Abstract
In mammals, the nuclear lamina interacts with hundreds of large genomic regions, termed lamina-associated domains (LADs) that are generally in a transcriptionally repressed state. Lamins form the major structural component of the lamina and have been reported to bind DNA and chromatin. Here, we systematically evaluate whether lamins are necessary for the LAD organization in murine embryonic stem cells. Surprisingly, removal of essentially all lamins does not have any detectable effect on the genome-wide interaction pattern of chromatin with emerin, a marker of the inner nuclear membrane. This suggests that other components of the lamina mediate these interactions.
Keywords: interphase chromosome organization, genome-wide mapping; nuclear architecture, nuclear lamina
Introduction
The nuclear lamina (NL) is thought to play a role in the spatial organization of the genome inside the interphase nucleus, by providing an interaction platform for hundreds of large genomic regions named lamina-associated domains (LADs). Genes in LADs are generally inactive, indicating that LADs form a transcriptionally repressive chromatin compartment 1. It is likely that specific proteins of the NL recognize DNA sequences or chromatin features in LADs and thereby anchor LADs to the periphery 2. However, the identity of these NL proteins is still largely unknown.
The primary components of the NL are lamins, which are intermediate filament-type proteins that form a fibrous network 3. Most mammalian somatic cells express four lamin proteins derived from three genes: lamins A and C (LmA/C), encoded by the LMNA gene, and B1 and B2, encoded by LMNB1 and LMNB2, respectively. In addition to lamins, the NL harbors a range of other proteins, several of which are anchored in the inner nuclear membrane 4.
Several reports suggest that lamins may be involved in chromosome organization. Both A- and B-type lamins can directly interact with genomic DNA and chromatin in vitro 5. In D. melanogaster and C. elegans, depletion of lamins can have effects on gene positioning and silencing 6. Human fibroblasts with a dominant-negative mutation in the LmA gene causing Hutchinson–Gilford progeria syndrome exhibit altered chromatin–LmA/C interactions, loss of peripheral heterochromatin, and a perturbed overall chromosome structure 7, 8. Depletion of lamin B1 (LmB1) in somatic mouse and human cells was found to lead to changes in interphase chromosome morphology and positioning, concomitant with genome-wide changes in gene expression 9, 10, 11, 12. In contrast, no change in cell behavior and gene expression was observed in keratinocytes and murine embryonic stem (mES) cells lacking lamins 13, 14, 15.
While most of these data point to a role for lamins in certain aspects of interphase chromosome architecture, they do not directly address whether lamins are needed for overall LAD organization. Here, we address this issue by generating a series of high-resolution maps of NL–genome interactions in mES cells upon systematic removal of all lamins. We correlate these results with microscopy observations and gene expression profiling. Surprisingly, these data indicate that lamins are dispensable for LAD organization of the genome, at least in mES cells.
Results
To address the role of lamins in the formation of LADs, we used the DNA adenine methyltransferase identification (DamID) technique 1 to construct genome-wide maps of NL interactions in wild-type (wt) and lamin B1/B2 double knockout (dKO) mES cells 13.
Naturally, we could not perform DamID with a Dam-lamin fusion protein, as this would reintroduce a lamin into the null background. We instead selected the lamin-interacting inner nuclear membrane protein emerin (Emd) as Dam-fusion partner, for several reasons. First, Emd is endogenously present in mES cells and it is expressed at similar levels in wt and dKO mES cells, as indicated by mRNA sequencing (Supplementary Fig S1A). Second, by immunofluorescence microscopy, we found that the localization of Emd at the nuclear envelope is not perturbed in dKO mES cells (Supplementary Fig S1B), which is consistent with recent reports 13, 16. Third, in human cells, the DamID profiles of Emd and LmB1 are virtually identical 1, indicating that Dam-Emd can be used instead of Dam-LmB1 to identify LADs. To verify that this is also the case in wt mES cells, we generated a genome-wide DamID profile for Emd and compared it to the one previously generated with LmB1 17. Indeed, the two profiles were similar, with an overall Pearson correlation coefficient of 0.87 and highly similar domain patterns along the chromosomes (Supplementary Fig S1C and D).
Having established that Emd can substitute for LmB1 to identify LADs, we then generated DamID profiles in wt and dKO mES cells. Dam-Emd was expressed at very low levels using the low leaky activity of a non-induced Drosophila heat-shock promoter 18 in order to avoid interference with endogenous NL components. Amplification of the DNA methylated by Dam-Emd did not show differences in yield between the cell lines (Supplementary Fig S1E), further indicating that in dKO cells, there is no major relocation of Dam-Emd from the inner nuclear membrane to a cytoplasmic compartment. This is consistent with previous observations that Emd is largely retained in the nuclear envelope of mES cells lacking lamins, which contrasts with Emd behavior in differentiated cell types lacking lamins 16, 19. For both wt and dKO mES cells, we obtained Emd interaction maps by combining the data from two independent DamID experiments.
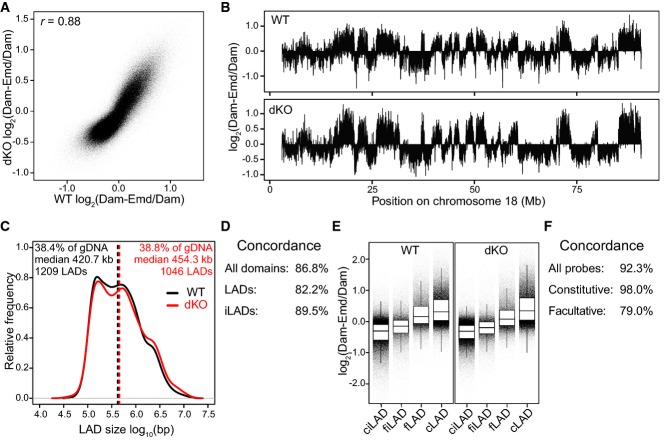
Strikingly, the Emd interaction patterns of wt and dKO mES cells were highly similar in genome-wide correlation, amplitude of the signals, and overall appearance (Fig1A and B). We used a domain detection algorithm 1 to determine for each cell line the number, size, and genome coverage of the LADs. While the total number of LADs was slightly reduced in dKO cells, their total coverage along the genome was nearly identical (38.4% vs 38.8%), and there was a strong concordance between their positions in wt and dKO cells (Fig1C and D). Taken together with a general lack of off-diagonal data points in the scatterplot analysis (Fig1A), these data indicate that overall LAD organization is largely retained in dKO cells.
Figure 1. No detectable changes in LADs organization in dKO mES cells.
- Scatterplot of log2 ratio of Dam-Emd over Dam for wt compared to dKO mES cells. Pearson correlation coefficient (r) is indicated.
- Emd interaction profile along chromosome 18 in wt and dKO mES cells.
- Distribution of LAD sizes in wt (black) and dKO (red) mES cells. Dashed lines mark the median LAD sizes. Percentage of total genomic DNA covered by LADs (% of gDNA), the median size of LADs, and the total number of LADs are listed for wt (black) and dKO (red) cells.
- Concordance between wt and dKO cells for LADs and iLADs as defined in (C).
- Distribution of Emd DamID probe values in wt and dKO mES cells, divided into facultative (f) and constitutive (c) LADs and iLADs. Horizontal lines of boxes depict percentiles 25, 50, and 75; vertical lines extend from the box edge to the highest or lowest value that is within 1.5× inter-quartile range of the edge.
- Concordance between wt and dKO cells for facultative (fLAD, fiLAD) and constitutive (cLAD, ciLAD) regions, as identified in (E).
Data information: Data in (A) and (B) were smoothed (median) with a running window of 11 probes. DamID data are averages of two independent experiments.
We then investigated whether specific subsets of LADs were affected, which may not be noticeable in the bulk analyses above. Specifically, we tested whether the previously identified facultative (cell-type specific) or constitutive (cell-type invariant) LADs and inter-LADs were affected 20. Given their different dynamics during cell differentiation, it was possible that they would respond differently to the loss of B-type lamins. However, these regions showed a high overall concordance, with almost identical interactions of the constitutive regions (Fig1E and F). A somewhat lower concordance was observed in facultative LADs, but this should be interpreted with caution because these regions have somewhat weaker DamID signals overall in wt cells, and therefore, the signal/noise ratio may be lower in these regions. Finally, we applied a specially designed statistical test to identify genes with significantly altered DamID signals 17. This test yielded no significant genes. We conclude that LADs remain largely unaffected in dKO mES cells.
Next, we investigated whether B-type lamins are involved in repressing genes at the NL. We generated mRNA expression profiles of wt and dKO mES cells and averaged two biological replicates for each cell line. In wt mES cells, the genes that interact with the NL (high DamID log2-ratios) generally exhibit low mRNA expression (Fig2A), as it was reported previously for various cell types 1, 17. This correlation was also observed in dKO mES cells, indicating that the NL remains a repressive environment regardless of the presence of B-type lamins (Fig2B). The wt and dKO mES cell mRNA profiles showed an overall Pearson correlation coefficient of 0.99, with only 94 genes changing expression (P ≤ 0.05 and fold change ≥ 2) out of 37,991 genes that were analyzed (Supplementary Fig S2A). Of the differentially expressed genes, 18 are in LADs and 76 in inter-LADs, a distribution that is not statistically different from what may be expected by chance (P = 0.7, Fisher's exact test). Indeed, gene expression changes between wt and dKO mES cells did not correlate with NL interaction changes (Fig2C) or with their constitutive or facultative status (Supplementary Fig 2B and C), again indicating that the rather modest changes in gene expression are generally not linked to altered NL interactions.
Figure 2. Gene repression is not detectably compromised in dKO mES cells.
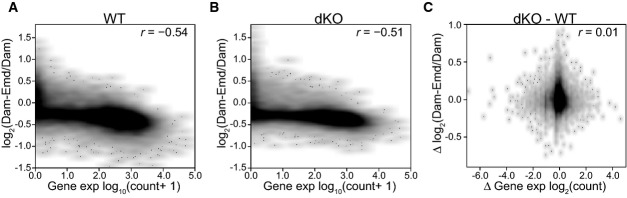
- A, B Density scatterplot of the mean Emd DamID score per gene versus expression levels based on RNA-seq for wt (A) and dKO (B) mES cells.
- C Density scatterplot of changes in Emd interaction versus changes in gene expression in dKO relative to wt cells.
Data information: Pearson correlation coefficients (r) are indicated. Gene expression data and DamID data are averages of two independent experiments.
One possible caveat to our analyses is that B-type lamins could be functionally compensated by LmA/C. While LmA/C was initially thought not to be present in mES cells 13, 21, it was later established with multiple antibodies that it is expressed in mES cells, including the cell lines studied here 15, 16, 22. In our cultures, we could detect moderate levels of LmA/C transcripts by RT–PCR and RNA sequencing and of LmA/C proteins by Western blot using two different antibodies, one detecting both isoforms and one specific for LmA (Supplementary Fig S3A–C). Co-staining with an Oct4 antibody confirmed that the cells are in an undifferentiated state (Supplementary Fig S3D).
Immunofluorescence microscopy of LmA/C showed the classic nuclear rim localization in wt mES cells. In dKO mES cells, LmA/C also localized to the nuclear rim, but it showed a more punctuated distribution. This we observed with the two different antibodies described above (Supplementary Figs S1B and S3E), and is in agreement with recently reported observations 16. In addition, we observed the patchy distribution of LmA/C in LmB1−/− mES cells, but not in LmB2−/− cells, pointing to LmB1 as being responsible for the even distribution of LmA/C along the nuclear envelope (Supplementary Fig S3F).
We could recapitulate the LmA patchy pattern by lentivirus-mediated expression of GFP-tagged LmA in dKO mES cells, while GFP-LmA exhibited a normal homogeneous rim staining in wt mES cells (Supplementary Fig S3G). Surprisingly, GFP-tagged LmC also accumulated in bright spots in the nuclear interior of dKO mES cells, while it was properly located at the nuclear periphery in wt mES cells (Supplementary Fig S3H), indicating that LmC requires one or both B-type lamins for its proper integration in the NL of mES cells.
Previous studies have shown that LmA and B-type lamins interact with the same LADs 20, 23, 24. To test whether LmA requires B-type lamins for its LAD interactions, we generated DamID maps using Dam-LmA in wt and dKO mES cells. Because the DamID protocol requires the Dam-LmA protein to be expressed at very low levels 18, it is unlikely that this small amount of fusion protein perturbs the distribution of endogenous LmA. In the rare cells in which the Dam-LmA is weakly detectable by immunofluorescence microscopy, we found it to show a similar patchy appearance at the nuclear rim as endogenous LmA in dKO cells (Supplementary Fig S3I).
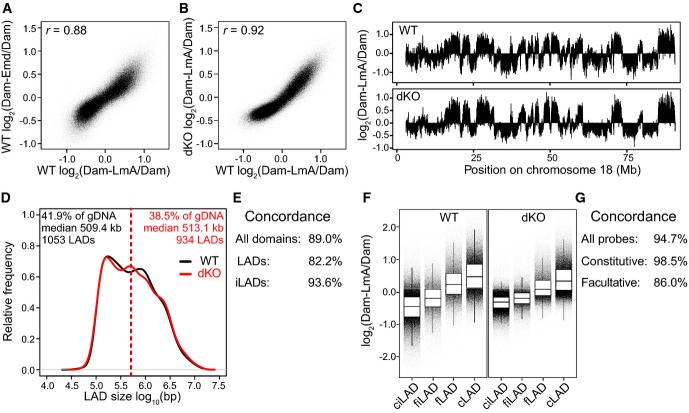
Inspection of the DamID profiles revealed that in wt mES cells, the detected LmA interactions were similar to the Emd interactions (Fig3A). Interestingly, LmA–genome interactions in dKO cells were highly similar to those in wt cells (Fig3B–G). Thus, even though B-type lamins are absent and LmA is not homogeneously distributed along the nuclear periphery, LmA in dKO mES cells can still interact with all LADs that are contacted by Emd in dKO mES cells and by LmA and Emd in wt mES cells. The difference in DamID signal between LADs and inter-LAD regions (iLADs) is lower in dKO mES cells (Fig3F), possibly since only a random subset of LADs in each single cell can interact with the patches of LmA.
Figure 3. LmA interactions with LADs in wt and dKO mES cells.
- A–C Comparisons of LmA and Emd interaction profiles in wt and dKO mES cells. Samples were smoothed (median) with a running window of 11 probes. Pearson correlation coefficients (r) are indicated.
- D–G Analysis of LADs as detected with Dam-LmA in wt and dKO cells, similar to Fig1C–F, respectively.
Data information: DamID data are averages of two independent experiments.
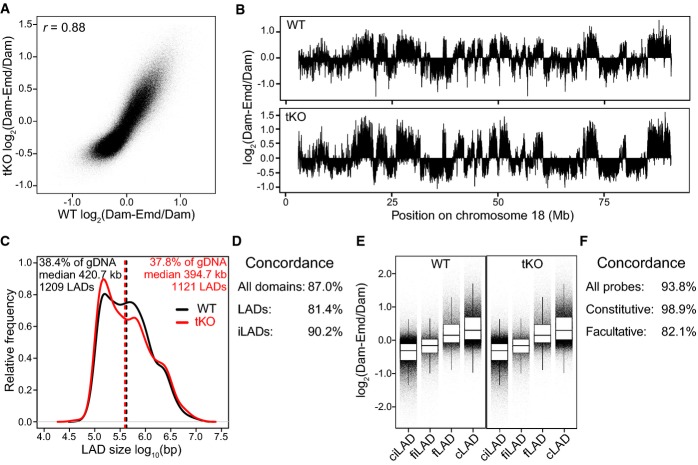
To address whether LmA/C are required for LAD formation in the absence of B-type lamins, we further reduced LmA/C expression in dKO mES cells by RNA interference using a short hairpin RNA (shRNA). By RT–qPCR, Western blot, and immunofluorescence labeling, we found that the LmA/C transcripts and proteins were substantially reduced (Supplementary Fig S4A–C). For simplicity, we will refer to the resulting cells as tKO mES cells. We confirmed that in the absence of all lamins, mES cells maintain their undifferentiated phenotype (Supplementary Fig S4D). To perform DamID, we again used Dam-Emd, since Emd is clearly present at the NE of mES cells also in the absence of LmA/C (Supplementary Fig S4E) 15, 16 and amplification of the DNA methylated by Dam-Emd did not show differences in yield between the cell lines (Supplementary Fig S4F). Again, the DamID maps show that chromatin organization in wt and tKO mES cells is virtually indistinguishable (Fig4A–F). Statistical analysis 17 did not detect any gene with a significant change in DamID log-ratios. Thus, removal of nearly the entire lamin meshwork has no detectable influence on the interaction pattern of the genome with the residual NL in mES cells.
Figure 4. No detectable changes in LADs organization in tKO mES cells.
- A Scatterplot of log2 ratio of Dam-Emd over Dam for wt against tKO mES cells. Pearson correlation coefficient (r) is indicated.
- B Emd interaction profile along chromosome 18 in wt and tKO mES cells.
- C–F Analysis of LADs as detected with Dam-Emd in wt and tKO cells, similar to Fig1C–F, respectively.
Data information: Data in (A) and (B) were smoothed (median) with a running window of 11 probes. DamID data are averages of two independent experiments. The wt dataset is identical to that in Fig1.
Complementary to this DamID mapping in tKO mES cells, we inspected the distribution of DNA inside the nucleus of dKO mES cells by confocal microscopy. We noticed that not only LmA has a patchy appearance, but also the density of DAPI staining is inhomogeneous along the nuclear rim (Fig5A–C). We reasoned that if LmA participates in the tethering of heterochromatin, then the LmA-rich patches might preferentially bind regions with more dense DAPI staining, compared to the intervening LmA-poor areas of the NL. We designed a quantitative image analysis approach to test this (Fig5B–D). Analysis of 54 nuclei revealed that there is no significant positive correlation between the presence of LmA and local DNA intensity (Fig5D), again suggesting that the presence of LmA does not detectably help in tethering DNA to the NL.
Figure 5. Random distribution of DNA relative to LmA patches in dKO cells.
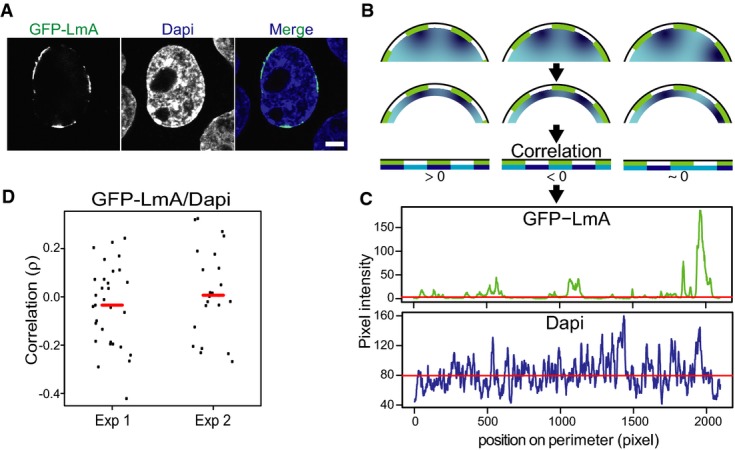
- Representative confocal image showing the patchy NL localization of GFP-LmA in dKO mES cells and the corresponding DAPI staining of DNA. Scale bar, 5 μm.
- Cartoon showing how the image analysis was performed. LmA patches are depicted in green, different DNA concentrations in shades of blue. The correlation between LmA and DAPI pixel intensity is determined in a 10-pixel shell, as shown in the bottom row.
- Pixel intensity values of GFP-LmnA and DAPI along the cell perimeter for the nucleus shown (A).
- Correlation analysis of DAPI and GFP-LmA signals along the nuclear perimeter. Graph shows the distribution of Spearman correlation coefficients for two independent experiments (Exp1, n = 31; Exp2, n = 23). These distributions are not significantly different from 0 (P > 0.2, Student's t-test). Red lines mark median correlation coefficients.
Discussion
Although lamins are able to interact with chromatin in vitro and in vivo 5, we report here that lamins are to a very large extent dispensable for the LAD organization of the genome in mES cells. Because Dam-Emd produces in wt cells essentially the same genome-wide DamID profile as Dam-LmB1, and because it has previously been demonstrated that Dam-LmnB1 methylation signals correlate with NL proximity in the nucleus 1, 17, 23, it is reasonable to interpret the DamID profiles obtained here with Dam-Emd as maps of NL contact probabilities in the nucleus. We found that this genome-wide NL interaction pattern remains virtually unchanged in the absence of LmB1, LmB2, and LmA/C. Moreover, only a handful of genes exhibit altered expression in the absence of LmB1 and LmB2, but these genes are not enriched in LADs, indicating that B-type lamins are not involved in silencing genes at the NL, which is in agreement with a previous study 13.
Our results contrast with results obtained in flies and worms, where depletion of lamins was found to affect the expression and the peripheral positioning of specific genomic loci 6, 25, 26. Loss of LmB1 has also been reported to result in changes in nuclear organization in differentiated mouse and human cells. For example, in mouse fibroblasts, the loss of LmB1 caused relocation of chromosome 18 from the periphery toward the nuclear interior 9. Depletion of LmB1 in a human colon cancer cell line resulted in overall decondensation of chromosome territories 10, while in HeLa cells, it was reported to lead either to an overall reduction in transcription followed by relocation of chromosomes toward the nuclear periphery 11, or to the formation of nuclear blebs that have a euchromatic appearance yet are devoid of transcription activity 12. These results were all based on microscopy observations. A study in human fibroblast expressing progerin, a dominant-negative form of lamin A, also indicates that the disruption of the nuclear lamina results in a loss of peripheral heterochromatin, change in H2K27me3 distribution, and a global loss of chromatin compartmentalization 7.
Our data suggest that mES cells do not require lamins for overall LAD organization. It is possible that mES cells employ a unique mechanism for LAD organization, because mES cells appear to have a more dynamic chromatin architecture 27, 28, exhibit an extremely rapid cell cycle 29, have a more plastic NL architecture in which LmB1 is less stably incorporated 30, and have a higher AT content in its LAD sequences compared to other differentiated cells 20.
It thus seems likely that one or more non-lamin components of the NL help to organize LADs. The lamin B receptor (Lbr) was reported to interact with chromatin in vitro 31, 32, 33 and recently to act redundantly with LmA/C in preventing pericentric heterochromatin from aggregating in the nuclear interior in mouse 34, 35. According to the latter studies, Lbr is important for organizing heterochromatin and for correct cell differentiation and gene expression. We note that this conclusion was based on microscopy analyses rather than on a genome-wide molecular contact assay such as DamID. We found that Lbr is expressed and localized at the NL in wt, dKO, and tKO mES cells (Supplementary Fig S5). It will thus be interesting to investigate the role of Lbr in genome-wide LAD organization in mES cells. Other candidates are the LEM-domain proteins such as Emd, which can interact with chromatin components 36, and other nuclear envelope transmembrane proteins, some of which were found to affect the nuclear localization of entire chromosomes in specific cell types 37. Systematic knockout of these candidate proteins combined with DamID mapping may help to identify NL proteins that govern LAD organization. Considering that ∽40% of the mouse genome consists of LADs, we anticipate that multiple, possibly redundant mechanisms exist.
Materials and Methods
Cell culture, lentivirus transductions, and DamID
mES cells were cultured as described in Supplementary Methods. The wt and dKO mES cells showed no major differences in proliferation rate, cell cycle distribution, and undifferentiated status (Supplementary Fig S6A–D). Comparison of our mRNA-seq data to published microarray-based RNA expression profiles from the same mES cells 13 showed good correspondence (within the limits of such a cross-platform comparison), with similar expression levels of LmA/C and various mES cell markers (Supplementary Fig S6E and F). The mRNA-seq data also confirmed a > 100 and > 30-fold reduction in LmB1 and LmB2 expression, respectively, in dKO cells compared to wt cells (Supplementary Fig S6E and F). Cloning, production, and transduction of lentiviral vectors (LVs) are described in the Supplementary Methods. DamID was performed as previously described 17. Briefly, mES cells were plated and transduced the following day with LV expressing the Dam-fusion protein of interest. gDNA was collected 2 days after transduction, and it was processed and hybridized to NimbleGen genomic arrays as previously described 17. The DamID data used in Figs4 are the average of two independent biological replicates for each condition, which in a previous study 17 was sufficient to detect changes in NL interactions at hundreds of sites throughput the genome.
Immunofluorescence
mES cells were plated on matrigel (BD Bioscience, 356231)-coated glass coverslips. The following day, cells were fixed, stained, and analyzed as described in Supplementary Methods.
Data analysis
All genome-wide analyses were performed using the R statistical software. DamID data normalization was done as previously described 17. LADs and iLADs were defined as described previously 1. Separately, we defined cLAD, fLAD, fiLAD, and ciLAD status for each array probe using a hidden Markov model approach as described 20, but using the Emd DamID profile obtained in wt mES cells in addition to the four cell types used previously. Facultative probes were subdivided in facultative iLAD (fiLAD) and facultative LAD (fLAD) if identified as iLAD or LAD, respectively, in Emd DamID performed in wt mES cells.
The concordance score is defined as: the amount of sequence overlap between LAD or iLAD domains of two cell types (domain defined as in 1) or the percentage of all calls in agreement (e.g., LAD vs.LAD or inter-LAD vs. inter-LAD) between two cell types (calls defined according to the HMM as in 20).
DamID values per gene were calculated by averaging the DamID scores of all probes overlapping with the gene. As a consequence, the comparison of gene expression levels to DamID scores (Fig2) focuses on 14,157 genes. A previously reported statistical test to identify genes with significant changes in DamID signals 17 was applied using default settings (minimum gene size 5 probes; FDR = 5%). For other analyses of gene expression where the position of genes relative to LADs is considered (but not the DamID scores of the genes), all 37,991 genes are included.
Data availability
All data are available from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62685) under accession number GSE62685.
Primary data
Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, and Zheng Y (2011) B-type lamins differentially associate with specific genes during differentiation without directly affecting their expression. Gene Expression Omnibus GSE24532.
Acknowledgments
We thank the NKI Genomics Core Facility for array hybridizations and RNA-seq; the Yixian Zheng lab for the wt and dKO mES cells; Harald Herrmann for the lamin B receptor antibody; Bram van den Broek, Sandra de Vries, and Ludo Pagie for help with, respectively, ImageJ analysis, mES cells culturing, and data analysis. This work was supported by ERC Advanced Grant 293662, NWO-ALW-VICI (BvS), and an EMBO Long-term Fellowship (MA).
Author contributions
MA performed the experiments and analyzed the data; BvS and MA conceived the study, designed the experiments, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Methods, Supplementary Table S1, Supplementary Figure Legends and Supplementary References
Supplementary Figures S1–S6
Review Process File
References
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Amendola M, van Steensel B. Mechanisms and dynamics of nuclear lamina-genome interactions. Curr Opin Cell Biol. 2014;28:61–68. doi: 10.1016/j.ceb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Zuela N, Bar DZ, Gruenbaum Y. Lamins in development, tissue maintenance and stress. EMBO Rep. 2012;13:1070–1078. doi: 10.1038/embor.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Lopez I, Worman HJ. Inner nuclear membrane proteins: impact on human disease. Chromosoma. 2012;121:153–167. doi: 10.1007/s00412-012-0360-2. [DOI] [PubMed] [Google Scholar]
- Zuleger N, Robson MI, Schirmer EC. The nuclear envelope as a chromatin organizer. Nucleus. 2011;2:339–349. doi: 10.4161/nucl.2.5.17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelyov YY, Nurminsky DI. The nuclear lamina as a gene-silencing hub. Curr Issues Mol Biol. 2012;14:27–38. [PubMed] [Google Scholar]
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23:260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LB, Rothman FG, Lopez-Otin C, Misteli T. Progeria: a paradigm for translational medicine. Cell. 2014;156:400–407. doi: 10.1016/j.cell.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J Cell Biol. 2007;176:593–603. doi: 10.1083/jcb.200607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Wangsa D, Falke M, Brown M, Case CM, Erdos MR, Ried T. Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB J. 2014;28:3423–3434. doi: 10.1096/fj.14-250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CW, Maya-Mendoza A, Martin C, Zeng K, Chen S, Feret D, Wilson SA, Jackson DA. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J Cell Sci. 2008;121:1014–1024. doi: 10.1242/jcs.020982. [DOI] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Chang SY, Yin L, Tu Y, Hu Y, Yoshinaga Y, de Jong PJ, Fong LG, Young SG. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum Mol Genet. 2011;20:3537–3544. doi: 10.1093/hmg/ddr266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Zheng Y. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res. 2013;23:1420–1423. doi: 10.1038/cr.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kim Y, Shimi T, Goldman RD, Zheng Y. Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol Biol Cell. 2014;25:1287–1297. doi: 10.1091/mbc.E13-11-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Vaughan A, Alvarez-Reyes M, Bridger JM, Broers JL, Ramaekers FC, Wehnert M, Morris GE, Whitfield WGF, Hutchison CJ. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J Cell Sci. 2001;114:2577–2590. doi: 10.1242/jcs.114.14.2577. [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Bergmann JH, Lazar Z, Spector DL. Lamin A/C is expressed in pluripotent mouse embryonic stem cells. Nucleus. 2013;4:53–60. doi: 10.4161/nucl.23384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. Stochastic genome-nuclear lamina interactions: modulating roles of Lamin A and BAF. Nucleus. 2014;5:124–130. doi: 10.4161/nucl.28825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Oldenburg AR, Collas P. Enriched domain detector: a program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res. 2014;42:e92. doi: 10.1093/nar/gku324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol. 2011;21:1603–1614. doi: 10.1016/j.cub.2011.08.030. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, Skoultchi A, Stewart CL, Bustin M, Meshorer E. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat Commun. 2012;3:910. doi: 10.1038/ncomms1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Talwar S, Mazumder A, Shivashankar GV. Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis. Biophys J. 2009;96:3832–3839. doi: 10.1016/j.bpj.2008.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, McLean K, Theodoropoulos PA, Singh PB, Georgatos SD. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J Biol Chem. 2004;279:25567–25573. doi: 10.1074/jbc.M313606200. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hizume K, Kimura H, Takeyasu K, Haraguchi T, Hiraoka Y. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J Biol Chem. 2012;287:42654–42663. doi: 10.1074/jbc.M112.397950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk JM, Tifft KE, Wilson KL. The nuclear envelope LEM-domain protein emerin. Nucleus. 2013;4:298–314. doi: 10.4161/nucl.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N, Boyle S, Kelly DA, de las Heras JI, Lazou V, Korfali N, Batrakou DG, Randles KN, Morris GE, Harrison DJ, et al. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol. 2013;14:R14. doi: 10.1186/gb-2013-14-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Supplementary Table S1, Supplementary Figure Legends and Supplementary References
Supplementary Figures S1–S6
Review Process File
Data Availability Statement
All data are available from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62685) under accession number GSE62685.