Abstract
In eukaryotic cells, covalent modifications to core histones contribute to the establishment and maintenance of cellular phenotype via regulation of gene expression. Histone acetyltransferases (HATs) cooperate with histone deacetylases (HDACs) to establish and maintain specific patterns of histone acetylation. HDAC inhibitors can cause pluripotent stem cells to cease proliferating and enter terminal differentiation pathways in culture. To better define the roles of individual HDACs in stem cell differentiation, we have constructed “dominant-negative” stem cell lines expressing mutant, Flag-tagged HDACs with reduced enzymatic activity. Replacement of a single residue (His → Ala) in the catalytic center reduced the activity of HDACs 1 and 2 by 80%, and abolished HDAC3 activity; the mutant HDACs were expressed at similar levels and in the same multiprotein complexes as wild-type HDACs. Hexamethylene bisacetamide-induced MEL cell differentiation was potentiated by the individual mutant HDACs, but only to 2%, versus 60% for an HDAC inhibitor, sodium butyrate, suggesting that inhibition of multiple HDACs is required for full potentiation. Cultured E14.5 cortical stem cells differentiate to neurons, astrocytes, and oligodendrocytes upon withdrawal of basic fibroblast growth factor. Transduction of stem cells with mutant HDACs 1, 2, or 3 shifted cell fate choice toward oligodendrocytes. Mutant HDAC2 also increased differentiation to astrocytes, while mutant HDAC1 reduced differentiation to neurons by 50%. These results indicate that HDAC activity inhibits differentiation to oligodendrocytes, and that HDAC2 activity specifically inhibits differentiation to astrocytes, while HDAC1 activity is required for differentiation to neurons.
Keywords: HDAC, dominant negative, transduction, cell fate, MEL, neural stem cell
Introduction
Pluripotent stem cells exhibit a stable phenotype in culture characterized by sustained proliferation and expression of specific protein markers. These cells can be induced to differentiate by either the addition or withdrawal of specific growth factors or drugs in the culture medium. Differentiation is characterized by a slowing or cessation of cell proliferation and the up- or down-regulation of gene expression at many loci, leading to a stable, differentiated cell phenotype.
Stable patterns of gene expression are maintained, in part, by covalent modification of DNA (methylation) and core histones (methylation and acetylation). The N-terminal domains of core histones contain specific patterns of covalent modification (i.e., acetylation, methylation, and phosphorylation) that is termed “the histone code” (Strahl and Allis, 2000; Turner, 2000; Jenuwein and Allis, 2001). Specific patterns of lysine acetylation are established and maintained by the coordinated activity of histone acetyl transferases (HATs) and histone deacetylases (HDACs). HDAC inhibitors can cause stem cells to cease proliferating and enter terminal differentiation pathways (Marks et al., 2000; Kramer et al., 2001; Hsieh et al., 2004), suggesting that reprogramming involves an increase in histone acetylation at specific loci.
Mammalian HDACs comprise a multiprotein family of zinc metallohydrolases sharing a conserved catalytic center (Finnin et al., 1999; Gray and Ekstrom, 2001; Khochbin et al., 2001). The quantitatively major class I HDACs (1, 2, 3, and 8), related to the yeast RPD3 protein, are ~500 residues long and localized to the nucleus. The class II HDACs (4–7, 9, 10), related to yeast HDA1 protein, are ~1,000 residues long, and shuttle between the nucleus and cytosol. HDACs exist in multiprotein complexes with transcription factors, DNA-binding proteins, and other chromatin-modifying enzymes; assembly into complexes is required for full deacetylase activity (Zhang et al., 1999). Transient transfection studies indicate that HDACs function as transcriptional corepressors in a variety of regulatory pathways. Class 1 HDACs 1, 2, and 3 interact with components of the p53 and RB tumor-suppressor pathways (Brehm et al., 1998; Magnaghi-Jaulin et al., 1998; Wade, 2001; Zhang and Dean, 2001), histone and DNA-methyltransferases (Fuks et al., 2000; Robertson et al., 2000; Fuks et al., 2001; Vaute et al., 2002), and mCpG-DNA-binding proteins (Ng et al., 1999; Wade et al., 1999), suggesting their direct involvement in growth control.
Existing HDAC inhibitors act on all class 1 and 2 HDACs by stereospecific binding to the catalytic center (Finnin et al., 1999). Thus, genetic approaches are required to assess the roles of individual HDACs in the maintenance of cell proliferation. Deletion of the hdac1 locus in mice causes an embryonic lethal cell proliferation defect, demonstrating that HDAC1 is required for cell proliferation (Lagger et al., 2002). Expressions of HDAC2 and HDAC3 are increased in HDAC1-null ES cells, but they do not fully compensate for the absence of HDAC1. Expression of cyclin-dependent kinase inhibitors p21WAF/CIP1 and p27KIP1 is also increased in HDAC1-null cells. These inhibitors can cause proliferation arrest by conversion of Rb protein to its active, hypophosphorylated form (Wade, 2001; Zhang and Dean, 2001).
In order to better define the roles of individual class 1 HDACs in cell proliferation and terminal differentiation, we have created retroviral vectors for stable expression of dominant-negative HDACs. These dominant-negative HDACs have reduced enzymatic activity due to an amino acid replacement (His → Ala) in the catalytic center, but retain the ability to form multiprotein complexes. We observed that mutant HDACs could potentiate hexamethylene bisacetamide (HMBA)-induced terminal differentiation of erythroleukemia (MEL) cells, albeit modestly, with a relative magnitude of HDAC3 > HDAC2 > HDAC1. We also examined the effect of dominant-negative HDACs on the fate of cultured central nervous system (CNS) cortical stem cells, which differentiate predominantly to neurons. More striking results were obtained in these studies. Mutant HDAC1, HDAC2, and HDAC3 shifted stem cell fate to oligodendrocytes while mutant HDAC2 shifted cell fate to astrocytes. Our findings delineate for the first time distinct roles for individual HDACs in neuronal differentiation.
Methods
Plasmid construction and mutagenesis
The construction of retroviral vectors (pOZ) for expression of Flag-tagged human HDAC1 and 2 is described in Humphrey et al. (2001). A pOZ expression construct for Flag-tagged human HDAC3 was prepared by the same procedure using a cDNA clone isolated from a CLONTECH Marathon cDNA library. Mutagenesis was performed by the overlap extension PCR method (Ho et al., 1989). All constructs were verified by DNA sequencing.
Isolation and analysis of HDAC complexes
Flag-tagged HDAC1, 2, and 3 were isolated from transduced Hela S3 cells using an anti-Flag immunoadsorbent as described in Humphrey et al. (2001). Histone deacetylase assays were performed as described in Humphrey et al. (2001). For sedimentation analysis of HDAC3 complexes, complexes eluted with Flag peptide were centrifuged in a 10%–35% glycerol gradient for 5.5 hr at 55,000 r.p.m. Marker proteins (ovalbumin 4.54S, aldolase 7.3S, catalase 11.3S) were centrifuged in parallel gradients. HDAC complex components were identified by Western blotting as described in Humphrey et al. (2001). Anti-ebi antibody was a gift from Dr. S. Lawrence Zipursky (University of California, Los Angeles, CA). Anti-NcoR and anti- SMRT were obtained from Upstate. Anti-HDAC3 antibodies were raised in rabbits to His-tagged proteins representing the N and C-terminal regions of human HDAC3. For peptide mass fingerprinting, tryptic peptides were prepared from excised gel bands as described in Humphrey et al. (2001), and mass spectra were collected using the Applied Biosystems (Foster City, CA) Voyager DE-STR MALDI-TOF instrument in the NICHD mass spectrometry facility. Protein identification was made by database searching using Protein Prospector and Mascot.
Murine erythroleukemia cell culture and transduction
MEL cells, kindly provided by Dr. Shoshana Segal (National Cancer Institute, Bethesda,MD), were cultured in R10 medium (Borre et al., 1996). For retroviral infection, cultures seeded at 5 × 105 cells/cm2 on retronectin-coated plates were exposed to an initial viral supernatant for 2 hr at 37°C, followed by a second supernatant for 22 hr at 37°C. Transduced cells were purified by magnetic affinity cell sorting for the co-expressed IL2R α subunit (Tac antigen) surface marker (Ogryzko et al., 1997; Humphrey et al., 2001). Differentiation was induced in 24-well plates (2 × 105 cells/well) in the presence of HMBA and/or sodium butyrate as indicated. In preliminary experiments (not shown), 0.9 mM HMBA was insufficient to induce measurable differentiation but potentiated the effect of butyrate. This HMBA concentration was used in plating MACS-purified cells transduced with HDAC1-3 mutants. Differentiation efficiency was quantified by benzidine staining.
Neural stem cell culture and transduction
Fetal stem cells were grown as described previously (Johe et al., 1996; Kim et al., 2003). Briefly, cortical tissue was dissected from embryonic day 14.5 rats, mechanically dissociated, and plated in serum-free DMEM/F12 medium with N2 supplements. Basic fibroblast growth factor (bFGF) was included at 10 ng/ml to promote stem cell proliferation. Cells were fed with bFGF daily and medium was replaced on alternate days during stem cell expansion. Passaging was performed nonenzymatically using Ca2+/Mg2+-free HBSS. Infections were performed as described above with the following modifications. Second- or third-passage stem cells were plated at 2 × 105 cells/well on 6-well plates. The next day, the medium was replaced with 2 ml of virus-conditioned medium (retrovirus collected in N2 medium supplemented with FGF2, centrifuged at 3,000 r.p.m. for 90 min) and cells were cultured for an additional 48 hr to allow expression of the retroviral construct. Cells were passaged, centrifuged, and resuspended in 2 ml volume, from which a 100 µl aliquot was taken for FACS analysis (Ogryzko et al., 1997). Cells were labeled with an anti-Tac IL2R with a fluorescent secondary antibody to visualize the number of cells expressing IL2R by multiparametric FACS analysis. After the procedures were optimized to limit spontaneous differentiation, the infection efficiency improved to 35%–65%.
Magnetic affinity cell sorting of neural stem cells
IL2R-expressing cells were isolated by MACS as described previously (Ogryzko et al., 1997) with the following modifications to suit the elution of neuroepithelial cells. The cells were eluted, centrifuged, and resuspended in Ca2+/Mg2+-free HBSS containing 0.1% BSA fraction V. A small number of cells were plated separately to verify IL2R expression. The remaining sorted cells were plated at 5–50 × 104 in 6 cm dishes in DMEM/F12 plus N2 supplements, expanded for two days with bFGF, and then differentiated by incubation in medium without bFGF for an additional 7 days.
Immunocytochemical analysis of neural cells
The antibodies used for detection of cell lineage-specific markers were: rabbit anti-nestin 130 from the McKay lab (Bethesda, MD), rabbit anti-βIII-tubulin from Covance (Berkeley, CA), mouse antiglial fibrillary acidic protein (GFAP) from ICN (Costa Mesa, CA), and mouse IgM anti-O4 from Chemicon (Temecula, CA) and mouse IgG1 anti-CNPase from Chemicon. Cells were fixed with 4% paraformaldehyde for 10–15 min at room temperature, blocked for 30 min in phosphate-buffered saline (PBS) with 0.1% triton and 5% normal goat serum, and incubated with the primary antibody for 2–4 hr at room temperature. Cells were immunostained as directed with anti-Tac to detect transduced IL2R-expressing cells, rabbit anti-nestin 130 to detect proliferating CNS precursors, mouse anti-βIII-tubulin to detect neurons, rabbit anti-GFAP to detect astrocytes, and mouse anti-O4 to detect oligodendrocytes. After two washes in PBS, the secondary antibody was applied at a 200-fold dilution for 1 hr at room temperature. Cultures were then washed with PBS and mounted in Vectashield containing DAPI (Vector Laboratories, Burlingame, CA). Images were photographed under fluorescent filters using a Zeiss Axioplan microscope (Thornwood, NY) and a Spot digital camera (Diagnostic Instruments Inc., Sterling Heights, MI). Statistical analysis and histogram illustration of cell numbers were performed using SigmaPlot 5.0 (SPSS, Inc.).
Results
Expression constructs for dominant-negative HDAC1, 2, and 3 mutants
Our laboratory has developed bicistronic retroviral vectors for stable expression of chromatin-modifying proteins in proliferating mammalian cells (Ogryzko et al., 1996). These vectors code for a cell surface tag, in addition to the protein of interest. Following transduction, the cells are sorted using the surface tag to obtain a pure population expressing both the surface tag and the protein of interest (see “Methods”). Previously, we used this system to express Flag-tagged class 1 HDACs in Hela cells (Humphrey et al., 2001). These tagged HDACs were expressed at comparable levels to the endogenous HDACs (1–5 × overexpression), were enzymatically active, and associated with the same proteins in complexes as the endogenous HDACs.
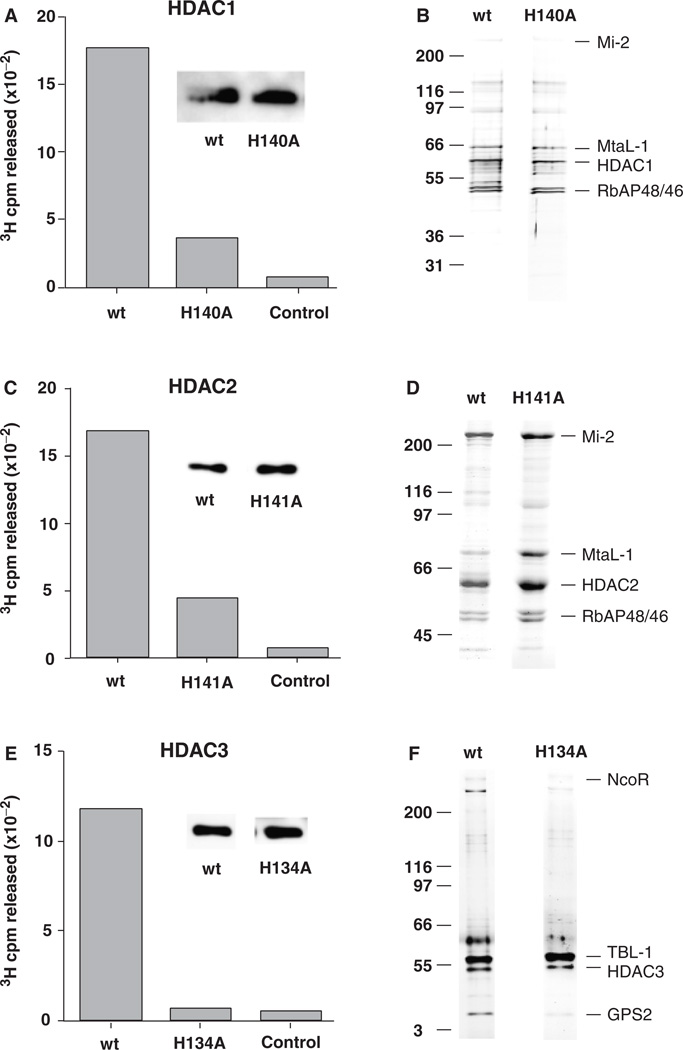
In order to assess the role(s) of individual HDACs in cell proliferation and differentiation, we created mutant HDACs with reduced enzymatic activity (i.e., dominant negatives). Our objective in creating these mutants was to reduce enzymatic activity while preserving the ability of the mutant HDACs to form multiprotein complexes (Hassig et al., 1998). Our strategy was guided by a proposed mechanism for HDAC catalysis based on the crystal structure of the Aquifex aeolicus HDAC-like protein (HDLP) (Finnin et al., 1999). The HDAC catalytic center contains an H2O molecule bound to a Zn2+ atom (Fig. 1). In the reaction scheme, this H2O nucleophile attacks the carbonyl carbon of the acetyl group in acetyl lysine. The nucleophilicity of the H2O is enhanced by hydrogen bonding via a His140-D174 charge relay system. The oxyanion reaction intermediate is stabilized by the Zn2+ atom and Tyr303. Following bond breakage, the ε-nitrogen of lysine acquires a proton from a second charge relay system, His141-D191. Replacement of His140 with alanine in HDAC1 (mutant H140A) reduced deacetylase activity by 80%, compared with wild-type HDAC1, in HDAC complexes immunopurified from Hela cells (Fig. 2A). In an effort to further reduce HDAC1 activity, we created double mutants: H140A-H141A and H140A-Y303A. These double mutants exhibited the same level of deacetylase activity as the single H140A mutant, indicating a critical role for His140 in HDAC1 catalysis. The H140A mutant was found in stable, multiprotein complexes with the same proteins as wild-type HDAC1, including NURD complex components Mi-2, MtaL-1, and RbAP46/48 (Fig. 2B), as we have described previously (Humphrey et al., 2001).
Fig. 1.
Conserved catalytic domain of type I histone deacetylases. Sequence alignment of Aquifex aeolicus HDLP (1C3R_B) with human HDAC1 (Q13547), HDAC2 (Q92769), and HDAC3 (O15379). Residues located in the catalytic center are in caps. Residues mutated for this study are in bold. Zinc-binding residues are indicated by “Z.” Horizontal lines connect the two His–Asp charge relays (see text). HDAC, histone deacetylases; HDLP, HDAC-like protein.
Fig. 2.
Wild-type (wt) and mutant HDACs form similar multiprotein complexes. Flag-tagged HDACs were expressed in Hela cells and isolated using an anti-Flag immunoaffinity adsorbent. HDAC1 enzymatic activity was determined by 3H cpm released from 3H acetate-labeled histones in a 30-min incubation (see “Methods”). (A) Wt, mutant (H140A) HDAC1 complexes isolated from transduced Hela cells; control anti-Flag adsorbent using normal (untransduced) Hela cells. Insets show that similar amounts of Flag-tagged protein were used for each assay. (C) Wt or mutant (H141A) HDAC2 complexes isolated from transduced Hela cells, control anti-Flag adsorbent using normal (untransduced) Hela cells. (E) Wt or mutant (H134A) HDAC3 complexes isolated from transduced Hela cells, control anti- Flag adsorbent using normal (untransduced) Hela cells. Wt and mutant HDACs form similar multiprotein complexes. (B) Comparison of immunopurified Flag-tagged HDAC1 wt and mutant (H140A) multiprotein complexes resolved by SDS-PAGE and visualized by silver staining. Right, principal components; left, size markers. (D) Comparison of immunopurified Flag-tagged HDAC2 wt and mutant (H141A) multiprotein complexes. (F) Comparison of wt and mutant (H134A) HDAC3 form similar multiprotein complexes. HDAC, histone deacetylases.
We created dominant-negative mutants of HDAC2 and HDAC3 by replacing the histidine residues homologous to His140 in HDAC1, i.e., His141 in HDAC2 (H141A) and His134 in HDAC3 (H134A). Replacement of His141 with alanine reduced deacetylase activity by 75% in HDAC2 complexes (Fig. 2C). Replacement of His134 by alanine completely eliminated detectable deacetylase activity in HDAC3 complexes (Fig. 2E). It is possible that the residual activity in mutant HDAC1 and HDAC2 complexes is due to the presence of endogenous HDAC1/2 in these complexes (Humphrey et al., 2001). HDAC2 complexes are similar in polypeptide composition to HDAC1 complexes. The HDAC2 H141A mutant also formed the same complexes as wild-type HDAC2 (Fig. 2D).
HDAC3 occurs in a different multiprotein complex than HDAC1/2. This complex sediments at 9S in a glycerol gradient and consists of four proteins in addition to HDAC3 (not shown), as described in other reports (Guenther et al., 2000; Li et al., 2000; Wen et al., 2000; Guenther et al., 2001). These proteins were identified as a silencing mediator for retinoid and thyroid receptors (SMRT), nuclear receptor corepressor (N-CoR), β transducin-like protein (TBL1), and G-protein pathway suppressor 2 (GPS2) by Western blotting and mass spectrometry. The HDAC3 H134A mutant formed the same complexes as wild-type HDAC3 (Fig. 2F).
The Flag-tagged HDACs, both wild type and mutants, were expressed at similar levels in the transduced cells (Fig. 2). As the level of HDAC expression from the pOZ constructs is at least equal to, and generally higher than the level of endogenous HDAC expression (Humphrey et al., 2001), we expect that ≥50% of the HDAC complexes would contain a mutant HDAC subunit that should be sufficient to observe a dominant-negative phenotype.
Dominant-negative HDACs induce MEL cell differentiation
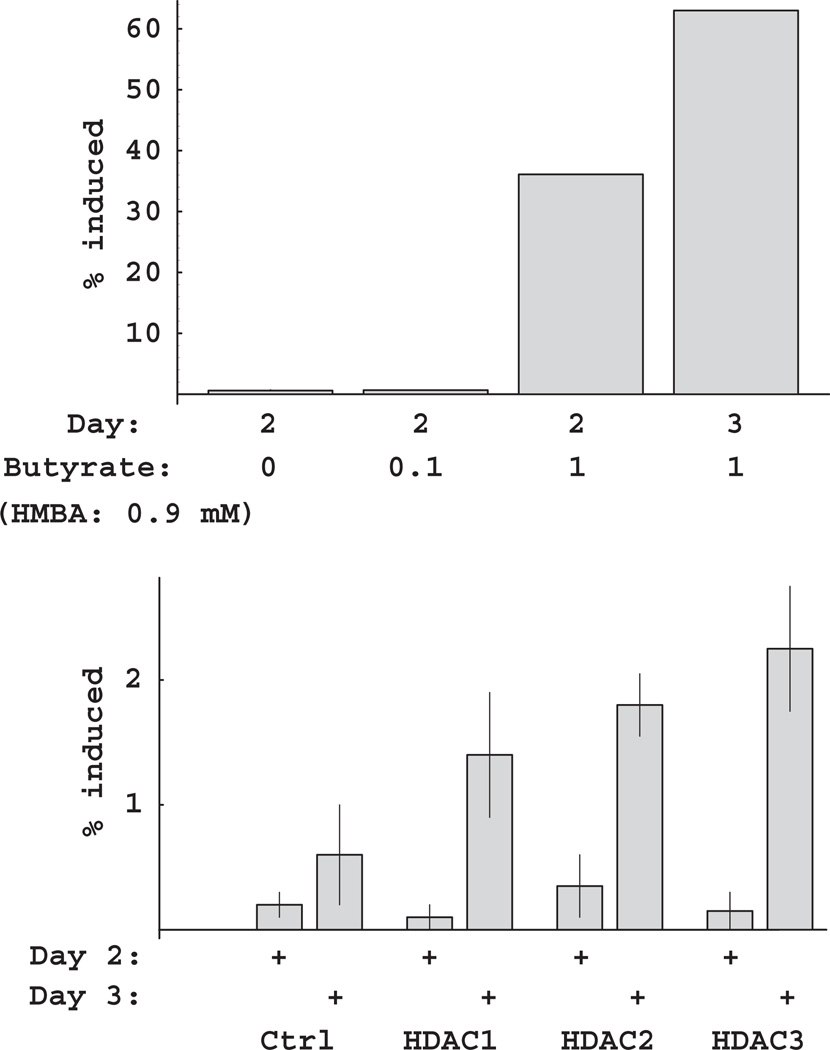
The murine erythroleukemia cell line (MEL) can be induced to cease proliferation in culture and undergo terminal differentiation by treatment with HMBA (Richon et al., 1998; Matushansky et al., 2000). HMBA-induced differentiation can be further potentiated by histone deacetylase inhibitors. For example, whereas HMBA at a sub-threshold level (0.9 mM) yielded few benzidine-positive cells, combined treatment with 0.9 mM HMBA and 1 mM sodium butyrate induced differentiation in 60% of the cultured cells after 3 days (Fig. 3A). We examined whether transduction of MEL cells with dominant-negative HDACs would have a similar effect on induction of differentiation. All three dominant-negative HDACs could potentiate a significant amount of induction above background levels; however, the percentage of cells induced (1–2%) was considerably lower than with butyrate (Fig. 3B). The relative effects of the three class I HDACs for potentiation of differentiation were the same as for growth inhibition: HDAC3 > HDAC2 > HDAC1 (Fig. 3B and unpublished results). The relatively modest effect of the dominant-negative HDACs in this assay may indicate a requirement to inhibit multiple HDACs in order to potentiate MEL cell differentiation (see “Discussion”).
Fig. 3.
Potentiation of HMBA-induced MEL differentiation by inhibition of HDAC activity. (A) Potentiation of HMBA-induced MEL differentiation by sodium butyrate. MEL cultures were treated with 0, 0.1, or 1.0 mM butyrate in the presence of 0.9 mM HMBA and assayed for differentiation on days 2 and 3. (B) MEL differentiation following retroviral transduction with HDAC1-3 mutants. MEL cultures were transduced with a retrovirus containing no insert (Ctrl), or dominant-negative HDAC constructs (HDAC1, 2, or 3). Following treatment with 0.9 mM HMBA, cultures were assayed for differentiation on days 2 and 3. HDAC, histone deacetylases; HMBA, Hexamethylene bisacetamide.
Dominant-negative HDACs alter the fate of CNS stem cells
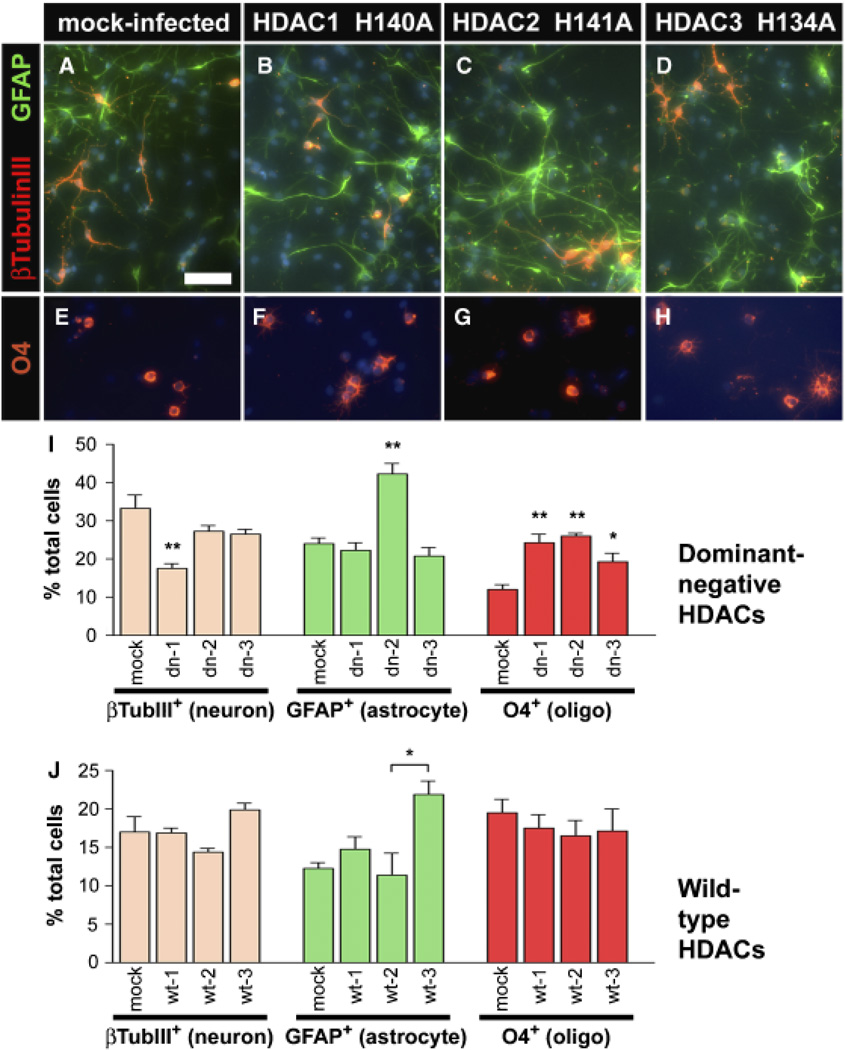
CNS stem cells are characterized by their ability to self-renew and to differentiate into neurons, astrocytes, and oligodendrocytes (Panchision and McKay, 2002). Selfrenewal in vitro requires the presence of a mitogen such as bFGF, while removal of this mitogen causes the cells to differentiate (Johe et al., 1996). We analyzed the effect of blocking HDAC function on E14.5 cortical stem cell proliferation and fate choice. Cells infected with a control pOZ construct expressing green fluorescent protein, and Tac-selected, showed no obvious difference in proliferation versus non-manipulated cells in the presence of bFGF (data not shown). By 7 days of differentiation after bFGF withdrawal, 69% of mock-infected cells expressed differentiation markers for neurons (βIII-tubulin, 33%), astrocytes (GFAP, 24%), or oligodendrocytes (O4, 12%), while more than 30% still expressed the stem cell marker nestin (Figs. 4A, 4E and 4I). These proportions are consistent with previous reports of CNS stem cell differentiation (Johe et al., 1996).
Fig. 4.
Loss of HDAC function causes changes in stem cell fate. Stem cells were cultured from E14.5 rat forebrain and infected with a retrovirus containing no insert (A, E) or dominant-negative HDAC constructs (B–D, F–H). Infected cells were purified based on the expression of an IL2R selectable marker, cultured in bFGF for 2 days, and then differentiated by bFGF withdrawal for 7 additional days. Cells were immunostained for β-III-Tubulin to mark neurons (A–D, red), GFAP to identify astrocytes (A–D, green) or O4 to mark oligodendrocytes (E–H, red) and DAPI to identify cell nuclei (blue). (I) Quantitation of differentiated cells after mock or dominant-negative construct infections of stem cells (mean SEM, n = 4–9). (J) Quantitation of differentiated cells after mock or wild-type construct infections of stem cells. CNPase is used instead of O4 to mark oligodendrocytes. Lower baseline values reflect slower differentiation due to lower initial plating densities (mean ± SEM, n = 2). *p < 0.05; **p < 0.001. Bar = 200 µm for all images. HDAC, histone deacetylases; bFGF, basic fibroblast growth factor; GFAP, glial fibrillary acidic protein.
In contrast, stem cells that were infected with mutant HDAC1 (H140A) proliferated more slowly in the presence of bFGF than mock-infected cells (not shown). After differentiation, HDAC1 (H140A) cells generated 50% fewer neurons and twice as many oligodendrocytes than normal or mock-infected stem cells (Figs. 4B, 4F and 4I). Stem cells that were infected with HDAC2 (H141A) also proliferated more slowly in the presence of bFGF (not shown). After differentiation, HDAC2 (H141A) cells differentiated preferentially into astrocytes and oligodendrocytes with only a modest decrease in neurons compared with mock-infected stem cells (Figs. 4C, 4G and 4I). Furthermore, the astrocytes generated from these infected cells tended to have longer processes than in any other condition (Fig. 4C), suggesting greater maturation. Stem cells that were infected with HDAC3 (H134A) also proliferated more slowly in the presence of bFGF (not shown). After differentiation, these cells generated greater numbers of oligodendrocytes, with little change in the percentage of neurons or astrocytes versus mock-infected stem cells (Figs. 4D, 4H and 4I). Infection with wild-type HDACs did not show these effects (Fig. 4J), indicating that the response was due to the dominant-negative loss of deacetylase activity effect rather than simply the overexpression of HDAC protein. In fact, the increase in GFAP+ cells in wild-type HDAC3 versus HDAC2-infected cells was in contrast to that seen with the dominant-negative constructs. These results support the conclusion that all three HDACs act to inhibit oligodendrocyte differentiation, while only HDAC2 acts to inhibit astrocyte differentiation. The results also suggest that HDAC1 is selectively required for neuronal differentiation.
Discussion
Studies using inhibitory drugs have indicated a critical role for histone deacetylase activity in stem cell differentiation. Two possible approaches to define the role(s) of individual HDACs in this process are (1) stable expression of a mutant (dominant negative) enzyme or (2) stable expression of siRNA (Sandy et al., 2005). Expression of siRNA would result in loss of the HDAC protein, similar to a deletion mutant, and the disruption of HDAC complexes. In this study, we wished to focus specifically on the requirement for HDAC enzymatic activity in stem cell differentiation, as distinct from a requirement for HDAC complexes, and so we chose to express dominant-negative HDAC proteins.
Role of HDAC3 in MEL cell differentiation
We found that expression of dominant-negative HDACs in murine erythroleukemia cells could potentiate MEL cell differentiation in response to HMBA, supporting the previous inhibitor studies (Fig. 3). Inhibition of HDAC3 activity had the greatest effect. Studies in other differentiating systems have also indicated that HDAC3 activity blocks differentiation. In mouse embryo fibroblasts, HDAC3 exists in a ternary complex with Rb protein and PPARγ, a nuclear receptor that promotes terminal differentiation to adipocytes in response to ligand binding. The ternary complex inhibits the PPARγ function and serves to maintain cells in an actively proliferating state. HDAC inhibitors cause dissociation of the complex, activation of PPARγ, and terminal differentiation to adipocytes (Fajas et al., 2002; Fajas et al., 2003). An analogous ternary complex may block differentiation in MEL cells. The degree of potentiation due to inhibiting individual HDACs is significantly less than when using a general inhibitor; thus, efficient potentiation likely requires inhibiting more than one HDAC.
Role of HDACs in CNS stem cell fate
While blocking HDAC function had a general effect on the proliferation of cortical stem cells, it had a specific effect on the differentiation of oligodendrocytes. Expression of all three dominant-negative HDAC constructs led to an increase in oligodendrocytes as a percentage generated from neural stem cells (Fig. 4I). Interestingly, triiodothyronine (T3), a factor known to promote the differentiation of oligodendrocytes and astrocytes from neural stem cells (Johe et al., 1996), is involved in HDAC recruitment leading to TSHβ repression (Sasaki et al., 1999). Our results suggest that in stem cells, HDACs may normally complex with repressors of oligodendrocyte or astrocyte differentiation and that this repression may be reversed by the actions of T3. Because HDAC activity is required for maturation of post-mitotic oligodendrocytes (Marin-Husstege et al., 2002), these repressor complexes may be displaced by glia promoting complexes shortly after mitotic arrest.
Interestingly, blocking HDAC2 increased the total percentage of cells expressing the differentiation markers β-III-tubulin, GFAP or O4 (95.2%), compared with blocking HDAC1 (64.0%), HDAC 3 (66.7%), or mock infection (69.2%). This included a twofold increase in both GFAP and O4 expression (Fig. 4I). GFAP expression is indicative of radial glial cells as well as astrocytes; radial glial cells are the likely multipotent descendants of early gestation neuroepithelial stem cells (Merkle and Alvarez-Buylla, 2006). This leads to the intriguing possibility that HDAC2 may repress the transition from “early” to “late” stem cells during development, or may repress mitotic arrest or quiescence of radial glia once they have formed.
Our results differ from previous studies (Hsieh et al., 2004; Siebzehnrubl et al., 2007), which found that exposure of postnatal CNS precursors to pharmacological HDAC inhibitors led to an increase in neuron numbers, rather than the decrease we see after HDAC1 inhibition. One explanation for this discrepancy is that the pharmacological agents may have non-selective effects in addition to HDAC inhibition. Valproate is known to inhibit multiple pathways including HDAC, Wnt, and PI3 kinase (Chuang, 2005; Wiltse, 2005). The broad HDAC inhibitors suberoylanilide hydroxamic acid (SAHA) and N-Hydroxy-7-(4-dimethylaminobenzoyl) aminoheptanamide (M344) and the class I HDAC inhibitor N-(2-aminophenyl)-4-[N-(pyridin-3-yl-methoxycarbonyl) aminomethyl] benzamide (MS-275) yield similar results (Siebzehnrubl et al., 2007), but may also affect pathways other than HDAC. In contrast, we used dominant-negative constructs that selectively inhibit distinct HDACs. Secondly, the stoichiometry of different HDACs within larger complexes may modulate the function of the HDACs. This effect would not be seen with a broad pharmacological inhibition. Thirdly, because our studies assayed HDAC inhibition in fetal stem cells rather than the adult stem cells of the previous study, the differences in outcomes may reflect the cellular context in which HDAC inhibition occurs. Finally, we used a paradigm in which HDAC inhibition acted during the expansion phase as well as the differentiation phase. In contrast, the other studies assayed HDAC inhibition after mitogen withdrawal, suggesting that HDACs may have distinct actions on the instruction or expansion of precursors compared with the later differentiation of these cells.
The transcription factor REST acts as a repressor of neuronal genes in non-neuronal cells through its interactions with HDACs (Huang et al., 1999; Roopra et al., 2000; Ballas et al., 2001). Specifically, HDAC2 was shown to be required for the repressive actions of REST (Ballas et al., 2001). While these previous studies used heterologous systems to characterize the function of HDACs on the neuronal phenotype, our studies specifically assayed the function of HDACs in the generation of differentiated progeny from neural stem cells. We show that inhibition of HDAC2 activity had minimal effect on the generation of neurons from cortical stem cells (Fig. 4). Thus, HDACs may interact with REST to repress neuronal differentiation or with other, as yet unknown factors to promote neuronal differentiation during development.
Acknowledgments
We thank Elisabeth Brinkmann for assistance with the cDNA cloning, and Justin Boyd for assistance with immunocytochemical analysis of the neural cells. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Child Health and Human Development, and National Institute of Neurological Disorders and Stroke. D.P. is supported by the NIH Mental Retardation and Developmental Disabilites Research Center Grant P30HD40677.
Contributor Information
Glen W. Humphrey, Laboratory of Molecular Growth Regulation, National Institute of Child Health and Human Development, Bethesda, MD 20892, USA, Tel: +301 496 9038, Fax: +301 480 9354.
Yong-Hong Wang, Laboratory of Molecular Growth Regulation, National Institute of Child Health and Human Development, Bethesda, MD 20892, USA, Tel: +301 496 9038, Fax: +301 480 9354.
Tazuko Hirai, Laboratory of Molecular Growth Regulation, National Institute of Child Health and Human Development, Bethesda, MD 20892, USA, Tel: +301 496 9038, Fax: +301 480 9354.
Raji Padmanabhan, Laboratory of Molecular Biology, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
David M. Panchision, Laboratory of Molecular Biology, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Laura F. Newell, Laboratory of Molecular Biology, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA
Ronald D. G. McKay, Laboratory of Molecular Biology, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA
Bruce H. Howard, Email: howard@helix.nih.gov, Laboratory of Molecular Growth Regulation, National Institute of Child Health and Human Development, Bethesda, MD 20892, USA, Tel: +301 496 9038, Fax: +301 480 9354.
References
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Borre A, Cultraro CM, Segal S. c-Myc inactivation by mutant Max alters growth and morphology of NCI-H-630 colon cancer cells. J Cell Physiol. 1996;169:200–208. doi: 10.1002/(SICI)1097-4652(199610)169:1<200::AID-JCP20>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Chuang DM. The antiapoptotic actions of mood stabilizers: molecular mechanisms and therapeutic potentials. Ann N Y Acad Sci. 2005;1053:195–204. doi: 10.1196/annals.1344.018. [DOI] [PubMed] [Google Scholar]
- Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell. 2002;3:903–910. doi: 10.1016/s1534-5807(02)00360-x. [DOI] [PubMed] [Google Scholar]
- Fajas L, Egler V, Reiter R, Miard S, Lefebvre AM, Auwerx J. PPARgamma controls cell proliferation and apoptosis in an RB-dependent manner. Oncogene. 2003;22:4186–4193. doi: 10.1038/sj.onc.1206530. [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequencespecific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D. Functional significance of histone deacetylase diversity. Curr Opin Genet Dev. 2001;11:162–166. doi: 10.1016/s0959-437x(00)00174-x. [DOI] [PubMed] [Google Scholar]
- Kim J-H, Panchision DM, Kittappa R, McKay RD. Generating CNS neurons from embryonic, fetal and adult stem cells. In: Wassarman PM, Keller GM, editors. Differentiation of embryonic stem cells. New York: Academic Press; 2003. pp. 303–327. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Gottlicher M, Heinzel T. Histone deacetylase as a therapeutic target. Trends Endocrinol Metab. 2001;12:294–300. doi: 10.1016/s1043-2760(01)00438-6. [DOI] [PubMed] [Google Scholar]
- Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- Matushansky I, Radparvar F, Rekhtman N, Skoultchi A. Reprogramming erythroleuekmia cells to terminal differentiation and terminal cell division. Front Biosci. 2000;5:D488–D492. doi: 10.2741/matushan. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Alvarez-Buylla A. Neural stem cells in mammalian development. Curr Opin Cell Biol. 2006;18:704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Wong P, Howard BH. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4877–4882. doi: 10.1128/mcb.17.8.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchision DM, McKay RD. The control of neural stem cells by morphogenic signals. Curr Opin Genet Dev. 2002;12:478–487. doi: 10.1016/s0959-437x(02)00329-5. [DOI] [PubMed] [Google Scholar]
- Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, Buckley NJ. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3- histone deacetylase complex. Mol Cell Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy P, Ventura A, Jacks T. Mammalian RNAi: a practical guide. Biotechniques. 2005;39:215–224. doi: 10.2144/05392RV01. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Lesoon-Wood LA, Dey A, Kuwata T, Weintraub BD, Humphrey G, Yang WM, Seto E, Yen PM, Howard BH, Ozato K. Ligand-induced recruitment of a histone deacetylase in the negative-feedback regulation of the thyrotropin beta gene. EMBO J. 1999;18:5389–5398. doi: 10.1093/emboj/18.19.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebzehnrubl FA, Buslei R, Eyupoglu IY, Seufert S, Hahnen E, Blumcke I. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Exp Brain Res. 2007;176:672–678. doi: 10.1007/s00221-006-0831-x. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA. Transcriptional control at regulatory checkpoints by histone deacetylases: molecular connections between cancer and chromatin. Hum Mol Genet. 2001;10:693–698. doi: 10.1093/hmg/10.7.693. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltse J. Mode of action: inhibition of histone deacetylase, altering WNT-dependent gene expression, and regulation of beta-catenin—developmental effects of valproic acid. Crit Rev Toxicol. 2005;35:727–738. doi: 10.1080/10408440591007403. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Dean DC. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene. 2001;20:3134–3138. doi: 10.1038/sj.onc.1204338. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]