Abstract
Arabidopsis flowers early under long days (LD) and late under short days (SD). The repressor of photomorphogenesis DE-ETIOLATED1 (DET1) delays flowering; det1-1 mutants flower early, especially under SD, but the molecular mechanism of DET1 regulation remains unknown. Here we examine the regulatory function of DET1 in repression of flowering. Under SD, the det1-1 mutation causes daytime expression of FKF1 and CO; however, their altered expression has only a small effect on early flowering in det1-1 mutants. Notably, DET1 interacts with GI and binding of GI to the FT promoter increases in det1-1 mutants, suggesting that DET1 mainly restricts GI function, directly promoting FT expression independent of CO expression. Moreover, DET1 interacts with MSI4/FVE, which epigenetically inhibits FLC expression, indicating that the lack of FLC expression in det1-1 mutants likely involves altered histone modifications at the FLC locus. These data demonstrate that DET1 acts in both photoperiod and autonomous pathways to inhibit expression of FT and SOC1. Consistent with this, the early flowering of det1-1 mutants disappears completely in the ft-1 soc1-2 double mutant background. Thus, we propose that DET1 is a strong repressor of flowering and has a pivotal role in maintaining photoperiod sensitivity in the regulation of flowering time.
The appropriate timing of flowering is tightly linked to the success of reproduction in higher plants. Intrinsic genetic programs and various environmental factors, mainly day length and temperature, determine the transition from vegetative to reproductive development. In particular, photoperiod provides a major cue for controlling flowering time, as perception of light enables plants to synchronize initiation of flowering with seasonal changes in photoperiod1.
In Arabidopsis thaliana, several signaling components participate in the regulatory circuit promoting photoperiodic flowering, including GIGANTEA (GI), CONSTANS (CO), and FLOWERING LOCUS T (FT)2,3,4. FT integrates multiple flowering pathways and FT protein is an essential component of florigen, which moves from the induced leaf to the shoot apex2,5. CO directly regulates expression of FT mRNA and CO mediates between the circadian clock and the control of flowering. CO is stable in the light, but is degraded in the dark by ubiquitin-mediated proteolysis4,6. GI and FLAVIN-BINDING, KELCH REPEAT, F-BOX PROTEIN 1 (FKF1) form a complex and regulate the timing of CO expression. The diurnal expression of GI and FKF1 has little overlap in SD, leading to minimal formation of the GI-FKF1 complex7. By contrast, in LD, the more extensive overlap of GI and FKF1 diurnal expression leads to formation of more GI-FKF1 complex. Thus, GI acts as a flowering inducer with FKF1 in the CO-FT pathway mainly in LD. In a CO-independent flowering pathway, GI can also directly activate FT expression by binding to its promoter region8, indicating that GI can directly or indirectly induce FT transcription in the photoperiod pathway.
In addition to regulation by the photoperiod pathway, genes involved in the autonomous and vernalization pathways also control FT expression. FLOWERING LOCUS C (FLC) has a central place in those two pathways and directly regulates FT and SOC1 expression by binding to their promoters9,10,11. Chromatin remodeling also affects FLC expression. For example, MULTICOPY SUPPRESSOR OF IRA1 4 (MSI4)/FVE, in the autonomous pathway, negatively regulates FLC expression via histone deacetylation of the FLC locus12. Furthermore, MSI4/FVE interacts with DDB1 and HDA6, and mediates transcriptional silencing by histone modification of H3K4me313 and H3K27me314. This indicates that MSI4/FVE plays a significant role in FLC expression by making a complex with various chromatin remodeling factors.
DET1, a repressor of photomorphogenesis, was first identified as a member of the CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) gene family15. DET1 forms a complex with COP10 and DAMAGED DNA BINDING PROTEIN 1 (DDB1) to promote the activity of ubiquitin-conjugating enzymes (E2) for repression of photomorphogenesis in the ubiquitination pathway16,17. DET1 also acts as a pacemaker to adjust the period length of the circadian rhythm18, possibly through interaction with LHY and CCA119. DET1 acts as a flowering repressor; det1-1 mutants flower slightly early in LD and extremely early in SD20. Despite recent advances in the understanding of DET1 function, the molecular mechanism causing early flowering in det1-1 mutants remains unknown.
Here we demonstrate that DET1 delays flowering time in SD, mainly by reducing the affinity of GI binding to the FT promoter in the photoperiod pathway. DET1 also contributes to upregulating FLC expression in the autonomous pathway, possibly by weakening the activity of MSI4/FVE in histone modification of the FLC locus. These effects, in turn, lead to reduced expression of FT and SOC1. These findings provide new insights into how DET1 dynamically suppresses flowering in SD and thus plays an important role in maintaining photoperiod sensitivity in Arabidopsis.
Results
The det1 mutation alters the expression of flowering-time genes
The det1 null mutants are lethal; to study the molecular mechanism by which DET1 functions in floral repression, we therefore used a weak allele, det1-1, and counted the rosette leaf number at bolting to measure flowering time (Fig. 1a, b). We found that det1-1 mutants flower early under LD and extremely early under SD, which shows that flowering in det1-1 mutants is photoperiod-insensitive. These results indicate that DET1 acts as a strong floral repressor in SD and has a key role in maintaining the photoperiod sensitivity of the regulation of flowering time in Arabidopsis.
Figure 1. Flowering-time phenotypes of det1-1 mutants.
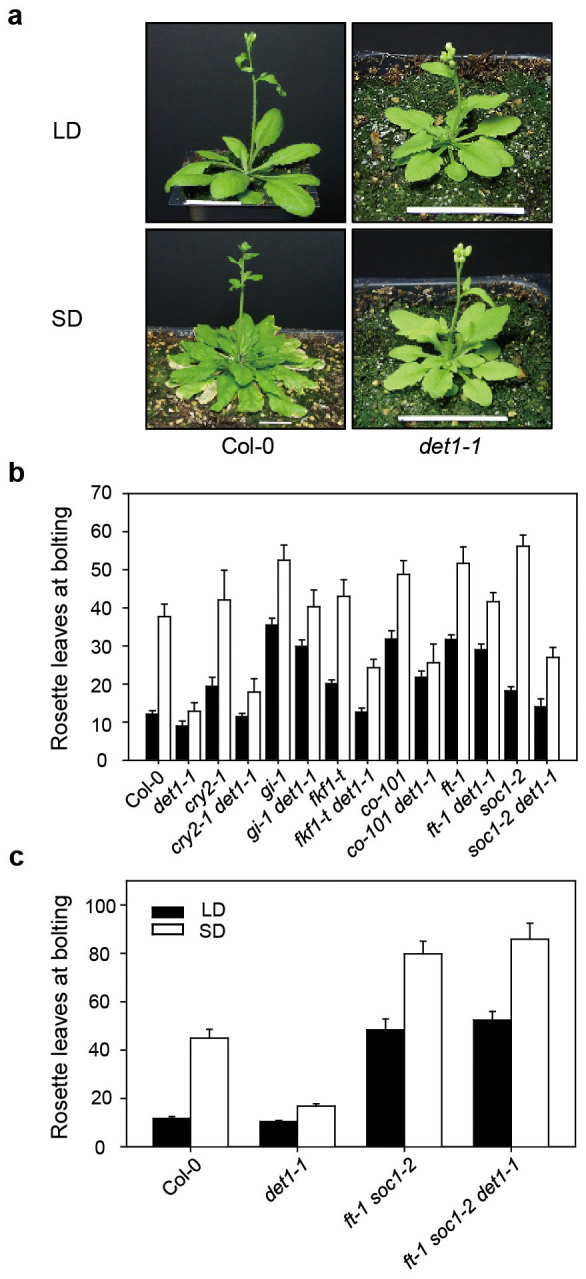
(a) Phenotypes of wild-type (WT, Col-0 ecotype) and det1-1 mutant plants. Plants were grown at 22°C under cool-white fluorescent light (90–100 μmol m−2s−1) in LD (16-h light:8-h dark) or SD (10-h light:14-h dark), and photographed at 2 to 4 days after bolting. Scale bars = 2 cm. (b–c) Genetic analysis to show epistasis between det1-1 and flowering mutants using double (b) and triple mutants (c). The number of rosette leaves of WT (Col-0) and flowering-time mutants grown under LD (16-h light:8-h dark) and SD (10-h light:14-h dark) in (b), and LD (16-h light:8-h dark) and SD (8-h light:16-h dark) conditions in (c) (see Table S1). Flowering time was measured as the number of rosette leaves at bolting. Means and standard deviations were obtained from more than 20 plants.
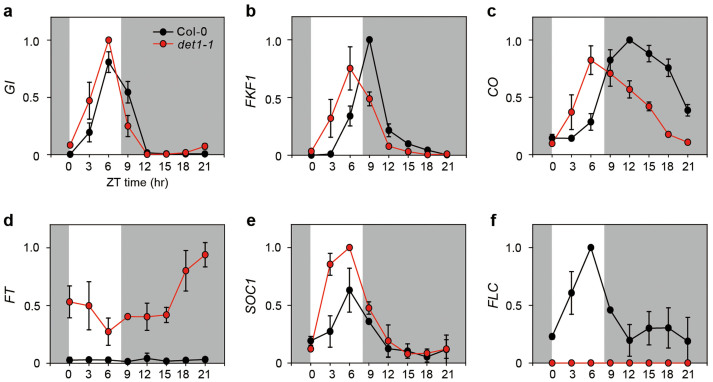
The det1-1 mutation causes period-shortening of clock-regulated gene expression; the internal circadian periods of CAB2:LUC (encoding a luciferase) expression in det1-1 mutants were approximately 18 h in continuous darkness and 21 h in continuous light conditions18. To investigate whether the circadian defect in det1-1 mutants causes extremely early flowering under SD (Fig. 1 and Table S1), we analyzed the expression modes of floral inducers by measuring the phases and amplitudes of GI, FKF1, CO, FT, and SOC1 mRNA abundance, in WT and det1-1 mutants grown in SD (Fig. 2). In WT, GI expression peaked at ZT6 (zeitgeber time; 6 h after dawn) during daytime, but the peaks of FKF1 and CO expression occurred at ZT9 and ZT12 during nighttime, respectively, resulting in no FT expression21. In det1-1 mutants, GI, FKF1, CO, FT, and SOC1 also showed rhythmic expression (Fig. 2a–e) and GI expression did not significantly differ compared with WT (Fig. 2a). However, the peaks of FKF1 and CO expression shifted 3 h and 6 h earlier than those in WT, respectively (Fig. 2b, c). Accordingly, the peaks of GI, FKF1, and CO expression occurred at ZT6 during daytime in det1-1 mutants under SD. Thus, it appears that the daytime expression of CO and light-stabilized CO (Fig. 2c) can activate FT expression in det1-1 mutants under SD (Fig. 2d). The waveform and peak time of SOC1 expression did not change in det1-1 mutants, but SOC1 mRNA abundance increased (Fig. 2e), possibly due to daytime expression of CO and/or increased expression of FT (Fig. 2c, d)9,22,23. Thus, we first speculated that circadian dysfunction might cause the early flowering in det1-1 mutants, as previously reported19.
Figure 2. Effect of det1-1 mutation on GI, FKF1, CO, FT, SOC1, and FLC expression under SD.
The expression of GI (a), FKF1 (b), CO (c), FT (d), SOC1 (e), and FLC (f) was analyzed in Col-0 and det1-1 mutants by real-time PCR using 3-week-old plants. Plants were grown at 22°C under SD (8-h light:16-h dark) conditions, and plant tissues were harvested every 3 h. ACT2 expression was used for normalization. Means and standard deviations were obtained from three biological replicates.
To test whether circadian-period shortening causes the extremely early flowering of det1-1 mutants in SD (Fig. 1 and Table S1), we examined whether the flowering-time defect can be recovered when det1-1 mutants were entrained in SD (light:dark = 1:2) under reduced diurnal cycles, i.e. environmental time periods (T) of 24 T (8-h light:16-h dark), 21 T (7-h light:14-h dark), and 18 T (6-h light:12-h dark). Although reduced diurnal cycles of 21 T and 18 T slightly delayed flowering compared to normal cycles of 24 T, the det1-1 mutants still flowered much earlier than WT under SD of 24 T (Fig. 3). To investigate the cause of early flowering in det1-1 mutants under reduced T cycles, we analyzed the phases and amplitudes of GI, FKF1, CO, FT, and SOC1 mRNA abundance in det1-1 mutants grown under SD of 18 T (Fig. S1). Unlike the SD of 24 T, the waveforms and peaks of GI, FKF1, and CO expression in det1-1 mutants were very similar to those of WT. However, FT and SOC1 expression was still upregulated in det1-1 mutants, suggesting that the internal period-shortening defect in det1-1 mutants cannot fully explain the extremely early flowering under SD of 24 T. The FKF1 and CO peak shifts likely produce a small effect on early flowering in det1-1 mutants, because fkf1-t and co-101 mutations delayed flowering in det1-1 mutants under SD whereas they were almost ineffective in WT21 (Fig. 1b and Table S1). Thus, these results strongly suggest that other defects in mechanisms of floral repression lead to photoperiod-insensitive early flowering in det1-1 mutants, rather than the circadian dysfunction in the FKF1-CO-FT pathway.
Figure 3. Flowering time of det1-1 mutants under reduced diurnal cycles.
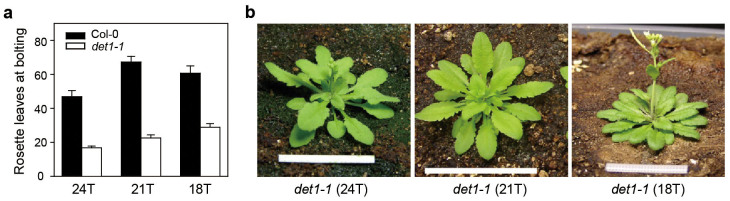
(a) Effect of reduced diurnal cycles on the flowering time of det1-1 mutants. Plants were entrained in SD (light [L]:dark [D] = 1:2) of 24 h (24 T = 8 L:16 D), 21 h (21 T = 7 L:14 D), and 18 h (18 T = 6 L:12 D). T represents environmental time period. Means and standard deviations were obtained from more than 20 plants. Col-0 means Columbia-0 ecotype (wild type). (b) Phenotypes of det1-1 mutants after bolting under SD of 24 T, 21 T, and 18 T. Plants were grown at 22–24°C under cool-white fluorescent light (90–100 μmol m−2 s−1). Scale bars = 2 cm.
DET1 mainly functions in the photoperiod and autonomous pathways
To test which genetic pathways of floral induction are responsible for the early flowering phenotype of det1-1 mutants, we examined the flowering-time phenotypes of double mutants of det1-1 and mutations with late-flowering phenotypes, specifically cry2-1, fkf1-t, gi-1, co-101, ft-1, and soc1-2 (Fig. 1b and Table S1). The cry2-1 det1-1 double mutants flowered much earlier than the cry2-1 single mutants in both LD and SD, suggesting that DET1 acts downstream of CRY2. The fkf1-t det1-1 and co-101 det1-1 double mutants exhibited intermediate flowering times compared with fkf1-t, co-101, and det1-1 single mutants in both LD and SD, suggesting that although daytime expression of FKF1 and CO contributes to early flowering in SD, det1-1 mutants can flower early in the absence of FKF1 and CO activity in both photoperiod conditions. In gi-1 det1-1 and ft-1 det1-1 mutants, the early-flowering effect of det1-1 was almost abolished by gi-1 or ft-1 in both LD and SD (Fig. 1b and Table S1), indicating that GI and FT play major roles in the DET1-mediated flowering pathway.
As both the photoperiod and autonomous pathways regulate SOC1 expression10, we further tested whether DET1 also participates in the autonomous pathway. We found that soc1-2 det1-1 double mutants showed intermediate flowering times in both LD and SD. Also, in ft-1 soc1-2 det1-1 triple mutants, the early flowering effect of det1-1 completely disappeared (Fig. 1b, c, and Table S1). These results indicate that the regulation of flowering time by DET1 does not entirely depend on the FT-mediated photoperiod pathway, but also depends on the SOC1-mediated autonomous pathway. Thus, we further examined the expression of FLC, a major gene in the autonomous pathway, in det1-1 mutants. We found that the det1-1 mutants under SD had very low levels of FLC mRNA (Fig. 2f), suggesting that DET1 induces FLC expression to repress FT and SOC1. Taking these results together, we concluded that DET1 mainly acts in the photoperiod and autonomous pathways as a strong floral repressor.
DET1 interacts with GI in vivo
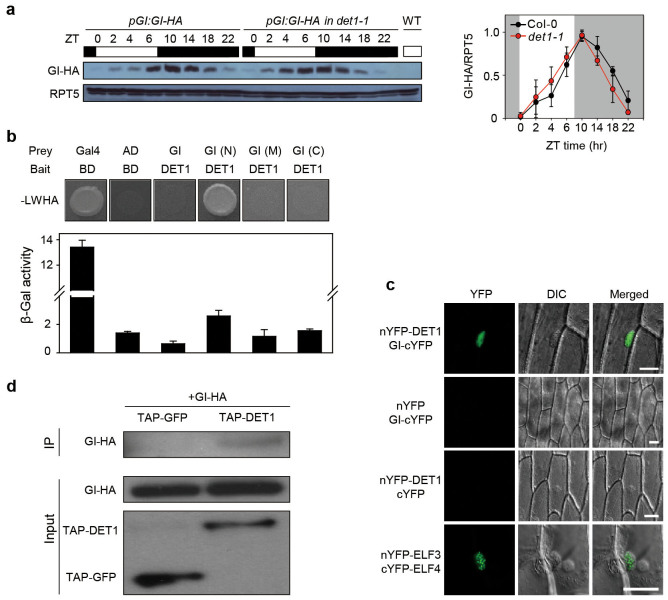
GI functions in the photoperiod pathway and det1-1 mutants did not show significant alterations in GI mRNA levels (Fig. 2a), but the gi-1 mutation nearly abolished the early flowering effect of det1-1 in gi-1 det1-1 double mutants (Table S1). Based on these observations, we postulated that DET1 mainly regulates GI at the post-translational level. Thus, we used transgenic plants expressing a tagged GI protein (pGI:GI-HA gi-2 and pGI:GI-HA gi-2 det1-1) to examine whether DET1 negatively regulates GI stability. We found that det1-1 mutants showed no significant alteration in the rhythmic accumulation of GI protein in SD (Fig. 4a). This indicates that the det1-1 mutation does not affect GI protein stability.
Figure 4. DET1 directly interacts with GI.
(a) Comparison of GI protein stability between pGI:GI-HA and pGI:GI-HA det1-1 plants under SD conditions. The plant tissues were collected every 2 h during the daytime and every 4 h during the nighttime, using 3-week-old seedlings. GI protein was detected with an anti-HA antibody. RFT5 expression was used for normalization. Means and standard deviations were obtained from three biological replicates. (b) Interaction of DET1-GI was tested by yeast 2-hybrid assay. The bait was full-length DET1. For prey, GI was divided into three pieces: N-terminal (N; 1–507), middle (M; 401–907), and C-terminal (C; 801–1173). Gal4 indicates a positive control. Empty pGBKT7 (BD) and pGADT7 (AD) vectors were used as the negative control. SD medium (-LWHA; lacking tryptophan, leucine, histidine, and adenine) was used to select for the interaction between bait and prey proteins. β-galactosidase (β-Gal) activity assays were performed according to the manufacturer's protocol. Means and standard deviations were obtained from three biological replicates. (c) BiFC analysis of the interaction of between DET1 and GI in the nucleus of an onion epidermal cell. nYFP-ELF3 and cYFP-ELF4 plasmids served as a positive control. For the negative control, empty nYFP/GI-cYFP and nYFP-DET1/cYFP were used. Scale bar = 50 μm. (d) Coimmunoprecipitation of DET1 and GI. Total protein was extracted from 2-week-old seedlings of p35S:TAP-DET1 pGI:GI-HA gi-2 and p35S:TAP-GFP pGI:GI-HA gi-2. IgG beads were used for the pull-down. An anti-HA antibody was used for GI-HA protein band. p35S:TAP-GFP pGI:GI-HA gi-2 plants served as a negative control. The upper panel is a coimmunoprecipitated sample, and the middle panel is the input sample for GI-HA protein. The lower panel shows input samples of p35S:TAP-GFP and p35S:TAP-DET1.
DET1 interacts with LHY and CCA1, which regulate the circadian rhythms of expression of clock-regulated genes19. This raises the possibility that DET1 could negatively regulate GI activity by protein-protein interaction. To examine this, we performed yeast 2-hybrid assays and found that DET1 interacts with the N-terminal region of GI (amino acids [aa] 1-507) (Fig. 4b). To test the in vivo interaction of DET1 and GI, we performed bimolecular fluorescence complementation (BiFC) assays. In the onion epidermal cells, we detected reconstituted YFP fluorescence in the nucleus when nYFP-DET1 and GI-cYFP plasmids were co-transformed (Fig. 4c). To further confirm their interaction, we tested whether GI and DET1 co-immunoprecipitate from transgenic plants expressing tagged proteins. To that end, we sampled the p35S:TAP-DET1 pGI:GI-HA gi-2 and p35S:TAP-GFP pGI:GI-HA gi-2 (a negative control) transgenic plants at ZT8 in SD, and used antibodies for the TAP tag to immunoprecipitate DET1. We found that HA-GI co-immunoprecipitated with TAP-DET1, but not with TAP-GFP (Fig. 4d). These results indicate that DET1 interacts directly with GI in the nucleus.
DET1 negatively regulates GI binding to the FT promoter
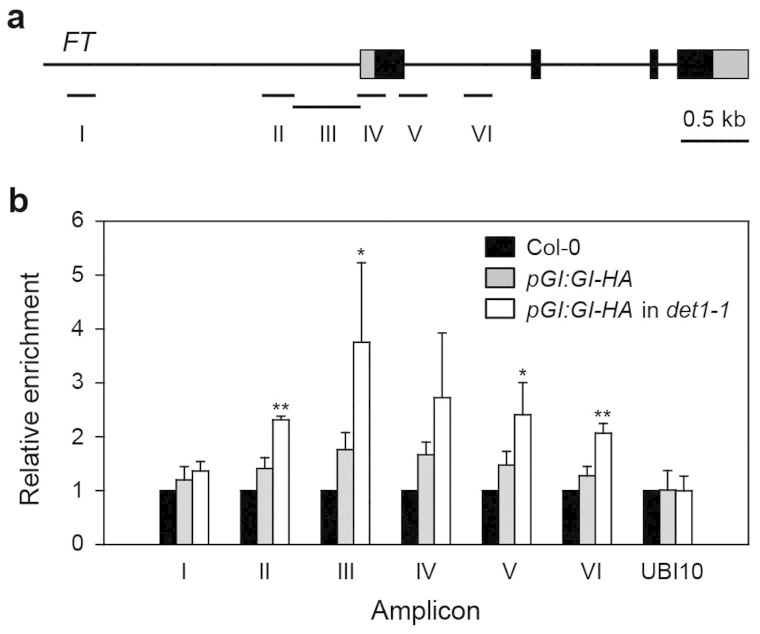
The det1-1 mutation does not alter GI mRNA expression (Fig. 2a) or GI protein levels (Fig. 4a) but gi-1 shows nearly complete epistasis to det1-1 in flowering time (Fig. 1b and Table S1). Based on this observation, we hypothesized that in the photoperiod pathway, DET1 negatively regulates the activity of GI, which directly upregulates FT expression through a CO-independent pathway8. To test whether det1-1 mutation affects the GI-FT module, we performed chromatin immunoprecipitation (ChIP) assays, using pGI:GI-HA gi-2 and pGI:GI-HA gi-2 det1-1 seedlings entrained in SD, to test whether det1-1 affects the ability of GI to bind to the FT promoter. We collected tissues from 10-day-old seedlings at ZT8 and detected relative enrichment of the promoter regions by PCR with primers for six regions of the FT promoter, as described previously8. When we compared GI binding affinity to the FT promoter regions, the amplicons close to the 5′ untranslated region (UTR) were significantly more enriched in ChIP from det1-1 mutants (Fig. 5b). This result strongly supports the notion that DET1 plays an important role in the suppression of FT transcription by preventing GI binding to the FT promoter, and thus contributing to late flowering in SD conditions.
Figure 5. DET1 affects GI binding to the FT promoter.
(a) Gene structure of FT and the amplicon regions for the ChIP assay. Six amplicon locations (I, II, III, IV, V and VI) are shown. (b) FT promoter binding affinity of GI in the det1-1 mutant, relative to the wild type. All samples were harvested at ZT8 under SD (8-h light:16-h dark) conditions. Chromatin isolated from these samples was immunoprecipitated with anti-HA. Relative enrichment in Col-0, pGI:GI-HA gi-2, and pGI:GI-HA gi-2 det1-1 are shown. Means and standard deviations were obtained from three biological replicates. This experiment was replicated at least three times with similar results. UBIQUITIN 10 (UBI10) was used as a negative control. Black, gray, and white boxes represent Col-0, pGI:GI-HA gi-2, and pGI:GI-HA gi-2 det1-1, respectively. Asterisks indicate statistically significant differences compared to pGI:GI-HA as determined by Student's t-test (*P < 0.05 and **P < 0.01, respectively).
DET1 positively regulates FLC expression to delay flowering time in SD
In the autonomous pathway, FLC functions as a key floral repressor and downregulates the transcription of FT and SOC124,25,26. As the transcript levels of FT and SOC1 were upregulated in det1-1 mutants under SD (Fig. 2d, e), and FLC expression was almost absent in det1-1 mutants entrained in SD (Fig. 2f), we reasoned that DET1 also functions to delay flowering in the autonomous pathway by upregulating expression of FLC. A previous report showed that the COP10-DET1-DDB1 complex interacts with CUL427 and the DDB1-CUL4 complex interacts with MSI4/FVE to induce FLC transcription14. Thus, we asked if DET1 interacts with MSI4 to form a DET1-MSI4 complex to regulate FLC mRNA levels. To test this, we examined the in vivo interaction of MSI4-DET1 by BiFC assays (Fig. 6a). We detected strong YFP fluorescence in the nuclei of cells co-transformed with plasmids expressing DET1-nYFP and cYFP-MSI4, indicating that DET1 interacts with MSI4, which directly binds to the FLC promoter to repress FLC transcription.
Figure 6. DET1 interacts with MSI4 and regulates histone methylation of the FLC locus.
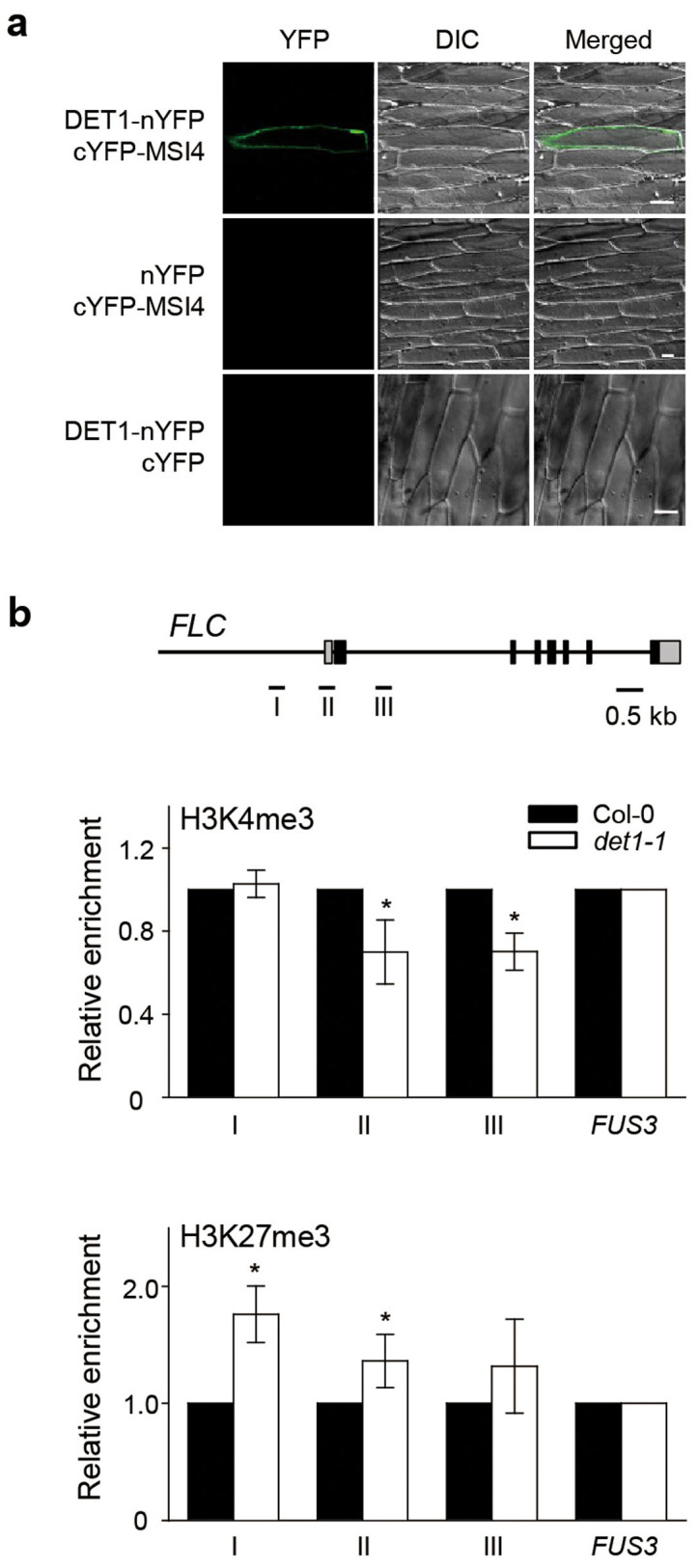
(a) BiFC analysis of the interaction between MSI4 and DET1 in onion epidermal cells. For negative controls, nYFP/cYFP-MSI4 and DET1-nYFP/cYFP were used. Scale bar = 50 μm. (b) Relative levels of histone modifications on the FLC locus were examined by ChIP analysis using H3K4me3 and H3K27me3 antibodies in Col-0 and det1-1 plants. The top of the panel represents the FLC gene structure and the region used for primers (I, II and III) in the ChIP-quantitative PCR analyses. Chromatin was prepared from 14-day-old seedlings grown under SD (8-h light:16-h dark). FUSCA 3 (FUS3) was used for the normalization of the quantitative PCR analysis. Means and standard deviations were obtained from three biological replicates. This experiment was replicated at least three times with similar results. Asterisks indicate statistically significant difference compared to Col-0 as determined by Student's t-test (*P < 0.05).
Since MSI4 binds to the FLC promoter and alters histone modification, specifically H3K27me3 and H3K4me3, at the FLC locus13,14, we further examined the histone methylation levels of the FLC locus, using anti-H3K27me3 and anti-H3K4me3 antibodies in WT and det1-1 mutants. The ChIP analysis revealed that det1-1 mutants maintained higher levels of H3K27me3 and lower levels of H3K4me3 at the FLC locus than did WT (Fig. 6b), consistent with the histone modification states observed in the early-flowering hos1-3 mutants28. Taking these results together, we suggest that the DET1-MSI4/FVE complex likely contributes to late flowering in SD by altering histone modification of the FLC locus in the autonomous pathway.
Discussion
DET1 is involved in repression of photomorphogenesis in the ubiquitination pathway16,17,29, light-response regulatory pathway20, and circadian period18,19. However, the function of DET1 in the regulation of flowering time remains unclear. In this study, we provide evidence showing how DET1 regulates photoperiod sensitivity by delaying flowering time in SD. For example, det1-1 mutants showed increased GI activity (Fig. 5) and epigenetic silencing of FLC expression (Fig. 6), resulting in upregulation of FT and SOC1. Thus, we propose a model for the regulatory role of DET1 in both photoperiod and autonomous pathways (Fig. 7).
Figure 7. Working model of DET1 function in floral repression in Arabidopsis.
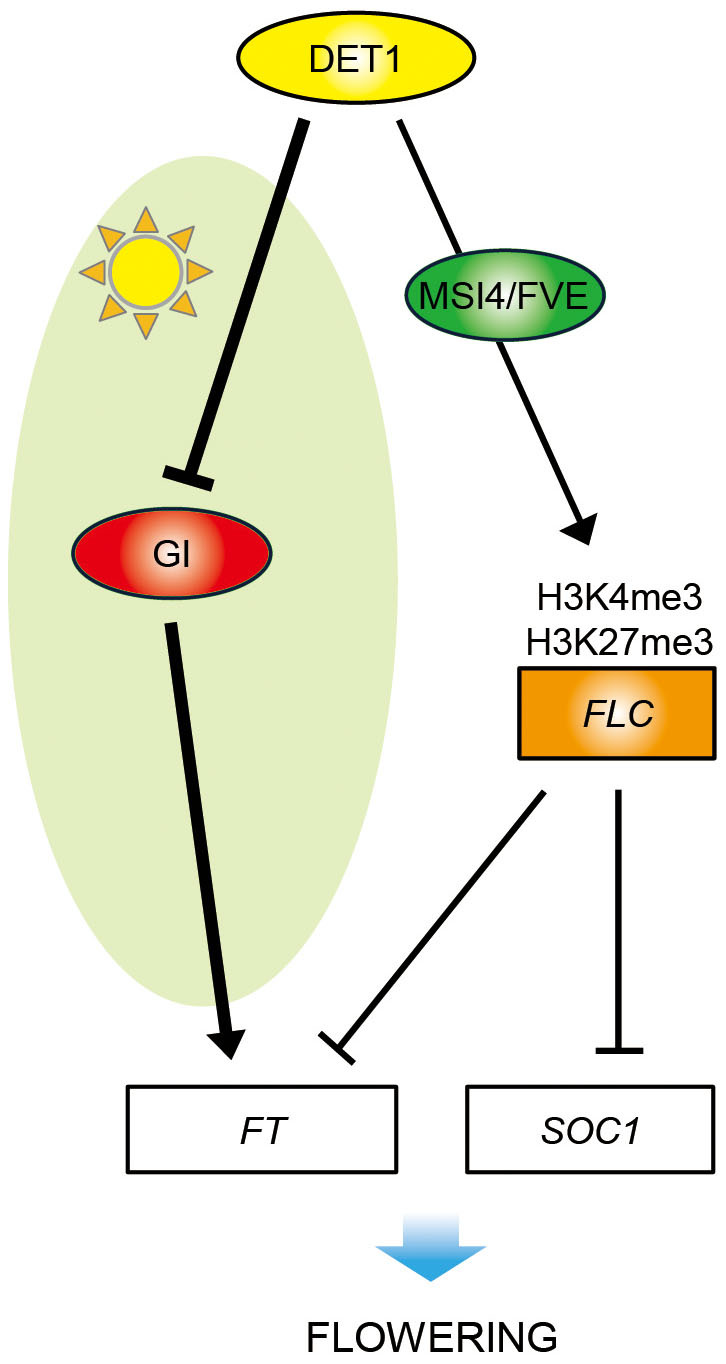
DET1 suppresses FT and SOC1 expression through the photoperiod and autonomous pathways of flowering. In the photoperiod pathway, DET1 mainly represses flowering by modulating GI-mediated floral induction at the transcriptional and post-translational levels during daytime under SD. DET1 represses the function of daytime-expressed GI by preventing GI from binding to the FT promoter in a CO-independent pathway. In the autonomous pathway, DET1 interacts with MSI4/FVE and possibly modulates trimethylation of FLC chromatin to epigenetically induce FLC expression. Genes and proteins are represented as rectangles and ovals, respectively.
In this study, we showed that gi-1 and ft-1 nearly completely suppressed the early flowering of det1-1 mutants and that DET1 directly interacts with GI in vitro and in vivo (Fig. 4). However, DET1 does not interact with the light-input components PHYA, PHYB, CRY1 C-terminus (CCT1), or CRY2 C-terminus (CCT2), or the floral inducers CO or FKF1 (Fig. S3), indicating that DET1 has a unique role in the posttranslational regulation of GI activity in the photoperiod pathway. A previous study revealed that EARLY FLOWERING4 (ELF4), one of the circadian-clock components30, acts upstream of GI31. ELF4 represses GI binding to the CO promoter to control flowering32. Our results revealed that co-101 det1-1 mutants showed intermediate flowering-time phenotypes, but in ft-1 det1-1 mutants, the early flowering phenotype of det1-1 almost completely disappeared under LD (Fig. 1b), indicating that DET1 function in the regulation of photoperiodic flowering mainly depends on FT expression. Thus, we hypothesized that DET1 regulates GI binding to the FT promoter to delay flowering time and showed that GI binding to the FT promoter significantly increased in the det1-1 mutant background (Fig. 5). This result indicates that DET1 represses FT expression via direct regulation of GI binding to the FT promoter.
DET1 functions as a repressor of photomorphogenesis in darkness by forming a complex with COP10 and DDB1 and promoting the activity of ubiquitin-conjugating E2 enzymes in the ubiquitination pathway16,17. The RING-type E3 ubiquitin ligase COP1, a member of the COP/DET/FUS family15, also represses photomorphogenesis in darkness; cop1-4 mutants display very similar phenotypes to det1-1 mutants, such as short hypocotyls and opened cotyledons34. This implies a potential functional connection between DET1 and COP1. Indeed, COP1 interacts with COP10, but not with DET116, suggesting that COP1 could interact with the COP10-DET1-DDB1 (CDD) complex to repress photomorphogenesis. In addition, cop1-4 mutants flower extremely early under SD, similar to det1-1 mutants33. Thus, the CDD complex may function with COP1 in regulation of flowering time, although we have no direct evidence because the det1-1 cop1-4 double mutant is lethal34. COP1 directly controls GI stability by interacting with GI in the presence of ELF3 for photoperiodic flowering33. However, DET1 does not regulate GI stability but does negatively affect GI binding to the FT promoter (Fig. 4a). Therefore, although DET1 and COP1 have very similar mutant phenotypes and post-translational behavior, they seem to regulate GI function independently through distinct molecular mechanisms.
Other negative regulators of FT transcription, including FLC, SVP, TEM1, and TEM2, bind to the regions near the 5′UTR of FT. In single mutants of these regulatory genes, FT mRNA expression increases to levels similar to those seen in det1-1 mutants11,35,36. Notably, SVP, TEM1, and TEM2 interact with GI to regulate FT expression, although the regulatory function of their interaction is not clearly understood8. Therefore, DET1 could be involved in the function of these FT repressors. To investigate this possibility, we examined the interaction of DET1 with these four FT repressors by yeast 2-hybrid assays, which revealed that DET1 does not interact with FLC, SVP, TEM1, or TEM2 (Fig. S4). This result strongly suggests that DET1 may regulate the GI-FT module independent of these known FT repressors.
In addition, we revealed that DET1 regulates the expression of FLC, a key component in the autonomous pathway. We found that the det1-1 mutants showed a remarkable decrease in FLC mRNA levels and had altered levels of H3K4me3 and H3K27me3 (Figs. 2f and 6b), as observed in the early-flowering hos1-3 mutants28. Furthermore, our examination of the components of the CDD complex showed that in addition to interacting with DDB1, DET1 also interacts with MSI4/FVE, which repress FLC expression in the autonomous pathway (Fig. 6a)14. This indicates that DET1 represses FLC expression possibly through direct interaction with MSI4/FVE. Meanwhile, FLC negatively regulates not only FT but also the downstream factor SOC1, which encodes a MADS box transcription factor37. In genetic analysis, ft-1 was completely epistatic to det1-1 in LD, but in SD the ft-1 det1-1 double mutants showed an intermediate phenotype, indicating incomplete epistasis. Consistent with this, SOC1 expression was upregulated in det1-1 mutants (Fig. 2e), but soc1-2 did not rescue the early flowering of det1-1 (Fig. 1b and Table S1). Notably, the ft-1 soc1-2 det1-1 triple mutants showed complete suppression of the early flowering of det1-1 in both photoperiods. This supports the idea that DET1 suppresses both FT and SOC1 via promoting FLC expression in the autonomous pathway.
DET1 interacts with LHY/CCA1 and is required for transcriptional repression of CCA1/LHY target genes such as TOC119. These observations indicate that DET1 functions with LHY/CCA1 to regulate the circadian rhythms of evening genes. Moreover, DET1 could act with LHY/CCA1 to negatively regulate GI binding to the FT promoter mainly in SD, because lhy cca1 double mutants also exhibit photoperiod-insensitive early flowering38. To prove this hypothesis will require further analysis, such as examination of the in vivo interaction of CCA1-GI or LHY-GI, and GI binding activity to the FT promoter in either lhy cca1 double mutants or LHY or CCA1 overexpressors.
Based on these data, we propose a model for the molecular mechanism by which DET1 represses flowering in non-inductive SD conditions (Fig. 7). In WT plants, the absence of FT expression under SD can be explained by the incongruity of peak expression of FKF1 and GI; GI peaks in the late afternoon but FKF1 peaks at night, leading to reduced expression of CO and FT during daytime21. As GI also directly induces FT expression in a CO-independent pathway8, we wondered why GI, which is expressed in the afternoon21 (Fig. 2a), is not capable of inducing FT expression under SD (Fig. 2a, d). In this study, we found that DET1 suppresses FT transcription by repressing GI binding activity to the FT promoter (Fig. 5b). This model is further supported by genetic analysis showing that gi-1 and ft-1 are almost completely epistatic to det1-1 (Fig. 1b and Table S1), indicating that DET1 mainly regulates flowering via GI.
In conclusion, we propose that DET1 functions as a strong repressor of flowering, acting in both photoperiodic and autonomous pathways (Fig. 7); DET1 suppresses flowering mainly by decreasing GI binding activity to the FT promoter in the photoperiod pathway and epigenetically upregulating FLC expression in the autonomous pathway. Whether DET1 acts in the CDD complex17 to delay flowering time under SD in Arabidopsis remains to be elucidated.
Methods
Plant materials and growth conditions
All the Arabidopsis thaliana lines used in this study are in the Columbia (Col-0) genetic background. Flowering-time mutants were obtained from the Arabidopsis Biological Resource Center (USA), except for det1-1 which was kindly provided by Joanne Chory. cry2-1 (CS3732), gi-1 (CS3123), soc1-2 and ft-139, fkf1-t40, and co-10141 were used for genetic analysis. To create double and triple mutants, F1 heterozygotes were obtained by crossing the det1-1 mutant as the female plant with other flowering-time mutants as pollen donors. To select correct transformants, the plants showing the det1-1 morphological phenotype were first isolated from F3 plants, and flowering-time mutations were finally confirmed by PCR-based genotyping. Plants were grown on soil at a constant 22°C under white fluorescent light (90-100 μmol m−2s−1) in LD (16 h light:8 h dark) and SD (10 h light:14 h dark) or SD (8 h light:16 h dark).
Analysis of flowering time
The bolting date was measured as the number of days from seed sowing to opening of the first flower and as the total number of rosette leaves at bolting. Data were obtained from three experimental replications (20 to 60 plants per replication).
RNA preparation and quantitative real-time PCR analysis
Tissue samples were collected every 3 h from 3-week-old seedlings. Total RNA was extracted with the plant RNA extraction kit (Macrogen). For each sample, 2 μg of total mRNA was reverse transcribed using M-MLV reverse transcriptase (Promega). The level of the transcripts was measured by real-time PCR, using GoTaq qPCR Master Mix (Promega) and the Light Cycler 2.0 instrument (Roche). Each PCR was repeated at least three times using biologically independent samples. The amount of each RNA level was determined using specific primers. The primers used for real-time PCR are listed in Table S2.
Yeast 2-hybrid assays
The full-length cDNAs of DET1, GI, PHYA, PHYB, CCT1, CCT2, CO, FKF1, FLC, SVP, TEM1, and TEM2 were amplified from wild-type total RNA using RT-PCR. GI was divided into three parts: GI N-terminal (aa 1-507), GI middle (aa 401-907), and GI C-terminal (aa 801-1173) regions. The PCR products were cloned into pGBKT7 and pGADT7 vectors (MATCHMAKER GAL4 TWO-hybrid system 3, Clontech) to get the bait and prey clones. For the interaction study, plasmids containing fusion proteins were transformed into Saccharomyces cerevisiae AH109 and grown on media lacking adenine, leucine, histidine, and tryptophan. Galactosidase activity assays were performed according to the manufacturer's protocol.
In vivo pull-down assays
TAP-DET1 and TAP-GFP were from Xing Wang Deng. pGI:GI-HA gi-2 det1-1 was obtained by crossing pGI:GI-HA gi-2 and det1-1. For DET1-GI binding assays, TAP-DET pGI:GI-HA gi-2 and TAP-GFP pGI:GI-HA gi-2 plants were grown on MS medium in SD (8 h light:16 h dark) for 10 days and then vacuum infiltrated for 7 ~ 10 min in 1X MS (Duchefa) liquid medium supplemented with 50 mM MG132 (Sigma) for proteasome inhibitor treatment. After that, plants were incubated for 10 h under light conditions. These plants were homogenized and total proteins were extracted in total protein extract buffer [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA (pH 8.0), 10% glycerol, 1 mM PMSF, 1 mM DTT]. These experiments were performed with IgG beads for TAP-IP. After washing, the immunoprecipitated fractions were analyzed by immunoblotting. The TAP-DET1 and GI fusion proteins were detected by using anti-HA antibody.
Bimolecular fluorescence complementation assays
Each cDNA of GI, ELF3, DET1, and MSI4 was cloned into the BiFC gateway vectors42 to examine their in vivo interactions. For partial YFP-tagged DET1, and MSI4 constructs, the cDNA of the gene was obtained by RT-PCR from wild-type (WT, Col-0) plants and fused into four BiFC plasmid sets, pSAT5-DEST-cEYFP(175-end)-C1(B) (pE3130), pSAT5(A)-DEST-cEYFP(175-end)-N1 (pE3132), pSAT4(A)-DEST-nEYFP(1-174)-N1(pE3134), and pSAT4-DEST-nEYFP(1-174)-C1 (pE3136). Partial YFP-tagged ELF3 and GI constructs were previously described33. Each pair of recombinant plasmids encoding nEYFP and cEYFP fusions was mixed 1:1 (w/w), co-bombarded into onion epidermal layers using a DNA particle delivery system (Biolistic PDS-1000/He, BioRad), and incubated on MS solid media with MG132 (50 mM) for 16–24 h at 22°C under light or dark incubation, followed by observation and image analysis using a confocal laser scanning microscope (Carl Zeiss LSM710).
Chromatin immunoprecipitation assay
For the ChIP assay, Col-0, pGI:GI-HA gi-2, and pGI:GI-HA gi-2 det1-1 plants were grown for 10 days under SD (8 h light:16 h dark) conditions and collected at ZT8. The samples were cross-linked with 1% formaldehyde, ground to powder in liquid nitrogen, and then sonicated43. The sonicated chromatin complexes were bound with anti-HA antibody (ab9110, Abcam) for immunoprecipitation. The amount of DNA fragment was analyzed by quantitative real-time PCR (qPCR) using specific primers. UBI10 was used as an internal standard for normalization. The primers used for qPCR are listed in Table S2. For another ChIP assay, Col-0 and det1-1 plants were grown for 14 days under SD (8 h light/16 h dark) conditions and collected at ZT8. For immunoprecipitation, we used the anti-trimethyl H3K4 (07-473, Millipore), and anti-trimethyl H3K27 (07-449, Millipore). FUS3 was used as an internal standard for normalization14. Experiments were performed with three biological repeats.
Author Contributions
M.-Y.K., S.-C.Y., Y.-S.N. and N.-C.P. conceived the study and designed the research. M.-Y.K., H.-Y.K., J.-N.C. and B.-D.L. performed experiments. M.-Y.K. and S.-C.Y. analyzed data with suggestions by Y.-S.N. and N.-C.P. M.-Y.K., S.-C.Y. and N.-C.P. wrote the article. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr. Xing Wang Deng at Yale University for 35S:TAP-DET1 seeds, and Dr. Woe-Yeon Kim at Gyeongsang National University for pGI:GI-HA/gi-2 seeds. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (NRF-2011-0017308), Republic of Korea.
References
- de Montaigu A., Toth R. & Coupland G. Plant development goes like clockwork. Trends Genet. 26, 296–306 (2010). [DOI] [PubMed] [Google Scholar]
- Kardailsky I. et al. Activation tagging of the floral inducer FT. Science 286, 1962–1965 (1999). [DOI] [PubMed] [Google Scholar]
- Park D. H. et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582 (1999). [DOI] [PubMed] [Google Scholar]
- Suárez-López P. et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120 (2001). [DOI] [PubMed] [Google Scholar]
- Corbesier L. et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 (2007). [DOI] [PubMed] [Google Scholar]
- Valverde F. et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006 (2004). [DOI] [PubMed] [Google Scholar]
- Sawa M., Kay S. A. & Imaizumi T. Photoperiodic flowering occurs under internal and external coincidence. Plant Signal. Behav. 3, 269–271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M. & Kay S. A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108, 11698–11703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A. et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616 (2000). [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Valverde F., Ravenscroft D., Mouradov A. & Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I. et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I., Alonso-Blanco C., Jarillo J. A., Ruiz-Garcia L. & Martinez-Zapater J. M. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166 (2004). [DOI] [PubMed] [Google Scholar]
- Gu X. et al. Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 7, e1002366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M., Molinier J., Berr A. & Genschik P. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 3430–3435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J., Peto C., Feinbaum R., Pratt L. & Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58, 991–999 (1989). [DOI] [PubMed] [Google Scholar]
- Suzuki G., Yanagawa Y., Kwok S. F., Matsui M. & Deng X. W. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16, 554–559 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y. et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18, 2172–2181 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. J., Straume M., Chory J., Chua N. H. & Kay S. A. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267, 1163–1166 (1995). [DOI] [PubMed] [Google Scholar]
- Lau O. S. et al. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 43, 703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A. E. & Chory J. Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics 145, 1125–1137 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D. A., Kay S. A. & Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K. et al. CONSTANS activates SUPPRESSOR OF OVEREXPRESSIONOF CONSTANS 1 through Flowering Locus T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D. & Amasino R. M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D. & Amasino R. M. Loss of Flowering Locus C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Himelblau E., Kim S. Y., Schomburg F. M. & Amasino R. M. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 137, 149–156 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Arabidopsis CULLIN4 forms an E3 ubiquitin Ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18, 1991–2004 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. H. et al. The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates Flowering Locus C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25, 4378–4390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixdorf M. & Hoecker U. SPA1 and DET1 act together to control photomorphogenesis throughout plant development. Planta 231, 825–833 (2010). [DOI] [PubMed] [Google Scholar]
- Doyle M. R. et al. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77 (2002). [DOI] [PubMed] [Google Scholar]
- Kim Y. et al. GIGANTEA and EARLY FLOWERING 4 in Arabidopsis exhibit differential phase-specific genetic influences over a diurnal cycle. Mol. Plant 5, 678–687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. et al. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 3, 671–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. W. et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32, 617–630 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L. H. & Deng X. W. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H. et al. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21, 397–402 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C. & Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343 (2008). [DOI] [PubMed] [Google Scholar]
- Lee J. & Lee I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254 (2010). [DOI] [PubMed] [Google Scholar]
- Mizoguchi T. et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641 (2002). [DOI] [PubMed] [Google Scholar]
- Moon J., Lee H., Kim M. & Lee I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46, 292–299 (2005). [DOI] [PubMed] [Google Scholar]
- Cheng X. F. & Wang Z. Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 43, 758–768 (2005). [DOI] [PubMed] [Google Scholar]
- Takada S. & Goto K. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V. et al. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362, 1120–1131 (2006). [DOI] [PubMed] [Google Scholar]
- Cho J. N. et al. Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 22, 736–748 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information