA products in the 50- to 100-base range. In the second step, fragmented product is labeled by enzymatic attachment of a biotin-labeled nucleotide to the 3-hydroxyl end of the fragmented cDNA.
Hybridization and Bioinformatic Analysis of Messenger RNA Expression Profiling
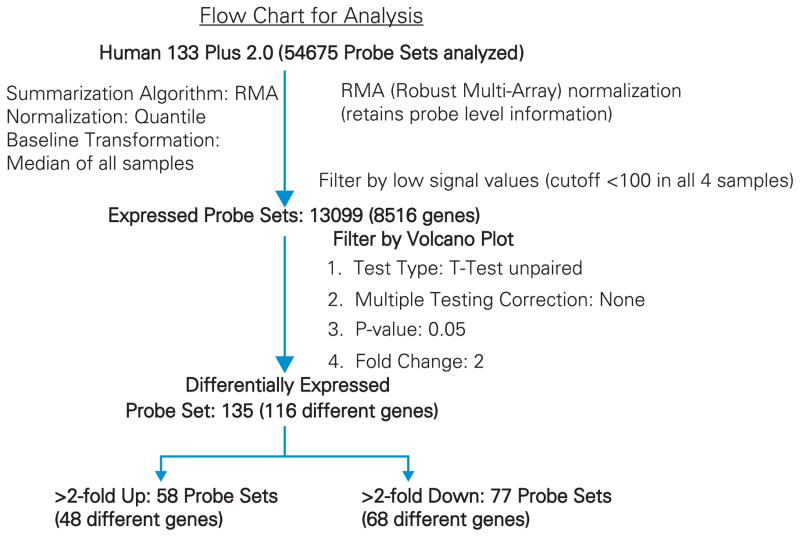
Fragmented and biotin-labeled target (3.75 μg) in 200 μL of hybridization cocktail was used for each Affymetrix HG U133 Plus 2.0 array (Affymetrix, Santa Clara, California, USA), which contains 56,000 probe sets representing 34,000 well-characterized human genes. Target denaturation was done at 99°C for 2 minutes, and hybridization was performed for 18 hours. Arrays were washed and stained using GeneChip Fluidic Station 450, and hybridization signals were amplified using antibody amplification with goat IgG (Sigma-Aldrich) and anti-streptavidin biotinylated antibody (Vector Laboratories, Burlin-game, California, USA). Chips were scanned on an Affymetrix GeneChip Scanner 3000 using GeneChip Operating Software version 3.0. A flow-chart analysis is shown in Figure 1. Normalization was accomplished using robust multi-array average to arrive at baseline transformation for the median of all samples using GeneSpring GX version 10.0 software (Agilent). The gene list was filtered by removing low expressers (<100 signal value in all four samples). A volcano plot was used to identify differentially expressed genes (DEGs) using unpaired t-test with no multiple testing correction (condition, P = 0.05, and twofold change).
Figure 1.
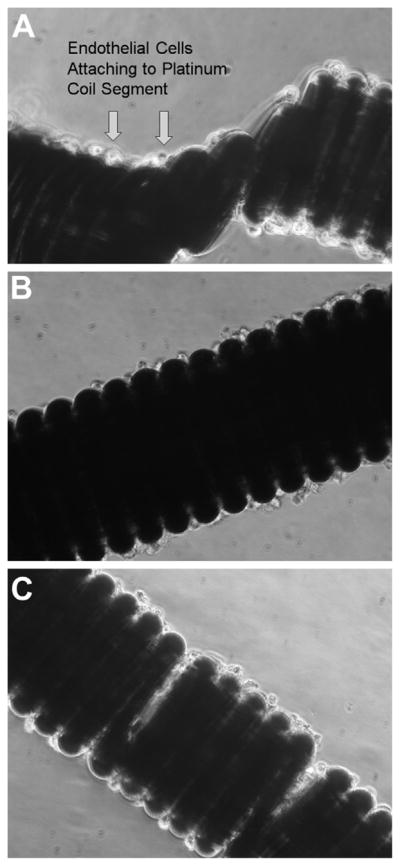
Phase contrast photomicrographs showing endothelial cells attached to coil at day 1 (A), day 4 (B), and day 7 (C).
Gene Annotation
Expressed and DEG lists were linked to the genome database NetAffx at the NetAffx Analysis Center (http://www.affymetrix.com) using Microsoft Excel and Microsoft Access. Gene ontology functions and pathway analysis were assigned using database for Annotation, Visualization, and Integrated Discovery (DAVID version 2.0; http://david.abcc.ncifcrf.gov/). The DEG list was used to perform biological network and functional analysis using Ingenuity Pathway Analysis (IPA) software version 5.0 (Ingenuity Systems Inc., Redwood City, California, USA).
RESULTS
EC Attachment and Proliferation on Platinum-Based Coils
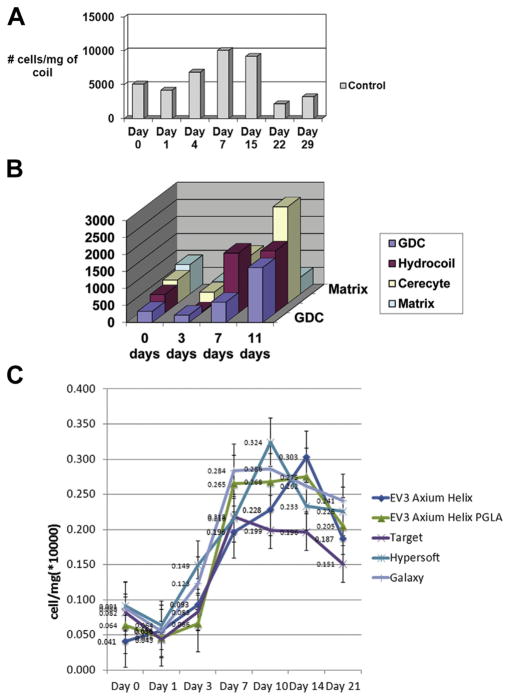
Upon incubation of ECs with platinum coil segments, anywhere from 2000 to 12,000 cells attached for every milligram of coil segment. The cells appeared to attach as clusters, and during the next 24 hours most cells within the clusters appeared to detach as aggregates. Within the first 24 hours, there was an approximate 20% loss of cells, but during the next 96 hours there was an approximate 60% increase in the number of cells associated with the coils, confirming EC proliferation on platinum coils (Figure 1A, B). This increase in cell numbers continued until days 10–15 when there were 7000–10,000 cells per milligram of coil (Figure 1C). After this time and during the next week, there was a reduction in the number of coil-bound cells (Figure 2A). The same phenomenon was observed for human brain EC growth on platinum-based coil surfaces (Figure 2C).
Figure 2.
Bar graphs illustrating human umbilical vein endothelial cell growth on platinum (A) and bioactive (B) coils. Human brain endothelial cell growth on platinum-based coil surfaces per mg of coil (C). GDC, Guglielmi detachable coil; PGLA, polyglycolic acid.
Cellular Attachment and Proliferation on Coated Coils
GDCs, HydroCoils, Matrix coils, and Cerecyte coils all had similar numbers of ECs attaching during the incubation period (300–850 cells/mg of coil). Within the first 72 hours, 200–400 ECs/mg of coil segment remained. During the next 4 days there was marked cell proliferation on the Cerecyte coils (~1400 cells/mg coil) and HydroCoils (~1750 cells/mg coil); however, cell numbers on the GDCs (600 cells/mg coil) and Matrix coils (100 cells/mg coil) were constant with no evidence of proliferation (Figure 2B). During the next week, cellular proliferation occurred on the Cerecyte coils (~2850 cells/mg coil) and the GDCs (~1600 cells/mg coil), remained stable on the HydroCoils (~1800 cells/mg coil), and showed no evidence of proliferation on the Matrix coil (~500 cells/mg coil).
Immunohistochemistry
Figure 1 shows EC attachment to coil segments, and evidence of cellular proliferation and their colonization within coil crevices. Immunohistochemical staining for EC adhesion/signaling receptors α1 integrin, α2 integrin, β1 integrin, and platelet EC adhesion molecule showed diffuse staining of cells when attached to coil segments, or when grown on tissue culture plastic in the absence of coils (Figure 3).
Figure 3.
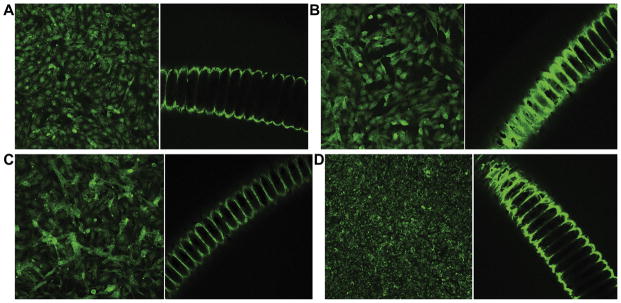
Control (left) and experimental (right) immunohistochemical staining for α1 integrin (A), α2 integrin (B), β1 integrin (C), and platelet endothelial cell adhesion molecule (D) showing endothelial cell localization on coils. Control images represent staining of cells attached to the surface of the wells rather than the coil segments.
Microarray Analyses of ECs Grown in Culture with or without a Coil
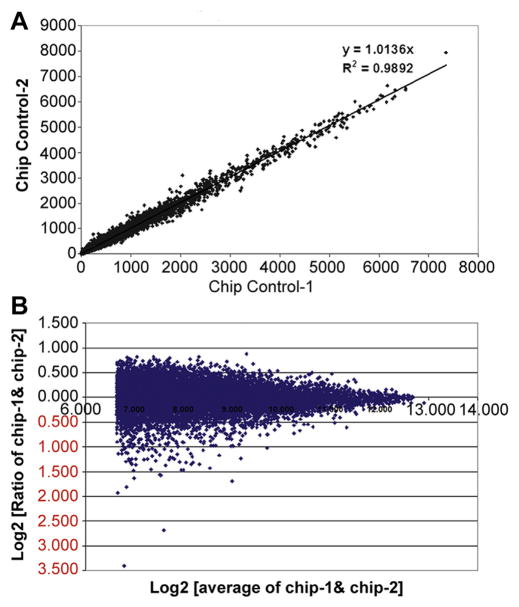
Transcriptome analyses were performed by genomewide microarray expression profiling to define expressed and DEGs after 24-hour growth of ECs in the presence versus absence of a coil. A flow chart for microarray analysis is shown in Figure 4. Array data reproducibility was documented by comparison of a linear scatter plot of spot intensities at each register on duplicate arrays using RNA derived from two replicate control cultures grown in the absence of a coil (Figure 5; slope =1.0136, R2 =0.9892). When average signal values <100 were excluded, 11,894 probe sets (7929 unique genes) were expressed from duplicate untreated cultures. This represents 21.8% of arrayed probe sets (11,894/54,675) and corresponds to 7929 unique genes. The number of expressed probe sets showed minimal change when comparing control to coil-grown cultures. Many of the highest values were messenger RNAs coding for ribosomal proteins, thymosin beta 10, β-actin, eukaryotic translation elongation factor 1 α1, ferritin light chain, cytochrome c oxidase subunit II, tubulin, α1B, and annexin A2.
Figure 4.
Flow chart analysis of gene expression.
Figure 5.
Scatter plots used to define array reproducibility. (A) Gene expression intensities from microarrays using duplicate control cultures (self versus self-experiment). (B) Log scatter plot (control self-experiment). Signal values are graphed from all probe sets on the two arrays, each hybridized with one of two duplicate untreated sample targets.
Identification of DEGs after 24-Hour Growth of ECs
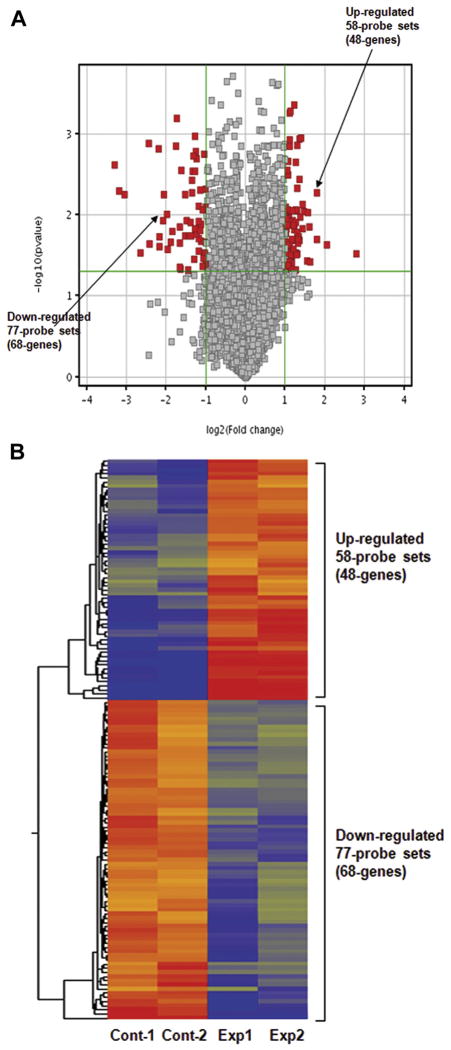
Using signal intensities >100 in at least one of the four samples and ≥2-fold up or down as the criterion for definition of a DEG, we next performed a volcano plot analysis to compare the two replicate controls against the two cultures grown on the coil (Figure 6, left). Such a plot defines fold change (x-axis) as a function of statistical significance (y-axis). This analysis identified 135 probe sets at a P value of 0.05 (red dots), which were differentially expressed as a result of EC growth on the coil. This included 58 probe sets (48 genes) that were up-regulated and 77 (68 genes) that were down-regulated in response to growth on the coil (Tables 1 and 2). A heat map shows up-regulated and down-regulated genes for duplicate cultures (Figure 6, right). Seventy-one of 77 down-regulated probe sets belong to functional categories. Functional categories include phosphoprotein metabolism [42], cell division [12], cell cycle [15], mitosis [10], DNA replication [4], cell cycle control [3], and metal binding [2]. Kyoto Encyclopedia of Genes and Genomes pathway analysis shows “cell cycle” as the major affected pathway containing four different genes (cyclin A1, cell division control protein 6 homolog, cell division cycle 25C splice variant 3, and extra spindle poles-like 1 protein) designated as down-regulated in ECs in response to growth on the coil.
Figure 6.
(A) Volcano plot showing differentially expressed genes (135 differentially expressed probe set; 116 genes). Relationship between fold-change (>2; magnitude of change; x-axis) and statistical significance (P = 0. 05; y-axis) is shown. (B) Heat map from microarray data reflecting gene expression comparing four different samples: two controls and two coil-treated cultures.
Table 1.
Down-Regulated Gene List
| Probe Set ID | Gene Symbol | Gene Title | Fold Change | Control | Experimental |
|---|---|---|---|---|---|
| 239331_at | Transcribed locus | 0.151 | 139 | 21 | |
| 232174_at | CDNA: FLJ21635 fis, clone COL08233, highly similar to AF131819 Homo sapiens clone 24838 mRNA sequence | 0.245 | 323 | 79 | |
| 222040_at | HNRNPA1///LOC728844 | Heterogeneous nuclear ribonucleoprotein A1///hypothetical LOC728844 | 0.288 | 125 | 36 |
| 230127_at | Transcribed locus | 0.293 | 167 | 49 | |
| 227165_at | C13orf3 | Chromosome 13 open reading frame 3 | 0.331 | 118 | 39 |
| 209990_s_at | GABBR2 | Gamma-aminobutyric acid (GABA) B receptor, 2 | 0.331 | 136 | 45 |
| 215100_at | C6orf105 | Chromosome 6 open reading frame 105 | 0.339 | 112 | 38 |
| 207526_s_at | IL1RL1 | Interleukin 1 receptor-like 1 | 0.341 | 464 | 158 |
| 204338_s_at | RGS4 | Regulator of G-protein signaling 4 | 0.361 | 108 | 39 |
| 221521_s_at | GINS2 | GINS complex subunit 2 (Psf2 homolog) | 0.370 | 211 | 78 |
| 205899_at | CCNA1 | Cyclin A1 | 0.371 | 510 | 189 |
| 205681_at | BCL2A1 | BCL2-related protein A1 | 0.378 | 172 | 65 |
| 204817_at | ESPL1 | Extra spindle pole bodies homolog 1 (S. cerevisiae) | 0.380 | 92 | 35 |
| 201506_at | TGFBI | Transforming growth factor, beta-induced, 68 kDa | 0.391 | 128 | 50 |
| 205576_at | SERPIND1 | Serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | 0.391 | 391 | 153 |
| 235425_at | SGOL2 | Shugoshin-like 2 (S. pombe) | 0.392 | 158 | 62 |
| 234066_at | IL1RL1 | Interleukin 1 receptor-like 1 | 0.395 | 233 | 92 |
| 226695_at | PRRX1 | Paired related homeobox 1 | 0.403 | 124 | 50 |
| 235236_at | Transcribed locus | 0.404 | 171 | 69 | |
| 207601_at | SULT1B1 | Sulfotransferase family, cytosolic, 1B, member 1 | 0.404 | 314 | 127 |
| 225239_at | CDNA FLJ26120 fis, clone SYN00419 | 0.407 | 280 | 114 | |
| 205046_at | CENPE | Centromere protein E, 312 kDa | 0.407 | 194 | 79 |
| 231403_at | TRIO | Triple functional domain (PTPRF interacting) | 0.407 | 108 | 44 |
| 220940_at | ANKRD36B | Ankyrin repeat domain 36B | 0.408 | 179 | 73 |
| 217165_x_at | MT1F | Metallothionein 1F | 0.408 | 218 | 89 |
| 220651_s_at | MCM10 | Minichromosome maintenance complex component 10 | 0.412 | 102 | 42 |
| 229610_at | CKAP2L | Cytoskeleton-associated protein 2-like | 0.415 | 224 | 93 |
| 204823_at | NAV3 | Neuron navigator 3 | 0.415 | 426 | 177 |
| 226419_s_at | FLJ44342 | Hypothetical LOC645460 | 0.416 | 377 | 157 |
| 204602_at | DKK1 | Dickkopf homolog 1 (Xenopus laevis ) | 0.422 | 446 | 188 |
| 213684_s_at | PDLIM5 | PDZ and LIM domain 5 | 0.424 | 125 | 53 |
| 212020_s_at | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 0.426 | 141 | 60 |
| 201195_s_at | SLC7A5 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 0.429 | 392 | 168 |
| 230082_at | LOC100133660 | Hypothetical LOC100133660 | 0.432 | 95 | 41 |
| 221436_s_at | CDCA3 | Cell division cycle associated 3 | 0.434 | 143 | 62 |
| 221685_s_at | CCDC99 | Coiled–coil domain containing 99 | 0.435 | 308 | 134 |
| 218009_s_at | PRC1 | Protein regulator of cytokinesis 1 | 0.437 | 421 | 184 |
| 242809_at | IL1RL1 | Interleukin 1 receptor-like 1 | 0.439 | 611 | 268 |
| 218115_at | ASF1B | ASF1 anti-silencing function 1 homolog B (S. cerevisiae) | 0.444 | 169 | 75 |
| 212021_s_at | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 0.447 | 132 | 59 |
| 220865_s_at | PDSS1 | Prenyl (decaprenyl) diphosphate synthase, subunit 1 | 0.448 | 125 | 56 |
| 203968_s_at | CDC6 | Cell division cycle 6 homolog (S. cerevisiae) | 0.451 | 122 | 55 |
| 219148_at | PBK | PDZ binding kinase | 0.455 | 321 | 146 |
| 212023_s_at | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 0.455 | 134 | 61 |
| 203108_at | GPRC5A | G protein-coupled receptor, family C, group 5, member A | 0.459 | 196 | 90 |
| 204326_x_at | MT1X | Metallothionein 1X | 0.461 | 206 | 95 |
| 210052_s_at | TPX2 | TPX2, microtubule-associated, homolog (Xenopus laevis) | 0.464 | 466 | 216 |
| 219557_s_at | NRIP3 | Nuclear receptor interacting protein 3 | 0.466 | 131 | 61 |
| 205167_s_at | CDC25C | Cell division cycle 25 homolog C (S. pombe) | 0.467 | 120 | 56 |
| 218663_at | NCAPG | Non-SMC condensin I complex, subunit G | 0.467 | 345 | 161 |
| 203622_s_at | PNO1 | Partner of NOB1 homolog (S. cerevisiae) | 0.470 | 117 | 55 |
| 218355_at | KIF4A | Kinesin family member 4A | 0.473 | 510 | 241 |
| 206343_s_at | NRG1 | Neuregulin 1 | 0.475 | 101 | 48 |
| 239202_at | CDNA FLJ34848 fis, clone NT2NE2011684, weakly similar to H. sapiens mRNA for plakophilin 2a and b | 0.477 | 239 | 114 | |
| 206224_at | CST1 | Cystatin SN | 0.477 | 1404 | 670 |
| 202016_at | MEST | Mesoderm specific transcript homolog (mouse) | 0.477 | 220 | 105 |
| 222958_s_at | DEPDC1 | DEP domain containing 1 | 0.478 | 301 | 144 |
| 239973_at | Transcribed locus | 0.479 | 119 | 57 | |
| 208600_s_at | GPR39 | G protein-coupled receptor 39 | 0.480 | 667 | 320 |
| 226210_s_at | MEG3 | Maternally expressed 3 | 0.480 | 175 | 84 |
| 227211_at | PHF19 | PHD finger protein 19 | 0.483 | 236 | 114 |
| 219918_s_at | ASPM | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) | 0.484 | 411 | 199 |
| 219493_at | SHCBP1 | SHC SH2-domain binding protein 1 | 0.486 | 144 | 70 |
| 213007_at | FANCI | Fanconi anemia, complementation group I | 0.487 | 388 | 189 |
| 229070_at | C6orf105 | Chromosome 6 open reading frame 105 | 0.487 | 956 | 466 |
| 203805_s_at | FANCA | Fanconi anemia, complementation group A | 0.489 | 94 | 46 |
| 203358_s_at | EZH2 | Enhancer of zeste homolog 2 (Drosophila) | 0.491 | 330 | 162 |
| 223381_at | NUF2 | NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) | 0.491 | 234 | 115 |
| 203438_at | STC2 | Stanniocalcin 2 | 0.495 | 186 | 92 |
| 209714_s_at | CDKN3 | Cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 0.497 | 290 | 144 |
| 212022_s_at | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 0.499 | 517 | 258 |
| 202533_s_at | DHFR | Dihydrofolate reductase | 0.500 | 134 | 67 |
| 203755_at | BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 0.500 | 416 | 208 |
| 206653_at | POLR3G | Polymerase (RNA) III (DNA directed) polypeptide G (32kD) | 0.500 | 326 | 163 |
| 221520_s_at | CDCA8 | Cell division cycle associated 8 | 0.500 | 170 | 85 |
| 205909_at | POLE2 | Polymerase (DNA directed), epsilon 2 (p59 subunit) | 0.502 | 231 | 116 |
| 216250_s_at | LPXN | Leupaxin | 0.503 | 163 | 82 |
Table 2.
Up-Regulated Gene List
| Probe Set ID | Gene Symbol | Gene Title | Fold Change | Control | Experimental |
|---|---|---|---|---|---|
| 227235_at | CDNA clone IMAGE:5302158 | 9.887 | 11 | 113 | |
| 222106_at | PRND | Pprion protein 2 (dublet) | 9.071 | 39 | 356 |
| 202350_s_at | MATN2 | Matrilin 2 | 8.284 | 18 | 149 |
| 209392_at | ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 6.284 | 12 | 80 |
| 214180_at | MAN1C1 | Mannosidase, alpha, class 1C, member 1 | 5.427 | 27 | 144 |
| 217757_at | A2M | Alpha-2-macroglobulin | 5.347 | 31 | 161 |
| 212344_at | SULF1 | Sulfatase 1 | 4.597 | 60 | 274 |
| 201852_x_at | COL3A1 | Collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) | 4.504 | 46 | 203 |
| 215076_s_at | COL3A1 | Collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) | 4.498 | 45 | 197 |
| 201150_s_at | TIMP3 | TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 4.227 | 43 | 182 |
| 209047_at | AQP1 | Aquaporin 1 (Colton blood group) | 4.158 | 84 | 346 |
| 201009_s_at | TXNIP | Thioredoxin interacting protein | 3.936 | 54 | 211 |
| 204904_at | GJA4 | Gap junction protein, alpha 4, 37kDa | 3.92 | 36 | 137 |
| 203423_at | RBP1 | Retinol binding protein 1, cellular | 3.696 | 75 | 274 |
| 211161_s_at | COL3A1 | Collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) | 3.605 | 159 | 582 |
| 201148_s_at | TIMP3 | TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 3.387 | 55 | 187 |
| 209543_s_at | CD34 | CD34 molecule | 3.339 | 103 | 343 |
| 40687_at | GJA4 | Gap junction protein, alpha 4, 37 kDa | 3.18 | 42 | 133 |
| 206638_at | HTR2B | 5-hydroxytryptamine (serotonin) receptor 2B | 3.177 | 104 | 330 |
| 212353_at | SULF1 | Sulfatase 1 | 3.177 | 177 | 566 |
| 201262_s_at | BGN | Biglycan | 3.125 | 114 | 345 |
| 209583_s_at | CD200 | CD200 molecule | 3.067 | 33 | 101 |
| 228770_at | GPR146 | G protein-coupled receptor 146 | 2.893 | 75 | 216 |
| 201010_s_at | TXNIP | Thioredoxin interacting protein | 2.792 | 160 | 447 |
| 205019_s_at | VIPR1 | Vasoactive intestinal peptide receptor 1 | 2.722 | 36 | 96 |
| 202688_at | TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | 2.693 | 197 | 535 |
| 209182_s_at | C10orf10 | Chromosome 10 open reading frame 10 | 2.657 | 189 | 499 |
| 209894_at | LEPR | Leptin receptor | 2.632 | 48 | 127 |
| 227243_s_at | EBF3 | Early B-cell factor 3 | 2.579 | 41 | 105 |
| 226237_at | mRNA full length insert cDNA clone EUROIMAGE 1913076 | 2.562 | 139 | 358 | |
| 205779_at | RAMP2 | Receptor (G protein-coupled) activity modifying protein 2 | 2.546 | 52 | 133 |
| 217897_at | FXYD6 | FXYD domain containing ion transport regulator 6 | 2.529 | 93 | 235 |
| 238673_at | SAMD12 | Sterile alpha motif domain containing 12 | 2.494 | 61 | 151 |
| 226621_at | Transcribed locus | 2.458 | 124 | 308 | |
| 216598_s_at | CCL2 | Chemokine (C-C motif) ligand 2 | 2.433 | 545 | 1326 |
| 214329_x_at | TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | 2.422 | 63 | 151 |
| 219304_s_at | PDGFD | Platelet derived growth factor D | 2.421 | 45 | 108 |
| 202838_at | FUCA1 | Fucosidase, alpha-L- 1, tissue | 2.391 | 59 | 141 |
| 202291_s_at | MGP | Matrix Gla protein | 2.376 | 543 | 1291 |
| 212354_at | SULF1 | Sulfatase 1 | 2.352 | 547 | 1292 |
| 230740_at | Transcribed locus | 2.284 | 92 | 210 | |
| 202947_s_at | GYPC | Glycophorin C (Gerbich blood group) | 2.278 | 62 | 142 |
| 224818_at | SORT1 | Sortilin 1 | 2.275 | 54 | 122 |
| 227306_at | CDNA: FLJ21245 fis, clone COL01184 | 2.26 | 44 | 99 | |
| 223449_at | SEMA6A | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 2.223 | 145 | 322 |
| 225511_at | GPRC5B | G protein-coupled receptor, family C, group 5, member B | 2.203 | 113 | 249 |
| 203498_at | RCAN2 | Regulator of calcineurin 2 | 2.192 | 85 | 186 |
| 202023_at | EFNA1 | Ephrin-A1 | 2.179 | 184 | 398 |
| 227040_at | NHLRC3 | NHL repeat containing 3 | 2.114 | 43 | 91 |
| 240890_at | LOC643733 | Hypothetical LOC643733 | 2.113 | 370 | 782 |
| 235887_at | Transcribed locus | 2.088 | 44 | 93 | |
| 204358_s_at | FLRT2 | Fibronectin leucine rich transmembrane protein 2 | 2.079 | 140 | 291 |
| 218999_at | TMEM140 | Transmembrane protein 140 | 2.039 | 130 | 265 |
| 212951_at | GPR116 | G protein-coupled receptor 116 | 2.023 | 107 | 217 |
| 213894_at | THSD7A | Thrombospondin, type I, domain containing 7A | 2.021 | 69 | 140 |
| 218966_at | MYO5C | Myosin VC | 2.021 | 259 | 526 |
| 212325_at | LIMCH1 | LIM and calponin homology domains 1 | 2.007 | 56 | 113 |
A functional annotation chart also is given for the 58 up-regulated probe sets demonstrating 50 belong to different functional categories: signal [26], glycoprotein [27], transmembrane [20], membrane [22], secreted [10], extracellular [4], and receptor [9]. KEGG pathway analysis shows “cytokine-cytokine receptor interaction” as the major affected pathway containing four different genes (C-C motif chemokine 2 precursor, oncostatin M receptor, tumor necrosis factor, and leptin receptor) up-regulated in response to growth on the coil.
Biological Network and Functional Analysis
To further refine the functional properties of the DEGs, all 134 genes were analyzed in the IPA tool for network, functional, and pathway analysis. IPA converts a list of genes with accompanying expression information into a set of relevant networks based on the Ingenuity Knowledge Base pathways. A core analysis is performed for the list of genes. The genes were categorized based on molecular function in the IPA software. The identified genes also were mapped to molecular networks in the IPA database and ranked by score. The score reflects the probability that a collection of genes equal to or greater than the number in a network could be achieved by chance alone. The top five (high score) networks are listed in Table 3. Network analysis revealed two important networks related to cellular function and cell-to-cell signaling (Figure 7). Two networks have high scores of 40 and 30, with 20 and 16 focus genes, respectively. Several important genes, such as CCNA1, BUB1B, CDC6, CDC25c, A2M, SULF1, TNFSF10, and TXN1P are related in the network. Further molecular and cellular functional classifications are listed in Table 4, showing that 26 genes are related to cell cycle (4 molecules are up-regulated and 22 genes are down-regulated) and 19 genes are related to cell-to-cell signaling interaction function (14 genes are up-regulated and 5 are down-regulated).
Table 3.
Selected Networks with High Scores for Differentially Expressed Genes
| Network | Molecules in Network | Score | Focus Molecules | Top Functions |
|---|---|---|---|---|
| 1 | A2M, Alp, Ap1, AQP1, BCL2A1, BGN, CCL2, CD34, CDKN3, Collagen type I, Collagen(s), EFNA1, FUCA1, IL1, IL1RL1, KIF4A, LDL, MKI67, Mmp, MT1F, NCAPG, NFkB, PBK, Pdgf, PDGF BB, PDGFD, PRC1, PRRX1, RBP1, SERPIND1, Tgf beta, TGFBI, TIMP3, TXNIP, Vegf | 48 | 23 | Connective tissue disorders, infectious disease, inflammatory disease |
| 2 | APC, ASF1B, BUB1B, Caspase, CCNA1, CDC6, CDC25C, CENPE, COL3A1, Cyclin A, DHFR, DKK1, E2f, ERK1/2, EZH2, hCG, Histone h3, Histone h4, HNRNPA1, IgG, Insulin, LEPR, Mapk, MCM10, NGF, NRG1, NUF2, P38 MAPK, PHF19, PNO1, Rb, RGS4, Tnf, TNFSF10, TPX2 | 40 | 20 | Cellular assembly and organization, cell cycle, cell morphology |
| 3 | AMBP, AQP1, ASPM, butyric acid, C10ORF10, CCDC99, CDC14B, DEPDC1, EHD1, EHD3, ENPP2, FANCI, FXYD6, GINS2, HMGN2, HNF4A, IFITM2, KNTC1, L-triiodothyronine, LIMCH1, LZTR1, MEG3, MIR124, PELO, PODXL, POLR3G, RCAN2, SULT1B1, TGFBI, THSD7A, TMEM140, TNF, TP53, WTAP, ZNF224 | 34 | 18 | Gene expression, infection mechanism, cellular development |
| 4 | 26s Proteasome, Akt, CALCRL, CD200, ERK, FANCA, Focal adhesion kinase, FSH, GABBR2, Gpcr, GPR39, GPR108, GPR116, GPR146, GPRC5A, GPRC5B, GPRC6A, HCRTR2, HTR2B, Interferon alpha, Jnk, LPXN, MT1X, NTSR2, OSMR, P2RY12, PI3K, Rac, RAMP2, RAMP3, Ras, STAT, SULF1, TRIO, VIPR1 | 30 | 16 | Cell signaling, nucleic acid metabolism, small molecule biochemistry |
| 5 | ADAM12, ADCY3, ANKRD36B, ARAP1, ASPM, BDKRB1, C5ORF13, Ca2+, CDCA3, CLTC, EBF3, EHD1, ESPL1, GLI1, GYPC, KDM5B, MAN1C1, MEST, MPP1, NAV3, PDLIM5, PLOD1, PRC1, PRND, PTH1R, RTKN2, SEMA6A, SERPINH1, SH3D19, SHCBP1, SLC2A4, SMAD2, TGFB1, TGFBR3, TUFM | 25 | 14 | Cardiovascular system development and function, organismal development, tissue morphology |
Figure 7.
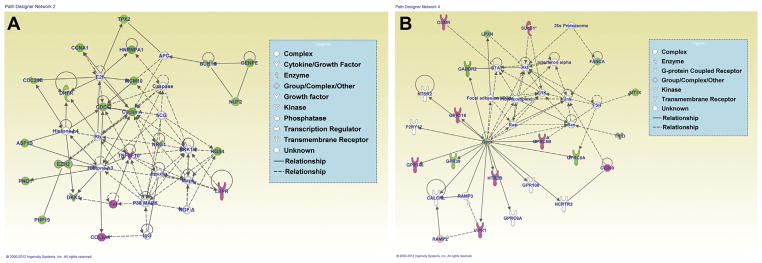
Network analysis using Ingenuity Pathway Analysis (IPA). IPA analysis from differentially expressed genes shows predominant networks affected. (A) Cellular assembly and organization, cell cycle, cell morphology (green indicates down-regulated, pink represents up-regulated genes). (B) Cell signaling, nucleic acid metabolism, small molecule biochemistry.
Table 4.
Molecular and Cellular Functions of Differentially Expressed Genes
| Functions | Number of Molecules |
|---|---|
| Cellular growth and proliferation | 46 |
| Cell cycle | 26 |
| Cellular assembly and organization | 18 |
| Cellular movement | 23 |
| Cell morphology | 13 |
| Cell-to-cell signaling and interaction | 19 |
DISCUSSION
Cerebral aneurysmal rupture leads to significant morbidity and mortality (21). Endovascular occlusion of aneurysms is a safe and efficacious treatment modality as supported by the ISAT; however, recurrence of previously coiled aneurysms occurs in 10%–30% of patients (15, 18). Endothelialization across the neck of an aneurysm is essential in preventing such recurrence and associated complications (5, 6, 22). The role of ECs in aneurysmal healing after coiling is essential, as these cells are involved in control of vascular tone, thrombosis, thrombolysis, and platelet activation regulation (2, 7, 28). To evaluate the interaction of ECs with platinum-based coils, we developed an in vitro model by seeding coils with HUVECs and examining their proliferation and gene expression profiles.
We observed cellular attachment to coil segments within hours of incubation. Most adherent cells then detached from the coils as aggregates. During the next 7–14 days, cellular proliferation continued until a confluent population of ECs was observed on coil segments. Cell populations could also be observed within the coil segment grooves—a potentially ideal location, as the cells were protected from shear stress associated with flow within the wells. Intercellular contact on the coil segment as well as EC differentiation could have led to inhibition of cellular proliferation as well as cellular death; thus, the observed decrease in number of attached cells in the third week after attachment (25).
With the introduction of different coil types to the clinical market, we evaluated HUVEC attachment and growth pattern on these modified coil fragments. GDCs are unmodified platinum coils and have been used as the control in these experiments. Matrix coils have a polyglycolic acid (PGLA) coating on the platinum surface as opposed to Cerecyte coils that have a PGLA coating within the platinum core (8, 11). HydroCoils have a hydrogel coating on the platinum surface, which allows for volumetric enlargement when the coils are hydrated within an in vivo system (14, 16). A similar number of cells attached to each coil type; however, the greatest proliferation occurred on Cerecyte coils, followed by HydroCoils and GDCs. Matrix coil segments supported the least proliferation, which could be secondary to the PGLA coating being in direct contact with the HUVECs. HydroCoil enlargement leads to an increase in surface area; thus, more potential space for cellular proliferation without intercellular inhibition, which could explain the larger number of HUVEC cells colonizing this coil type.
Immunohistochemical analysis with antibody labeling for α1, α2, and β1 integrins revealed similar labeling of cells to the coil segments compared with unattached cells (14). Thus, these represent functional HUVECs, as they do not seem to change their cellular membrane integrity when attaching and proliferating on GDC segments (10, 26). In addition, labeling revealed the confluence of cellular proliferation on the coil segments as well as within the crevices of the coils. We analyzed the gene expression of cells attached to the platinum coils versus those grown on tissue culture plastic in the absence of coils, and found that 48 genes were up-regulated whereas 68 genes were down-regulated. The dominating cellular function of down-regulated genes involved cell cycle regulation, whereas genes up-regulated mostly involved cytokine-to-cytokine interaction. The platinum surface most certainly decreased cellular proliferation compared with cells grown in wells. Thus, down-regulation of cell cycle-associated genes was expected and confirmed by our microarray expression studies. It is feasible that EC differentiation could be occurring sooner on the platinum surfaces, leading to a down-regulation of proliferation-associated genes and decreased growth rates on the platinum-based surfaces. Coil segments could also initiate an inflammatory response from the HUVECs, as cytokine-related genes were differentially up-regulated within the attached cellular population (20).
Ozawa et al. (17) and Tamatani et al. (23, 24) reported that canine ECs require extracellular matrix (ECM; fibronectin, laminin, collagen) coating on embolic material for cellular proliferation. They did not observe EC proliferation on uncoated embolic materials. They concluded that ECs require ECM for attachment, growth, and proliferation. Our in vitro model uses HUVECs and achieves cellular attachment, growth, and proliferation on bare platinum as well as PGLA-coated coils. However, it is possible that culture media-derived growth and differentiation factors (e.g., plasma fibronectin) may adhere to the coil surfaces in our model system, creating an ECM-like surface.
Most investigators have reported that collagen type I represents the most ideal substrate for EC proliferation (13). Although collagen provides the appropriate extracellular support for attachment and proliferation, it is also thrombogenic and thus could pose significant thromboembolic risk in the event that coils are coated with this material. In theory, allowing for cellular proliferation on uncoated material would be ideal in reducing the probability of thrombogenicity, as well as allowing for permanent healing within the aneurysm.
We also evaluated the growth of human brain ECs (hCMEC/D3) on different coil types to understand whether brain ECs would have different growth curves on the platinum surfaces compared with HUVECs (27). The overall growth curve is similar to what we observed for HUVECs, as there is exponential growth during the first 7–10 days after cellular attachment. After confluency was reached in the first 10 days, there was a decrease in cell numbers at 2–3 weeks after attachment secondary to cellular growth inhibition, secondary to a lack of surface area. We were not able to detect a statistically significant difference in the number of cells present on the different coil types.
The in vitro model for EC interaction with coils reported here could be used to identify factors to enhance cellular attachment and proliferation in an in vivo situation. In addition, it could serve as the initial step in tissue engineering, as a mechanism for permanent healing within cerebral aneurysms. Tissue engineering requires cellular harvest, attachment to a matrix, and proliferation. The platinum coil could serve as the scaffold, with the goal of attaching smooth muscle cells to endothelium-coated coils with the aim of introducing functional endothelial and smooth muscle cells into the aneurysm lumen (29). Although these cells were able to attach and proliferate when placed on a rocker causing continuous motion, we do aim to modify our in vitro design to introduce calculated flow rates as would be present in an in vivo situation.
Based on our results, we propose a model for the endothelialization of coils: 1) cells attach loosely to the outermost surface of the coil; 2) loosely adherent cells detach; and 3) strongly adherent cells then migrate or compact within flow-protected area of the coil surface (i.e., in the grooves between coil wires). Our results suggest that coils may have the innate capacity to foster EC growth and interactions. These results could be used for development of coils optimized for EC seeding and growth with their surface profile configured with grooves to facilitate attachment and growth of parallel lines of ECs (as has been done for vascular stents) (19). Increasing the surface area-to-volume ratio, as well as decreasing the distance between metallic surfaces within the coil grooves to 10–20 μm (the diameter of ECs), could optimize cellular migration and proliferation. Optimizing EC colonization of coils with biological as well as mechanical modifications should result in endogenous endothelialization within weeks of coil introduction and thus, prevent recurrence as well as need for long-term anticoagulation.
CONCLUSIONS
EC proliferation across the neck of an aneurysm plays a critical role in the formation of permanent healing and prevention of recurrence in previously coiled aneurysms. Our in vitro model of HUVEC and human brain EC attachment and proliferation on coil segments advances toward the goal of rational design of platinum-based coils to support optimal growth and differentiation of ECs, thus potentially enhancing coil efficacy in endovascular surgeries.
Acknowledgments
The authors wish to thank Kari Habursky for technical assistance.
Abbreviations and Acronyms
- cDNA
Complementary DNA
- DEG
Differentially expressed gene
- EC
Endothelial cell
- ECM
Extracellular matrix
- GDC
Guglielmi detachable coil
- HUVEC
Human umbilical vein endothelial cell
- IPA
Ingenuity Pathway Analysis
- ISAT
International Subarachnoid Aneurysm Trial
- PGLA
Polyglycolic acid
Footnotes
Conflict of interest statement: This work was supported in part by a grant from the National Institutes of Health within the Department of Cancer Biology (SA and PF) at the Kimmel Cancer Center, Thomas Jefferson University, the Cardeza Foundation for Hematologic Research at Jefferson Medical College (SS), the Department of Neurosurgery at Thomas Jefferson Hospital, and Covidien, Mansfield, MA.
References
- 1.Babiker MH, Gonzalez LF, Albuquerque F, Collins D, Elvikis A, Frakes DH. Quantitative effects of coil packing density on cerebral aneurysm fluid dynamics: an in vitro steady flow study. Ann Biomed Eng. 2010;38:2293–2301. doi: 10.1007/s10439-010-9995-4. [DOI] [PubMed] [Google Scholar]
- 2.Barry OP, Pratico D, Lawson JA, FitzGerald GA. Transcellular activation of platelets and endothelial cells by bioactive lipids in platelet microparticles. J Clin Invest. 1997;99:2118–2127. doi: 10.1172/JCI119385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bavinzski G, Richling B, Binder BR, Gruber A, Talazoglu V, Dietrich W, Schwendtenwein I, Plenk H., Jr Histopathological findings in experimental aneurysms embolized with conventional and thrombogenic/antithrombolytic Guglielmi coils. Minim Invasive Neurosurg. 1999;42:167–174. doi: 10.1055/s-2008-1053392. [DOI] [PubMed] [Google Scholar]
- 4.Bavinzski G, Talazoglu V, Killer M, Richling B, Gruber A, Gross CE, Plenk H., Jr Gross and microscopic histopathological findings in aneurysms of the human brain treated with Guglielmi detachable coils. J Neurosurg. 1999;91:284–293. doi: 10.3171/jns.1999.91.2.0284. [DOI] [PubMed] [Google Scholar]
- 5.Cognard C, Weill A, Spelle L, Piotin M, Castaings L, Rey A, Moret J. Long-term angiographic follow-up of 169 intracranial berry aneurysms occluded with detachable coils. Radiology. 1999;212:348–356. doi: 10.1148/radiology.212.2.r99jl47348. [DOI] [PubMed] [Google Scholar]
- 6.Frosen J, Piippo A, Paetau A, Kangasniemi M, Niemela M, Hernesniemi J, Jaaskelainen J. Remodeling of saccular cerebral artery aneurysm wall is associated with rupture: histological analysis of 24 unruptured and 42 ruptured cases. Stroke. 2004;35:2287–2293. doi: 10.1161/01.STR.0000140636.30204.da. [DOI] [PubMed] [Google Scholar]
- 7.Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–376. doi: 10.1038/288373a0. [DOI] [PubMed] [Google Scholar]
- 8.Geyik S, Yavuz K, Ergun O, Koc O, Cekirge S, Saatci I. Endovascular treatment of intracranial aneurysms with bioactive Cerecyte coils: effects on treatment stability. Neuroradiology. 2008;50:787–793. doi: 10.1007/s00234-008-0399-1. [DOI] [PubMed] [Google Scholar]
- 9.Hebert CA, Luscinskas FW, Kiely JM, Luis EA, Darbonne WC, Bennett GL, Liu CC, Obin MS, Gimbrone MA, Jr, Baker JB. Endothelial and leukocyte forms of IL-8. Conversion by thrombin and interactions with neutrophils. J Immunol. 1990;145:3033–3040. [PubMed] [Google Scholar]
- 10.Kubota Y, Kawa Y, Mizoguchi M. CDw49b/CD29 integrin complex mediates the differentiation of human endothelial cells into capillary-like structures in vitro. J Dermatol Sci. 1996;12:36–43. doi: 10.1016/0923-1811(95)00462-9. [DOI] [PubMed] [Google Scholar]
- 11.Linfante I, Akkawi NM, Perlow A, Andreone V, Wakhloo AK. Polyglycolide/polylactide-coated platinum coils for patients with ruptured and unruptured cerebral aneurysms: a single-center experience. Stroke. 2005;36:1948–1953. doi: 10.1161/01.STR.0000177532.94736.85. [DOI] [PubMed] [Google Scholar]
- 12.Maciag T, Cerundolo J, Ilsley S, Kelley PR, Forand R. An endothelial cell growth factor from bovine hypothalamus: identification and partial characterization. Proc Natl Acad Sci U S A. 1979;76:5674–5678. doi: 10.1073/pnas.76.11.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuire PG, Orkin RW. Isolation of rat aortic endothelial cells by primary explant techniques and their phenotypic modulation by defined substrata. Lab Invest. 1987;57:94–105. [PubMed] [Google Scholar]
- 14.Mechtersheimer G, Barth T, Hartschuh W, Lehnert T, Moller P. In situ expression of beta 1, beta 3 and beta 4 integrin subunits in non-neoplastic endothelium and vascular tumours. Virchows Arch. 1994;425:375–384. doi: 10.1007/BF00189575. [DOI] [PubMed] [Google Scholar]
- 15.Molyneux A, Kerr R, Stratton I, Sandercock P, Clarke M, Shrimpton J, Holman R. International Subarachnoid Aneurysm Trial (ISAT) of neurosurgical clipping versus endovascular coiling in 2143 patients with ruptured intracranial aneurysms: a randomised trial. Lancet. 2002;360:1267–1274. doi: 10.1016/s0140-6736(02)11314-6. [DOI] [PubMed] [Google Scholar]
- 16.O’Hare AM, Fanning NF, Ti JP, Dunne R, Brennan PR, Thornton JM. HydroCoils, occlusion rates, and outcomes: a large single-center study. AJNR Am J Neuroradiol. 2010;31:1917–1922. doi: 10.3174/ajnr.A2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozawa T, Tamatani S, Koike T, Abe H, Ito Y, Soga Y, Hasegawa H, Morita K, Tanaka R. Histological evaluation of endothelial reactions after endovascular coil embolization for intracranial aneurysm. Clinical and experimental studies and review of the literature. Interv Neuroradiol. 2003;9:69–82. doi: 10.1177/15910199030090S109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey AS, Koebbe C, Rosenwasser RH, Veznedaroglu E. Endovascular coil embolization of ruptured and unruptured posterior circulation aneurysms: review of a 10-year experience. Neurosurgery. 2007;60:626–637. doi: 10.1227/01.NEU.0000255433.47044.8F. [DOI] [PubMed] [Google Scholar]
- 19.Panetta CJ, Miyauchi K, Berry D, Simari RD, Holmes DR, Schwartz RS, Caplice NM. A tissue-engineered stent for cell-based vascular gene transfer. Hum Gene Ther. 2002;13:433–441. doi: 10.1089/10430340252792567. [DOI] [PubMed] [Google Scholar]
- 20.Plachokova A, Link D, van den Dolder J, van den Beucken J, Jansen J. Bone regenerative properties of injectable PGLA-CaP composite with TGF-beta1 in a rat augmentation model. J Tissue Eng Regen Med. 2007;1:457–464. doi: 10.1002/term.59. [DOI] [PubMed] [Google Scholar]
- 21.Sarrafzadeh A, Haux D, Kuchler I, Lanksch WR, Unterberg AW. Poor-grade aneurysmal subarachnoid hemorrhage: relationship of cerebral metabolism to outcome. J Neurosurg. 2004;100:400–406. doi: 10.3171/jns.2004.100.3.0400. [DOI] [PubMed] [Google Scholar]
- 22.Stiver SI, Porter PJ, Willinsky RA, Wallace MC. Acute human histopathology of an intracranial aneurysm treated using Guglielmi detachable coils: case report and review of the literature. Neurosurgery. 1998;43:1203–1208. doi: 10.1097/00006123-199811000-00106. [DOI] [PubMed] [Google Scholar]
- 23.Tamatani S, Ozawa T, Minakawa T, Takeuchi S, Koike T, Tanaka R. Histological interaction of cultured endothelial cells and endovascular embolic materials coated with extracellular matrix. J Neurosurg. 1997;86:109–112. doi: 10.3171/jns.1997.86.1.0109. [DOI] [PubMed] [Google Scholar]
- 24.Tamatani S, Ozawa T, Minakawa T, Takeuchi S, Koike T, Tanaka R. Radiologic and histopathologic evaluation of canine artery occlusion after collagen-coated platinum microcoil delivery. AJNR Am J Neuroradiol. 1999;20:541–545. [PMC free article] [PubMed] [Google Scholar]
- 25.Vance MM, Wiley LM. Gap junction intercellular communication mediates the competitive cell proliferation disadvantage of irradiated mouse preimplantation embryos in aggregation chimeras. Radiat Res. 1999;152:544–551. [PubMed] [Google Scholar]
- 26.Vittet D, Prandini MH, Berthier R, Schweitzer A, Martin-Sisteron H, Uzan G, Dejana E. Embryonic stem cells differentiate in vitro to endothelial cells through successive maturation steps. Blood. 1996;88:3424–3431. [PubMed] [Google Scholar]
- 27.Vu K, Weksler B, Romero I, Couraud PO, Gelli A. Immortalized human brain endothelial cell line HCMEC/D3 as a model of the blood-brain barrier facilitates in vitro studies of central nervous system infection by Cryptococcus neoformans. Eukaryot Cell. 2009;8:1803–1807. doi: 10.1128/EC.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu KK, Thiagarajan P. Role of endothelium in thrombosis and hemostasis. Annu Rev Med. 1996;47:315–331. doi: 10.1146/annurev.med.47.1.315. [DOI] [PubMed] [Google Scholar]
- 29.Yang D, Guo T, Nie C, Morris SF. Tissue-engineered blood vessel graft produced by self-derived cells and allogenic acellular matrix: a functional performance and histologic study. Ann Plast Surg. 2009;62:297–303. doi: 10.1097/SAP.0b013e318197eb19. [DOI] [PubMed] [Google Scholar]