Abstract
The thymus provides a specialized microenvironment in which distinct subsets of thymic epithelial cells (TECs) support T-cell development. Here, we describe the significance of cortical TECs (cTECs) in T-cell development, using a newly established mouse model of cTEC deficiency. The deficiency of mature cTECs caused a massive loss of thymic cellularity and impaired the development of αβT cells and invariant natural killer T cells. Unexpectedly, the differentiation of certain γδT-cell subpopulations—interleukin-17-producing Vγ4 and Vγ6 cells—was strongly dysregulated, resulting in the perturbation of γδT-mediated inflammatory responses in peripheral tissues. These findings show that cTECs contribute to the shaping of the TCR repertoire, not only of “conventional” αβT cells but also of inflammatory “innate” γδT cells.
Keywords: IL-17, thymic epithelial cell, thymus, β5t, γδT
Introduction
The thymic microenvironment is a three-dimensional cellular architecture composed of a set of stromal cells, including thymic epithelial cells (TECs), which guide the development and repertoire formation of T cells 1, 2. In the thymus, there are two anatomically discrete environments, the cortex and medulla, that contain phenotypically and functionally different TEC subsets called cortical TECs (cTECs) and medullary TECs (mTECs), respectively. cTECs form a cortical meshwork densely filled with CD4+CD8+ (double positive, DP) thymocytes undergoing repertoire selection. Positively selected CD4+CD8− (CD4 single positive, CD4SP) and CD4−CD8+ (CD8SP) thymocytes migrate into the medulla, where mTECs filter out self-reactive SP thymocytes by negative selection. This stepwise developmental process across the distinct microenvironments achieves the development of αβT cells with a diverse yet self-tolerant T-cell receptor (TCR) repertoire.
A series of studies has revealed the importance of mTECs in the establishment of T-cell tolerance. More than 10 strains of mice defective in mTEC development, due to the mutation of cytokines, cytokine receptors, signaling molecules, or transcription factors, have been reported, all of which showed organ-specific, T-cell-dependent autoimmunity 3. mTECs express a variety of peripheral tissue-specific antigens, at least partly by virtue of the nuclear factor Aire, such that SP thymocytes reactive to self-antigens are deleted 4. A subset of mTECs produce chemokines CCL19 and CCL21, which attract SP thymocytes from the cortex to the medulla, ensuring deletion of self-reactive cells 5. Studies with mTEC-deficient mice revealed that mTECs are also required for development of regulatory T cells (Tregs) 6 and maturation of invariant natural killer T (iNKT) cells 7.
On the other hand, the thymic cortex has been studied mainly as a place for positive selection of αβT cells owing to the unique proteolytic and antigen processing capabilities of cTECs that are essential for positive selection. A proteasome subunit β5t, which is specifically expressed in cTECs 8, 9, regulates positive selection of CD8 T cells by producing unique MHC class I-bound peptides in cTECs 10, 11. Furthermore, cTECs highly produce endosomal/lysosomal proteases, cathepsin L 12 and thymus-specific serine protease 13, which contribute to the generation of MHC class II-bound peptides and positive selection of certain repertoire of CD4 T cells. However, when compared with mTECs, the physiological significance of cTECs has been less addressed, partly because of a few reports on cTEC-deficient mice 14, 15. Thus, whether and how cTECs contribute to the development and function of the immune system, other than in the context of positive selection of αβT cells, remains to be elucidated.
In addition to the conventional and unconventional αβT-cell lineages described above, the thymus also supports the development of γδT cells 16. In fact, development of Vγ5+ γδT cells was shown to be dependent on Skint1 expressed on mTECs 17. Recent studies have paid particular attention to a subpopulation of γδT cells as an important source of interleukin (IL)-17 in various infections or inflammatory disorders 18, 19, 20, although regulation of the development of IL-17-producing γδT cells in the thymus remains largely unclear.
In the present study, we describe a novel mutant mouse strain harboring a spontaneous point mutation resulting in substantial loss of mature cTECs. We utilized these mice to study the significance of cTECs in T-cell development in vivo. Our results demonstrated that cTECs are required for the increase in thymic volume and optimum development of conventional αβT cells and iNKT cells. We also found that cTECs control the TCR repertoire of IL-17-producing γδT cells and thereby contribute to γδT cell-dependent inflammatory responses.
Results
Thymic hypoplasia and impaired thymocyte development in TN mice
A spontaneous mutant mouse line that exhibited a T lymphopenia was found in our in-house breeding colony of C57BL/6 mice. These mice showed a significant reduction of CD3+CD44lo naïve T cells in peripheral blood (Fig1A), with no apparent defects in growth or reproduction. We named this mouse strain “T lymphopenia of naïve population (TN)”. After several generations of in- and outbreeding, we found that heterozygous mutant mice (+/tn) showed a mild reduction of naïve T cells (25.7% of wild-type) and homozygous mutant mice (tn/tn) showed a severe naïve T-cell reduction (6.0% of wild-type). Mating tests indicated that the T lymphopenia was inherited as an autosomal dominant trait (Supplementary Fig S1).
Figure 1. Thymus hypoplasia and impaired T-cell development in TN mice.
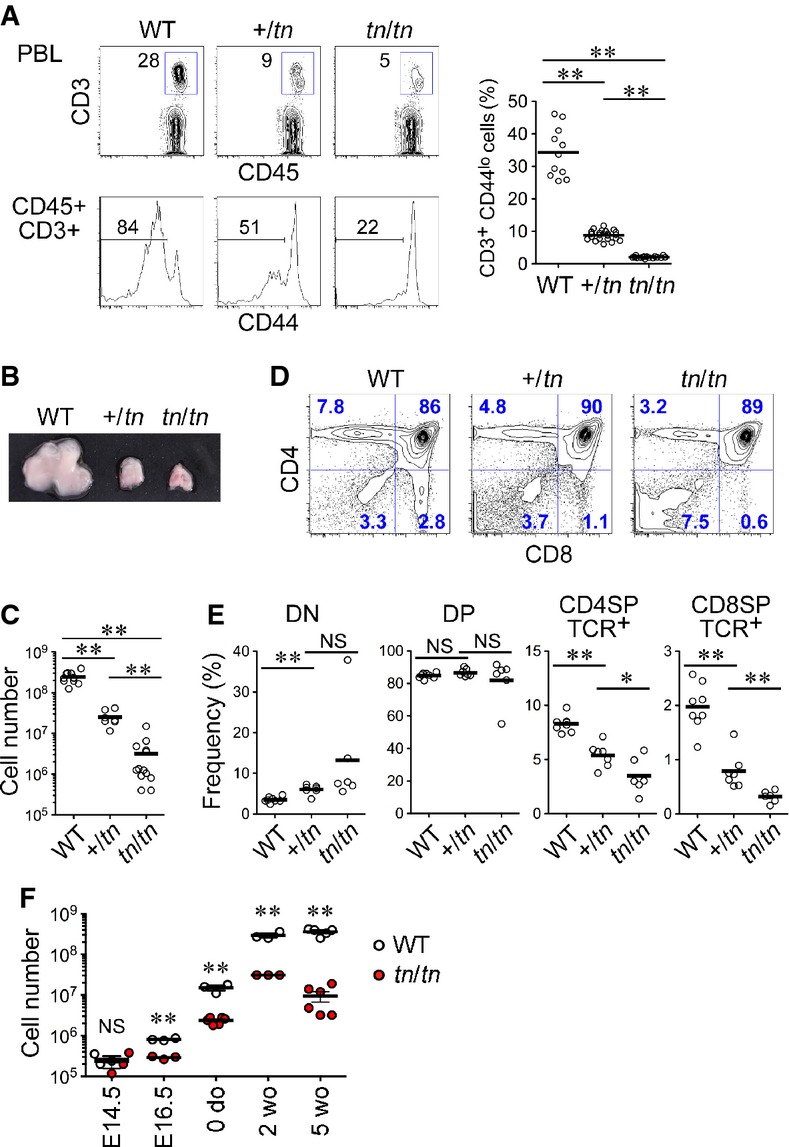
- Frequency of naïve T cells in peripheral blood leukocytes (PBL). PBL from wild-type (WT) (n = 11), +/tn (n = 22), or tn/tn (n = 16) mice were analyzed by flow cytometry for CD45, CD3ε, and CD44 (left). Numbers indicate percentage of cells within indicated areas. Graph indicates percentages of CD3+CD44lo naïve T cells in total CD45+ leukocytes (right).
- Representative photograph of thymi from 5-week-old mice.
- Total numbers of thymocytes from 5-week-old mice (n = 7–13).
- Flow cytometry profiles for CD4 and CD8 of total thymocytes from 5-week-old mice.
- Frequency of indicated thymocyte populations.
- Numbers of whole thymic cells prepared from indicated ages of WT (n = 3–5) or tn/tn (n = 3–6) mice.
Data information: Data represent more than three independent experiments. Each circle represents an individual mouse, and horizontal bars indicate the mean. The statistical significance between indicated groups was calculated with an unpaired one-tailed Student's t-test. *P < 0.05; **P < 0.01; NS, not significant.
Compared with wild-type, TN mice had strikingly smaller thymi and markedly reduced numbers of total thymocytes (Fig1B and C). The frequency of CD4SP and CD8SP mature thymocytes was significantly reduced in TN mice (Fig1D and E), whereas the frequency of DP thymocytes was unchanged. Bone marrow cells from tn/tn mice readily reconstituted thymocyte development in irradiated wild-type mice, whereas +/tn and tn/tn host mice did not support thymocyte development of wild-type bone marrow cells (Supplementary Fig S2), indicating that non-hematopoietic stromal cells, likely thymic stromal cells, are responsible for the impaired T-cell development in TN mice.
TN mice lack mature cTECs
In the thymus from tn/tn mice, the contrast and boundary between cortex (wherein DP thymocytes localize) and medulla (wherein CD4SP and CD8SP thymocytes localize) were clearly detectable as seen in wild-type thymus (Fig2A). However, the expression of cTEC markers such as CD205, Ly51, and keratin 8 was almost undetectable in tn/tn thymus, whereas mTEC markers such as UEA1, keratin 5, Aire, and CCL21 were detectable (Fig2A and Supplementary Fig S3A). The tn/tn cortex that hosted DP thymocytes was composed of keratin+ TECs without expression of cTEC and mTEC markers (likely immature TECs as described later). Electron microscopy showed that the cortical epithelial network that was characteristic in wild-type thymus was poorly developed in tn/tn thymus (Fig2B). Flow cytometric analysis of collagenase-digested thymic stromal cells from adult mice confirmed the nearly complete loss of CD205hiUEA1− cTECs in tn/tn mice (Fig2C). During thymic ontogeny in wild-type mice, CD205hiUEA1− cTECs were detected by embryonic day (E) 16.5 and their number increased exponentially until birth and was maintained in postnatal thymus until young adult age. However, this same cTEC population was negligible throughout embryogenesis and postnatal development in tn/tn mice (Fig2D and Supplementary Fig S3C). Development of cTECs also failed in in vitro organ culture of E14.5 fetal thymus, indicating that this defect was thymus-intrinsic (Supplementary Fig S3D).
Figure 2. TN mice lack mature cTECs.
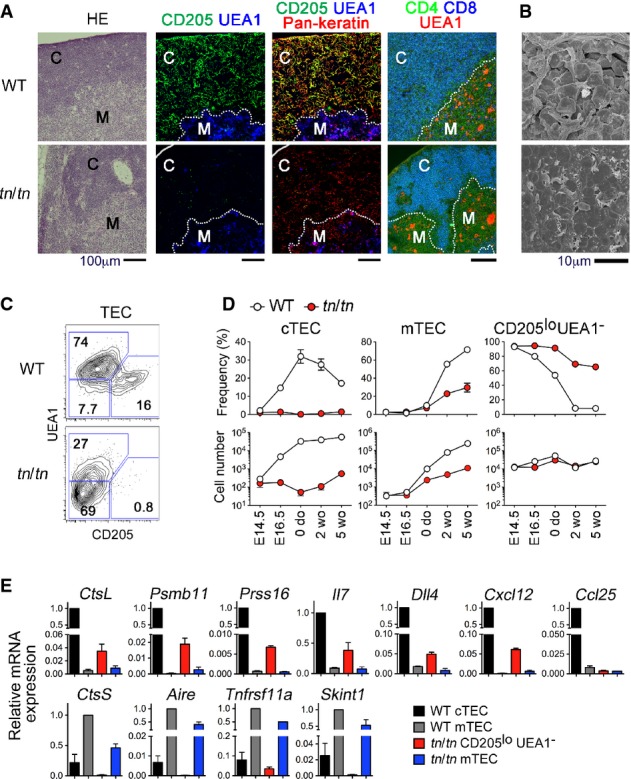
- Thymus sections from 5-week-old WT or tn/tn mice were stained with hematoxylin and eosin (HE), or for CD205, UEA1, pan-keratin, CD4, and CD8. “C” denotes cortex and “M” denotes medulla. Dotted lines indicate cortex/medulla boundary. Scale bars indicate 100 μm.
- Scanning electron micrographs of thymic cortex from WT or tn/tn mice. Scale bar indicates 10 μm.
- Flow cytometry profiles for CD205 and UEA1 of EpCAM+Keratin+ TECs prepared from 5-week-old WT or tn/tn mice.
- Frequency (% of EpCAM+Keratin+ cells) (top) and numbers (per mouse) (bottom) of CD205hiUEA1− (cTEC), CD205loUEA1+ (mTEC), or CD205loUEA1− TECs from the indicated ages of WT or tn/tn mice (n = 3–5, mean ± SEM).
- Quantitative RT–PCR analysis of the indicated genes in isolated TECs (n = 3, mean ± SE). mRNA expression was normalized to EpCAM mRNA, and those in WT cTECs or WT mTECs were arbitrarily set to 1.
Data information: Data represent more than three independent experiments (A, C, D, E) or one experiment with three mice per group (B).
In the postnatal thymus from tn/tn mice, the frequency of CD205loUEA1+ mTECs was partially reduced (Fig2C and D). Despite the reduced frequency of mTECs in tn/tn mice, treatment with RANKL, an mTEC-promoting cytokine 21, successfully induced expansion of mTECs in organ culture of tn/tn thymus (Supplementary Fig S3E), indicating that the developmental potential of mTECs was not aberrant in tn/tn mice.
The most prominent population of TECs from tn/tn mice was CD205loUEA1− cells that showed low surface expression of MHC class II (Supplementary Fig S3F). As the expression of MHC class II gradually increases along the maturation process of TECs 9, 22, our results indicate that CD205loUEA1− cells in tn/tn mice are immature TECs. Expression of cTEC-associated genes, including CtsL, Psmb11, Prss16, Il7, Dll4, and Cxcl12, was detected at low levels in CD205loUEA1− TECs from tn/tn mice, while mTEC-associated genes, including CtsS, Aire, and Skint1, were almost undetectable (Fig3E). These results, along with recent reports that cTECs and mTECs are derived from TEC progenitors expressing cTEC-associated genes 23, 24, 25, indicate that CD205loUEA1− TECs that reside in the thymic cortex of tn/tn mice are immature TEC progenitors and that tn/tn mice are defective in the generation of mature cTECs from immature TEC progenitors.
Figure 3. A missense mutation of the Psmb11 gene causes cTEC deficiency in TN mice.
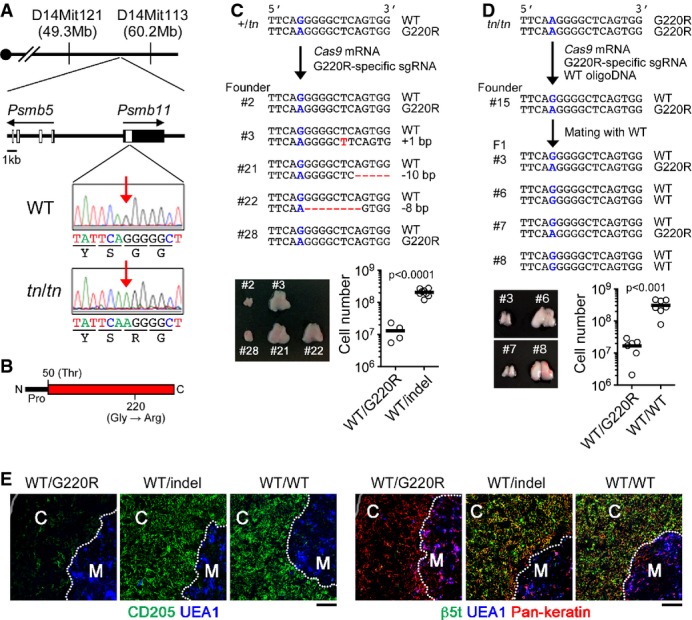
- A Genetic map of the tn locus on chromosome 14 and representative sequencing results of Psmb11 from WT or tn/tn mice. Red arrows indicate the position of the mutation.
- B The structure of the β5t protein. The positions of N-terminal propeptide (Pro), catalytically active site (Thr50), and G220R mutation are shown.
- C, D Cas9mRNA and Psmb11G220R-specific single-guide (sg) RNA were injected into +/tn fertilized eggs, and generated founder mice were genotyped and analyzed (C). Cas9mRNA, Psmb11G220R-specific sgRNA, and WT oligonucleotide were injected into tn/tn fertilized eggs, and a generated founder mouse (#15) was crossed with WT mice (D). The sequences of both alleles of Psmb11 in representative mice are shown (top). G220R mutation site is labeled in blue. Insertion and deletion (indels) are labeled in red. Representative photographs of thymi and total numbers of thymocytes from mice of the indicated genotype are shown (bottom). Each circle represents an individual mouse, and horizontal bars indicate the mean. The statistical significance was calculated with an unpaired one-tailed Student's t-test.
- E Thymus sections from mice with the indicated Psmb11 genotypes (see C, D) were stained for CD205, UEA1, β5t, and pan-keratin. Scale bars indicate 100 μm. “C” denotes cortex and “M” denotes medulla. Dotted lines indicate cortex/medulla boundary.
Data information: Data represent more than three (C, D) or two (E) experiment(s).
A missense mutation of β5t impairs cTEC development
By linkage analysis followed by deep sequencing of the entire 11-Mb candidate region on chromosome 14, we identified a homozygous missense mutation in tn/tn mice in the Psmb11 gene, the gene that encodes the cTEC-specific proteasome subunit β5t (Fig3A). This mutation is a G to A nucleotide substitution, which causes a Gly 220 to Arg codon change (G220R) in β5t (Fig3B). To confirm that the Psmb11G220R mutation was primarily responsible for the TN phenotypes, we performed CRISPR/Cas9-mediated genome editing in mice. Targeted disruption of the Psmb11G220R allele or single nucleotide reversion from Psmb11G220R to Psmb11WT in +/tn mice completely restored thymocyte cellularity and development of mature cTECs up to wild-type levels (Fig3C–E). These results clearly indicate that the Psmb11G220R is the causative mutation responsible for the TN phenotype.
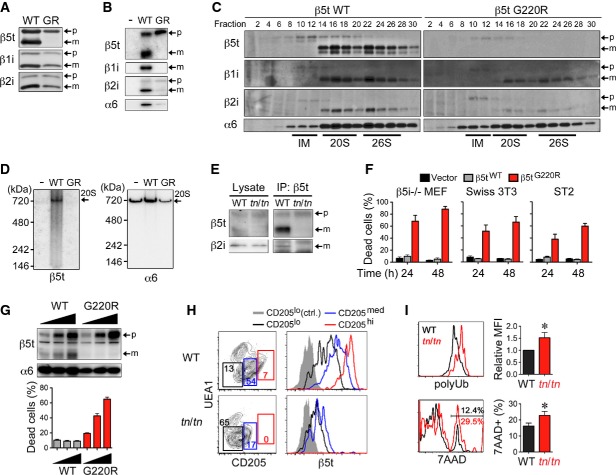
β5t is a proteasome subunit exclusively expressed in cTECs that forms an atypical type of proteasome, termed the “thymoproteasome” 8, 9, that is required for positive selection of CD8+ T cells 10, 11. It was quite unexpected that the single amino acid substitution of β5t could account for such severe defects, because mice lacking β5t showed normal cTEC development by the redundant complementation with another subunit β5i 10, 11. Along with the fact that heterozygous +/tn mice also exhibited the TN phenotype, it is evident that the G220R mutation of β5t has a dominant-negative effect, instead of a loss-of-function effect. To examine the effect of the G220R mutant on thymoproteasomes containing β5t, β1i, and β2i subunits, we utilized retroviral expression of β5t in β5i-deficient (β5i−/−) mouse embryonic fibroblast (MEF) cells, wherein β1i and β2i could be induced by interferon-γ (IFNγ) stimulation. The mutant β5tG220R protein was not proteolytically processed to its mature form (Fig4A), nor was it assembled with other subunits (Fig4B). Its expression resulted in the accumulation of proteasome assembly intermediates (Fig4C), and correlated with reduced amounts of mature β1i and β2i subunits (Fig4A) and of total 20S proteasomes (Fig4D). These results indicate that the β5tG220R mutant protein disturbs normal proteasome assembly in cTECs. In fact, impaired thymoproteasome assembly was observed in vivo in fetal thymus from tn/tn mice (Fig4E). Consistent with an absolute requirement of proteasome activity for cell survival 26, 27, exogenous expression of β5tG220R induced cell death in different cell types in a dose-dependent manner (Fig4F and G). Indeed, in tn/tn neonatal thymus, mature β5thi cTECs were specifically absent, while immature β5tmed/lo TEC populations remained (Fig4H). Finally, residual cTECs in fetal thymus from tn/tn mice exhibited higher accumulation of polyubiquitinated proteins and a higher occurrence of cell death than those from wild-type mice (Fig4I). Collectively, we concluded that β5tG220R mutant protein inhibits normal proteasome assembly and thus induces cell death in mature cTECs that highly express β5t. The moderate defects of mTECs in tn/tn mice may be due to an effect of low level expression of β5tG220R on bipotent TEC progenitors and a reduction of SP thymocytes that provide RANKL. Thus, TN mice represent a β5t-driven, novel mouse model of cTEC deficiency.
Figure 4. β5tG220R impairs proteasome assembly and cell survival in cTECs.
- A–D β5i−/− mouse embryonic fibroblasts (MEFs) were infected with retroviruses expressing C-terminally FLAG-tagged β5tWT or β5tG220R (GR) and treated with IFNγ. Forty hours after infection, total cell lysates (A) and immunoprecipitates with anti-FLAG antibody (B) were analyzed by immunoblotting. Cell lysates were fractionated by glycerol density gradient, and each fraction was used for immunoblotting and measurement of chymotrypsin-like proteasomal activity (C). Fractions 8–12, 14–18, and 22–26 contained assembly intermediates (IM), 20S proteasomes, and 26S proteasomes, respectively. Cell lysates were subjected to native PAGE followed by immunoblotting (D). Arrows indicate precursor (p) and mature (m) forms of β-subunits (A–C) or 20S proteasomes (20S) (D).
- E Cell lysates from E15.5 fetal thymus were immunoprecipitated with anti-β5t antibody followed by immunoblotting for β5t and β2i.
- F β5i−/− MEFs, Swiss 3T3 fibroblast cells, and ST2 bone marrow stromal cells were infected with retroviruses expressing β5tWT or β5tG220R or with empty retroviruses (Vector). β5i−/− MEFs were treated with IFNγ. Thirty hours after infection, EGFP+ cells were sorted and re-plated for additional culture for 24 or 48 h. Cells were detached and their viability was assessed by the trypan blue exclusion method (n = 3).
- G β5i−/− MEFs were retrovirally infected and treated with IFNγ. Thirty hours after infection, EGFPlo, EGFPmed, and EGFPhi cells were sorted and then immunoblotted for expression of β5t and α6 or re-plated for additional 24-h culture. Cell viability was assessed by the trypan blue exclusion method (n = 3).
- H Flow cytometry profiles for CD205 and UEA1 of gated EpCAM+Keratin+ TECs prepared from 1-week-old mice (left). Numbers indicate percentage of cells within indicated areas. Histograms show intracellular β5t expression in CD205lo (black), CD205med (blue), and CD205hi (red) cells and control staining of CD205lo cells (gray shaded).
- I Gated CD205med/hiUEA1− cTECs from E16.5 fetal thymus were further analyzed for cellular accumulation of polyubiquitinated proteins (polyUb) (top) and dead-cell staining by 7-aminoactinomycin D (7AAD) (bottom). Graphs show polyUb mean fluorescence intensity (MFI) and frequency of 7AAD+ dead cells (n = 3–5). The statistical significance was calculated with an unpaired one-tailed Student's t-test. *P < 0.05.
Data information: Data represent three (A–F, I) or two (G, H) independent experiments (error bars, SEM).
Impaired αβT-cell development in TN mice
The findings described above prompted us to utilize TN mice to study the significance of cTECs in T-cell development and the immune system in vivo. First, we observed that thymic cellularity was associated with the expansion of mature cTECs during ontogeny. In E14.5 fetal thymus, there were no differences in thymic cellularity (Fig1F) or epithelial architecture (Supplementary Fig S3B) between wild-type and tn/tn mice. After E16.5, tn/tn mice exhibited reduced thymic cellularity, which coincided with the impaired maturation of cTECs (Fig2D). These results indicate the requirement of mature cTECs for maximizing the size of the thymus.
We next examined αβT-cell development. Among CD4−CD8− (double negative, DN) thymocytes, the proportion of c-Kit+CD25− (DN1) and c-Kit+CD25+ (DN2) cells was significantly reduced in postnatal tn/tn mice (Fig5A). tn/tn mice also showed a markedly reduced frequency of c-Kit−CD25+CD27hi (DN3b) cells, which represent post-β-selection DN3 thymocytes, whereas c-Kit−CD25+CD27lo (DN3a) and c-Kit−CD25− (DN4) cells were unaffected. These results indicate that optimal development of DN thymocytes requires mature cTECs.
Figure 5. Impaired development of conventional αβT cells in TN mice.
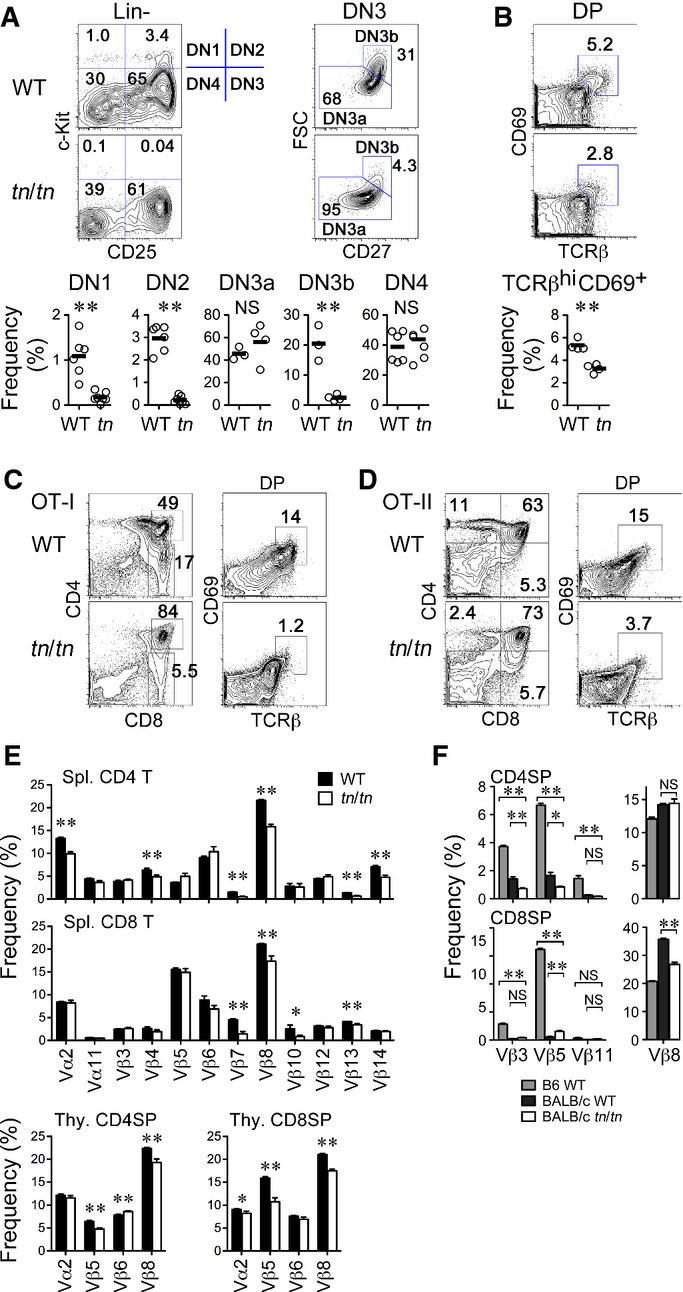
Thymocytes from 5-week-old mice were analyzed by flow cytometry.
- A Flow cytometry profiles of gated Lin− (CD3−CD4−CD8−CD11b−CD11c−CD19−NK1.1−) or DN3 (Lin−c-Kit−CD25+) thymocytes (top) and frequency of DN1 (c-Kit+CD25−), DN2 (c-Kit+CD25+), DN3a (c-Kit−CD25+CD27loFSClo), DN3b (c-Kit−CD25+CD27hiFSChi), or DN4 (c-Kit−CD25−) populations (bottom) (n = 4–6) are shown.
- B TCRβ and CD69 expression profiles of DP (CD4+CD8+) thymocytes (top) and their frequency (bottom) (n = 4).
- C, D Flow cytometry profiles of total and gated thymocytes from OT-I (C) or OT-II TCR transgenic (D) mice.
- E Distribution of TCR-Vα and TCR-Vβ in spleen CD4 T or CD8 T cells or CD4SP or CD8SP thymocytes (n = 6–12).
- F Distribution of TCR-Vβ3, TCR-Vβ5, TCR-Vβ11, and TCR-Vβ8 in CD4SP or CD8SP thymocytes from WT mice of C57BL/6 (B6) background (n = 2–5) and WT (n = 3–4) or tn/tn mice (n = 3–4) of BALB/c background.
Data information: Each circle represents an individual mouse, and horizontal bars indicate the mean (A, B). Mean ± SEM (E, F). *P < 0.05; **P < 0.01; NS, not significant (unpaired t-test). Data in (F) were analyzed by one-way ANOVA. Data represent more than three independent experiments.
In mice expressing either polyclonal or monoclonal TCRs (OT-I and OT-II), TCRβhiCD69+ post-selected DP thymocytes and CD4SP or CD8SP thymocytes were markedly reduced, indicating that positive selection of αβT cells was impaired in tn/tn mice (Fig5B–D). The αβT cells that developed in tn/tn mice carried an altered TCR repertoire, as the distribution of TCR-Vα and TCR-Vβ in splenic T cells and SP thymocytes was significantly different between wild-type and tn/tn mice (Fig5E). By contrast, negative selection mediated by mammary tumor virus (mtv)-encoded superantigen was not affected in tn/tn mice (Fig5F). These results indicate that mature cTECs are required for positive selection and repertoire formation, but dispensable for negative selection, of αβT cells, which is consistent with previous findings 10, 11.
Reduced development of iNKT cells in TN mice
We then asked whether the cTEC deficiency influenced the development of unconventional αβT-cell lineages such as iNKT cells, Foxp3+ Tregs, and CD8αα+ TCRαβ intraepithelial lymphocytes (IELs), all of which develop from cortical DP thymocytes. We found a significant reduction of iNKT cells that are reactive to CD1d tetramer loaded with α-galactosylceramide (αGalCer) in the thymus, liver, and spleen from tn/tn mice (Fig6A and B). The ratio of thymic iNKT to DP cells was markedly reduced in tn/tn mice, indicating that development of iNKT cells from DP thymocytes is impaired in the absence of mature cTECs (Fig6C). iNKT cell development depends on positive selection by CD1d/glycolipid complexes expressed on the surface of other DP thymocytes 28. However, no significant reduction was observed in either CD1d expression or TCR-Vα14-Jα18 rearrangement in DP thymocytes from tn/tn mice (Fig6D and E), suggesting the possibility that the reduced development of iNKT cells is due to impaired interaction between CD1d-expressing “selector” DP thymocytes and TCR-Vα14-Jα18-expressing “iNKT precursor” DP thymocytes. It could be an effect of the marked reduction of absolute numbers of DP thymocytes in tn/tn thymus. It is also possible that the cTEC-dependent cortical epithelial network is important for optimal cell–cell contact among DP thymocytes, thereby supporting positive selection of iNKT cells. iNKT cells from tn/tn thymus normally expressed TCR-Vβ2, TCR-Vβ7, or TCR-Vβ8 (Fig6F), indicating that the TCR repertoire of iNKT cells was unaffected by cTEC deficiency. The majority of iNKT cells from tn/tn thymus were mature CD44+NK1.1+ (stage 3) cells (Fig6G), but the frequency of CD44+NK1.1− (stage 2) cells was significantly reduced. Intracellular staining for key transcription factors revealed that PLZF+ NKT2 cells, but not RORγt+ NKT17 cells, were markedly (P < 0.01) reduced (Fig6H). Reduction of stage 2 iNKT cells could be due to the reduction of NKT2 cells, because most of the NKT2 cell falls into stage 2 29. Although further analysis of the molecular mechanisms of cTEC-mediated selection and differentiation, as well as mTEC-mediated maturation 7, of iNKT cells should be required, our present finding that differential requirement of cTEC for the differentiation of NKT2 and NKT17 cells is intriguing.
Figure 6. Reduced development of iNKT cells in TN mice.
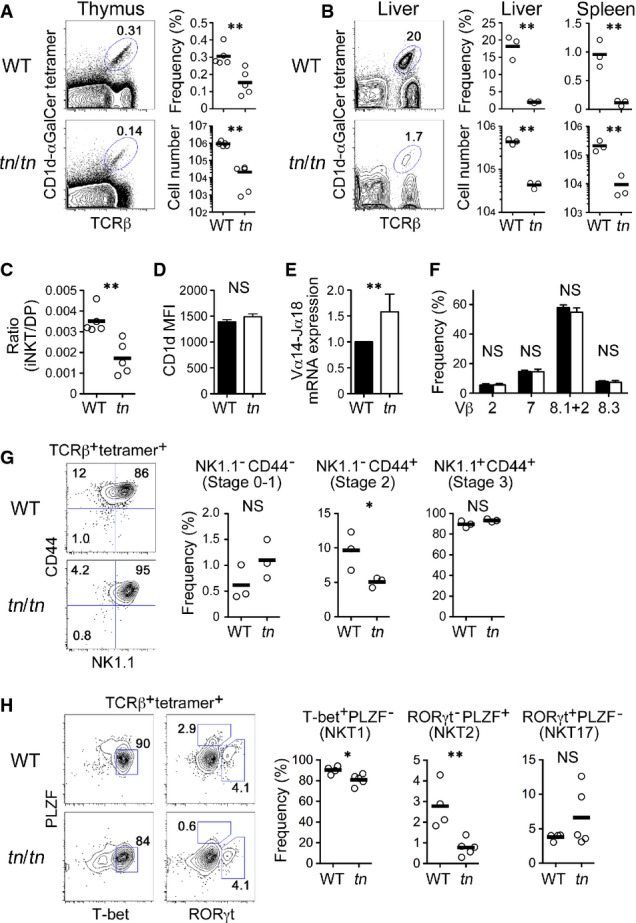
- A, B Thymocytes (A) or liver cells (B) from WT or tn/tn mice were stained with αGal-Cer/CD1d tetramer and anti-TCRβ antibody (left). The frequency and numbers of TCRβ+ αGal-Cer/CD1d tetramer+ cells from thymus (A), liver, or spleen (B) are shown (right) (n = 3–5).
- C The ratio of TCRβ+ tetramer+ iNKT cells to DP thymocytes (n = 5).
- D CD1d expression on DP thymocytes was equivalent between WT and tn/tn mice (n = 3–5).
- E DP thymocytes were isolated from WT or tn/tn mice and analyzed by quantitative RT–PCR (n = 3). The level of Vα14-Jα18 transcript was normalized to TCR-Cα mRNA, and those in WT were arbitrarily set to 1.
- F TCRβ+ tetramer+ thymic iNKT cells were further stained for Vβ2, Vβ7, Vβ8.1 + 2, or Vβ8.3 (n = 3–4).
- G TCRβ+ tetramer+ thymic iNKT cells were further stained for NK1.1 and CD44. Representative flow cytometry profiles (left) and the frequency among total iNKT cells (right) of NK1.1−CD44− (stage 0–1), NK1.1−CD44+ (stage 2), or NK1.1+CD44+ (stage 3) cells are shown (n = 3).
- H TCRβ+ tetramer+ thymic iNKT cells were intracellularly stained for PLZF, T-bet, and RORγt. Representative flow cytometry profiles (left) and the frequency among total iNKT cells (right) of T-bet+PLZF− (NKT1), RORγt−PLZF+ (NKT2), or RORγt+PLZF− (NKT17) cells are shown (WT, n = 4; tn/tn, n = 5).
Data information: Data represent more than three independent experiments. Each circle represents an individual mouse, and horizontal bars indicate the mean (A, B, C, G, H). Mean ± SEM (D, E, F). The statistical significance was calculated with an unpaired one-tailed Student's t-test. *P < 0.05; **P < 0.01; NS, not significant.
The frequency of Foxp3+CD25+ Tregs in CD4SP thymocytes and their distribution in the thymic medulla were similar between wild-type and tn/tn mice (Supplementary Fig S4A and B). We also found no significant reduction in the frequencies of TCRβ+CD5+ thymic CD8αα+ IEL precursors or IEL subpopulations, including CD8αα+TCRαβ IELs, isolated from small intestine (Supplementary Fig S4C and D). These results indicate that the thymic development of Tregs and IELs does not require mature cTECs.
Altered γδT17 cell repertoire in TN mice
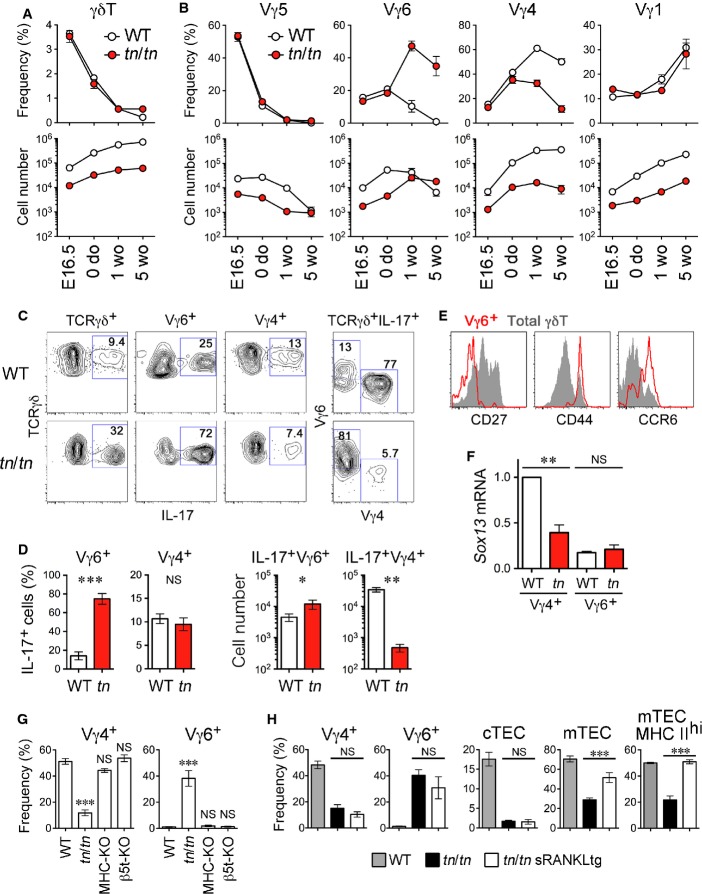
Lastly, we addressed γδT-cell development in tn/tn mice. The frequency of γδT cells in the thymus showed no remarkable difference between wild-type and tn/tn mice during thymic ontogeny (Fig7A). Absolute numbers of thymic γδT cells were reduced in tn/tn mice, because of the severe decrease in total thymic cellularity (Fig1F).
Figure 7. Altered γδT17 TCR repertoire in TN mice.
- Frequency (% of total thymocytes) and numbers (per mouse) of thymic γδT cells from indicated ages of WT or tn/tn mice (n = 4–9).
- Frequency (% of total γδT cells) of Vγ5+, Vγ6+ (17D1+ Vγ5−), Vγ4+, and Vγ1+ cells and their numbers (per mouse) from indicated ages of WT or tn/tn mice (n = 3–8).
- Thymocytes from 5-week-old mice were cultured for 4 h with PMA, ionomycin, and brefeldin A and analyzed by flow cytometry.
- Frequency of IL-17-producing cells in Vγ6+ or Vγ4+ cells and absolute numbers of IL-17-producing Vγ6+ and Vγ4+ cells in individual mice (n = 4–5).
- Histograms show expression of CD27, CD44, and CCR6 in total γδT (shaded) and Vγ6+ (red) cells from adult tn/tn mice.
- Quantitative RT–PCR analysis of Sox13 in isolated Vγ4+ or Vγ6+ cells from WT or tn/tn mice (n = 4). Sox13mRNA expression was normalized to GapdhmRNA, and those in WT Vγ4+ cells were arbitrarily set to 1.
- Thymic γδT cells from 4- to 5-week-old WT, tn/tn, B6-H-2Ab−/−B2m−/− (MHC-KO), or Psmb11−/− (β5t-KO) mice were analyzed for expression of Vγ4 and Vγ6 (n = 3–7).
- Frequency of Vγ4+ or Vγ6+ γδT cells (% of total thymic γδT cells), CD205hiUEA1− cTECs or CD205loUEA1+ mTECs (% of total TECs), and MHC class IIhi mTECs (% of total mTECs) from 4- to 7-week-old WT, tn/tn, or tn/tnsRANKL transgenic (tg) mice (n = 3–12).
Data information: Data represent more than two independent experiments. Graphs indicate mean ± SEM. The statistical significance between indicated groups was calculated with an unpaired one-tailed Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
It is known that the generation of murine γδT-cell subsets expressing distinct TCR-Vγ chains is developmentally regulated during ontogeny 30. Vγ5+ cells develop during the embryonic period, Vγ6+ cells around birth, Vγ4+ cells at the neonatal period, and Vγ1+ cells at adult stages (according to the nomenclature of Heilig and Tonegawa 31). These γδT-cell subsets distribute to different tissues in adult mice 30: Vγ5+ cells to epidermis, Vγ6+ and Vγ4+ cells to mucosal epithelia such as dermis, lung, tongue, and uterus, and Vγ1+ cells to peripheral lymphoid organs. Although the proportion of Vγ5+ and Vγ1+ cells among thymic γδT cells was largely unchanged between wild-type and tn/tn mice throughout ontogeny (Fig7B), development of Vγ6+ and Vγ4+ cells was dramatically altered in postnatal thymus from tn/tn mice. Vγ6+ cells continued to expand after birth and their numbers exceeded those from wild-type thymus at 5 weeks old. In contrast, Vγ4+ cells in tn/tn thymus did not show the robust postnatal increase that was apparent in wild-type thymus. TCR sequencing analysis demonstrated that Vγ6+ cells expressed an invariant TCR-Vγ6-Jγ1 chain and Vγ4+ γδT cells expressed a diverse TCR-Vγ4-Jγ1 repertoire, in both wild-type and tn/tn mice, representing qualitatively equivalent subpopulations (Supplementary Table S1).
Vγ6+ and Vγ4+ cells include an IL-17-producing subset, termed γδT17, whose development is restricted to the thymus during late embryonic and neonatal stages 32, 33; they also contribute to the pathogenesis of inflammation in the periphery 18, 19, 20. In adult tn/tn mice, the proportion of IL-17-producing cells among γδT cells was greatly increased (Fig5C). Among these γδT17 cells, the Vγ6+ subset vigorously increased, whereas the Vγ4+ subset substantially decreased, resulting in the marked skewing from Vγ4 to Vγ6 in TCR repertoire of γδT17 cells from tn/tn mice (Fig7C and D). The majority of thymic Vγ6+ cells in tn/tn mice were CD27− and expressed CD44 and CCR6, indicating that these are functionally mature cells (called γδ27−) 34 (Fig7E). Vγ4+ cells from tn/tn mice exhibited reduced mRNA expression of Sox13 (Fig7F), a key transcription factor for development of Vγ4+ γδT17 cells 35, 36. These results indicate increased and prolonged production of functional Vγ6+ γδT17 cells and restrained development of Vγ4+ γδT17 cells in the thymus from tn/tn mice.
Proportions of Vγ4+ and Vγ6+ cells among thymic γδT cells was unaffected in both β5t-deficient mice and MHC-deficient mice that exhibit defective mTEC expansion (Fig7G). Therefore, the altered repertoire of γδT cells was due to neither β5t deficiency nor reduced mTEC development. To further examine the effect of mTECs on γδT-cell repertoire, we utilized soluble RANKL (sRANKL) transgenic mice, causing enhanced expansion of mTECs 37. Although introduction of sRANKL transgene rescued substantial number of MHC class IIhi mature mTECs in the thymus of tn/tn mice, altered repertoire of γδT cells was not rescued in mTEC-sufficient tn/tn sRANKL transgenic mice (Fig7H and Supplementary Fig S5). These results clearly exclude the responsibility of β5t deficiency and reduced mTEC development to the altered repertoire of γδT cells observed in tn/tn thymus.
To explore the significance of the alteration in thymic γδT cells, we examined the distribution and function of peripheral γδT cells. In adult tn/tn mice, Vγ6+ cells in lung were robustly increased, whereas Vγ4+ cells were markedly reduced in lung, skin, spleen, and lymph nodes (Fig8A). Lung Vγ6+ γδT17 cells have been reported to trigger the inflammation induced by trehalose 6,6′-dimycolate (TDM), a cord factor of Mycobacterium tuberculosis 38. Indeed, in response to TDM administration, tn/tn mice exhibited enhanced acute inflammatory responses in lung, including the production of IL-17 mRNA, organ swelling, and granuloma formation when compared with wild-type mice (Fig8B–D), indicating that increased lung Vγ6+ γδT17 cells are functionally normal. To the contrary, in the skin, psoriasis-like dermatitis induced by imiquimod (IMQ), in which Vγ4+ γδT17 cells that expand in lymph node and home to inflamed skin play a crucial role 35, 39, 40, was significantly attenuated in tn/tn mice (Fig8E and F). These results indicate that the γδT cells generated in the absence of mature cTECs express an altered TCR repertoire that fails to properly mount inflammatory responses.
Figure 8. TN mice exhibited altered γδT-cell-mediated inflammatory responses.
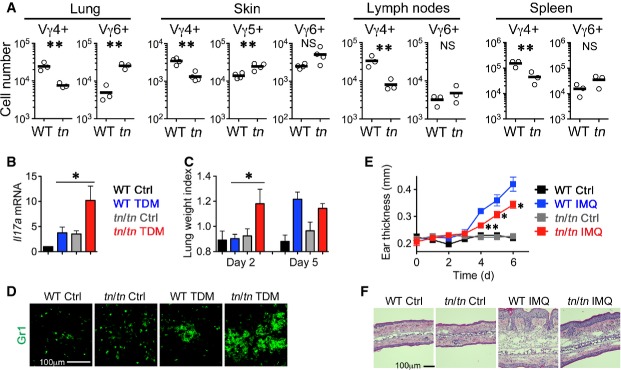
- A Absolute numbers (per mouse) of indicated γδT-cell subsets in lung, skin, lymph nodes (inguinal, axillary, and submandibular), and spleen from 5-week-old WT or tn/tn mice (n = 3). Each circle represents an individual mouse, and horizontal bars indicate the mean. The statistical significance was calculated with an unpaired one-tailed Student's t-test. *P < 0.05; **P < 0.01; NS, not significant.
- B–D WT or tn/tn mice were injected intravenously with an oil-in-water emulsion containing TDM (15 μg). Emulsion without TDM was injected as a vehicle control. Il17amRNA levels in lungs at day 2 were examined by quantitative RT–PCR and normalized to GapdhmRNA (n = 3–4, mean ± SEM) (B). Lung inflammation intensity was measured by calculating the lung weight index (n = 3–5, mean ± SEM) (C). The statistical significance between TDM-treated WT and tn/tn mice was calculated with an unpaired one-tailed Student's t-test. *P < 0.05. Lung sections at day 2 were stained for granulocyte marker Gr1 (D).
- E, F WT or tn/tn mice were treated daily for 6 days with IMQ cream or control cream on the ears. Ear skin thickness at the days indicated (n = 3, mean ± SEM) (E). The statistical significance between IMQ-treated WT and tn/tn mice was calculated with an unpaired one-tailed Student's t-test. *P < 0.05; **P < 0.01. HE staining of the ear section at day 6 (F).
Discussion
In the current study, we established a novel mouse model, designated TN mice, which intrinsically lacks mature cTECs. We identified the G220R mutation of β5t as being primarily responsible for cTEC deficiency in TN mice. Thus, TN is a β5t-driven, mature cTEC-deficient mouse strain, which will be useful to investigate the physiological significance of cTECs.
It is interesting to note that in the thymus from tn/tn mice, the compartmentalization of cortex and medulla is readily detectable, despite the loss of mature cTECs. It was reported that outward migration of developing thymocytes from the corticomedullary junction to the cortex requires CXCL12-CXCR4 and CCL25-CCR9 chemokine signals 41, 42. However, the expression of CXCL12 and CCL25 in CD205loUEA1− TECs from tn/tn mice is very low, ∽6 and 0.3%, respectively, compared with that in wild-type mature cTECs. Another thymic chemokine CCL21, which attracts mature CD4SP and CD8SP thymocytes from the cortex to the medulla 5, is normally produced by mTECs in tn/tn mice. These results demonstrate that the cortex/medulla compartmentalization in the thymus does not require mature cTECs and can be mediated by CD205loUEA1− immature TECs along with CCL21-expressing mTECs. Further studies will be needed to dissect developmental processes of cTECs and to better understand their nature and function.
Although the thymic cortex is apparently formed, cortical T-cell development without mature cTECs is obviously abnormal, as tn/tn mice showed impairment of positive selection of αβT cells. tn/tn mouse thymi showed diminished expression of cathepsin L, Prss16 (also called thymus-specific serine protease), and complete loss of β5t-containing thymoproteasomes. Cathepsin L and Prss16 are endosomal/lysosomal proteases highly expressed by cTECs that regulate positive selection of MHC class II-restricted CD4 T cells 12, 13. Thymoproteasomes are essential for positive selection of MHC class I-restricted CD8 T cells, likely by producing cTEC-specific MHC class I-bound peptides 8, 10, 11. Our results that tn/tn mice have an altered TCR-Vα and TCR-Vβ distribution of both CD4 and CD8 T cells indicate that cTECs contribute not only to generate the optimum cellularity of T cells but also to shape their αβTCR repertoire, as suggested by previous reports. In addition to the lack of such functional molecules in the cortex, it is also possible that a loss of the cortical epithelial network per se impairs thymocyte development through physical interference with intracortical migration or cell–cell interactions of cortical thymocytes. Taken together, cortex-resident, immature TECs in tn/tn thymus are incompetent for αβT-cell repertoire formation, even though they are functional in forming the thymic cortex and supporting development of a small fraction of αβT cells, confirming the essential role for cTECs in development and repertoire formation of conventional αβT cells.
To the contrary, negative selection of mtv superantigen-reactive thymocytes occurred normally in tn/tn mice. Indeed, no signs of systemic autoimmunity were detected in tn/tn mice (unpublished observation), indicating that self-tolerance is established in the absence of mature cTECs. Although a recent report indicated that three quarters of negative selection occurs in the cortex 43, negative selection was not affected in the absence of mature cTECs. Therefore, cortex-resident, immature TECs and/or dendritic cells 44 may induce negative selection of self-reactive thymocytes.
The most unexpected finding from the current study was the influence of mature cTEC deficiency on γδT-cell development. γδT cells generated in the absence of mature cTECs were skewed toward IL-17-producing (γδT17) lineage that expressed an altered γδTCR repertoire. As the development of Vγ5+ and Vγ1+ γδT cells (mostly non-γδT17 cells) was normal in tn/tn thymus, the influence of mature cTEC deficiency is specific to γδT17 lineage. It was shown that development of γδT cells in the thymus is dependent on IL-7 45, and expansion and development of γδT17 cells requires IL-7 46 and Dll4-Notch signals 47, respectively. As tn/tn mice lack mature cTECs that express high levels of IL-7 and Dll4, the increase of γδT17 cells seen in the tn/tn thymus could not be directly attributed to signaling on these molecules. It may be that relatively low levels of expression of IL-7 and Dll4 by immature TECs and mTECs, and also likely by other thymic stromal cells, are sufficient for inducing development of γδT17 cells. Most interestingly, two major subsets of γδT17 cells, Vγ6+ cells and Vγ4+ cells, are reciprocally differentiated in tn/tn mice. Increased and prolonged production of Vγ6+ cells resulted in boosted Vγ6+ γδT17-mediated inflammatory responses, and impaired production of Vγ4+ cells, which in turn provided protection from Vγ4+ γδT17-dependent dermatitis. The altered repertoire of γδT cells in tn/tn mice was due to neither β5t deficiency nor reduced mTEC development, as the distribution of Vγ4+ and Vγ6+ cells was unaffected in β5t-deficient mice and mTEC-reduced, MHC-deficient mice, and the altered γδT-cell repertoire in tn/tn mice was not restored by sRANKL-mediated enhancement of mTEC differentiation. These results suggest that mature cTECs play roles in repressing the development of Vγ6+ cells and promoting that of Vγ4+ cells, although it is also possible that cortex-resident, immature TECs in tn/tn mice have reciprocal functions and that partly defective development of (likely RANKL-independent) mTECs in tn/tn mice caused the γδT17 repertoire alteration. A recent study showed a differential developmental requirement for Vγ4+ and Vγ6+ γδT17 cells; Vγ6+ γδT17 cells absolutely require fetal thymus for their development, whereas Vγ4+ γδT17 cells can be generated in adult thymus 48. Thus, it is suggested that the thymus from adult tn/tn mice provides fetal type microenvironment that supports predominant development of Vγ6+ γδT17 cells.
Our results first reveal that cTECs determine the balance between Vγ4+ and Vγ6+ γδT-cell development, which is important for properly mounting inflammatory responses. The mechanisms by which cTECs regulate the γδT17 TCR repertoire remain to be elucidated. Mature cTECs and immature TECs may express different cell-surface proteins, such as Skint family proteins 49, or as of yet unidentified selecting ligand molecule(s), that mediate differentiation or deletion of Vγ4+ or Vγ6+ γδT cells; for example, as mTECs support maturation of Vγ5+ cells via expression of Skint1 17, 50. The conversion of the Vγ repertoire in tn/tn mice could be attributed to the alternation of interaction between the γδTCRs and its putative ligands on TECs, because recent studies proposed that strength of ligand–γδTCR interaction controls lineage commitment and specification of effector fate of γδT cells 16, 20, 51. It is also possible that restricted access of developing γδT cells to cTECs 52 or reduced negative feedback of γδT17 development by αβT cells 33 may be responsible for the repertoire conversion in tn/tn mice. We observed reduced expression of the transcription factor Sox13 in Vγ4+ γδT cells from cTEC-deficient tn/tn mice. Interestingly, deficiency of Sox13 in mice resulted in the loss of Vγ4+ γδT17 cell development but a partial reduction of Vγ4− (likely to be Vγ6+) γδT17 cells 35, 36. Therefore, the conversion of the γδT17 repertoire in tn/tn mice could be partly explained by the decrease of Sox13, caused by the absence of proper interaction with cTECs.
Taken together, the present study, utilizing a newly established mouse strain that lacks mature cTECs, provides novel evidence for γδT-cell repertoire determination by thymic epithelium. cTECs are required for the shaping of the TCR repertoire, not only of conventional αβT-cell subsets, but also of an “innate-type” γδT-cell subset programmed for IL-17 production. Future studies to clarify the molecular mechanisms should unveil developmental rules for γδT cells that can be applied to γδT-cell-based immunotherapies.
Materials and Methods
Mice
C57BL/6, BALB/c, and C57BL/6× DBA/2F1 (BDF1) mice were purchased from SLC Japan (Shizuoka, Japan). TN mice were derived from an in-house breeding colony of a C57BL/6NCrSlc strain originally derived from SLC Japan. B6.SJL-Ptprca (B6-CD45.1) mice were purchased from Taconic Farm (NY, USA). Psmb11−/− mice 8, OT-I TCR transgenic mice 53, OT-II TCR transgenic mice 54, Rag2−/− mice 55, H-2Ab−/− mice 56, B2m−/− mice 57, and sRANKL transgenic mice 58 were described previously. All mice were bred and maintained under specific pathogen-free conditions in our animal facility. All animal experiments were approved by the Animal Care and Use Committee of the National Center for Global Health Medicine (NCGM) Research Institute (number: 23-Tg-31) and conducted in accordance with institutional procedures.
Flow cytometry and cell sorting
Flow cytometry analysis and cell sorting were performed with FACSCantoII and FACSAriaIII (BD Bioscience) as previously described 59. Thymic stromal cells were prepared by digesting thymic fragments with collagenase D (Roche) and DNase I (Roche), as described 60. For intracellular staining, cells were fixed with 2% paraformaldehyde for 30 min, permeabilized with 0.1% saponin, and stained with antibodies. Foxp3 staining was performed using the Foxp3 staining kit (eBioscience) according to the manufacturer's protocol.
Antibodies
Monoclonal antibodies specific for B220 (RA3-6B2), CD4 (GK1.5), CD5 (53-7.3), CD8α (53-6.7), CD8β (CT-CD8b), CD25 (PC61.5), CD69 (H1.2F3), CD205 (205yekta), NK1.1 (PK136), TCRβ (H57-597), Vα11 (RR8-1), Aire (5H12), PLZF (Mags.21F7), and RORγt (B2D) were purchased from eBioscience. Monoclonal antibodies against CD3ε (2C11), CD44 (IM7), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), EpCAM (G8.8), Ly51 (6C3), CCR6 (29-2L17), TCRγδ (GL3), TCR-Vγ1 (2.11), TCR-Vγ4 (UC3-10A6), TCR-Vγ5 (536), IL-17 (TC11-18H10.1), and T-bet (4B10) were purchased from BioLegend. Mouse Vβ TCR screening panel and anti-Vα2 (B20.1) monoclonal antibody were purchased from BD PharMingen. Anti-pan-keratin (C-11) and anti-FLAG (M2) monoclonal antibodies were purchased from Sigma-Aldrich. UEA1 was purchased from Vector Laboratories. Polyclonal antibodies for keratin 5 (Covance), keratin 8 (Progen), keratin 14 (Covance), and CCL21 (R&D Systems) were used. Monoclonal antibody 17D1 specific for TCR-Vγ6/Vδ1 and TCR-Vγ5/Vδ1 was provided by Dr. Robert E. Tigelaar (Yale University) and used as described previously 61. Polyclonal antibody for β5t was described previously 8. Anti-polyubiquitin monoclonal antibody (FK2) was purchased from MBL.
Bone marrow chimera
Bone marrow cells obtained from donor mice were depleted of T cells using biotin-conjugated anti-CD4, anti-CD8, and anti-Thy1.2 antibodies and streptavidin-conjugated magnetic beads (Miltenyi Biotec). Recipient mice were X-ray-irradiated (9.25 Gy) and injected with T-cell-depleted bone marrow cells (4 × 107 cells) 1 day after irradiation. Mice were analyzed 5 weeks after the injection.
Histological analysis
Frozen tissues embedded in OCT compound (Sakura Finetek) were sliced into 5-μm-thick sections by cryostat (Leica), air-dried, fixed with acetone, and stained with hematoxylin and eosin, or with specific antibodies as described previously 21. Multi-color images were obtained by BZ-9000 fluorescent microscope (Keyence).
Electron microscopy
Thymus lobes from 5-week-old mice were fixed and analyzed by scanning electron microscopy at Tokai Electron Microscopy Analysis (Aichi, Japan).
Fetal thymic organ culture
Thymic lobes isolated from E14.5 fetuses were cultured as previously described 62. To induce mTEC development, recombinant soluble RANKL (1 μg/ml) (Wako) was added to the medium.
Quantitative mRNA analysis
Total cellular RNA was reverse-transcribed with Superscript III first-strand synthesis supermix for qRT–PCR (Life Technologies). Real-time PCR was performed with SYBR Premix ExTaq (TaKaRa) and StepOne real-time PCR system (Life Technologies). Primer sequences are available upon request.
Identification of the responsible mutation of TN mice
For linkage analysis, TN mice originated from C57BL/6 background were crossed with BALB/c mice. Seventy-seven mice (55%) out of 139 (TN × BALB/c) N2 mice exhibited thymic hypoplasia and naïve T lymphopenia, which confirmed that the trait of this mutation was dominant. Genomic DNA was prepared from tail biopsies by the standard method of SDS/proteinase K lysis followed by phenol/chloroform. Information on microsatellite markers was obtained from Mouse Genome Informatics (MGI; http://www.informatics.jax.org/). For sequencing the candidate region, the SureSelect Target Enrichment System (Agilent Technologies) was used to enrich for 11.2-Mb genomic region between D14Mit121 and D14Mit113. Enriched DNA fragments were sequenced by HiSeq 2000 (Illumina).
CRISPR/Cas9-mediated genome editing in mice
Single-guide (sg) RNA was designed for the target sequence (5′-TGCTTATTCAAGGGGCTCAGTGG-3′) and prepared as described previously 63 with some modifications. Briefly, sgRNA expression vector for target sequence with a T7 promoter was synthesized (Operon Biotechnologies, Tokyo, Japan), and transcribed in vitro using MEGAshortscript kit (Life Technologies). hCas9 sequence was removed from pX330 (http://www.addgene.org/42330) and placed under the T7 promoter. hCas9 mRNA was synthesized using mMESSAGE mMACHINE T7 kit (Life Technologies) and was polyadenylated with polyA tailing kit (Life Technologies). The sgRNAs and hCas9 mRNAs were purified by phenol/chloroform/isoamylalchol extraction and concentrated by ethanol precipitation. For injection into one-cell embryos, fertilized eggs were obtained by in vitro fertilization as described previously 64. hCas9 mRNAs (100 ng/μl) and sgRNAs (50 ng/μl) were co-injected into the cytoplasm of the pronuclear stage eggs. For generating targeted mutant mice, WT oligonucleotide (5′-GAAGCCTACACCCTGGCCCGCTGTGCTGTGGCCCATGCCACCCACCGTGATGCTTATTCAGGGGGCTCAGTGGACCTCTTCCACGTTCGGGAGAGCGGATGGGAGTATGTATCCCGCAGT-3′) (100 ng/μl) was co-injected. The eggs were transferred into the oviducts of pseudopregnant ICR female mice. The efficiency of targeted gene disruption and potential off-target effects are shown in Supplementary Tables S2 and S3.
Cell culture and retrovirus infection
Mouse embryonic fibroblasts isolated from Psmb8−/− (β5i−/−) mice 65, Swiss 3T3 cells (ATCC), and ST2 cells (kindly provided by Dr. Meinrad Busslinger) were cultured in DMEM medium supplemented with 10% fetal calf serum (FCS), penicillin, and streptomycin (Life Technologies). DNA fragments encoding an open-reading frame of mouse β5tWT or β5tG220R were cloned into the retrovirus vector pMRX-IRES-EGFP 66. Retrovirus supernatant was prepared as described previously 62.
Proteasomal analysis
Glycerol density gradient was described previously 67. For immunoprecipitation, cells were lysed in an ice-cold lysis buffer (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.5% NP-40, and protease inhibitor cocktail (Thermo)). Cell lysates were incubated with agarose-bead-conjugated anti-FLAG M2 antibody or anti-β5t antibody followed by sepharose bead-conjugated protein G (GE Healthcare). The beads were then washed and boiled in Laemmli gel-loading buffer. The proteins were separated by SDS–PAGE [15% Bis-Tris gel (Life Technologies)] or native PAGE [7% Tris-acetate gel (Life Technologies)], transferred onto PVDF membrane, and detected with indicated antibodies.
Preparation of tissue-resident lymphocytes
IELs were prepared as described elsewhere with some modifications 59. Briefly, small intestine was isolated and Peyer's patches were completely removed prior to opening longitudinally. After six quick rinses in PBS, the intestine was cut into 5-mm pieces, followed by incubating twice in calcium/magnesium-free HBSS containing 2% FCS and 2 mM EDTA at 37°C for 20 min. The cell suspension was filtered through glass wool and the filtrate was centrifuged in 30% Percoll (GE Healthcare) to remove mucus. Cell pellets were then resuspended with 40% Percoll and further separated through a 40–70% Percoll step gradient. The cells at the interface of the Percoll gradient were collected and used for further analysis. For isolation of lymphocytes from skin or lung, ears or lungs from adult mice were cut into small pieces and digested with 0.2% collagenase D (Roche) and DNase I (Roche) at 37°C for 30 min. After digestion, tissue fragments were mechanically disrupted using a syringe and 18-G needle, and cells were passed through 100-μm nylon mesh to remove tissue clumps, collected by centrifugation, and resuspended in PBS containing 2 mM EDTA and 2% FCS.
Inflammatory responses
Adult mice (6 to 12 week old) were injected intravenously with 100 μl of emulsion containing 15 μg TDM (Sigma) prepared as described previously 68. Lung weight index was calculated as described previously 69. Emulsion without TDM was injected as a control. For testing psoriasis-like dermatitis, a daily dose of 5 mg of Beselna cream (5% imiquimod; Mochida Pharmaceutical) or control vaseline cream (Wako) was applied to each ear for 6 days. At the days indicated, the ear thickness was measured using a micrometer.
Sequencing analysis of TCRγ
Vγ4+ or Vγ6+ (17D1+ Vγ5−) thymic γδT cells were isolated from 3- to 4-week-old wild-type or tn/tn mice (pooled from three mice per group). Total cellular RNA was extracted using RNeasy micro kit (QIAGEN) and reverse-transcribed with oligo-dT primer and Superscript III reverse transcriptase (Life Technologies). Vγ4-Cγ1 and Vγ6-Cγ1 cDNA fragments were PCR-amplified and cloned in the pBluescript vector. DNA sequences of randomly picked clones were determined. Primer sequences are as follows: Vγ4 forward, 5-CTTGCAACCCCTACCCATAT-3′; Vγ6 forward, 5-TCAAGAGGAAAGGAAATACGGC-3′; and Cγ1 reverse, 5-CTGGGAGTCCAGGATAGTATTG-3′.
Statistical analysis
The statistical significance was calculated with an unpaired one-tailed Student's t-test. When indicated, one-way ANOVA was used. All calculations and justifications were done using the GraphPad Prism software. No randomization or exclusion of samples was used. For animal experiments, sample size was chosen based on previous experience and preliminary experiments. No blinding of investigators was done.
Acknowledgments
We thank Dr. Robert E. Tigelaar for providing 17D1 antibody; Drs. Kensuke Shibata and Yasunobu Yoshikai for helpful discussion; and Dr. Michael S. Patrick for reading the manuscript. This study was supported by Grants-in-Aid for Research from the Ministry of Education, Culture, Sports, and Technology in Japan (25111516), the grant for National Center for Global Health and Medicine (24-112), Astellas Foundation, Inamori Foundation, and Kanae Foundation.
Author contributions
TNi and HS designed the project. TNi, RM, SN, HO, TNa, and HT performed experiments on mouse, tissue, and cell analysis. YS, MG, RY, and TO worked on the linkage analysis and CRISPR/Cas9-mediated genome editing; YO and SM worked on the proteasome analysis. HY analyzed sRANKLtg mice. TNit and HS wrote the manuscript with help from the other authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figures S1–S5, Supplementary Tables S1–S3
Review Process File
References
- Anderson G, Takahama Y. Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 2012;33:256–263. doi: 10.1016/j.it.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, Shinzawa M, Qin J, Akiyama N. Regulations of gene expression in medullary thymic epithelial cells required for preventing the onset of autoimmune diseases. Front Immunol. 2013;4:249–249. doi: 10.3389/fimmu.2013.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tykocinski L-O, Sinemus A, Kyewski B. The thymus medulla slowly yields its secrets. Ann NY Acad Sci. 2008;1143:105–122. doi: 10.1196/annals.1443.018. [DOI] [PubMed] [Google Scholar]
- Ueno T, Saito F, Gray DHD, Kuse S, Hieshima K, Nakano H, Kakiuchi T, Lipp M, Boyd RL, Takahama Y. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan JE, Parnell SM, Nakamura K, Caamano JH, Lane PJL, Jenkinson EJ, Jenkinson WE, Anderson G. The thymic medulla is required for Foxp3(+) regulatory but not conventional CD4(+) thymocyte development. J Exp Med. 2013;210:675–681. doi: 10.1084/jem.20122070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, Jenkinson EJ, Anderson G. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J Immunol. 2014;192:2659–2666. doi: 10.4049/jimmunol.1303057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S-I, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- Ripen AM, Nitta T, Murata S, Tanaka K, Takahama Y. Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit β5t. Eur J Immunol. 2011;41:1278–1287. doi: 10.1002/eji.201041375. [DOI] [PubMed] [Google Scholar]
- Nitta T, Murata S, Sasaki K, Fujii H, Ripen AM, Ishimaru N, Koyasu S, Tanaka K, Takahama Y. Thymoproteasome shapes immunocompetent repertoire of CD8(+) T cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Xing Y, Jameson SC, Hogquist KA. Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proc Natl Acad Sci USA. 2013;110:6979–6984. doi: 10.1073/pnas.1222244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey K, Nakagawa T, Peters C, Rudensky A. Cathepsin L regulates CD4(+) T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands. J Exp Med. 2002;195:1349–1358. doi: 10.1084/jem.20011904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommeaux J, Gregoire C, Nguessan P, Richelme M, Malissen M, Guerder S, Malissen B, Carrier A. Thymus-specific serine protease regulates positive selection of a subset of CD4(+) thymocytes. Eur J Immunol. 2009;39:956–964. doi: 10.1002/eji.200839175. [DOI] [PubMed] [Google Scholar]
- Assarsson E, Chambers BJ, Hogstrand K, Berntman E, Lundmark C, Fedorova L, Imreh S, Grandien A, Cardell S, Rozell B, et al. Severe defect in thymic development in an insertional mutant mouse model. J Immunol. 2007;178:5018–5027. doi: 10.4049/jimmunol.178.8.5018. [DOI] [PubMed] [Google Scholar]
- Rode I, Boehm T. Regenerative capacity of adult cortical thymic epithelial cells. Proc Natl Acad Sci USA. 2012;109:3463–3468. doi: 10.1073/pnas.1118823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz I, Silva-Santos B, Pennington DJ. Functional development of γδT cells. Eur J Immunol. 2013;43:1988–1994. doi: 10.1002/eji.201343759. [DOI] [PubMed] [Google Scholar]
- Roberts NA, White AJ, Jenkinson WE, Turchinovich G, Nakamura K, Withers DR, McConnell FM, Desanti GE, Benezech C, Parnell SM, et al. Rank signaling links the development of invariant γδT cell progenitors and Aire(+) medullary epithelium. Immunity. 2012;36:427–437. doi: 10.1016/j.immuni.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher B, Pantelyushin S. Hiding under the skin Interleukin-17-producing γδT cells go under the skin? Nat Med. 2012;18:1748–1750. doi: 10.1038/nm.3016. [DOI] [PubMed] [Google Scholar]
- Chien Y-h, Zeng X, Prinz I. The natural and the inducible interleukin (IL)-17-producing γδT cells. Trends Immunol. 2013;34:151–154. doi: 10.1016/j.it.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδT cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka Y, Nitta T, Ohigashi I, Yano K, Ishimaru N, Hayashi Y, Matsumoto M, Matsuo K, Penninger JM, Takayanagi H, et al. The cytokine RANKL produced by positively selected thymocytes Fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 2008;29:438–450. doi: 10.1016/j.immuni.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Shakib S, Desanti GE, Jenkinson WE, Parnell SM, Jenkinson EJ, Anderson G. Checkpoints in the development of thymic cortical epithelial cells. J Immunol. 2009;182:130–137. doi: 10.4049/jimmunol.182.1.130. [DOI] [PubMed] [Google Scholar]
- Ohigashi I, Zuklys S, Sakata M, Mayer CE, Zhanybekova S, Murata S, Tanaka K, Hollaender GA, Takahama Y. Aire-expressing thymic medullary epithelial cells originate from β5t-expressing progenitor cells. Proc Natl Acad Sci USA. 2013;110:9885–9890. doi: 10.1073/pnas.1301799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik S, Jenkinson EJ, Lane PJL, Anderson G, Jenkinson WE. Generation of both cortical and Aire+ medullary thymic epithelial compartments from CD205+ progenitors. Eur J Immunol. 2013;43:589–594. doi: 10.1002/eji.201243209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves NL, Takahama Y, Ohigashi I, Ribeiro AR, Baik S, Anderson G, Jenkinson WE. Serial progression of cortical and medullary thymic epithelial microenvironments. Eur J Immunol. 2014;44:16–22. doi: 10.1002/eji.201344110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi D, Tahiliani P, Kumar A, Chandu D. The ubiquitin-proteasome system. J Biosci. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroeter F, Prozorovski T, Lange N, Steffen J, Rieger M, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carding SR, Egan PJ. γδT cells: functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- Heilig JS, Tonegawa S. Diversity of murine gamma genes and expression in fetal and adult lymphocytes-T. Nature. 1986;322:836–840. doi: 10.1038/322836a0. [DOI] [PubMed] [Google Scholar]
- Shibata K, Yamada H, Nakamura R, Sun X, Itsumi M, Yoshikai Y. Identification of CD25+ γδT cells as fetal thymus-derived naturally occurring IL-17 producers. J Immunol. 2008;181:5940–5947. doi: 10.4049/jimmunol.181.9.5940. [DOI] [PubMed] [Google Scholar]
- Haas JD, Ravens S, Dueber S, Sandrock I, Oberdoerfer L, Kashani E, Chennupati V, Foehse L, Naumann R, Weiss S, et al. Development of interleukin-17-producing γδT cells is restricted to a functional embryonic wave. Immunity. 2012;37:48–59. doi: 10.1016/j.immuni.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, Girardi M, Borst J, Hayday AC, Pennington DJ, et al. CD27 is a thymic determinant of the balance between Interferon-γ-and Interleukin 17-producing γδT cell subsets. Nat Immunol. 2009;10:427–436. doi: 10.1038/ni.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Ramirez-Valle F, Xu Y, Wu S, Wu Z, Karjalainen KE, Cyster JG. Deficiency in IL-17-committed Vγ4(+) γδT cells in a spontaneous Sox13-mutant CD45.1(+) congenic mouse substrain provides protection from dermatitis. Nat Immunol. 2013;14:584–592. doi: 10.1038/ni.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N, Narayan K, Cho OH, Sylvia KE, Yin C, Melichar H, Rashighi M, Lefebvre V, Harris JE, Berg LJ, et al. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity. 2013;38:681–693. doi: 10.1016/j.immuni.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohigashi I, Nitta T, Lkhagvasuren E, Yasuda H, Takahama Y. Effects of RANKL on the thymic medulla. Eur J Immunol. 2011;41:1822–1827. doi: 10.1002/eji.201141480. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Yano I, Kumazawa Y, Takimoto H. Pulmonary TCR gamma delta T cells induce the early inflammation of granuloma formation by a glycolipid trehalose 6,6′-dimycolate (TDM) isolated from Mycobacterium tuberculosis. Immunopharmacol Immunotoxicol. 2012;34:815–823. doi: 10.3109/08923973.2012.658922. [DOI] [PubMed] [Google Scholar]
- Cai Y, Shen X, Ding C, Qi C, Li K, Li X, Jala VR, Zhang H-G, Wang T, Zheng J, et al. Pivotal role of dermal IL-17-producing γδT cells in skin inflammation. Immunity. 2011;35:649–649. doi: 10.1016/j.immuni.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelyushin S, Haak S, Ingold B, Kulig P, Heppner FL, Navarini AA, Becher B. Ror γt(+) innate lymphocytes and γδT cells initiate psoriasiform plaque formation in mice. J Clin Invest. 2012;122:2252–2256. doi: 10.1172/JCI61862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J, Prockop SE, Lepique A, Petrie HT. Critical role for CXCR4 signaling in progenitor localization and T cell differentiation in the postnatal thymus. J Immunol. 2003;171:4521–4527. doi: 10.4049/jimmunol.171.9.4521. [DOI] [PubMed] [Google Scholar]
- Benz C, Heinzel K, Bleul CC. Homing of immature thymocytes to the subcapsular microenvironment within the thymus is not an absolute requirement for T cell development. Eur J Immunol. 2004;34:3652–3663. doi: 10.1002/eji.200425248. [DOI] [PubMed] [Google Scholar]
- Stritesky GL, Xing Y, Erickson JR, Kalekar LA, Wang X, Mueller DL, Jameson SC, Hogquist KA. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc Natl Acad Sci USA. 2013;110:4679–4684. doi: 10.1073/pnas.1217532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med. 2008;205:2575–2584. doi: 10.1084/jem.20080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitara S, Hara T, Liang B, Wagatsuma K, Zuklys S, Hollaender GA, Nakase H, Chiba T, Tani-ichi S, Ikuta K. IL-7 produced by thymic epithelial cells plays a major role in the development of thymocytes and TCRγδ(+) intraepithelial lymphocytes. J Immunol. 2013;190:6173–6179. doi: 10.4049/jimmunol.1202573. [DOI] [PubMed] [Google Scholar]
- Michel M-L, Pang DJ, Haque SFY, Potocnik AJ, Pennington DJ, Hayday AC. Interleukin 7 (IL-7) selectively promotes mouse and human IL-17-producing γδ cells. Proc Natl Acad Sci USA. 2012;109:17549–17554. doi: 10.1073/pnas.1204327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K, Yamada H, Sato T, Dejima T, Nakamura M, Ikawa T, Hara H, Yamasaki S, Kageyama R, Iwakura Y, et al. Notch-Hes1 pathway is required for the development of IL-17-producing γδT cells. Blood. 2011;118:586–593. doi: 10.1182/blood-2011-02-334995. [DOI] [PubMed] [Google Scholar]
- Cai Y, Xue F, Fleming C, Yang J, Ding C, Ma Y, Liu M, Zhang HG, Zheng J, Xiong N, et al. Differential developmental requirement and peripheral regulation for dermal Vγ4 and Vγ6T17 cells in health and inflammation. Nat Commun. 2014;5:3986. doi: 10.1038/ncomms4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden LM, Lewis JM, Barbee SD, Bas A, Girardi M, Hayday AC, Tigelaar RE, Lifton RP. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal γδT cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SD, Woodward MJ, Turchinovich G, Mention JJ, Lewis JM, Boyden LM, Lifton RP, Tigelaar R, Hayday AC. Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc Natl Acad Sci USA. 2011;108:3330–3335. doi: 10.1073/pnas.1010890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KDC, Su X, Shin S, Li L, Youssef S, Yarnasaki S, Steinman L, Saito T, Locksley RM, Davis MM, et al. Thymic selection determines γδ cell effector fate: antigen-naive cells make Interleukin-17 and antigen-experienced cells make interferon γ. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt A, Ravens S, Fleige H, Haas JD, Oberdörfer L, Łyszkiewicz M, Förster R, Prinz I. CCR7-mediated migration in the thymus controls γδ T-cell development. Eur J Immunol. 2014;44:1320–1329. doi: 10.1002/eji.201344330. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T-cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Cosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D. Mice lacking MHC class-II molecules. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- Koller BH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in beta-2-m, MHC class I proteins, and CD8+ T-cells. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- Mizuno A, Kanno T, Hoshi M, Shibata O, Yano K, Fujise N, Kinosaki M, Yamaguchi K, Tsuda E, Murakami A, et al. Transgenic mice overexpressing soluble osteoclast differentiation factor (sODF) exhibit severe osteoporosis. J Bone Miner Metab. 2002;20:337–344. doi: 10.1007/s007740200049. [DOI] [PubMed] [Google Scholar]
- Oda H, Tamehiro N, Patrick MS, Hayakawa K, Suzuki H. Differential requirement for RhoH in development of TCRαβ CD8αα IELs and other types of T cells. Immunol Lett. 2013;151:1–9. doi: 10.1016/j.imlet.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Gray DHD, Chidgey AP, Boyd RL. Analysis of thymic stromal cell populations using flow cytometry. J Immunol Methods. 2002;260:15–28. doi: 10.1016/s0022-1759(01)00493-8. [DOI] [PubMed] [Google Scholar]
- Roark CL, Aydintug MK, Lewis J, Yin X, Lahn M, Hahn YS, Born WK, Tigelaar RE, O'Brien RL. Subset-specific, uniform activation among Vγ6/Vδ1(+) γδT cells elicited by inflammation. J Leukoc Biol. 2004;75:68–75. doi: 10.1189/jlb.0703326. [DOI] [PubMed] [Google Scholar]
- Nitta T, Ohigashi I, Takahama Y. The development of T lymphocytes in fetal thymus organ culture. Methods Mol Biol. 2013;946:85–102. doi: 10.1007/978-1-62703-128-8_6. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo T, Kaneko T, Haruguchi Y, Fukumoto K, Machida H, Koga M, Nakagawa Y, Takeshita Y, Matsuguma T, Tsuchiyama S, et al. Birth of mice from vitrified/warmed 2-cell embryos transported at a cold temperature. Cryobiology. 2009;58:196–202. doi: 10.1016/j.cryobiol.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Fehling HJ, Swat W, Laplace C, Kuhn R, Rajewsky K, Muller U, Vonboehmer H. MHC class-I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Nakano H, Yamamoto N, Yamaoka S. Lymphotoxin-β receptor mediates NEMO-independent NF-κB activation. FEBS Lett. 2002;532:45–51. doi: 10.1016/s0014-5793(02)03622-0. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K, Kato K, Tanaka K, Murata S. Dissecting β-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27:2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez RL, Roman J, Staton GW, Hunter RL. Extravascular coagulation and fibrinolysis in murine lung inflammation-induced by the mycobacterial cord factor trehalose-6,6′-dimycolate. Am J Respir Crit Care Med. 1994;149:510–518. doi: 10.1164/ajrccm.149.2.8306054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S5, Supplementary Tables S1–S3
Review Process File