Abstract
The control of cell fate decisions is vital to build functional organs and maintain normal tissue homeostasis, and many pathways and processes cooperate to direct cells to an appropriate final identity. Because of its continuously renewing state and its carefully organised hierarchy, the mammalian intestine has become a powerful model to dissect these pathways in health and disease. One of the signalling pathways that is key to maintaining the balance between proliferation and differentiation in the intestinal epithelium is the Notch pathway, most famous for specifying distinct cell fates in adjacent cells via the evolutionarily conserved process of lateral inhibition. Here, we will review recent discoveries that advance our understanding of how cell fate in the mammalian intestine is decided by Notch and lateral inhibition, focusing on the molecular determinants that regulate protein turnover, transcriptional control and epigenetic regulation.
Keywords: intestinal stem cells, lateral inhibition, Notch
Introduction
Tissue stem cells (SCs) exist in most adult organisms and are generally defined as cells having self-renewing capacity, as well as the ability to generate all cell types of a given organ. The cell types generated must be tailored to the needs of the tissue, and thus cell progeny are influenced to adopt particular cell fates depending on a complex interplay of internal and external signals. One of the regulatory mechanisms controlling cell fate is lateral inhibition, which enables differential activation of Notch signalling in neighbouring cells to generate different cell types. Notch signalling and lateral inhibition were initially described in Drosophila melanogaster, but are used throughout metazoan development as well as in adult organisms to specify cell fate in many tissues (for a historical perspective, see 1).
A widely used model system to investigate the signals controlling cell fate decisions in mammals is the murine intestine. The organisation of the intestinal epithelium into proliferative crypts that constantly renew the differentiated cells in the villi presents researchers with a repeating array of the complete set of stem, progenitor and differentiated cell types, the majority of which turn over within a matter of days due to the constant cell death of differentiated cells at the tip of the villi (Fig1A). This continuous production of multiple cell lineages facilitates genetic investigation of the system, since abnormal proliferation/differentiation phenotypes rapidly manifest as altered cellular compositions of the crypts and villi. The balance between self-renewal and differentiation is under stringent control to allow proper development and avoid uncontrolled growth, which can lead to intestinal hyperplasia, inflammatory processes and cancer. In this review, we will focus on how Notch signalling is regulated by protein turnover of signalling pathway components, as well as by transcriptional and epigenetic mechanisms, to achieve correct specification of cell fates in the mammalian intestine.
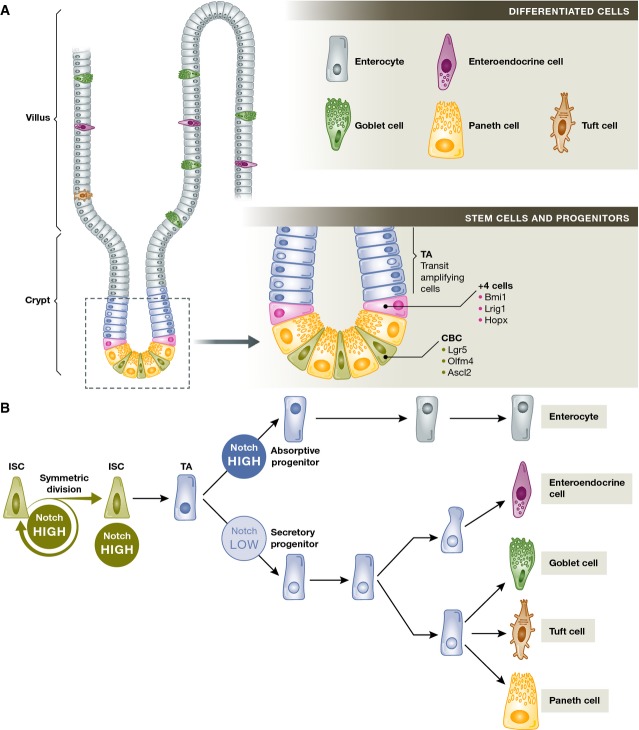
Figure 1. Intestinal homeostasis and Notch signalling in cell fate decisions.
(A) Schematic diagram of the different cell populations in the mouse small intestinal epithelium. In the mammalian small intestine, a crypt and villus represents the fundamental repetitive unit. Intestinal stem cells (CBCs and +4 cells) and TA (transit amplifying) cells are located in the crypt, and differentiated cells (enterocytes, goblet, enteroendocrine and tuft cells) are located along the villus. Paneth cells are a special case of differentiated cells, which migrate down to the crypt base and reside amongst the CBC stem cells. (B) Simplified scheme of intestinal cell lineages and differentiation. Notch signalling plays a crucial role in intestinal stem cell and progenitor differentiation. Notch signalling directly targets the intestinal stem cells (ISCs) to maintain proliferation and promote cell survival. Notch also acts to promote differentiation to the absorptive lineage, while the Notch-low state allows differentiation of secretory cells.
Stem cells and cell fate decisions in the mammalian intestine
The cellular hierarchy of the intestinal epithelium begins at the crypt base, the main stem cell niche, which contains the crypt base columnar (CBC) stem cells (Fig1A). CBC cells divide daily, producing rapidly proliferating daughter cells known as transit amplifying (TA) cells, which fill the crypts and gradually lose their progenitor identity as they move upwards towards the intestinal lumen. Upon reaching the crypt–villus junction, TA cells differentiate into the two main epithelial lineages: the absorptive lineage, which comprises all enterocytes, and the secretory lineage, which is composed of goblet cells (secreting protective mucins) and enteroendocrine cells (secreting hormones like serotonin or secretin) located within the villi, as well as Paneth cells that are restricted to the bottom of the crypt in the small intestine (reviewed in 2) (Fig1A). Other cell types with distinct ultrastructures, including tuft cells, M cells and cup cells, are present in the mature epithelium, but their lineages and functions are less well understood 3 (Fig1B).
CBC stem cells express the markers Lgr5 (one of a family of 7-transmembrane receptors containing a large leucine-rich extracellular domain) and Olfm4, an extracellular matrix glycoprotein that is a direct Notch target gene 4, 5, 6. In conditions of normal epithelial turnover, all cell types of the intestinal epithelium can be lineage traced back to a CBC cell (5 and reviewed in 7). Following injury and loss of CBC stem cells, a “reserve” population (or populations) of cells that reside outside the crypt base may act as facultative stem cells, moving down to the crypt niche to regenerate the Lgr5 stem cell pool and repopulate the entire tissue 8, 9 (Fig1A). Originally identified as Bmi1 positive 9, the unique identity of this population remains under debate. Unambiguously identifying non-CBC stem cell populations has been especially difficult because markers including Lgr5, Bmi1, Lrig1 and HopX are not exclusive to stem cells 10, 11 (Fig1A). Recent reports suggest that in the absence of injury, the cells of the “reserve” population continue as secretory or Paneth cell progenitors, rather than multipotent stem cells 12, 13. However, the characteristics of non-CBC stem cells and their behaviour in conditions of normal homeostasis, disease and injury are still very active areas of research 12, 13, 14, 15, 16.
Unlike stem cells in other tissues such as skin, which divide asymmetrically to maintain one stem cell at the same time as creating a progenitor, the CBC stem cells divide symmetrically and only become TA progenitors when they leave the niche 17, 18. The signals for maintaining the stem cell fate thus include a cell-extrinsic component, although the exact sources of these molecular stem cell niche signals are still not entirely clear. Direct cell–cell contact with a Paneth cell has been proposed as the stem-cell-maintaining niche signal 19, but more recent reports show that correct intestinal epithelial homeostasis can be maintained in the absence of Paneth cells 20, 21. However, it remains possible that Paneth cells do provide niche signals in the intact intestine and that alternative sources of signalling ligands, such as mesenchymal cells, may compensate when Paneth cells are missing 22.
Progenitor cells outside the crypt base niche occupy the proliferative transit-amplifying compartment. TA cells continuously migrate upwards towards the intestinal lumen, forming a dynamic epithelial “conveyor belt” that replenishes the short-lived enterocytes, goblet and enteroendocrine cells. On this journey, cells must rapidly decide whether to continue to proliferate, or to differentiate; differentiating cells will join the absorptive or the secretory lineage; and differentiating secretory cells can become goblet or enteroendocrine cells (Fig1B). Since differentiated Paneth cells reside at the crypt base and their turnover is very slow compared to other intestinal cell types, they constitute a special case. Presumptive Paneth cell progenitors begin to express EphB3 receptors so that they are repelled by the ephrin ligands further up the crypt and so Paneth cells remain in the crypt base where they are needed 23. For the majority of cells, however, their distance from the crypt base, measured by a decreasing gradient of Wnt and increasing gradients of BMP/ephrin ligands 24, is the major factor in controlling progressive migration and differentiation. Although the early stages of differentiation towards different lineages begin just a couple of cells above the stem cell niche (the “+ 5” position 13 or “Common origin of differentiation” described by Bjerknes and Cheng 25), the TA cells retain a degree of plasticity at lower levels in the crypt and may not fully commit until they reach the crypt–villus junction (Fig1A).
Molecular mechanisms controlling intestinal stem cell fate decisions
The key developmental signalling pathways Wnt and Notch, conserved throughout multicellular evolution to regulate cell patterning in many contexts, are used in the adult mammalian intestine to control the proliferation of stem and progenitor cells and differentiation of the various cell lineages. Though not the focus of this review, the Wnt signalling pathway is particularly important in maintaining the stem and progenitor cell compartments within the intestinal crypts 26, 27. Activating mutations in this pathway, frequently truncations of the APC gene that indirectly stabilise the Wnt effector β-catenin, have been found in over 90% of colorectal cancers 28 and APCmin/+ mice harbouring mutant APC serve as the most widely used intestinal tumour model in mammals 29, 30. Activation of the stem cell marker Lgr5 by R-spondins promotes Wnt signalling 31, 32, 33, which activates transcription of Lgr5 as well as the stem cell transcription factor Ascl2 5, 34.
There is much crosstalk between the Notch and Wnt pathways (recently reviewed by Collu et al 35), and in the intestine this manifests in different ways in the different compartments. In the stem cell niche, a combination of Wnt and Notch signals is required for maintenance of the stem cell pool, since without either one of these the stem cells are lost 36, 37. However, amongst progenitors, Wnt and Notch activation is more polarised: secretory progenitors are Wnt high and Notch low, whereas absorptive progenitors are Wnt low and Notch high (Fig1B).
Lateral inhibition and the Notch pathway in cell fate decisions in the intestine
The Notch cascade has a unique mode of action and has been recognised as one of a few signalling pathways that are repeatedly used in multiple developmental processes in embryonic and adult tissues. The canonical Notch pathway uses juxtacrine cell-to-cell contact and converts this interaction directly into changes of gene expression, frequently resulting in opposite fate determinations in adjacent cells (lateral inhibition). The Notch pathway has been extensively reviewed elsewhere 38, 39, 40, 41 and so will be only briefly introduced here. Upon binding of the Notch ligand (in mammals, Delta-like or Jagged) to the Notch receptor at the cell surface, the receptor undergoes a series of proteolytic cleavages, notably the shedding of the extracellular portion of the receptor by the metalloprotease ADAM10 and the release of a cytoplasmic portion by the gamma-secretase complex. This active fragment, the Notch intracellular domain (NICD), subsequently translocates to the nucleus and alters gene expression in complex with several cofactors, notably RBPJk. One of the best characterised groups of NICD target genes is the Hes (hairy enhancer of split) family that is upregulated in many different tissue types. The Hes family of transcriptional repressors comprises Hes1, Hes5 and Hes7 proteins and the related family of the Herp/Hey proteins including Hey1, Hey2 and HeyL 42. Hes and Hey transcription factors are responsible for the initiation of an extensive genetic program upon Notch activation. This program of altered gene expression determines the final fate of the cell 43: proliferation in the case of stem/progenitor cells or differentiation to an absorptive phenotype in the case of TA cells (Fig2).
Figure 2. Notch and lateral inhibition in ISCs and TA cells.
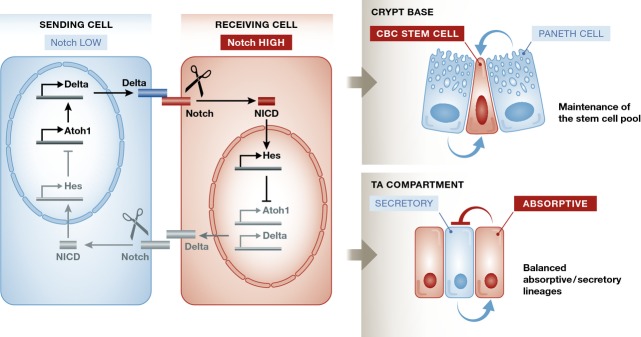
Notch signalling is initiated when a cell-surface-expressed Delta ligand binds to the Notch receptor expressed on an opposing cell surface. The membrane-tethered Notch is then cleaved by ADAM10 and then by the γ-secretase complex to release the intracellular fragment of Notch (NICD). This translocates to the nucleus and assembles into a transcriptional activation complex that relieves repression of Notch target genes such as the Hes family. The Hes family of transcriptional repressors controls Delta and a variety of differentiation/proliferation genes. An important function of the Notch pathway is in lateral inhibition—an interaction between equal adjacent cells that serves to drive them towards different final states. The basic principle of lateral inhibition is that activation of Notch represses production of the Notch ligand (Delta). Consequently, the cell with lower Notch activity produces more ligand (a status reinforced by derepression of the transcription factor Atoh1, which directly activates Delta transcription). More ligand at the cell surface activates Notch signalling in the neighbouring cell which results in reduced ligand production in that cell. This in turn enables the cell with lower Notch activity to increase its ligand production even further, because it receives a weakened inhibitory signal back from its neighbours. The effect of this feedback loop is that any initial difference in Notch activity between them, whether stochastic or genetically controlled, is amplified to drive the neighbouring cells into opposite Notch-level status and hence into different developmental pathways. Notch plays an important role in maintaining the intestinal stem cell pool: Paneth cells, and perhaps other sources, provide a constant Notch ligand stimulus to ISCs. In the TA compartment, Notch-high progenitors will differentiate to enterocytes while they will push neighbouring cells to commit to a secretory fate.
The fate of the cell also depends on the strength of the Notch signal it receives. The NICD transcriptional program represses genes encoding the Notch ligands (Delta-like, Jagged), so strong Notch activation in the receiving cell reduces its ability to activate its neighbouring cell. Because fate specification is controlled by cell-to-cell signalling between adjacent cells, this process is referred to as “lateral cell fate specification” or “lateral inhibition”. At its most basic, lateral inhibition amplifies and stabilises the stochastic initial differences in Notch signalling between two equivalent adjacent cells, rapidly pushing them towards opposite fates 1.
In the intestine, the Notch pathway uses two different mechanisms to achieve its two major roles: (i) negative regulation prevents the differentiation of stem cells, thereby maintaining the stem cell pool; and (ii) in binary cell decisions, Notch promotes differentiation in one direction while suppressing the other possible outcome, thereby controlling the balance between absorptive and secretory lineages (Fig2).
Maintenance of the stem cell pool
Notch signalling generally promotes proliferation, and Notch-high cells include the rapidly cycling CBC stem cells and the absorptive lineage progenitors, which are more proliferative than those of the secretory lineage. Mechanistically, Notch activation upregulates Hes transcription factors, which suppress CDK inhibitor expression 44. CBC stem cells receive Notch signalling input from the Delta-like ligands Dll1 and Dll4, which are partly redundant but together are crucial for stem cell proliferation 45 (Fig2). Paneth cells within the stem cell niche express Dll4 and also transiently Dll1 19, 46, although the finding that Paneth cells are dispensable in vivo 20, 21 implies that other sources of Dll1 and Dll4 may be available. Lineage tracing of Notch-expressing cells results in the labelling of stem cells followed by labelling of entire crypt–villus units 45, 47. Inhibiting Notch signalling by using a gamma-secretase inhibitor, by deleting the intermediate Notch protease ADAM10 or by combined genetic inactivation of the ligands Dll1 and Dll4 results in downregulation of the stem cell markers Olfm4 and Lgr5 and loss of CBC stem cells 6, 45, 48. As a result, inhibition of Notch signalling leads to rapid weight loss and death consistent with a failure of tissue replenishment and lack of nutrient absorption, demonstrating the essential role of Notch signalling in maintaining the stem cell pool.
Balance between absorptive and secretory lineages
While Notch inhibition is lethal, constitutive activation of Notch signalling in the intestinal epithelium using a Villin-driven NICD transgene is also lethal, due to loss of secretory cells 49. A balance is clearly necessary to ensure that stem cells are maintained and absorptive cells produced, while allowing the emergence of Notch-low cells that adopt a secretory fate. The transcription factor responsible for secretory cell fate is Atoh1 (also known as Math1 or Hath1) 50, 51, 52. Hes factors inhibit Atoh1, and so Notch-high cells are directed away from the secretory and towards the absorptive lineage 49. Suppressing Notch signalling results in an increase in secretory goblet cells at the expense of proliferating cells, as shown by deletion of ADAM10 48, inhibiting gamma-secretase 53, inactivating RBPJ 37 or knocking out the Hes genes 43. These phenotypes are dependent on Atoh1, since Atoh1 loss restores crypt cell proliferation and reduces accumulation of secretory cells in a Notch null background or when Notch signalling is inhibited 54, 55, 56.
The capacity of the Notch pathway to rapidly induce different and mutually exclusive fates in adjacent cells makes it ideally suited for the division of intestinal progenitor cells into absorptive and secretory lineages (Fig2). Committing to the absorptive lineage does not immediately halt proliferation, while cells committed to the secretory lineage no longer proliferate, leading to fewer secretory cells overall 46. The use of Notch signalling and lateral inhibition to differentiate absorptive and secretory progenitors is broadly conserved from zebrafish to mammals 57. Lateral inhibition ensures that a Notch-high cell (which is skewed towards the absorptive lineage) limits activation of Notch in its neighbouring cells, which promote secretory differentiation. Lineage tracing of strongly Dll1-positive (Notch-low) cells 13 and mathematical modelling 46 support this lateral inhibition pattern of secretory differentiation adjacent to neighbouring absorptive progenitors. Despite the apparent simplicity of the lateral inhibition model, the underlying complexity of this conserved process requires exquisite control in order to maintain the proper homeostasis of the intestine.
Molecular regulation of Notch and lateral inhibition
Lateral inhibition via the Delta-like and Jagged transmembrane ligands forms the core of cell-extrinsic Notch pathway regulation and lays the basis for a balanced distribution of absorptive and secretory cells. Expression of ligand in a neighbouring cell trans-activates Notch, while co-expression of ligand and receptor in cis inhibits Notch signalling via Fringe proteins 58, 59. However, there are also many other cell-intrinsic mechanisms that combine to determine the level of Notch activation within individual cells (Fig3).
Figure 3. Molecular regulation of Notch and lateral inhibition in health and disease.
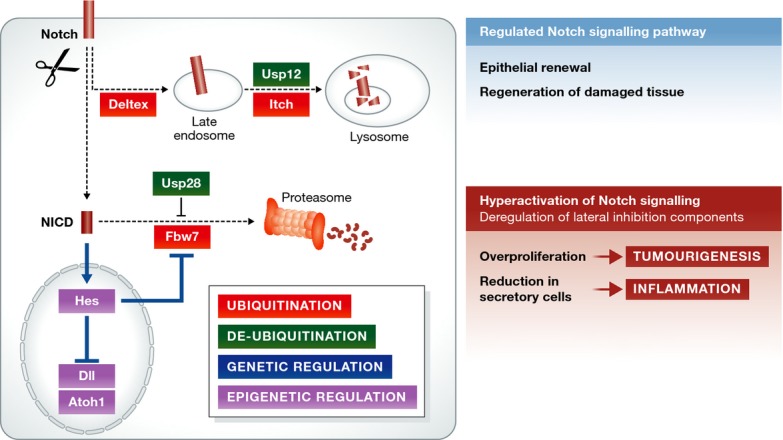
Cell-intrinsic mechanisms regulate the level of Notch activation within individual cells: ubiquitination (by Deltex/Itch/Fbw7) and deubiquitinating enzymes (Usp28 and Usp12) control the intracellular levels of Notch; genetic and epigenetic regulation ensures a correct stoichiometry of the Notch signalling components. The regulation of the Notch pathway maintains proper intestinal homeostasis. When Notch signalling regulators are altered, this can result in an aberrant hyperactivation of the pathway with severe complications such as intestinal inflammation (due to loss of secretory cells) or overproliferation/tumourigenesis.
Ubiquitination
The stability and trafficking of both inactive and active Notch receptors are regulated by ubiquitination. The availability of Notch at the cell surface is a key determinant of the cell's capacity for Notch signalling, and the pathway output also relies on the levels of active Notch intracellular domain (NICD) available to control transcription in the nucleus. Notch may also be activated within cells in an endocytic compartment 60, further sensitising the signalling output to subtle changes in the localisation and protein levels of Notch pathway components. Ubiquitin-mediated regulation therefore plays a major role in the levels of Notch signalling in each cell and hence its fate. Many of the molecular mechanisms involved were initially characterised in other systems, and their roles in the intestine are still uncharacterised. Itch (acting together with Numb) and Fbw7 are the best characterised E3 ligases regulating Notch in the mammalian intestine. Itch regulates trafficking and degradation of the membrane-bound Notch receptor via the lysosomal pathway, whereas Fbw7 regulates degradation of cleaved NICD via the proteasome (Fig3).
Itch, Numb and Deltex
Deltex is a RING-finger E3 ubiquitin ligase that in Drosophila promotes the late-endosomal activation of Notch in a ligand-independent manner, probably by mediating its internalisation 61. However, in both Drosophila and mammals, Deltex and Notch also form a complex with beta-arrestin, which modulates the ubiquitination and trafficking of the Notch receptor, leading to its degradation in the lysosome 62, 63. Thus, Deltex can regulate Notch signalling in either a positive or a negative manner, depending on its interactions with other regulatory factors.
The HECT family E3 ligase Itch (suppressor of Deltex in Drosophila; AIP4 in humans) ubiquitinates membrane-bound inactive Notch receptor, targeting it for lysosomal degradation 64. Itch interacts with the endocytic sorting protein Numb, a well-known cell fate determinant that segregates asymmetrically in dividing cells and antagonises Notch signalling 65, 66. In human colon cancer cell lines, Numb promotes the goblet cell phenotype, consistent with its Notch-antagonising effects 67. Interestingly, however, Numb was also reported to be ubiquitously expressed throughout the murine intestinal epithelium 67, suggesting that there is a further layer of regulation that can mute this antagonism in Notch-high cells. The regulation of Notch signalling output by intracellular trafficking is still a subject of intense research (reviewed in 60), and the effects of most mammalian components of these pathways on intestinal homeostasis are yet to be clarified.
Fbw7
The F-box protein Fbw7 (also known as Fbxw7, Cdc4, Sel10, Ago) is part of a multisubunit SCF (Skp1, Cullin1, F-box)-type E3 ubiquitin ligase that targets many oncoproteins for proteasomal degradation (recently reviewed in 68). Many of these oncoproteins are also cell fate determinants that affect the balance between proliferation and differentiation within tissues as within tumours. NICD1 was identified as an Fbw7 target more than a decade ago 69, 70, 71, and the phenotype of Fbw7 deficiency often reflects that of increased Notch signalling. Notably, in the intestine, we and others have shown that complete inactivation of Fbw7 results in a decrease in the numbers of goblet cells and an increase in crypt cell proliferation 72, 73, 74. Interestingly, loss of a single Fbw7 allele also increases NICD levels and reduces goblet cell numbers 75. It was found that Fbw7 is haploinsufficient for Notch degradation in the intestine (and nervous system) as a consequence of an additional positive feedback loop between Notch and Fbw7. The Notch downstream target Hes5 inhibits Fbw7 transcription, thus limiting Notch degradation by Fbw7 when Notch is active 75. This additional level of cell-intrinsic regulation ensures that the initially small differences between a Notch-high progenitor and a Notch-low progenitor established by lateral inhibition are drastically augmented, which accelerates the differentiation of the two neighbouring progenitors towards different fates.
Deubiquitination: Usp12 and Usp28
Ubiquitination is a reversible process that is counter-regulated by the deubiquitinating enzymes (DUBs). There are nearly 100 encoded DUBs in humans 76. Of those, the ubiquitin-specific proteases Usp28 and Usp12 have been shown to regulate Notch 77, 78 (Fig3). Usp28 counteracts the action of Fbw7 and reduces ubiquitin-mediated proteasomal degradation of activated Notch (NICD), resulting in higher NICD levels 77, 79. Consistent with this modulation of Notch signalling, Usp28 activity regulates the balance of cell fates within the intestine. Deletion of Usp28 results in increased numbers of goblet cells and a corresponding decrease in proliferation 77. Although Usp28, like Fbw7, also targets other proteins involved in intestinal epithelial proliferation such as Myc 80, 81, the goblet cell phenotype highly resembles that of Notch inhibition and is likely due to the stabilising effect of Usp28 on NICD.
On the other hand, deubiquitination of Notch can also promote its degradation. Deubiquitination of the inactive, uncleaved Notch receptor by the ubiquitin-specific protease Usp12 promotes its trafficking away from the cell membrane and towards lysosomal degradation 78. This step is thought to occur after Itch-mediated polyubiquitination as part of the same trafficking pathway. Loss of Usp12, part of a family of deubiquitinating enzymes that act together with the Usp-activating factor UAF1, resulted in increased Notch activity 78.
Genetic regulation of Notch and lateral inhibition
The correct stoichiometric ratio of the different components of the lateral inhibition network is important for proper signalling, making it sensitive to variations in gene dosage and expression. Although the basic mechanism of lateral inhibition relies on positive feedback to promote strongly divergent signalling outcomes in adjacent cells, negative feedback mechanisms are also at play to ensure that the precise level of Notch signalling can be modulated spatially and temporally. For example, the Hes family of transcription factors negatively regulate their own transcription, leading to an intrinsic oscillation of signalling within cells 82. In the intestine, Hes factors also seem to be inhibited by their downstream target Atoh1 independently of Notch, since Hes expression was rescued in Atoh1/Notch double mutants 54. This rescue of upstream transcription factor expression as well as secretory cell overproduction with a single transcription factor change suggests that the secretory lineage “commitment”, although robustly patterned at the outset, is actually fairly easily reversible. Combined with the observed capacity of committed progenitors for reacquiring stem cell capability 12, 13, this adds a surprising amount of plasticity in lineage selection (an idea discussed in greater detail by Philpott and Winton 15). As will be discussed below, even within a Notch-high state, considerable variation in transcriptional output is possible via epigenetic regulation.
Epigenetic regulation of Notch and lateral inhibition
Notch activity depends on the chromatin status of its target genes. The chromatin serves as a platform to integrate different signals and enable interplay with other pathways. In a non-activated state, RBPJ transcriptional complexes are associated with histone demethylases 83, histone deacetylases and histone chaperones that collectively repress target gene expression. Upon binding of active NICD, these corepressors are displaced and histone acetylases, methylases and ubiquitinases are recruited to modulate chromatin accessibility to the transcriptional machinery. The histone acetyltransferases p300/CBP and PCAF act synergistically together with NICD and RBPJ to acetylate different residues within the histone tails, resulting in a transcriptionally activated chromatin status 84. Overlaid on this basic on–off switch is a complex network of epigenetic regulatory mechanisms that modulate gene expression depending on context. Although there has been relatively little investigation of these regulatory networks in the mammalian intestine, it was expected that epigenetic regulation of Notch signalling would involve selective chromatin accessibility depending on the transcriptional program of the committed cell type, similar to other systems. However, recent work from Kim and colleagues 85 has suggested that in the case of the intestinal crypt, a broadly open chromatin structure in most progenitor cells allows flexibility in cell fate assignment based on the rapidly changing needs of the tissue. They found comparable levels of H3K4me2 and H3K27Ac histone marks, indicating a permissive chromatin status, at most cis-transcriptional enhancer loci in both secretory and absorptive progenitors. Enhancers acting uniquely in progenitors were already marked in Lgr5-positive stem cells, suggesting early priming of chromatin for divergent transcriptional programs, and the marks were retained after lineage specification. On this chromatin background, the secretory-specific transcription factor Atoh1 was sufficient to determine two different final fates of “equally chromatinised” progenitors: differentiation of some of the progenitor cells towards the secretory fate by activating transcription of secretory genes and diversion of neighbouring cells from this fate by lateral inhibition via upregulation of Notch ligands 85. The direct transcriptional upregulation of Notch ligands by Atoh1 in Notch-low cells adds to the previously known repression of ligand genes in Notch-high cells and thus reinforces lateral inhibition.
Lateral inhibition deregulation in intestinal inflammation and cancer
Inflammation
Pathological inflammation in the intestine typically results from damage either to the mucus barrier or to the integrity of the underlying intestinal epithelium, leading to inappropriate contact with micro-organisms and immune response. This damage can be caused by acute infection, radiation injury or inflammatory bowel disorders such as Crohn's disease or ulcerative colitis. Because of its twin roles in secretory cell production and proliferation of intestinal epithelial cells, Notch signalling affects both the susceptibility to inflammation and the recovery from it (Fig3). Mutations or deregulation in Notch pathway components can cause insufficient or immature secretory cell production, reducing the effectiveness of the mucous barrier and increasing the vulnerability to inflammation. For example, abnormal expression of Hes1 and repression of Atoh1 are associated with goblet cell depletion in ulcerative colitis 86. In murine models of colitis [typically treatment with dextran sodium sulphate (DSS)], Notch is activated in the inflamed mucosa to stimulate cellular proliferation and regeneration of the tissue. When this process is disrupted, for example by deleting RBPJ in the intestinal epithelium, mice develop chronic colitis 87. Abnormally activated Notch leading to insufficient mucus production can also impair recovery from induced colitis 88. Interestingly, tight junctions seem to link the barrier function of the epithelium and Notch activation: overexpression of Claudin-1, a structural tight junction component, in the intestinal epithelium activated Notch. The molecular mechanism of this activation is not entirely clear but was found to rely on the activity of the matrix metallopeptidase MMP9 88. Since Notch is also thought to induce MMP9 89, this may be an example of a positive feedback loop. A recent study has found that deletion of Dclk1 (Dcamkl1), a marker of tuft cells 90, reduces expression of both Claudin-1 and Notch1 and impairs epithelial repair after radiation injury, which could fit with this link 91. It will be interesting to discover whether endogenous Claudin-1 is also upregulated in models of induced inflammation, as a mechanism of activating Notch signalling in response to damage. Although many of the details are still to be worked out, it is clear that tight control of Notch signalling is important to prevent and manage inflammation in the intestine, ensuring proper secretory cell production via lateral inhibition, while stimulating tissue regeneration via Notch activation.
Cancer
Unsurprisingly, given its crucial functions in both differentiation and proliferation, inappropriate activation of the Notch signalling pathway has been associated with the pathogenesis of colorectal cancer (CRC) (Fig3). Upregulation of Notch signalling pathway components (Notch, Hes1 and downstream targets) has been detected in intestinal adenomas in both human and mouse 37, 92, 93, 94. Although activation of Wnt signalling by mutation of the APC gene (APCmin/+) is sufficient to initiate intestinal adenomas 29, 30, Notch signalling promotes the development of adenomas in APCmin/+ mice 95 and is essential for the self-renewal of human colorectal tumour-initiating cells 96. Furthermore, inhibiting Notch cleavage by treatment with gamma-secretase inhibitors dramatically decreases APCmin/+-induced tumour formation by promoting differentiation of intestinal progenitors and intestinal tumour cells towards a secretory fate 37, 97. This finding highlights the importance of Notch signalling and lateral inhibition in tumorigenic processes. However, because of the pleiotropic functions of Notch, loss or inhibition of Notch signalling can be pro-tumorigenic in other tissues such as skin and vasculature and result in serious side effects 98, 99, precluding the systemic use of gamma-secretase inhibitors for colorectal cancer treatment.
Several examples demonstrate that disrupting or enhancing the activity of regulators of the lateral inhibition network is key in intestinal tumourigenesis. As described above, one of the mechanisms regulating Notch levels relies on its ubiquitination by different E3 ligases and subsequent degradation. Fbw7 is the best characterised E3 ligase regulating Notch in the intestinal tissue. Fbw7 loss-of-function mutations are observed in 10% of human CRC 28. Furthermore, in an APCmin/+ background, loss of Fbw7 causes very aggressive adenocarcinomas by promoting self-renewal in the crypt cells and by inhibiting differentiation towards a secretory fate 72. Interestingly, Fbw7 is haploinsufficient for APCmin/+-induced tumourigenesis and NICD1 protein degradation, suggesting that a small deregulation in Notch regulators can be amplified by the multiple regulatory loops of lateral inhibition 75.
While restoring the activity of Fbw7 in tumours would be difficult, an alternative could be to inhibit the function of its associated deubiquitinase, Usp28. Genetic deletion of Usp28 in the intestinal epithelium reduces NICD levels and increases goblet cell differentiation in APCmin/+ tumours, consistent with an inhibition of Notch signalling 77. Moreover, inducible deletion of Usp28 in established APCmin/+ tumours slows their progression and increases the lifespan of affected mice 77. Further work will be needed to determine whether chemical inhibition of Usp28 is possible and whether the desired tumour-suppressive outcomes can be achieved in vivo. Once again, a balance must be achieved in disrupting the excessive activation of Notch that leads to intestinal overproliferation, while preserving the essential and sometimes tumour-suppressive functions of Notch signalling within normal tissues.
Conclusion and outlook
The multiple uses of Notch signalling within metazoan tissues continue to be revealed, more than 70 years after the “notched” wing phenotype was first noted in Drosophila melanogaster. Although the relative simplicity of the canonical Notch pathway compared with other cell signalling mechanisms was striking (described in 1998 as a “short cut to the nucleus” 100), the capacity of Notch signalling for intercellular communication and sensitivity to multiple levels of regulation and feedback inevitably mean that its outputs are varied and complex. In the mammalian intestine, Notch signalling is essential for the self-renewal and proper differentiation of the intestinal epithelium. Its characteristic property of lateral inhibition has been co-opted to regulate the arrangement of mixed cell populations: stem cells within the Paneth cell niche at the crypt base, and secretory cells amongst the absorptive lineage cells of the transit-amplifying compartment and villus. Disruption of these cell fate-determining and regenerative mechanisms can lead to inflammatory disorders and cancer. Although modulating such a fundamental pathway is never straightforward, understanding the more subtle regulatory mechanisms that influence Notch signalling should help us to identify more precise therapeutic targets. The recent findings in the gut system described here may also facilitate the elucidation of Notch-mediated mechanisms in other species and tissues.
Acknowledgments
The authors wish to dedicate this review to the memory of Julian Lewis. We thank Vivian Li for critical reading of the manuscript and apologise to the many colleagues whose work could not be cited due to space considerations. This work was supported by ERC grant 281661 and MRC grant G0901677. RS was funded by a Marie Curie Intra-European Fellowship, MEIF-CT-2006-041119. The London Research Institute is funded by Cancer Research UK.
Glossary
- ADAM10
a disintegrin and metallopeptidase domain-containing protein 10
- APC
adenomatous polyposis coli
- ATOH1
atonal homologue 1
- BMP
bone morphogenetic protein
- CBC
crypt base columnar cells
- CBP
CREB-binding protein
- CDK
cyclin-dependent kinase
- DLL4
delta-like 4
- DSS
dextran sodium sulphate
- DUB
deubiquitinating enzyme
- EphB3
ephrin type-B receptor 3
- Fbw7
F-box and WD repeat domain-containing 7
- Hes
hairy and enhancer of split
- Hey
hairy/enhancer-of-split related with YRPW motif protein
- ISC
intestinal stem cell
- Itch
Itchy E3 ubiquitin protein ligase
- Lgr5
leucine-rich repeat-containing G-protein-coupled receptor 5
- NICD
Notch intracellular domain
- Olfm4
olfactomedin 4
- PCAF
P300/CBP-associated factor
- RBPJ-k
recombining binding protein suppressor of hairless
- SCF
Skp, Cullin, F-box containing complex
- TA
transit amplifying
- Usp12
ubiquitin-specific protease 12
- Usp28
ubiquitin-specific protease 28
- Wnt
wingless-related MMTV integration site 1
Conflict of interest
The authors declare that they have no conflict of interest.
Sidebar A: Some important unanswered questions in the field
-
What are the sources of Notch ligand in the crypt base stem cell niche?
The Notch ligands Dll1 and Dll4 have been shown to be crucial for maintenance of the stem cell niche 45, but the Paneth cells, a known source of these ligands, have been shown to be dispensable in vivo 20, 21. However, in these studies, the need for Notch signalling is largely bypassed by Atoh1 deletion, and others have argued that Paneth cells do contribute to the stem cell niche in wild-type animals 19, 22. Secretory progenitors located immediately above the niche that express Dll1 13, 46, or possibly underlying mesenchymal cells, are potential alternative sources of Notch ligand.
-
Is Notch signalling required for facultative as well as CBC stem cell function?
Two of the cell types proposed as facultative stem cells, which can move down into the crypt base and take over the function of CBC stem cells during regeneration, are secretory progenitors—a Notch-low state 12, 13. It has been shown that these re-express Lgr5 when “recalled” to the crypt base, but do they also return to a Notch-high state and re-express Olfm4? If so, how is Notch activated? To what extent is the change in location to the crypt base “niche” required for plasticity?
-
Does Notch signalling in the intestine provide new clues for other tissues/systems?
As reviewed here, the Notch signalling pathway is a widely conserved pathway that regulates cellular identity, proliferation and differentiation via the process of lateral inhibition. The core components of Notch signalling have been shown to be conserved and essential in different tissues such as the central nervous system, lung and haematopoietic system as well as the intestine 101, 102, 103. The intestinal model has recently provided new examples of Notch signalling regulators (Fig3). Whether these regulatory mechanisms are also shared in the different tissues or are specific to the intestine remains to be elucidated.
-
Is Notch stability suitable for therapeutic targeting?
Increased Notch signalling is associated with different intestinal disease conditions (see Fig3), thus making Notch a potential therapeutic target. So far, the only clinically available drugs are gamma-secretase inhibitors, which inhibit Notch cleavage. However, the new regulatory steps in the Notch pathway identified in the intestine (Fig3) provide additional druggable targets (e.g. DUBs) that could be useful not only in the intestine but also in other systems. It remains to be seen whether modulation of protein stability can overcome some of the side effects associated with systemic Notch inhibition.
References
- Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- Scoville DH, Sato T, He XC, Li L. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- Gerbe F, Legraverend C, Jay P. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci. 2012;69:2907–2917. doi: 10.1007/s00018-012-0984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud A, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, van Oudenaarden A. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2012;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J, Stange DE, Schepers AG, de van Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent “+4” cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- van Es JH, Sato T, de van Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Philpott A, Winton DJ. Lineage selection and plasticity in the intestinal crypt. Curr Opin Cell Biol. 2014;31C:39–45. doi: 10.1016/j.ceb.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Escudero S, Shivdasani RA. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc Natl Acad Sci USA. 2012;109:3932–3937. doi: 10.1073/pnas.1113890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand A, Donahue B, Peignon G, Letourneur F, Cagnard N, Slomianny C, Perret C, Shroyer NF, Romagnolo B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc Natl Acad Sci USA. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518–1529. doi: 10.1053/j.gastro.2012.08.031. .e7. [DOI] [PubMed] [Google Scholar]
- Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci USA. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 2006;419:337–383. doi: 10.1016/S0076-6879(06)19014-X. [DOI] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26:570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Mori H. Multistep carcinogenesis of the colon in Apc(Min/+) mouse. Cancer Sci. 2007;98:6–10. doi: 10.1111/j.1349-7006.2006.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Collu GM, Hidalgo-Sastre A, Brennan K. Wnt-Notch signalling crosstalk in development and disease. Cell Mol Life Sci. 2014;71:3553–3567. doi: 10.1007/s00018-014-1644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Fischer A, Gessler M. Delta-Notch–and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueo T, Imayoshi I, Kobayashi T, Ohtsuka T, Seno H, Nakase H, Chiba T, Kageyama R. The role of Hes genes in intestinal development, homeostasis and tumor formation. Development. 2012;139:1071–1082. doi: 10.1242/dev.069070. [DOI] [PubMed] [Google Scholar]
- Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, Kaestner KH, Kopan R, Lewis J, Radtke F. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240. doi: 10.1053/j.gastro.2011.01.005. .e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamataki D, Holder M, Hodgetts C, Jeffery R, Nye E, Spencer-Dene B, Winton DJ, Lewis J. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS ONE. 2011;6:e24484. doi: 10.1371/journal.pone.0024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Hannezo E, Sale S, Huyghe M, Lafkas D, Kissel H, Louvi A, Greve J, Louvard D, Artavanis-Tsakonas S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE. 2011;6:e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YH, VanDussen KL, Sawey ET, Wade AW, Kasper C, Rakshit S, Bhatt RG, Stoeck A, Maillard I, Crawford HC, et al. ADAM10 regulates Notch function in intestinal stem cells of mice. Gastroenterology. 2014;147:822–834. doi: 10.1053/j.gastro.2014.07.003. .e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Samuelson LC. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev Biol. 2010;346:215–223. doi: 10.1016/j.ydbio.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Milano J, McKay J, Dagenais C, Foster-Brown L, Pognan F, Gadient R, Jacobs RT, Zacco A, Greenberg B, Ciaccio PJ. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82:341–358. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- Kim TH, Shivdasani RA. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J Biol Chem. 2011;286:11427–11433. doi: 10.1074/jbc.M110.188797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanjian A, Noah T, Brown D, Burkart J, Shroyer NF. Atonal homolog 1 is required for growth and differentiation effects of notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology. 2010;139:918–928. doi: 10.1053/j.gastro.2010.05.081. .e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1:18. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development. 2005;132:1093–1104. doi: 10.1242/dev.01644. [DOI] [PubMed] [Google Scholar]
- LeBon L, Lee TV, Sprinzak D, Jafar-Nejad H, Elowitz MB. Fringe proteins modulate Notch-ligand cis and trans interactions to specify signaling states. Elife. 2014;3:e02950. doi: 10.7554/eLife.02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M. Endocytic routes to Notch activation. Semin Cell Dev Biol. 2012;23:437–442. doi: 10.1016/j.semcdb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- Puca L, Chastagner P, Meas-Yedid V, Israel A, Brou C. Alpha-arrestin 1 (ARRDC1) and beta-arrestins cooperate to mediate Notch degradation in mammals. J Cell Sci. 2013;126:4457–4468. doi: 10.1242/jcs.130500. [DOI] [PubMed] [Google Scholar]
- Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem. 2009;284:26427–26438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhu R, Bai J, Zhang X, Tian Y, Li X, Peng Z, He Y, Chen L, Ji Q, et al. Numb modulates intestinal epithelial cells toward goblet cell phenotype by inhibiting the Notch signaling pathway. Exp Cell Res. 2011;317:1640–1648. doi: 10.1016/j.yexcr.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R, Abuzinadah M, Davis H, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J, Delrow JJ, Small T, Grady WM, Nakayama KI, et al. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol Cell Biol. 2012;32:2160–2167. doi: 10.1128/MCB.00305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R, Blake SM, Tendeng C, Clurman BE, Lewis J, Behrens A. Fbw7 repression by hes5 creates a feedback loop that modulates Notch-mediated intestinal and neural stem cell fate decisions. PLoS Biol. 2013;11:e1001586. doi: 10.1371/journal.pbio.1001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbacher ME, Popov N, Blake SM, Schulein-Volk C, Nye E, Spencer-Dene B, Jaenicke LA, Eilers M, Behrens A. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest. 2014;124:3407–3418. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti J, Chastagner P, Liang CC, Cohn MA, Israel A, Brou C. The ubiquitin-specific protease 12 (USP12) is a negative regulator of notch signaling acting on notch receptor trafficking toward degradation. J Biol Chem. 2012;287:29429–29441. doi: 10.1074/jbc.M112.366807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbacher ME, Chakraborty A, Blake SM, Mitter R, Popov N, Eilers M, Behrens A. Usp28 counteracts Fbw7 in intestinal homeostasis and cancer. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-1726. doi: 10.1158/0008-5472.CAN-14-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- Sansom OJ. Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, Rodriguez P, Dominguez M, Borggrefe T. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Li F, Ferreiro-Neira I, Ho LL, Luyten A, Nalapareddy K, Long H, Verzi M, Shivdasani RA. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Tsuchiya K, Okamoto R, Iwasaki M, Kano Y, Sakamoto N, Nakamura T, Watanabe M. Suppression of hath1 gene expression directly regulated by hes1 via notch signaling is associated with goblet cell depletion in ulcerative colitis. Inflamm Bowel Dis. 2011;17:2251–2260. doi: 10.1002/ibd.21611. [DOI] [PubMed] [Google Scholar]
- Obata Y, Takahashi D, Ebisawa M, Kakiguchi K, Yonemura S, Jinnohara T, Kanaya T, Fujimura Y, Ohmae M, Hase K, et al. Epithelial cell-intrinsic Notch signaling plays an essential role in the maintenance of gut immune homeostasis. J Immunol. 2012;188:2427–2436. doi: 10.4049/jimmunol.1101128. [DOI] [PubMed] [Google Scholar]
- Pope JL, Bhat AA, Sharma A, Ahmad R, Krishnan M, Washington MK, Beauchamp RD, Singh AB, Dhawan P. Claudin-1 regulates intestinal epithelial homeostasis through the modulation of Notch-signalling. Gut. 2014;63:622–634. doi: 10.1136/gutjnl-2012-304241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tan J, Zhang Y, Han N, Di X, Xiao T, Cheng S, Gao Y, Liu Y. DLK1 promotes lung cancer cell invasion through upregulation of MMP9 expression depending on Notch signaling. PLoS ONE. 2014;9:e91509. doi: 10.1371/journal.pone.0091509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology. 2009;137:2179–2180. doi: 10.1053/j.gastro.2009.06.072. ; author reply 2180-2171. [DOI] [PubMed] [Google Scholar]
- May R, Qu D, Weygant N, Chandrakesan P, Ali N, Lightfoot SA, Li L, Sureban SM, Houchen CW. Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells. 2014;32:822–827. doi: 10.1002/stem.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Majada V, Aguilera C, Villanueva A, Vilardell F, Robert-Moreno A, Aytes A, Real FX, Capella G, Mayo MW, Espinosa L, et al. Nuclear IKK activity leads to dysregulated notch-dependent gene expression in colorectal cancer. Proc Natl Acad Sci USA. 2007;104:276–281. doi: 10.1073/pnas.0606476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, Artavanis-Tsakonas S, Louvard D. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedijk M, Odorcic S, Zhang H, Chetty R, Tennert C, Dickson BC, Lockwood G, Gallinger S, Egan SE. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncol. 2008;33:1223–1229. doi: 10.3892/ijo_00000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, Lopez-Bigas N, Bellora N, Alba MM, Torres F, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci USA. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, Lipkin SM. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaleb AM, Aggarwal G, Bialkowska AB, Nandan MO, Yang VW. Notch inhibits expression of the Kruppel-like factor 4 tumor suppressor in the intestinal epithelium. Mol Cancer Res. 2008;6:1920–1927. doi: 10.1158/1541-7786.MCR-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Liu Z, Turkoz A, Jackson EN, Corbo JC, Engelbach JA, Garbow JR, Piwnica-Worms DR, Kopan R. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest. 2011;121:800–808. doi: 10.1172/JCI43114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. Notch signalling. A short cut to the nucleus. Nature. 1998;393:304–305. doi: 10.1038/30597. [DOI] [PubMed] [Google Scholar]
- Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BJ, Kleeberger W, Ball DW. Notch in lung development and lung cancer. Semin Cancer Biol. 2004;14:357–364. doi: 10.1016/j.semcancer.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Bigas A, Robert-Moreno A, Espinosa L. The Notch pathway in the developing hematopoietic system. Int J Dev Biol. 2010;54:1175–1188. doi: 10.1387/ijdb.093049ab. [DOI] [PubMed] [Google Scholar]