Abstract
Background
Engineering dynamic, environmentally- and temporally-responsive control of gene expression is one of the principle objectives in the field of synthetic biology. Dynamic regulation is desirable because many engineered functions conflict with endogenous processes which have evolved to facilitate growth and survival, and minimising conflict between growth and production phases can improve product titres in microbial cell factories. There are a limited number of mechanisms that enable dynamic regulation in yeast, and fewer still that are appropriate for application in an industrial setting.
Results
To address this problem we have identified promoters that are repressed during growth on glucose, and activated during growth on sucrose. Catabolite repression and preferential glucose utilisation allows active growth on glucose before switching to production on sucrose. Using sucrose as an activator of gene expression circumvents the need for expensive inducer compounds and enables gene expression to be triggered during growth on a fermentable, high energy-yield carbon source. The ability to fine-tune the timing and population density at which gene expression is activated from the SUC2 promoter was demonstrated by varying the ratio of glucose to sucrose in the growth medium. Finally, we demonstrated that the system could also be used to repress gene expression (a process also required for many engineering projects). We used the glucose/sucrose system to control a heterologous RNA interference module and dynamically repress the expression of a constitutively regulated GFP gene.
Conclusions
The low noise levels and high dynamic range of the SUC2 promoter make it a promising option for implementing dynamic regulation in yeast. The capacity to repress gene expression using RNA interference makes the system highly versatile, with great potential for metabolic engineering applications.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0223-7) contains supplementary material, which is available to authorized users.
Keywords: Yeast, Sucrose, GFP, Metabolic engineering, Synthetic biology, Promoter, SUC2, TEF1, Diauxic shift
Background
Microorganisms possess a variety of mechanisms for modulating gene expression levels according to changes in their environment. The ability to engineer dynamic regulation is highly desirable in microbial cell factories because many of the most productive genetic modifications used for optimising engineering objectives are also detrimental to cell growth and survival. Engineered pathways compete for the carbon flux, redox potential, and ATP required by the cell for normal growth [1]. Engineered pathways can also result in accumulation of intermediates, side-products or end-products that are highly toxic to the host cell [2]. If a production strain cannot reach a high population density due to the metabolic burden imposed by an engineered pathway, then product titers are inherently limited. For genetic modifications that impose the most severe limits on growth, it is essential that they be implemented near or after the completion of a growth phase. One of the most promising techniques to optimise cell factory performance is to control the expression/repression of relevant genes using dynamic regulation.
S. cerevisiae (yeast) is an industrial microorganism which has been employed for the production of fuels, chemicals, and pharmaceuticals [3]. There are a limited number of molecular tools available for implementing dynamic regulation in yeast, with most being inadequate for industrial application. For example, the commonly used galactose induction system [4] has a noise level (expression in the absence of inducer) which is unacceptable for some applications such as controlling RNA interference [5]. Furthermore, galactose is prohibitively expensive for use at an industrial scale [6]. Previous efforts to implement dynamic regulation under industrially relevant conditions required the knockout of galactose utilization genes so that a small amount of galactose can be added to fermentations as a gratuitous inducer for gene expression [6]. High concentrations of fermentable carbon sources repress gene expression from galactose promoters (GAL) via a carbon catabolite repression mechanism [7], and galactose utilisation genes are not activated during growth on sucrose [8]. This means that cheap, fermentable sugars such as glucose or sucrose cannot be used during a production phase when the GAL promoters are used to achieve dynamic regulation. To circumvent this limitation, ethanol has been used as a carbon source for fed-batch cultures with the GAL promoters [6]. However, this is also problematic because ethanol is more expensive than commonly used sugar feedstocks, and is in fact a common commercial product of industrial yeast fermentations. Other induction systems such as the doxycycline inducible promoters have the advantage of being completely orthologous to native yeast regulation [9], but are also too expensive to be employed at a large scale. There are many other carbon source regulated promoters in yeast that could potentially be used for dynamic regulation [10], including the ADH2 promoter that is activated during growth on ethanol, the low-oxygen regulated DAN1 promoter, and the low phosphate activated PHO5 promoter [11]. These options all rely on the absence of a growth component that enables maximal metabolic flux and gene expression capacity (such as glucose or sucrose). There is therefore a significant unmet need for control systems that enable dynamic regulation and have suitable properties for industrial use of cell factories such as yeast. An ideal promoter would have low noise, high dynamic range, switch-like activation, high absolute expression levels, sustained induction, and high activity on carbon sources that support high metabolic flux.
To expand the toolbox of dynamic regulatory systems in yeast we have explored the use of promoters regulated by glucose de-repression in the presence of sucrose. Sucrose is a preferred feedstock for industrial scale fermentations due to sugarcane being a more environmentally friendly source of sugar than glucose which has been derived from corn [12,13]. Promoters that can regulate high levels of gene expression during a sucrose-fed production phase are therefore highly desirable. We sought to identify and characterise the spatiotemporal expression dynamics of promoters which are repressed during growth on glucose, and up-regulated during growth on sucrose. We applied these promoters to dynamic over-expression, and also to dynamic repression via a heterologous RNA interference module that can be used to destroy target mRNA according to base pair complementarity of expressed antisense RNA [5].
Results and discussion
Screening for sucrose responsive promoters
The most important properties to consider when assessing mechanisms for dynamic regulation are the dynamic range (induced minus non-induced expression levels) and noise levels (non-induced minus background). Four different promoters were screened with the aim of achieving low expression levels in the presence of glucose, and high expression in the presence of sucrose. Many promoters are known to be de-repressed in the presence of low glucose in yeast [10], little or no published data is available describing how these promoters behave on sucrose as the sole carbon source. It therefore could not be assumed that promoters known to be de-repressed by low glucose would be active on sucrose. For metabolic engineering applications in yeast it is desirable to have a production phase with a cheap carbon source. Promoters which are not only de-repressed by low glucose, but that are activated during growth on the cheap industrial carbon source sucrose are therefore highly desirable for industrial yeast fermentations.
Four promoters were selected based on literature which suggested they may be differentially regulated by sucrose. SUC2p is an invertase enzyme that hydrolyses sucrose into glucose and fructose; expression of SUC2p is strongly repressed by glucose [10,14]. Transcription of SUC2 is repressed around 200-fold when glucose levels are high and is activated only after glucose levels fall below 0.1% w/v [15-18]. The MAL12 gene encodes a disaccharide transporter which is highly induced in the presence of sucrose in non-laboratory strains of S. cerevisiae [19,20]. Glc3p is a glycogen branching enzyme essential for glycogen accumulation, and the Gph1p enzyme breaks down glycogen during stationary phase [21-23]. Both GLC3 and GPH1 transcripts have been observed to be up-regulated during growth on sucrose [24].
Yeast strains were generated with promoter-GFP fusion constructs for the promoters PSUC2, PMAL12, PGLC3, or PGPH1 integrated into the genome at the URA3 locus in single copy. Each strain was grown in glucose- or sucrose-containing medium with GFP fluorescence measured in the mid exponential phase (OD660nm of ~1.5) after 8 hours of growth (Figure 1).
Figure 1.
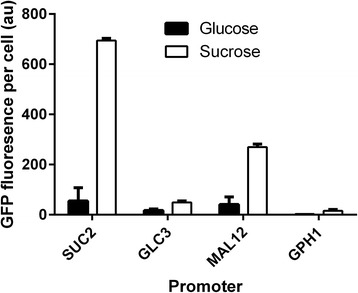
Testing promoters for differential expression on sucrose. SUC2, GLC3, MAL12, and GPH1 promoters were used to control GFP expression. Each strain was grown to mid-exponential phase on minimal medium containing 10 g/L glucose or 10 g/L sucrose prior to flow cytometry-based GFP measurement. Mean GFP fluorescence levels in arbitrary units (au) from duplicate fermentations with error bars representing the standard deviation are shown.
Each promoter had a significantly higher expression level on sucrose compared to on glucose. The SUC2 promoter had both the highest dynamic range (12.5-fold induction), and the highest absolute expression level. These properties made the SUC2 promoter an attractive choice for sucrose-mediated dynamic regulation in S. cerevisiae. Although this experiment was successful in identifying the best promoter, it only provided a ‘snapshot’ of expression levels during the exponential growth phase. In order to gain a deeper understanding of noise levels and dynamic range for comparison with other published data it is necessary to understand the spatiotemporal dynamics of gene expression throughout a fermentation.
Fine-tuning temporal gene expression dynamics using different glucose:sucrose ratios
Implementation of sucrose-mediated dynamic regulation would involve use of a growth medium which contains a known ratio of glucose to sucrose. In theory this would result in PSUC2-regulated genes being tightly repressed during a growth phase where glucose is preferentially consumed, then switched ‘on’ during a sucrose-consuming phase. When growth media with different ratios of glucose to sucrose were used to grow a PSUC2-GFP expressing strain, the timing of GFP induction responded according to glucose concentration (Figure 2a-d). A destabilized version of the GFP gene with a half-life of ~20 minutes (yEGFPCLN2PEST [25]) was used so that any decreases in gene expression level would not be obscured by the high stability (half-life of ~ 7 hours) of the regular yEGFP gene.
Figure 2.
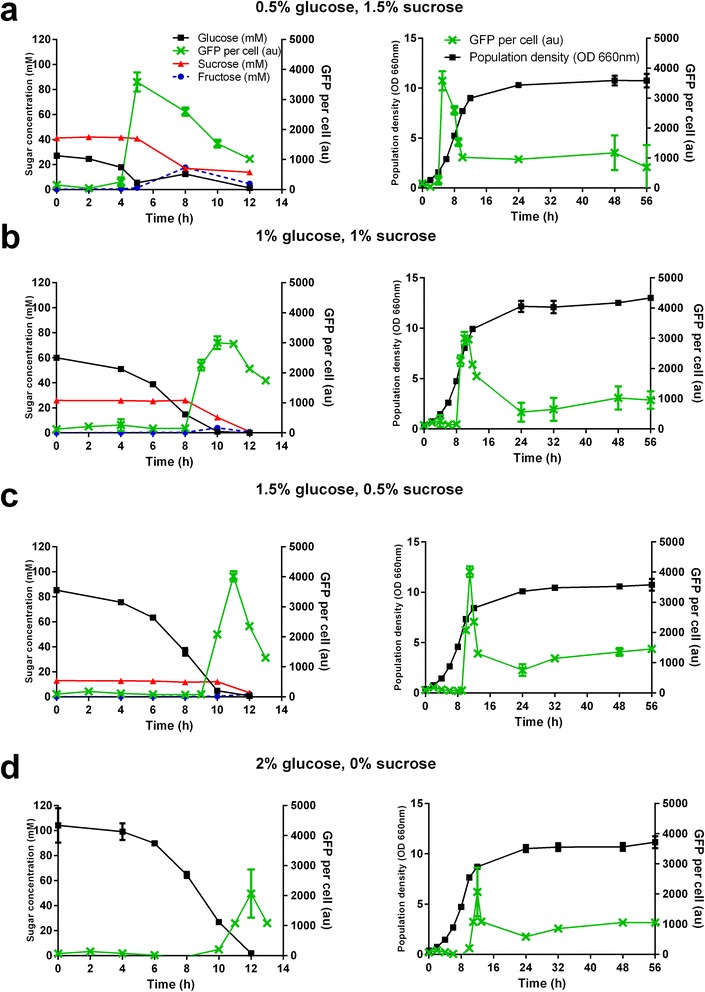
Tuning the timing of gene expression using different ratios of glucose to sucrose. A strain expressing the destabilized GFP gene driven by the SUC2 promoter was grown in media containing the indicated concentrations of glucose and sucrose. Extracellular glucose, sucrose, and fructose concentrations were measured using HPLC during the initial growth phase alongside GFP expression levels. Population density (OD660nm) and GFP expression levels were measured up to 56 hours of shake-flask cultivation. All data points and error bars represent the mean and standard deviation from triplicate cultivations.
In addition to the fine-tuning of gene expression, the time-course analysis of SUC2 promoter activity was necessary to gain more accurate insight into expression levels and dynamic range. As expected, GFP expression from the SUC2 promoter was strongly repressed during growth on glucose under all conditions. GFP expression levels during the glucose consumption phase were usually indistinguishable from autofluorescence, indicating tight regulation and a lack of undesirable leakiness. Once glucose concentrations dropped below about 5 mM, there was a rapid induction of GFP expression as the SUC2 promoter was activated. GFP levels from the SUC2 promoter peaked immediately after the switch from glucose to sucrose at around 4000 au before decreasing to 700–1000 au (Figure 2). This is consistent with the SUC2 expression level and dynamic range seen in the promoter screen, where populations had been growing on sucrose-only medium for 8 hours and would have progressed past the initial induction phase (Figure 1). As previously reported, the SUC2 promoter was activated after glucose was depleted even in the absence of sucrose [26]. When sucrose was present in the growth medium (Figure 2a-c) there was a 2 fold higher GFP expression level (~4000 au) (Figure 2a-c) compared to when sucrose was absent (~2000 au) (Figure 2d). As far as we are aware there is no known regulatory mechanism that can account for the increased SUC2 mediated gene expression in the presence of sucrose compared to in the absence of glucose (during the diauxic shift). It is possible that the phenomenon shown in Figure 2d could simply reflect a greater capacity for gene expression during exponential growth on fermentable carbon sources compared to during the diauxic shift and entry into the stationary phase [27].
It was possible to implement a very fine level of control over the timing and population density at which GFP expression was activated by varying glucose to sucrose ratios in the growth media. For example, with 0.5% glucose and 1.5% sucrose, GFP was fully induced after 5 hours of growth, at an OD660nm of ~2 (Figure 2a). With 1% glucose and 1% sucrose the switch occurred after 9 hours, at an OD660nm of ~6.4 (Figure 2b). When grown in medium containing 1.5% glucose and 0.5% sucrose, GFP expression was activated at 10 hours with a population density of ~7.3 (OD660nm), peaking after 11 hours at OD 8 (Figure 2c). With 2% glucose, and no sucrose in the growth medium, GFP expression was activated after 12 hours at an OD660nm of ~9 (Figure 2d). These variations demonstrate the fine-tuning of gene expression afforded by this fully autonomous and cheap induction system.
For application to metabolic engineering scenarios, it is important that any dynamic regulatory system has a sustained level of induction throughout cultivation. Measurement of GFP expression levels over 56 hours of shake-flask cultivation showed that expression from the SUC2 promoter remains at approximately 800–1000 fold higher than during the glucose consumption phase (Figure 2). This is a high expression level and after de-repression has occurred, is comparable to the commonly used TEF1 promoter (Figure 3). As a point of reference, the GAL10 promoter has a dynamic range of over 1000 [28] and an expression level similar to the TEF1 promoter [29]. The TEF1 promoter has previously been noted for its stable, high level of expression [29] and it was interesting that we observed a ~10 fold decrease in TEF1 promoter mediated GFP expression between the early exponential phase and the diauxic shift (Figure 3). This difference could possibly be due to the fact that previous observations relied on the beta-galactosidase reporter gene, which has a half-life of around 20 hours in S. cerevisiae [30]. In contrast, our system utilised a destabilized version of the yEGFP gene that has a half-life of ~ 20 minutes [25], making it far more sensitive to any decreases in gene expression levels. The TEF1 gene encodes a translational elongation factor that coordinates the positioning of aminoacylated tRNAs at ribosomes for the elongation of polypeptides [31]. Decreased expression from the TEF1 promoter is consistent with the overall decrease in gene expression during and after the diauxic shift in S. cerevisiae [32,33].
Figure 3.
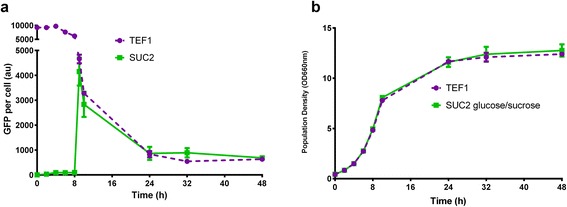
Comparison of TEF1 and SUC2 promoter strengths. GFP fluorescence (a) and population density (b) were measured for P TEF1-GFP and P SUC2-GFP expressing strains in 1% glucose, 1% sucrose containing medium over 48 hours. Mean and standard deviation for triplicate cultivations are shown.
Testing sucrose mediated regulation of RNA interference
Many of the most useful applications for dynamic regulatory systems require both the up-regulation and down-regulation of gene expression. To address this issue we sought to integrate the sucrose mediated induction system with a recently developed S. cerevisiae RNA interference (RNAi) system [5,34,35]. S. cerevisiae does not have a native system for RNA interference, but when the Argonaute and Dicer genes from Saccharomyces castelii are expressed, mRNAs can be specifically targeted for degradation by expressing complementary full-length antisense RNA [35] (Figure 4a), or double-stranded hairpin RNA [5,34,35].
Figure 4.
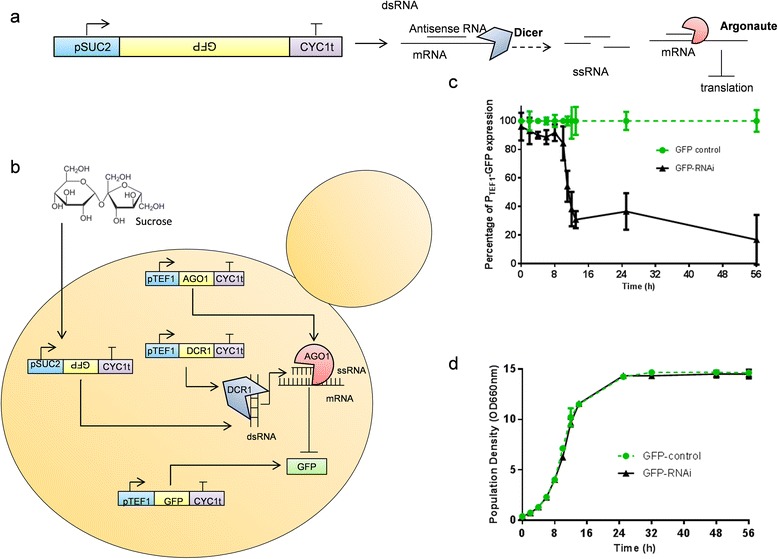
Dynamic repression of GFP expression using sucrose mediated RNAi. (a) The expression of an antisense RNA results in the destruction of complementary mRNA via Dicer and Argonaute enzymes. (b) Expression of the full GFP ORF in the antisense direction is triggered during growth on sucrose using the SUC2 promoter, causing constitutively regulated (TEF1 promoter) GFP expression to be repressed via Dicer/Argonaute-mediated RNA interference. GFP expression levels (c), and population density (d) were measured for P TEF1-GFP expressing strains both with (‘RNAi’, black triangles) and without (‘control’, green circles) a P SUC2-GFP antisense construct. The GFP expression level from the control strain was set to ‘100%’, with GFP expression values from the RNAi strain being normalised to this value. Means ± standard deviations are shown from triplicate cultures. Figure 4b was adapted from [42]. Non-normalised GFP expression values are shown in Additional file 1: Figure S1.
Previous efforts at implementing dynamic control of RNAi resulted in significant leakiness of target-gene repression [5]. With the tight repression of expression from the SUC2 promoter in the presence of glucose (Figure 2), we hypothesised that sucrose regulation would enable dynamic control of RNAi without non-induced repression occurring. The system was tested by expressing a destabilized GFP gene from the strong constitutive TEF1 promoter, and targeting it for repression by expressing the entire GFP ORF in the antisense direction from the SUC2 promoter in an RNAi capable strain (Argonaute and Dicer genes present) (Figure 4b). This strain was grown on medium containing a mixture of 1% glucose and 1% sucrose so that the SUC2 promoter would activate GFP repression only after the glucose in the growth medium was consumed as previously demonstrated (Figure 2). GFP expression levels were normalised to a ‘control’ strain which was identical except for the absence of the GFP antisense construct that facilitates RNAi. During the glucose consumption phase (0–8 hr; Figure 2b) there was no significant difference between RNAi and control strains, suggesting that little or no non-specific repression was occurring. Between 11 and 13 hours, GFP levels dropped sharply, indicating rapid activation of the repression module, with fluorescence levels reduced to 30% of the control strain (Figure 4c). Repression was maintained with expression levels being further reduced to around 17% of the control strain by 56 hours. The two strains were cultivated in the same media and there was no difference in growth dynamics (Figure 4d).
Conclusions
The dynamic control of gene expression is a core component of synthetic biology, and has great potential for the improvement of microbial cell factories. Dynamic control can be used to optimise a trade-off between growth based physiology and an organism’s engineered function [36]. By using dynamic regulation, any engineered functions that compete with normal growth-based physiology can be delayed until a population has reached a high density. Such functions are common, as even the most basic applications such as heterologous protein production can draw resources such as ATP, translation machinery, and amino acids away from native processes. In metabolic engineering the diversion of carbon flux, redox power, and ATP towards the production of target metabolites can reduce or potentially eliminate growth [1,2]. In support of this concept the SUC2 promoter has previously been used for high-level expression of amylase enzymes [37,38].
Although purified sucrose is not commonly used as an industrial carbon source, it is often hydrolysed into its constituent monosaccharides glucose and fructose. It is possible that the use of the SUC2 induction system would reduce the need for pre-hydrolysation of sucrose in industrial processes. For future directions it would also be interesting to test the sucrose induction system on an industrial carbon source such as sugarcane syrup at a much higher population density.
We present a simple dynamic regulatory system which does not require the addition of any expensive inducer compounds to the media, has a high dynamic range, low noise levels, maintains expression for prolonged periods, and can be coupled to RNAi to enable the repression of gene expression. We predict that this system will be of great utility for a diverse array of future applications in yeast cell factories.
Plasmids PTEF1-yEGFPCLN2PEST-pRS406, PSUC2-yEGFPCLN2PEST-pRS406, and PSUC2-GFPantisense-CYC1t-pRS413 will be made available from AddGene (www.addgene.org/).
Materials and methods
Media
Strain pre-cultures were grown in chemically defined liquid CBS medium with 5 g/L ammonium sulfate, 20 g/L glucose, vitamins and trace elements as described previously [39]. For GFP expression experiments CBS media with the indicated concentrations of glucose and/or sucrose was used. During strain construction purified amino acids (Sigma) were used to complement appropriate auxotrophies in agar plates (same composition as chemically defined media above) while YPD supplemented with appropriate antibiotics was used during gene deletion procedures. E. coli DH5α was used for plasmid propagation/storage and was grown in LB medium with kanamycin.
Strains and plasmids
Primers, plasmids, and strains used in this study are shown in (Table 1, Table 2, and Table 3). respectively. Annotated vector maps and sequences can be found in ‘Additional file 2’. DNA manipulation and propagation were carried out using standard techniques [40] unless stated otherwise. All S. cerevisiae transformations were carried out using the lithium acetate method [41]. BAR1 and FUS1 genes were deleted in the base RNAi strain ‘S01’ as part of another related project which utilises pheromone quorum sensing for dynamic regulation (reference [42]), and are not important design features for this study. Strains transformed with yeast integrating plasmids were screened for correct single copy integration using PCR as previously described [43] except using primers LEU2A2 and LEU2D2 to screen to LEU2 locus.
Table 1.
Primers
| Primer name | 5′ to 3′ sequence |
|---|---|
| MAL12F | TATTATctcgagACCAACCCGAAAATTCTTC |
| MAL12R | TATTATgaattcTTATGTAATTTAGTTACGCTTGAC |
| GPH1F | TATTATGatcgatTAGTTATCCGACTAGCAAG |
| GPH1R | TATTATgaattcTGTTCAAAATTAAATTAAGTTG |
| GLC3F | TATTATctcgagCGGTGATTTACAAGAAGAGG |
| GLC3R | TATTATatcgatTTTATTCTTGACGGTTCTTTATAC |
| SUC2F | TATTATctcgagACATACTAAGACATTTACCG |
| SUC2R | TATTATgaattcCATATACGTTAGTGAAAAGAAAAG |
| XhoI-pTEF1F | TATTATctcgagGCACACACCATAG |
| EcoRI-pTEF1R | TATTATgaattcTTGTAATTAAAACTTAGATTAGATTG |
| GFP1F | TATTATgaattcCTATATTACTTGGGTATTGCCC |
| GFP1R | TATTATcccgggTCTAAAGGTGAAGAATTATTCAC |
| LEU2A2 | ATAGAATTGTGTAGAATTGCAG |
| LEU2D2 | ATGAAATGAACATTGATTTACTATC |
| CYC1tF | TATAATtctagaACAGGCCCCTTTTCCTTTGT |
| CYC1tR | TATTATgagctcACGATGAGAGTGTAAACTGC |
| ApaI-SUC2F | TATTATgggcccACATACTAAGACATTTACCG |
| ClaI-SUC2R | TATTATatcgatCATATACGTTAGTGAAAAGAAAAG |
Restriction enzyme sites are shown in lower case.
Table 2.
Plasmids
| Name | Details | Origin |
|---|---|---|
| pRS406 | URA3 integrating vector | [45], Euroscarf |
| pSF019 | TEF1 promoter containing vector | [29] |
| yEGFPCLN2PEST-pRS406 | Destabilized GFP base plasmid | [36] |
| PMAL12- yEGFPCLN2PEST-pRS406 | MAL12 promoter driven GFP expression | This study |
| PGLC3- yEGFPCLN2PEST-pRS406 | GLC3 promoter driven GFP expression | This study |
| PGPH1- yEGFPCLN2PEST-pRS406 | GPH1 promoter driven GFP expression | This study |
| PSUC2- yEGFPCLN2PEST-pRS406 | SUC2 promoter driven GFP expression | This study |
| PTEF1-yEGFPCLN2PEST-406 | Constitutive TEF1 promoter driven GFP expression | This study |
| pRS413 | Yeast centromeric plasmid with HIS3 selection marker | [45], Euroscarf |
| CYC1t-pRS413 | CYC1 terminator | This study |
| PSUC2-CYC1t-pRS413 | SUC2 promoter, CYC1 terminator | This study |
| PSUC2-GFPantisense-CYC1t-pRS413 | SUC2 regulated expression of an antisense RNAi construct for GFP | This study |
| pRS404-PTEF -Ago1-CYC1t | TRP1 integrating vector with constitutive Argonaute expression | [5] |
| pRS405-PTEF-Dcr1-CYC1t | LEU2 integrating vector with constitutive Dicer expression | [5] |
| pUG6 | LoxP-KanMX-LoxP cassette | Euroscarf |
| pUG66 | LoxP-Ble-LoxP cassette | Euroscarf |
Table 3.
S. cerevisiae strains used in this study
| Name | Genotype | Notes | Origin |
|---|---|---|---|
| CEN.PK113-5D | MATa; ura3-52; MAL2-8C; SUC2 | Haploid MATa lab strain with uracil auxotrophy | Euroscarf |
| CEN.PK113-7D | MATa; MAL2-8C; SUC2 | Prototrophic haploid MATa lab strain | Euroscarf |
| S01 | CEN.PK2-1c, bar1Δ | BAR1 gene deleted | This study |
| S02 | CEN.PK2-1c, bar1Δ, fus1::KanMX, trp1::pRS404-PTEF-Ago1 | BAR1 and FUS1 deleted, Argonaute gene integrated at TRP1 | This study |
| S03 | CEN.PK2-1c, bar1Δ, fus1::KanMX, trp1::pRS404-PTEF-Ago1, leu2::pRS405-PTEF-Dcr1 | RNAi capable base strain with Argonaute and Dicer integration | This study |
| GFP01 | CEN.PK113-5D, bar1::phleo, fus1::KanMX, ura3-52::PTEF1-GFPCLN2PEST-ADH1t-pRS406 | constitutive destabilized GFP expression | This study |
| GFP02 | CEN.PK113-5D, ura3::PMAL12-GFPCLN2PEST-ADH1t-pRS406 | MAL12 regulated GFP expression | This study |
| GFP03 | CEN.PK113-5D, ura3::PGLC3-GFPCLN2PEST-ADH1t-pRS406 | GLC3 regulated GFP expression | This study |
| GFP04 | CEN.PK113-5D, ura3::PGPH1-GFPCLN2PEST-ADH1t-pRS406 | GPH1 regulated GFP expression | This study |
| GFP05 | CEN.PK113-5D, ura3::PSUC2-GFPCLN2PEST-ADH1t-pRS406 | SUC2 regulated GFP expression | This study |
| GFP06 | CEN.PK2-1c, bar1Δ, fus1::KanMX, ura3:: PTEF1-yEGFPCLN2PEST-406, trp1::pRS404-PTEF-Ago1, leu2::pRS405-PTEF-Dcr1, pRS413 | RNAi capable, constitutive GFP expressing control strain | This study |
| GFP07 | CEN.PK2-1c, bar1Δ, fus1::KanMX, ura3:: PTEF1-yEGFPCLN2PEST-406, trp1::pRS404-PTEF-Ago1, leu2::pRS405-PTEF-Dcr1, PSUC2-GFPantisense-CYC1t-pRS413 | SUC2 promoter mediated repression of GFP using RNAi | This study |
All putative sucrose-responsive promoters were PCR amplified from CENP.K2-1c genomic DNA, and contain ~700 bp upstream of the start codon of the native gene. MAL12 (XhoI/EcoRI), GPH1 (ClaI/EcoRI), GLC3 (XhoI/ClaI), and SUC2 (XhoI/EcoRI) promoter regions were cloned 5’ of the start codon in the GFPCLN2PEST-pRS406 plasmid using the indicated restriction enzyme combinations. The PTEF1-yEGFPCLN2PEST-pRS406 plasmid was made by inserting the TEF1 promoter amplified from pSF019 using primers 9 and 10 into the yEGFPCLN2PEST-pRS406 plasmid with XhoI/EcoRI.
The GFP silencing construct was expressed using the SUC2 promoter on the low copy number pRS413 plasmid. To construct this expression cassette a CYC1 terminator region was PCR amplified from genomic DNA using CYC1tF and CYC1tR primers and cloned into pRS413 using XbaI and SacI. The SUC2 promoter was then amplified using ApaI-SUC2F and ClaI-SUC2R and inserted 5’ of CYC1t in pRS413 to create PSUC2-CYC1t-pRS413. The GFP silencing construct was made by cloning the full length GFPCLN2PEST ORF sequence in the antisense direction between the SUC2 promoter and CYC1 terminator in PSUC2-CYC1t-pRS413 using GFP1F/GFPR primers and EcoRI/XmaI restriction enzymes to create the PSUC2-GFP1i-CYC1t-pRS413 plasmid.
Fermentation conditions
All growth experiments were carried out in baffled screw top flasks shaking at 200 rpm, 30°C with medium comprising 10% of the total flask volume. Individual colonies were transferred to liquid media and pre-cultured for approximately 15 hours. Cultures were then passaged into a second pre-culture and grown to mid exponential phase (OD660nm between 1 and 6) prior to inoculation of experimental cultures at an OD660nm of 0.1 for the initial promoter comparison (Figure 1) and 0.4 for the time course experiments (Figure 2).
Analytics
GFP fluorescence was measured using an Accuri C6 flow cytometer (BD Biosciences) as in [36]. HPLC was used to measure sucrose, fructose, and glucose as previously described [44].
Statistical analysis
Mean GFP fluorescence values from triplicate experiments were compared using a two-sided, unpaired student’s t-test with equal variance. Null hypotheses were rejected when p-values were ≤ 0.05.
Additional files
Supplementary material.
Annotated plasmid maps and sequences.
Acknowledgements
TCW was supported by an Australian Postgraduate Award and the AIBN Top Up Scholarship. We thank Manuel Plan of the Metabolomics Australia Queensland node for technical assistance with HPLC.
Footnotes
Competing interests
The authors declare no competing interests.
Authors’ contributions
TCW conceived of the study and designed experiments. TCW and MIE performed experiments. TCW, MIE, LKN, and CEV wrote and edited the manuscript. LKN and CEV participated in the design and coordination of the project. All authors read and approved the final manuscript.
Contributor Information
Thomas C Williams, Email: t.williams1@uq.edu.au.
Monica I Espinosa, Email: monica.espinosa@uq.net.au.
Lars K Nielsen, Email: lars.nielsen@uq.edu.au.
Claudia E Vickers, Email: c.vickers@uq.edu.au.
References
- 1.Lee SK, et al. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol. 2008;19(6):556–63. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Keasling JD. Synthetic Biology for Synthetic Chemistry. ACS Chem Biol. 2008;3(1):64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 3.Hong K-K, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci. 2012;69(16):2671–90. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9(9):777–87. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 5.Drinnenberg IA, et al. RNAi in Budding Yeast. Science. 2009;326(5952):544–50. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci U S A. 2012;109(3):E111–8. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gancedo JM. Yeast Carbon Catabolite Repression. Microbiol Mol Biol Rev. 1998;62(2):334–61. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto H, et al. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. Mol Gen Genet MGG. 1983;191(1):31–8. doi: 10.1007/BF00330886. [DOI] [PubMed] [Google Scholar]
- 9.Wishart JA, et al. Doxycycline, the drug used to control the tet-regulatable promoter system, has no effect on global gene expression in Saccharomyces cerevisiae. Yeast. 2005;22(7):565–9. doi: 10.1002/yea.1225. [DOI] [PubMed] [Google Scholar]
- 10.Weinhandl K, et al. Carbon source dependent promoters in yeasts. Microb Cell Fact. 2014;13(1):5. doi: 10.1186/1475-2859-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubmann G, Thevelein J, Nevoigt E. Natural and Modified Promoters for Tailored Metabolic Engineering of the Yeast Saccharomyces cerevisiae. In: Mapelli V, editor. Yeast Metabolic Engineering. New York: Springer; 2014. pp. 17–42. [DOI] [PubMed] [Google Scholar]
- 12.Bruschi M, et al. A transferable sucrose utilization approach for non-sucrose-utilizing Escherichia coli strains. Biotechnol Adv. 2012;30(5):1001–10. doi: 10.1016/j.biotechadv.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Sabri S, Nielsen LK, Vickers CE. Molecular Control of Sucrose Utilization in Escherichia coli W, an Efficient Sucrose-Utilizing Strain. Appl Environ Microbiol. 2013;79(2):478–87. doi: 10.1128/AEM.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2(2):202–7. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 15.Bu Y, Schmidt MC. Identification of cis-acting elements in the SUC2 promoter of Saccharomyces cerevisiae required for activation of transcription. Nucleic Acids Res. 1998;26(4):1002–9. doi: 10.1093/nar/26.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleming AB, Pennings S. Tup1-Ssn6 and Swi-Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene. Nucleic Acids Res. 2007;35(16):5520–31. doi: 10.1093/nar/gkm573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutfiyya LL, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16(9):4790–7. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozcan S, et al. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast. 1997;13(2):127–37. doi: 10.1002/(SICI)1097-0061(199702)13:2<127::AID-YEA68>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Q, Michels CA. MAL11 and MAL61 encode the inducible high-affinity maltose transporter of Saccharomyces cerevisiae. J Bacteriol. 1991;173(5):1817–20. doi: 10.1128/jb.173.5.1817-1820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao B, Marmur J, Sollitti P. Construction of glucose-repressible yeast expression vectors. Gene. 1993;137(2):223–6. doi: 10.1016/0378-1119(93)90010-Z. [DOI] [PubMed] [Google Scholar]
- 21.Francois J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25(1):125–45. doi: 10.1016/S0168-6445(00)00059-0. [DOI] [PubMed] [Google Scholar]
- 22.Sunnarborg SW, et al. Expression of the yeast glycogen phosphorylase gene is regulated by stress-response elements and by the HOG MAP kinase pathway. Yeast. 2001;18(16):1505–14. doi: 10.1002/yea.752. [DOI] [PubMed] [Google Scholar]
- 23.Thon VJ, et al. Coordinate regulation of glycogen metabolism in the yeast Saccharomyces cerevisiae. Induction of glycogen branching enzyme. J Biol Chem. 1992;267(21):15224–8. [PubMed] [Google Scholar]
- 24.Basso TO, et al. Engineering topology and kinetics of sucrose metabolism in Saccharomyces cerevisiae for improved ethanol yield. Metab Eng. 2011;13(6):694–703. doi: 10.1016/j.ymben.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Mateus C, Avery SV. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16(14):1313–23. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 26.Hua Lin K, et al. Control of gene expression from the SUC2 promoter of Saccharomyces cerevisiae with the aid of a glucose analyser. Appl Microbiol Biotechnol. 1989;32(3):313–6. [Google Scholar]
- 27.Galdieri L, et al. Transcriptional Regulation in Yeast during Diauxic Shift and Stationary Phase. OMICS: A Journal of Integrative Biology. 2010;14(6):629–38. doi: 10.1089/omi.2010.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St John TP, Davis RW. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- 29.Partow S, et al. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27(11):955–64. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- 30.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234(4773):179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 31.Schirmaier F, Philippsen P. Identification of two genes coding for the translation elongation factor EF-1 alpha of S. cerevisiae. EMBO J. 1984;3(13):3311–5. doi: 10.1002/j.1460-2075.1984.tb02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner JR, Vilardell J, Sohn JH. Economics of Ribosome Biosynthesis. Cold Spring Harb Symp Quant Biol. 2001;66:567–74. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 33.Werner-Washburne M, et al. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1993;57(2):383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crook NC, Schmitz AC, Alper HS. Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synth Biol. 2014;3(5):307–13. doi: 10.1021/sb4001432. [DOI] [PubMed] [Google Scholar]
- 35.Si T et al.. RNAi-Assisted Genome Evolution in Saccharomyces cerevisiae for Complex Phenotype Engineering. ACS Synthetic Biology, 2014. (http://pubs.acs.org/doi/abs/10.1021/sb500074a). The DOI is 10.1021/sb500074a. [DOI] [PubMed]
- 36.Williams TC, Nielsen LK, Vickers CE. Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth Biol. 2013;2(3):136–49. doi: 10.1021/sb300110b. [DOI] [PubMed] [Google Scholar]
- 37.Cha HJ, et al. Enhancement, by succinate addition, of the production of cloned glucoamylase from recombinant yeast using a SUC2 promoter. Process Biochem. 1998;33(3):257–61. doi: 10.1016/S0032-9592(97)00042-3. [DOI] [Google Scholar]
- 38.Zhang X, et al. Enhancement of production of cloned α-amylase by lactic acid feeding from recombinant Saccharomyces cerevisiae using a SUC2 promoter. Biotechnol Lett. 2001;23(4):259–62. doi: 10.1023/A:1005641926474. [DOI] [Google Scholar]
- 39.Verduyn C, et al. Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8(7):501–17. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Russell DW. Molecular cloning, a laboratory manual. New York: Cold Spring Harbour Laboratory Press; 2001. [Google Scholar]
- 41.Gietz RD, Schiestl RH. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protocols. 2007;2:35–7. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- 42.Stansfield I, Stark MJR. 2 Yeast Genetics and Strain Construction. In: Ian S, Michael JRS, editors. Methods in Microbiology. London, UK: Academic Press; 2007. pp. 23–43. [Google Scholar]
- 43.Arifin Y, et al. Deletion of cscR in Escherichia coli W improves growth and poly-3-hydroxybutyrate (PHB) production from sucrose in fed batch culture. J Biotechnol. 2011;156(4):275–8. doi: 10.1016/j.jbiotec.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Williams TC, Averesch NJH, Winter G, Plan MR, Vickers CE, Nielsen LK, et al. Quorum-Sensing Linked RNA interference for Dynamic Metabolic Pathway Control in Saccharomyces cerevisiae. Metab Eng. 2015. (http://www.sciencedirect.com/science/article/pii/S1096717615000348). The DOI is 10.1016/j.ymben.2015.03.008. [DOI] [PubMed]
- 45.Sikorski RS, Hieter P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Annotated plasmid maps and sequences.


