Summary
Clathrin triskelions form the lattice that organizes recruitment of proteins to coated pits and helps drive vesiculation of the lipid bilayer. We report the crystal structure at 2.6 Å resolution of a 55 kDa N-terminal fragment from the 190 kDa clathrin heavy chain. The structure comprises the globular “terminal domain” and the linker that joins it to the end of a triskelion leg. The terminal domain is a seven-blade β propeller, a structure well adapted to interaction with multiple partners, such as the AP-1 and AP-2 sorting adaptor complexes and the nonvisual arrestins. The linker is an α-helical zigzag emanating from the propeller domain. We propose that this simple motif may extend into the rest of the clathrin leg.
Introduction
Clathrin-coated vesicles transport lipids and proteins between intracellular membrane compartments. They are the most prominent of the carriers in the endocytic pathway, between the plasma membrane and early endosomes, and in the secretory pathway, between the trans-Golgi network and late endosomes (for recent reviews, see Robinson, 1994; Kirchhausen et al., 1997).
Clathrin is the scaffold protein of the basket-like coat that surrounds coated vesicles (Pearse, 1976). The soluble assembly unit, a “triskelion” (Ungewickell and Branton, 1981), contains three 190 kDa subunits (“heavy chains”) and three 24–27 kDa subunits (“light chains”) in an extended three-legged structure (Figure 1) (Kirchhausen and Harrison, 1981; Ungewickell and Branton, 1981). The triskelion legs, about 500 Å in length, radiate with a clockwise swirl when viewed from the cytosolic face (Kirchhausen et al., 1986). Each leg contains one heavy chain, with a light chain tightly bound near the vertex (Kirchhausen and Harrison, 1981; Ungewickell and Branton, 1981; Kirchhausen et al., 1983; Ungewickell, 1983). The N terminus of the heavy chain is in the globular “terminal domain” at the distal end of the leg; the C terminus is near the vertex of the triskelion (Kirchhausen and Harrison, 1984; Kirchhausen et al., 1987a; Lemmon et al., 1991; Nathke et al., 1992). Other positions in the heavy chain have been mapped to locations along the leg, particularly within the distal leg and in the linker segment that joins it to the terminal domain (Figure 1) (Kirchhausen and Harrison, 1984; Kirchhausen et al., 1987a). Sequence analysis of the heavy chain and comparisons with sequences of other proteins have not revealed internal repeats, nor have they shown recognizable motifs or domains (Kirchhausen et al., 1987a).
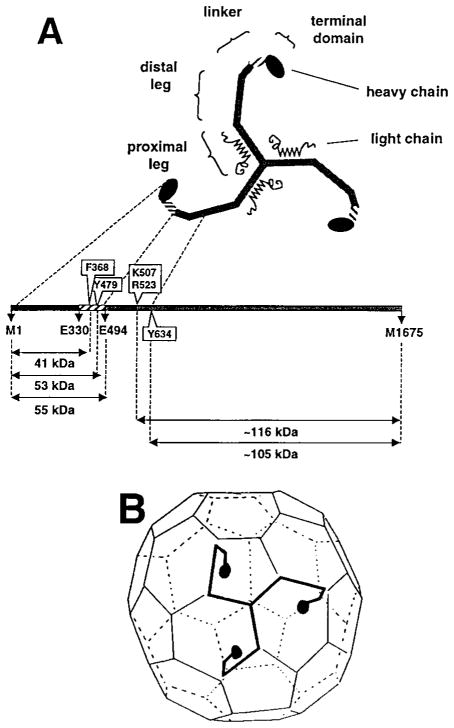
Figure 1. Schematic Representations of a Clathrin Triskelion, Showing Structural Subdivisions of a Leg and the Packing of a Triskelion in the Lattice of a Coat.
(A) Correspondence between molecular morphology and positions in the polypeptide chain. The diagram is based on results from single-molecule electron microscopy, limited proteolysis, sizing chromatography, and primary structure determination (Schmid et al., 1982; Kirchhausen et al., 1983; Ungewickell, 1983; Kirchhausen and Harrison, 1984; Kirchhausen et al., 1987a). Selective proteolytic cleavage sites in the native molecule are boxed.
(B) A typical lattice for a clathrin coat, with a single triskelion superimposed. Each leg of a triskelion contributes to two adjacent edges. The linker and terminal domain project inwards. The figure is based on results from electron microscopy (Crowther and Pearse, 1981; Heuser and Kirchhausen, 1985; Vigers et al., 1986).
Rapid assembly and disassembly of coats are central to the functions of clathrin in vesicular transport. Engulfment of receptors and budding of a vesicle require coat assembly; delivery of cargo and fusion with a target membrane require uncoating. In the absence of other proteins and membranes, clathrin trimers can self-assemble into hollow structures (“cages”) whose lattices resemble those of a normal coat (Keen et al., 1979). The proximal and distal legs participate in the interactions that hold the lattice together (Crowther and Pearse, 1981; Schmid et al., 1982; Ungewickell et al., 1982); the globular terminal domain projects inward toward the membrane (Kirchhausen and Harrison, 1984; Heuser and Kirchhausen, 1985; Vigers et al., 1986; Smith et al., 1998). Proteolysis of assembled cages leads to release of an amino-terminal fragment of about 52–59 kDa, but not to disassembly of the lattice (Schmid et al., 1982; Ungewickell et al., 1982). The released fragment includes the terminal domain and part of the linker connecting it to the distal leg (Kirchhausen and Harrison, 1984).
Other proteins in addition to clathrin are required for assembly under physiological conditions (Keen et al., 1979) and for recruiting cargo to a coated pit (Pearse, 1988). The heterotetrameric clathrin adaptor complexes AP-1 and AP-2 (Zaremba and Keen, 1983; Pearse and Robinson, 1984) and the monomeric AP180 (Murphy et al., 1991; Morris et al., 1993) stimulate coat assembly. AP-1 and AP-2 interact with the clathrin terminal domain (Contreras and Kirchhausen, unpublished results) through a contact that involves the hinge of their β chains (Gallusser and Kirchhausen, 1993; Shih et al., 1995; Wilde and Brodsky, 1996; Clairmont et al., 1997). The clathrin adaptors recognize sorting signals in the cytoplasmic domains of many membrane proteins (Glickman et al., 1989; Sorkin and Carpenter, 1993; Sosa et al., 1993)—for example, the Yppϕ motif (Trowbridge et al., 1993; Ohno et al., 1995; Boll et al., 1996; Rapoport et al., 1997; Rapoport et al., 1998) and LL motifs (Heilker et al., 1996; Rapoport et al., 1998), which direct endocytosis of various receptors and sorting from the trans-Golgi network.
In contrast to these “generic” adaptors, certain specialized adaptors mediate recruitment of specific membrane proteins to coated pits. The best-characterized examples are the “nonvisual” arrestins, proteins that associate with the clathrin terminal domain and bind the phosphorylated cytoplasmic face of activated G protein–coupled receptors, displacing the heterotrimeric G protein and directing receptor downregulation (Lohse et al., 1990; Lefkowitz et al., 1992; Goodman et al., 1996; Goodman et al., 1997). Arrestin3 and β-arrestin associate with α1b-, β1-, and β2-adrenergic receptors and odorant receptors (Dawson et al., 1993; Pippig et al., 1993; Freedman et al., 1995; Diviani et al., 1996). These nonvisual arrestins interact with terminal domain through a short sequence found near their C termini (Krupnick et al., 1997). Candidates for other receptor-specific adaptors that interact with clathrin include AP180 (Murphy et al., 1991; Morris et al., 1993) and amphyphysin (Ramjaun and McPherson, 1998).
The importance of the clathrin terminal domain for recruitment of cargo and for regulation of assembly motivates study of its structure. We report the crystal structure, determined to 2.6 Å resolution, of a 55 kDa fragment from the N terminus of the rat brain clathrin heavy chain. This fragment contains the complete terminal domain as well as part of the linker that joins it to the distal leg. The structure reveals two unexpected features. The cyclic structure of the terminal domain, a β propeller, creates around its outer surface a set of grooves that may provide multiple interaction sites for the variety of observed adaptor proteins. The linker has a different sort of repeating structure—a zigzag of α helices (“α zigzag”). We suggest that other parts of the clathrin leg may have this simple molecular architecture for building extended interactions within the lattice of a coat.
Results
Expression and Structure Determination
A 55 kDa fragment of clathrin, residues 1–494 of the rat heavy chain (Kirchhausen et al., 1987a), was expressed in E. coli and purified as described in Experimental Procedures. The crystal structure was determined by a combination of multiple isomorphous replacement with anomalous scattering and 3-fold noncrystallographic symmetry averaging. The current model has been refined at a resolution of 2.6 Å, with good R factors and stereochemical parameters. Details of the structure determination are reported in Experimental Procedures.
Overview of the Structure
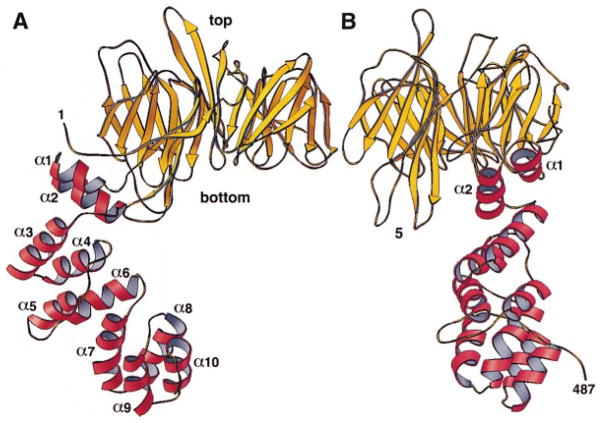
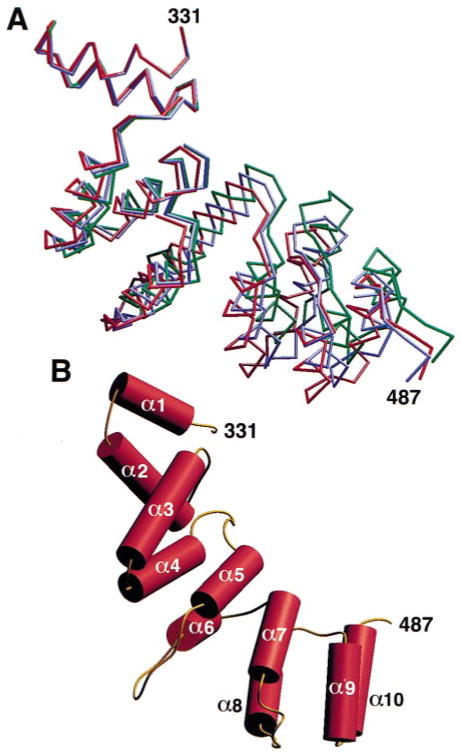
The 55 kDa fragment contains two distinct substructures (Figure 2). The N-terminal, globular part (Figure 3) is an all β-sheet domain; the C-terminal, extended part is entirely α helical. The globular domain is a seven-blade β propeller resembling those found in the β subunit of the heterotrimeric GTP-binding proteins, such as β-transducin (Wall et al., 1995; Lambright et al., 1996; Sondek et al., 1996), and in the regulator of chromosome condensation (RCC1) (Renault et al., 1998). The helical region is a zigzag of ten relatively short α helices (an eleventh is present in one of the three molecules in the asymmetric unit) that pack on each other in successive pairs to form a bent, stem-like structure. The β propeller corresponds to the globular terminal domain, and the α zigzag represents part of the linker that connects the terminal domain to the distal leg.
Figure 2. Overall Structure of the Terminal Domain and Part of the Linker from the Rat Clathrin Heavy Chain The ribbon diagram shows the structure of the 55 kDa fragment of clathrin.
The terminal domain (yellow) is a β propeller. The linker (red) is an α zigzag. The views in (A) and (B) include the first 487 amino acid residues of the clathrin heavy chain, which are visible in the electron density map in all the monomers in the crystallographic asymmetric unit. The view in (B) represents a 90° counterclockwise rotation about the vertical axis of the view in (A). The α helices are numbered α1 to α10. Blade 5 of the β propeller is labeled for orientation; loops 5a, 5b, 5c, and 5d project from the “bottom” surface of this blade. Figure was made with RIBBONS (Carson, 1991).
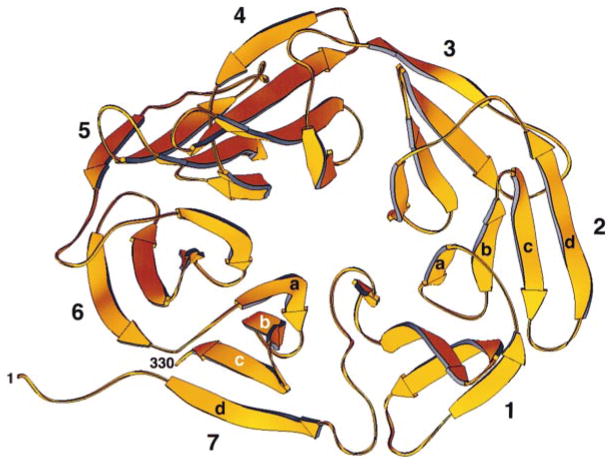
Figure 3. Ribbon Diagram of the Clathrin β Propeller Terminal Domain.
The β propeller spans amino acid residues M1–E330. It has seven blades (1–7) organized sequentially in a counterclockwise direction and viewed here looking from the “top” to the “bottom” surface along the pseudo-7-fold axis. Within each blade, the orientations of the four β strands (a, b, c, and d) alternate, starting with the innermost strand, “a,” which runs from top to bottom, and ending with the outermost strand, “d,” which runs from bottom to top. Closure is achieved in blade 7 by the contributions of β strand 7d (residues P6 to H12) derived from the N terminus of the polypeptide chain. Figure was made with RIBBONS (Carson, 1991).
Figure 4 shows an alignment of amino acid sequence and secondary structure elements. The sequence of clathrin is remarkably invariant among species as distant as yeast and man. Those portions in the 55 kDa fragment at which differences occur in the known clathrins do not correlate with any particular structural features. In general, they occur as frequently within strands and helices as they do in loops between secondary structures. This conservation suggests that almost all surfaces of the heavy chain engage in contacts, either with different parts of clathrin in a coat or with other proteins.
Figure 4. Structure-Based Sequence Alignment of Rat Clathrin Heavy Chain.
(A) The sequence of the heavy chain (residues M1–E494) is shown with the elements of secondary structure indicated above the alignment. Yellow arrows mark β strands in the propeller; red cylinders, α helices in the linker.
(B) Alignment of the seven blades within the β propeller of the terminal domain. The approximate boundaries of the strands are highlighted. ϕ designates conserved hydrophobic residues; W and D designate Trp and Asp. For comparison, the position of the WD repeat in bovine rod β transducin is shown (Sondek et al., 1996).
The β Propeller
An axial view of the β propeller, shown in Figure 3, illustrates the characteristics of this fold. There are seven β sheets, each with four antiparallel strands; the sheets are packed on each other around a central axis. The twist of each sheet, conforming to standard β sheet geometry, imparts a distinctly propeller-like character (Murzin, 1992). The sheets are numbered 1 to 7, and the strands within each sheet are denoted a to d in the order from axis to perimeter. We follow previous convention in using “top” to refer to the face of the domain in which the outer β strand (d) of one blade connects to the inner β strand (a) of the next blade. The polypeptide chain enters the propeller as strand 7d, crosses to strand 1a, continues through each blade in order, and ultimately exits the domain at the C terminus of strand 7c. Thus, it enters and leaves the domain at adjacent positions on the “bottom” face, and the β interaction of strands 7d and 7c stabilizes closure of the cyclic structure.
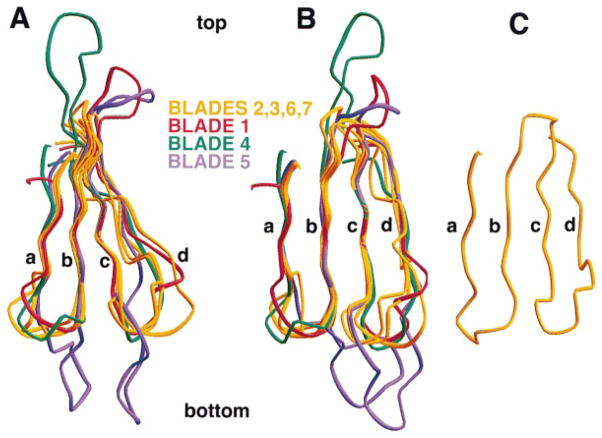
The core of each blade in the clathrin propeller has an essentially invariant structure (Figures 5A and 5B), particularly in the β strands b and c. The blades of β-transducin can be superposed almost equally well on those of clathrin (Figure 5C). The building blocks of the two structures are thus essentially the same. The β subunits of heterotrimeric G proteins contain recognizable “WD40 repeats,” each of which corresponds to a blade, but structural alignment of the amino acid sequence of the clathrin terminal domain (Figure 4B) reveals no striking repetitive features. The WD40-like characteristics of the clathrin sequence are restricted to conservation of a set of hydrophobic residues at positions of contact between successive sheets.
Figure 5. Structural Alignment of the Seven Blades in the Clathrin β Propeller.
The figure shows a superposition of Cα traces for the seven blades of the clathrin β propeller, aligned using strands βb and βc of each blade. Blades 2, 3, 6, and 7 (yellow) have the shortest loops between β strands. Blades 1, 4, and 5 (red, green, and blue) have at least one long loop projecting from the top or the bottom surface of the β propeller. (A) and (B) correspond to views rotated by about 50° with respect to each other. (C) shows blade 1 of the β propeller from β-transducin (Sondek et al., 1996).
The clathrin propeller has a roughly elliptical cross section (principal axes 47 and 40 Å). The elongation is due to increased spacings between blades 7 and 1 and between blades 3 and 4. The blade packing does not by itself create an irregularly grooved exterior, however, because the gaps at the outside of the domain between 7 and 1 and 3 and 4 are occupied by short helical segments in the connecting d–a loops. Indeed, the two most significant grooves are between blades 1 and 2 and blades 4 and 5. The former, created in part by an outward displacement of blade 2, appears to be a site for interaction with the adaptor molecules β-arrestin and arrestin3 (see below).
The number of residues in each blade of the clathrin propeller is quite variable because of significant differences in the interstrand loops. As a result, the clathrin terminal domain has some noteworthy surface features not present in the more regular transducin propeller. There are protrusions from the top and bottom surfaces of the clathrin terminal domain emanating as loops from blades 1, 4, and 5. In the case of blade 5, the proline-rich c–d loop has an unusual structure that opens the interface between blades 4 and 5. We suggest that this groove may also be an interaction site, perhaps for a component of the AP-1 and AP-2 adaptor complexes.
The β-Arrestin Binding Site
Mutational analysis of clathrin has identified the determinants of an interaction between the nonvisual arrestins (β-arrestin and arrestin3) and the clathrin terminal domain (Goodman et al., 1997). The first 100 residues of clathrin are sufficient for detection of binding in vitro. This fragment corresponds precisely to blades 1 and 2 of the propeller (Figures 6A and 6B). Site-directed alterations in the complete terminal domain show that Q89, F91, K96, and K98 are critical for arrestin binding. These residues lie near the margins of the groove between blades 1 and 2 (Figure 6C). Exposed hydrophobic side chains lie within the groove, and the two lysine residues impart a strong positive charge to its edge. The four residues identified by mutation form a coherent patch (red in Figures 6A and 6B), consistent with a localized contact between arrestin and clathrin. The part of the nonvisual arrestins required for clathrin binding is a short segment near the C terminus (Krupnick et al., 1997). A recently determined crystal structure of a related visual arrestin (Granzin et al., 1998), which is likely to be a general model for members of the family, shows that the C-terminal part of the polypeptide chain is disordered in free arrestins and that it projects away from the globular body of the protein. Arrestin3 and β-arrestin contain three hydrophobic residues that are required for their interaction with clathrin, as well as several acidic residues (Krupnick et al., 1997). The characteristics of this segment are thus precisely those that one would expect for a ligand that contacts the groove between blades 1 and 2 of clathrin. The corresponding sequence is absent from the visual arrestins, which do not direct photoreceptor proteins to coated pits.
Figure 6. Mapping of Interaction Sites in Clathrin Terminal Domain Required for the Association with Nonvisual Arrestins.
Residues of clathrin that participate in contacts with the C-terminal arm of arrestin3 and β-arrestin (Goodman et al., 1997) are shown in red in the space-filling representations (A and B) and as ball and stick models in the ribbon diagram (C). In (A) and (B), residues M1 to K100 are highlighted in gray; these correspond to blades 1 and 2 and to strand 7a. This segment by itself can bind to arrestin3 and β-arrestin (Goodman et al., 1997). The rest of the domain (residues A101 to E330) is yellow. The views in (A) and (B) are similar, respectively, to the top view in Figure 3 and to the side view in Figure 2A. The amino acid sequence of the segment of arrestin3 that binds this site is TNLIEFETN (Krupnick et al., 1997). Figure was made with RIBBONS (Carson, 1991).
The Helical Linker
The α zigzag fold in the linker contains helices of variable length (2–4 turns) connected by short loops (Figures 2 and 7). The overall length of the linker segment in the ordered part of this structure is about 45 Å. The first helical hairpin (helices 1 and 2) packs tightly against a hydrophobic surface of the propeller. It is flanked on one side by two long loops from blade 5, but it has no strong contacts with them. The remaining four helical hairpins in our structure pack into two four-helix bundles with a partially open interface between them. We can find no obvious sequence relationship between residues in the two bundles, and a spatial superposition also gives a rather poor fit. Thus, aside from the α zigzag fold itself and an apparent tendency for the hairpins to pair, there are no recognizable repetitive structural elements.
Figure 7. Structure of the α zigzag Region in the Clathrin Linker.
(A) Superposition of Cα traces from the helical region of the three monomers (red, blue, green) in the crystallographic asymmetric unit. The superposition was obtained by alignment of the β propeller domains (not shown).
(B) Cylinder representation of the α zigzag region of a monomer to show the overall right-handed twist in the linker region. Figure was made with Setor (Evans, 1993).
The packing of successive hairpins generates a consistent but irregular right-handed twist, with respect to the overall axis of the linker (Figure 7B). The α-helical interfaces are strongly hydrophobic, and even the partly open surfaces between the first and second and between the third and fourth hairpins contain exposed hydrophobic residues. The contacts evidently allow for some flexibility, since comparison of the three independent structures in the asymmetric unit shows somewhat different overall bends (Figure 7A), generated by the weak asymmetry of packing forces in the crystal. The gap between four-helix bundles does not appear to be a more significant hinge than any of the other interhairpin contacts.
Discussion
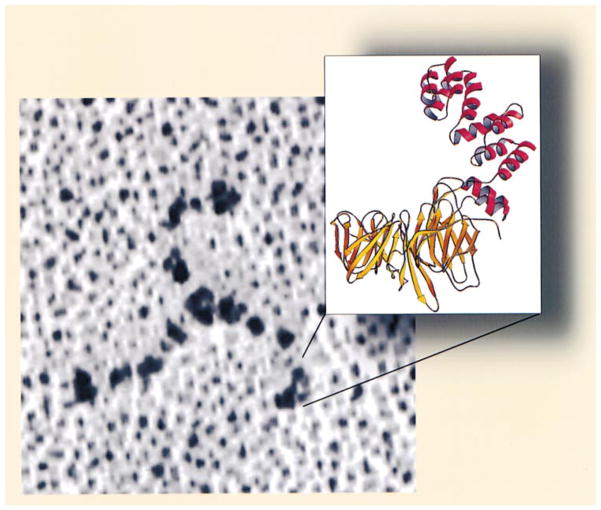
The relationship between the high-resolution structure described in this paper and the familiar electron-microscopic view of a clathrin triskelion is shown in Figure 8. The structure validates early conclusions from electron microscopy, limited proteolysis, and size-exclusion chromatograpy that the 55 kDa fragment comprises both the globular terminal domain and an extended linker segment (Schmid et al., 1982; Kirchhausen and Harrison, 1984; Kirchhausen et al., 1986). The unanticipated repeating substructures in these two elements are closely related to their distinct functions in a clathrin lattice.
Figure 8. Relationship between the Clathrin Triskelion and the Structure of the Terminal Domain and Linker.
Clathrin was deposited onto freshly cleaved mica by spraying in 50% glycerol, rotary shadowed with Pt, and visualized by electron microscopy (see Heuser and Kirchhausen, 1985). The ribbon diagram corresponds to the terminal domain and linker.
The Terminal Domain
The terminal domain has none of the sequence hallmarks known to form β propellers, such as the WD40 (Wall et al., 1995; Lambright et al., 1996; Sondek et al., 1996) or RCC1 repeats (Renault et al., 1998), but the same fold also occurs in proteins with no detectable internal repeat and with no sequence similarity to the signaling molecules that contain these motifs. We find that the individual blades of the clathrin propeller superpose well, both on each other and on a typical WD40 repeat. These observations are consistent with an analysis of the packing of sheets in β propeller structures, which has suggested that the driving forces leading to their formation are no different from those leading to β sheets in general and that the fold is therefore compatible with very different sequence patterns (Murzin, 1992).
The β propeller proteins do not have a common function. Some have enzymatic activity (e.g., methylamine dehydrogenase, methanol dehydrogenase, galactose oxidase, prolyl oligopeptidase), while others serve as scaffolds for protein associations in signal transduction and vesicular trafficking (e.g., heterotrimeric G proteins, a domain in the β subunit of COPI, Sec13 of COPII); for a recent summary, see Springer (1998). The β subunit of heterotrimeric G proteins is both a connector for the GTPase and a relay to many downstream effectors. RCC1 is a guanine nucleotide exchange factor for Ran, a small GTPase, and it also links Ran to DNA-binding proteins (Dasso et al., 1994; Renault et al., 1998). The clathrin terminal domain clearly resembles the G protein β subunit in one important respect: the diversity of its partners. Clathrin is a scaffold for association of a variety of adaptor molecules.
Several proteins known to interact with the terminal domain do so through short peptide segments that appear not to be part of a well-ordered, prefolded structure. Association with clathrin involves fitting a previously unstructured stretch of peptide into the appropriate site on the propeller. In the case of β-arrestin and arrestin3, the segment is part of a C-terminal tail, disordered in the visual arrestin crystal structure (Granzin et al., 1998), and its site in clathrin is the groove between blades 1 and 2. In the case of AP-1 and AP-2, the segment is part of the protease-sensitive “hinge” that connects the main part of the β chain to a projecting C-terminal “ear” (Shih et al., 1995). A targeted disruption of the interaction between clathrin and arrestin does not prevent association of clathrin with the AP-2 complex (Goodman et al., 1997), and we therefore suggest that the binding site for the AP β hinge may be another of the propeller grooves. Amphiphysin is also another potential clathrin adaptor. There is a sequence in amphiphysin that is required for interaction with clathrin and that is clearly related to a segment in the β hinge (Ramjaun and McPherson, 1998).
Binding of arrestin by a fragment of the terminal domain (Goodman et al., 1997) is at first surprising, since the cyclic character of the structure suggests that its stability might depend on a complete propeller. The fragment, residues 1–100, turns out to correspond precisely to the first two blades and an N-terminal extension (Figure 7), and the two sheets probably have sufficient stability, in association with the arrestin segment, to produce a detectable interaction. Even within the intact propeller, sequence constraints on individual pairs of blades may be relatively independent of the rest of the domain. Because the propeller fold does not require a particular sequence pattern, pairs of blades within it can specialize to recognize particular partners, subject only to conservation of packing of their β sheets. Thus, the design of the terminal domain is particularly adapted to independent evolution of multiple interactions—especially interactions with peptide segments that can fit a local contact, rather than with extended surfaces that may overlap adjacent sites.
The β subunit of a heterotrimeric G protein associates with a Gα through two distinct contacts (Wall et al., 1995; Lambright et al., 1996). In one, an N-terminal helix of Gα, disordered in the free subunit, runs along the groove between blades 7 and 1 of Gβ, and side chains from one face of the helix contact several blade-1 residues. In the other, the GTPase domain of Gα lies against the top surface of the Gβ propeller, forming an extended interface. The interaction of the clathrin terminal domain with the C-terminal peptide of nonvisual arrestins (and perhaps that with the β hinge of AP complexes) appears to resemble the contact between Gβ and the N-terminal segment of Gα. A groove between two blades serves as a docking site for a single, short stretch of polypeptide chain from the partner. The clathrin propeller may also use its top and bottom surfaces for more extended interactions. These surfaces are noticeably rough and irregular, due to the variable lengths of the loops that project from them.
Coats of vesicles in the ER and Golgi formed by the so-called “coatomer” complexes COPI and COPII contain WD40-repeat proteins that presumably have β propeller domains (Saxena et al., 1996; Csukai et al., 1997). Parallels between clathrin coats and those formed by coatomers are not well understood, but it is possible that the propeller domains have related functions in the three classes of transport vesicles.
The Clathrin Leg
Each leg of a triskelion has an overall contour length (including the terminal domain) of 500 Å (Crowther and Pearse, 1981; Kirchhausen et al., 1986) and spans two edges of a coated-vesicle lattice (Crowther and Pearse, 1981). The length of an edge is about 185 Å, and the diameter of the propeller is about 50 Å, leaving roughly 80 Å for the linker. The segment of α zigzag (residues N331–A493) in the fragment we have crystallized has a contour length of about 50 Å; it thus represents somewhat more than half of the complete linker joining terminal domain and distal leg.
Correlation of proteolytic cleavage points with distances along the leg from electron microscopy (Kirchhausen and Harrison, 1984; Kirchhausen et al., 1987a) yields estimates for the number of residues per unit length in various fragments (see Figure 1). Allowing roughly 75 residues for a region at the hub that mediates trimerization (Kirchhausen and Harrison, 1984; Liu et al., 1995; Huang et al., 1997), and excluding the 330-residue terminal domain (the β propeller), we can calculate an overall linear density for the two edges (approximately 185 Å each) and the linker (approximately 80 Å) of about 2.8 residues/Å. A cleavage point at 315 Å from the vertex corresponds to residue Y634 (Kirchhausen et al., 1987a). With the same 75-residue allowance for a trimerization element, the 966 residues in the 315 Å fragment give an average of 3 residues/Å. A simple calculation based on the crystal structure presented here indicates that the linear density in the linker region is again approximately 3 residues/Å, in good agreement with the values calculated from electron microscopy. Thus, the entire leg, including the linker, is of relatively uniform linear density. Indeed, electron micrographs of triskelions, contrasted either by rotary shadowing (Ungewickell and Branton, 1981; Kirchhausen and Harrison, 1984) or by negative staining (Crowther and Pearse, 1981; Kirchhausen and Harrison, 1981), reveal no bulges, knobs, or other striking variations in apparent thickness. Likewise, a recent three-dimensional reconstruction of a clathrin cage from cryo-electron microscopy at 21 Å resolution shows relatively uniform, rod-like elements within each edge (Smith et al., 1998). Circular dichroism measurements indicate a high (59%) α-helical content for the clathrin heavy chain (Winkler and Stanley, 1983), and the paucity of prolines and glycines is also consistent with a largely helical conformation. We therefore suggest that an α zigzag may be the fundamental structural motif of more than just the linker part of the leg.
The structural repeat in the linker is irregular—that is, the individual helical hairpins are of variable length, and the relationship of one to the next is not uniform—and the entire stack is somewhat flexible. The flexibility may be necessary to allow the terminal domain itself to interact with a variety of membrane-attached proteins. If the same α zigzag motif continues into the distal and proximal segments of the leg, we anticipate that the structure might be more regular, both in the length of the repeats and in the packing of one upon the next. We cannot, however, extrapolate to suggest whether the tendency to cluster into four-helix bundles will continue or not.
At least two other significantly more uniform motifs form helical zigzag structures. The “tetratricopeptide repeats” (TPR) are 34-residue, usually tandem motifs that form a zigzag with a right-handed twist (Das et al., 1998), rather like the clathrin linker. The ankyrin repeats are also about 33–34 residues in length: they form helical hairpins with a β loop between the two members, and these hairpins stack upon each other with a left-handed twist (Gorina and Pavletich, 1996). In the TPR structure, the axis of the stack is displaced from the center of the individual repeats, generating a deep, right-handed groove in which an α helix from another protein could bind (Das et al., 1998). It is possible that a groove in the proximal leg of clathrin could be the site of interaction of a light chain. The central one-third of a clathrin light chain, essential for heavy-chain association (Scarmato and Kirchhausen, 1990), contains ten amphipathic heptad repeats, and it is therefore likely that this part of the light chain forms an extended α helix (Kirchhausen et al., 1987b). The α zigzag structure in clathrin is also reminiscent of the armadillo repeats, which contain three α helices per unit (Huber et al., 1997). As in the TPR structure, individual units are displaced from the axis and tilted with respect to it, producing a groove that serves as a binding site. In karyopherin-α, which contains ten armadillo repeats, a recent structure shows that the groove accepts a nuclear localization signal in an extended conformation (Conti et al., 1998).
The α zigzag appears to be a new addition to the motifs that form extended, rod-like structures in the cytoplasm. For example, myosin, tropomyosin, and numerous other fibrous proteins contain α-helical coiled coils, and spectrin and dystrophin contain repeated anti-parallel three-helix bundles. Electron microscopy of free clathrin has indicated that the legs are relatively stiff, with hinge points at the connection between proximal and distal segments and likewise greater flexibility in the linker (Kirchhausen et al., 1986). The α zigzag segment in the linker part of our structure is rather like a stiff spring, with limited flexibility distributed uniformly along its length. This property may be important for presenting the terminal domain to its partners.
Experimental Procedures
Protein Expression and Purification
Rat clathrin heavy chain, residues 1–494, was expressed as a recombinant protein in E. coli BL21(DE3) using the vector pBAT (Peranen et al., 1996). The cells from a 2 l culture were harvested by centrifugation 4 hr postinduction with 0.1 mM IPTG, and the pellets were resuspended in 50 ml of buffer A (20 mM Tris [pH 9.0], 50 mM NaCI, 1 mM EDTA, 1 mM DTT). The cells were lysed by sonication, and the debris was removed by a 2 hr centrifugation at 4°C and 40,000 rpm using a Beckmann 60 Ti rotor. The supernatant was applied at 3 ml/min to a 50 ml HP-Q-Sepharose column (Pharmacia) equilibrated with buffer A, and the 55 kDa N-terminal portion was recovered in the flowthrough. The sample was concentrated ~25× by ultrafiltration (Amicon Centriprep) and loaded 1 ml at a time onto a Superdex 75 (16/60) size exclusion column (Pharmacia), run at 0.8 ml/min, and equilibrated with buffer B (20 mM MES [pH 6.5], 50 mM NaCl, 1 mM EDTA, 1 mM DTT). The recombinant fragment eluted as a single species slightly after bovine serum albumin. Fractions containing the recombinant fragment were pooled and concentrated by ultrafiltration to about 25 mg/ml and stored at 4°C. Monodispersity was verified by dynamic light scattering (Dyna Pro 801, Protein Solutions, Inc.).
Selenomethione-labeled clathrin was prepared by diluting 1.5 ml of an overnight culture of BL21(DE3) carrying the expression vector with 1.5 ml of defined LeMaster medium containing DL-selenomethionine (Sigma). The cells were grown at 37°C to an OD600 = 0.6. A 1 ml sample was then diluted with 100 ml of LeMaster medium plus selenomethionine and grown to an OD600 = 0.6, followed by further addition of 2 liters of the same medium. When the OD600 reached 0.6, protein expression was induced for 4 hr by addition of 0.1 mM IPTG. All the purification steps for this material were done in the presence of 5 mM DTT.
Crystallization and Structure Determination
Crystals were obtained by hanging-drop vapor diffusion in 7 days at room temperature by equilibrating against buffer C (21%–23% PEG 4000, 400–600 mM NaCl, 100 mM PIPES [pH 6.5], 10 mM DTT), a solution containing 2 μl of the sample and 2 μl of buffer C. The crystals belong to space group P3221 (a= b= 206.8 Å, c= 87.8 Å) with three molecules in the asymmetric unit. All native and heavy atom derivative diffraction data were collected at 160 K. Prior to freezing, crystals were dialyzed in two steps, first against buffer D (25% PEG 4000, 500 mM NaCl, 100 mM PIPES [pH 6.5], 15% glycerol, 10 mM DTT) and then against buffer E (25% PEG 4000, 100 mM PIPES [pH 6.5], 200 mM Na2SO4, 15% glycerol). Heavy atoms soaks were carried out for 7 days in buffer D with uranyl formate (0.5 mM) or for 1 day in buffer F (25% PEG 4000, 100 mM HEPES [pH 7.5], 200 mM Na2SO4, 15% glycerol) with p-hydroxymercuribenzoate (0.5 mM). Excess p-hydroxymercuribenzoate was removed by dialysis against buffer E prior to freezing.
The crystals were flash frozen for data collection. High-resolution data from a native crystal (to 2.6 Å resolution) and from a selenomethionyl derivative (to 3 Å) were collected on beamline X25 at the National Synchrotron Light Source at a wavelength of 0.9785 Å (to maximize the selenium anomalous signal) using a 34.5 cm image plate scanner (MAR Research, Hamburg, Germany). Data from the uranyl and mercury derivatives were recorded to 5 Å resolution using an 18 cm MAR image plate scanner on a GX13 rotating-anode source (Elliott, London). Oscillation images (0.5°) were processed using DENZO (Otwinowski, 1993), and data reduction was carried out using SCALEPACK (Otwinowski, 1991). Subsequent computation used CCP4 (Collaborative Computational Project, 1994) and X-PLOR (Brünger, 1992). The 62,848 unique reflections to a spacing of 2.6 Å in the native data set (96% complete) had an overall Rsym of 5.2% (26.1% in the outer shell, which was 70% complete). SCALEIT (CCP4) was used to scale derivative to native data, which had been put on an approximate absolute scale using TRUNCATE (CCP4).
Heavy atom positions for the mercury derivative were determined by inspection of difference Patterson maps calculated with resolution limits of 13 and 5 Å. Refinement of the heavy atom parameters was carried out with MLPHARE (CCP4), using data in the same resolution range, and the resulting phases were used to locate the uranium atoms in the uranyl formate derivative by difference Fourier techniques. Corefinement of the heavy atom parameters from the mercury and uranyl derivatives (13 to 5 Å resolution) provided a set of phases that readily identified 26 selenium atoms (out of a total of 42 such atoms in the asymmetric unit) from both isomorphous and anomalous difference maps. The refinement of strong and positive anomalous occupancies allowed unequivocal identification of the correct hand of the heavy atom structure and indicated that the space group was P3221. Both the anomalous and isomorphous signal from the selenomethionyl derivative extended to at least 3 Å resolution. Initial protein electron density maps (13 to 3 Å) were improved by solvent flattening and histogram matching using SOLO-MON (CCP4). The resulting map was skeletonized using MAPMAN (Kleywegt and Jones, 1994), and the skeleton was edited in “O” (Jones et al., 1991). This map allowed identification of several elements of secondary structure and definition of monomer boundaries. The initial skeleton was used to create a mask (Kleywegt and Jones, 1994) that covered a substantial fraction of one monomer. The 32 heavy atom positions were used to find the set of transformations relating the three monomers in the asymmetric unit. The combined application of solvent flattening, histogram matching, and 3-fold noncrystallographic symmetry (NCS) averaging led to an electron density map that could be interpreted readily.
A model corresponding to one monomer was built with the “baton” command in “O,” and the remaining monomers were built by rotating the coordinates of the first monomer according to the NCS operators. Refinement was carried out with X-PLOR (Brünger, 1992), with 5% of the data set aside for calculating Rfree. In order to account for some flexibility in the orientation of subdomains in the helical region, refinement was started with the rigid-body fitting of nine domains, roughly corresponding to the propeller and two four-helix bundles from each monomer. In subsequent steps, several attempts were made to define a set of protein segments and weights that would allow for application of NCS restraints while accounting for the observed flexibility of the helical domain. The best results were obtained when relatively high weights (300) were used and when each helix was taken as a separate NCS group. Many cycles of positional and B-factor refinement were followed by extensive rebuilding. Refinement was concluded by several runs of torsion-angle dynamics using simulated annealing with initial temperatures of 7500 K. The final model (Rcryst = 22.6%; Rfree = 29.2%, using data with F > 2σ between 6 and 2.6 Å) contains residues 1–487 (monomers A and C), 1–493 (monomer B), and 34 water molecules. The root-mean-squared deviation in bond lengths is 0.019 Å and in bond angles, 2.1°. Residues E46–Q47, Q173–N175, and A406–T411 are in poor density. The temperature factor for the C-terminal region of monomer C (E331–K487) is significantly higher (80.9 Å2) than for the corresponding regions of monomer A and B (47.1 and 55.1 Å2, respectively).
Supplementary Material
Acknowledgments
We thank the staff of beamline X25 at National Synchrotron Light Source laboratories for support and Jennifer Poitras for secretarial help. The work was supported in part by NIH grant GM36548 (T. K.) and CBR funds (T. K.). A. M. is an Armenise Fellow (Harvard Medical School Center for Structural Biology) and a Senior Postdoctoral Fellow of the American Cancer Society (Mass. division). S. C. H. is an Investigator in the Howard Hughes Medical Institute. This paper is a contribution from the Harvard Medical School Center for Structural Biology, with support from the Harvard-Armenise Foundation for Advanced Medical Research.
Footnotes
Brookhaven Protein Data Bank ID Code
The coordinates for clathrin have been deposited in the Brookhaven Protein Data Bank under identification code 1bpo.
References
- Boll W, Ohno H, Songyang Z, Rapoport I, Cantley LC, Bonifacino JS, Kirchhausen T. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 1996;15:5789–5795. [PMC free article] [PubMed] [Google Scholar]
- Brünger AT. X-PLOR Version 3.1: A System for X-Ray Crystallography and NMR. New Haven, Connecticut: Yale University Press; 1992. [Google Scholar]
- Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- Clairmont KB, Boll W, Ericsson M, Kirchhausen T. A role for the hinge/ear domain of the beta chains in the incorporation of AP complexes into clathrin-coated pits and coated vesicles. Cell Mol Life Sci. 1997;53:611–619. doi: 10.1007/s000180050080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Pearse BM. Assembly and packing of clathrin into coats. J Cell Biol. 1981;91:790–797. doi: 10.1083/jcb.91.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csukai M, Chen CH, Dematteis MA, Mochlyrosen D. The coatomer protein beta′-COP, a selective binding protein (RACK) for protein kinase cepsilon. J Biol Chem. 1997;272:29200–29206. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M, Seki T, Azuma Y, Ohba T, Nishimoto T. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 1994;13:5732–5744. doi: 10.1002/j.1460-2075.1994.tb06911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Arriza JL, Jaworsky DE, Borisy FF, Attramadal H, Lefkowitz RJ, Ronnett GV. Beta-adrenergic receptor kinase-2 and beta-arrestin-2 as mediators of odorant-induced desensitization. Science. 1993;259:825–829. doi: 10.1126/science.8381559. [DOI] [PubMed] [Google Scholar]
- Diviani D, Lattion AL, Larbi N, Kunapuli P, Pronin A, Benovic JL, Cotecchia S. Effect of different G protein-coupled receptor kinases on phosphorylation and desensitization of the alpha(1b)-adrenergic receptor. J Biol Chem. 1996;271:5049–5058. doi: 10.1074/jbc.271.9.5049. [DOI] [PubMed] [Google Scholar]
- Evans SV. SETOR: hardware lighted three-dimensional solid model representation of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Freedman NJ, Liggett SB, Drachman DE, Pei G, Caron MG, Lefkowitz RJ. Phosphorylation and desensitization of the human beta(1)-adrenergic receptor—involvement of G protein-coupled receptor kinases and CAMP-dependent protein kinase. J Biol Chem. 1995;270:17953–17961. doi: 10.1074/jbc.270.30.17953. [DOI] [PubMed] [Google Scholar]
- Gallusser A, Kirchhausen T. The β1 and β2 subunits of the AP complexes are the clathrin coat assembly components. EMBO J. 1993;12:5237–5244. doi: 10.1002/j.1460-2075.1993.tb06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman JN, Conibear E, Pearse BMF. Specificity of binding of clathrin adaptors to signals on the mannose-6-phosphate/insulin-like growth factor II receptor. EMBO J. 1989;8:1041–1047. doi: 10.1002/j.1460-2075.1989.tb03471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. β-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Gurevich VV, Benovic JL, Keen JH. Arrestin/clathrin interaction. Localization of the arrestin binding locus to the clathrin terminal domain. J Biol Chem. 1997;272:15017–15022. doi: 10.1074/jbc.272.23.15017. [DOI] [PubMed] [Google Scholar]
- Gorina S, Pavletich NP. Structure of the P53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- Heilker R, Manningkrieg U, Zuber JF, Spiess M. In vitro binding of clathrin adaptors to sorting signals correlates with endocytosis and basolateral sorting. EMBO J. 1996;15:2893–2899. [PMC free article] [PubMed] [Google Scholar]
- Heuser J, Kirchhausen T. Deep-etch views of clathrin assemblies. J Ultrastruct Res. 1985;92:1–27. doi: 10.1016/0889-1605(85)90123-5. [DOI] [PubMed] [Google Scholar]
- Huang KM, Gullberg L, Nelson KK, Stefan CJ, Blumer K, Lemmon SK. Novel functions of clathrin light chains: clathrin heavy chain trimerization is defective in light chain-deficient yeast. J Cell Sci. 1997;110:899–910. doi: 10.1242/jcs.110.7.899. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Keen JH, Willingham MC, Pastan IH. Clathrin-coated vesicles: isolation, dissociation and factor-dependent reassociation of clathrin baskets. Cell. 1979;16:303–312. doi: 10.1016/0092-8674(79)90007-2. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC. Protein organization in clathrin trimers. Cell. 1981;23:755–761. doi: 10.1016/0092-8674(81)90439-6. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC. Structural domains of clathrin heavy chains. J Cell Biol. 1984;99:1725–1734. doi: 10.1083/jcb.99.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC, Parham P, Brodsky FM. Location and distribution of the light chains in clathrin trimers. Proc Natl Acad Sci USA. 1983;80:2481–2485. doi: 10.1073/pnas.80.9.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC, Heuser J. Configuration of clathrin trimers: evidence from electron microscopy. J Ultrastruct Res. 1986;94:199–208. doi: 10.1016/0889-1605(86)90067-4. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC, Chow EP, Mattaliano RJ, Ramachandran KL, Smart J, Brosius J. Clathrin heavy chain: molecular cloning and complete primary structure. Proc Natl Acad Sci USA. 1987a;84:8805–8809. doi: 10.1073/pnas.84.24.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Scarmato P, Harrison SC, Monroe JJ, Chow EP, Mattaliano RJ, Ramachandran KL, Smart JE, Ahn AH, Brosius J. Clathrin light chains LCA and LCB are similar, polymorphic and share repeated heptad motifs. Science. 1987b;236:320–324. doi: 10.1126/science.3563513. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Bonifacino JS, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. From first map to final model. In: Bailey S, Hubbard R, Waller D, editors. Proceedings of the CCP4 Study Weekend. Daresbury, UK: Daresbury Laboratory; 1994. pp. 59–66. [Google Scholar]
- Krupnick JG, Goodman OB, Jr, Keen JH, Benovic JL. Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxy terminus. J Biol Chem. 1997;272:15011–15016. doi: 10.1074/jbc.272.23.15011. [DOI] [PubMed] [Google Scholar]
- Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Inglese J, Koch WJ, Pitcher J, Attramadal H, Caron MG. G-protein-coupled receptors: regulatory role of receptor kinases and arrestin proteins. Cold Spring Harbor Symp Quant Biol. 1992;57:127–133. doi: 10.1101/sqb.1992.057.01.016. [DOI] [PubMed] [Google Scholar]
- Lemmon SK, Pellicena Palle A, Conley K, Freund CL. Sequence of the clathrin heavy chain from Saccharomyces cerevisiae and requirement of the COOH terminus for clathrin function. J Cell Biol. 1991;112:65–80. doi: 10.1083/jcb.112.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SH, Wong ML, Craik CS, Brodsky FM. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell. 1995;83:257–267. doi: 10.1016/0092-8674(95)90167-1. [DOI] [PubMed] [Google Scholar]
- Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- Morris SA, Schröder S, Plessmann U, Weber K, Ungewickell E. Clathrin assembly protein AP180: primary structure, domain organization and identification of a clathrin binding site. EMBO J. 1993;12:667–675. doi: 10.1002/j.1460-2075.1993.tb05700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JE, Pleasure IT, Puszkin S, Prasad K, Keen JH. Clathrin assembly protein AP-3. The identity of the 155K protein, AP 180, and NP185 and demonstration of a clathrin binding domain. J Biol Chem. 1991;266:4401–4408. [PubMed] [Google Scholar]
- Murzin AG. Structural principles for the propeller assembly of beta-sheets: the preference for seven-fold symmetry. Proteins. 1992;14:191–201. doi: 10.1002/prot.340140206. [DOI] [PubMed] [Google Scholar]
- Näthke IS, Heuser J, Lupas A, Stock J, Turck CW, Brodsky FM. Folding and trimerization of clathrin subunits at the triskelion hub. Cell. 1992;68:899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. Maximum likelihood refinement of heavy atom parameters. In: Wolf W, Evans PR, Leslie AGW, editors. Isomorphous Replacement and Anomalous Scattering. Warrington, UK: Science and Engineering Council/Daresbury Laboratory; 1991. pp. 80–86. [Google Scholar]
- Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs N, Bailey S, editors. Data Collection and Processing. Warrington, UK: Science and Engineering Research Council/Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- Pearse BM. Clathrin: a unique protein associated with intracellular transfer of membrane by coated vesicles. Proc Natl Acad Sci USA. 1976;73:1255–1259. doi: 10.1073/pnas.73.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BMF. Receptors compete for adaptors found in plasma membrane coated pits. EMBO J. 1988;7:3331–3336. doi: 10.1002/j.1460-2075.1988.tb03204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BM, Robinson MS. Purification and properties of 100-kd proteins from coated vesicles and their reconstitution with clathrin. EMBO J. 1984;3:1951–1957. doi: 10.1002/j.1460-2075.1984.tb02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranen J, Rikkonen M, Hyvonen M, Kaariainen L. T7 vectors with modified T7 lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Pippig S, Andexinger S, Daniel K, Puzicha M, Caron MG, Lefkowitz RJ, Lohse MJ. Overexpression of beta-arrestin and beta-adrenergic receptor kinase augment desensitization of beta 2-adrenergic receptors. J Biol Chem. 1993;268:3201–3208. [PubMed] [Google Scholar]
- Ramjaun AR, McPherson PS. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J Neurochem. 1998;70:2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- Rapoport I, Miyazaki M, Boll W, Duckworth B, Cantley LC, Shoelson S, Kirchhausen T. Regulatory interactions in the recognition of endocytic sorting signals by AP-2 complexes. EMBO J. 1997;9:2240–2250. doi: 10.1093/emboj/16.9.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport I, Chen YC, Cupers P, Shoelson SE, Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- Robinson MS. The role of clathrin, adaptors and dynamin in endocytosis. Curr Opin Cell Biol. 1994;6:538–544. doi: 10.1016/0955-0674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Saxena K, Gaitatzes C, Walsh MT, Eck M, Neer EJ, Smith TF. Analysis of the physical properties and molecular modeling of Sec13—a WD repeat protein involved in vesicular traffic. Biochemistry. 1996;35:15215–15221. doi: 10.1021/bi961616x. [DOI] [PubMed] [Google Scholar]
- Scarmato P, Kirchhausen T. Analysis of clathrin light chain-heavy chain interactions using truncated mutants of rat liver light chain LCB3. J Biol Chem. 1990;265:3661–3668. [PubMed] [Google Scholar]
- Schmid SL, Matsumoto AK, Rothman JE. A domain of clathrin that forms coats. Proc Natl Acad Sci USA. 1982;79:91–95. doi: 10.1073/pnas.79.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih W, Gallusser A, Kirchhausen T. A clathrin-binding site in the hinge of the β-2 chain of mammalian AP-2 complexes. J Biol Chem. 1995;270:31083–31090. doi: 10.1074/jbc.270.52.31083. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Grigorieff N, Pearse BM. Clathrin coats at 21 Å resolution: a cellular assembly designed to recycle multiple membrane receptors. EMBO J. 1998;17:4943–4953. doi: 10.1093/emboj/17.17.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein beta gamma dimer at 2.1 Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Carpenter G. Interaction of activated EGF receptors with coated pit adaptins. Science. 1993;261:612–615. doi: 10.1126/science.8342026. [DOI] [PubMed] [Google Scholar]
- Sosa MA, Schmidt B, von Figura K, Hille-Rehfeld A. In vitro binding of plasma membrane-coated vesicle adaptors to the cytoplasmic domain of lysosomal acid phosphatase. J Biol Chem. 1993;268:12537–12543. [PubMed] [Google Scholar]
- Springer TA. Extracellular β-propeller module predicted in lipoprotein scavenger receptors, tyrosine kinases, epidermal growth gactor precursor and extracellular matrix components. J Mol Biol. 1998 doi: 10.1006/jmbi.1998.2115. in press. [DOI] [PubMed] [Google Scholar]
- Trowbridge IS, Collawn JF, Hopkins CR. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- Ungewickell E. Biochemical and immunological studies on clathrin light chains and their binding sites on clathrin triskelions. EMBO J. 1983;8:1401–1408. doi: 10.1002/j.1460-2075.1983.tb01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Branton D. Assembly units of clathrin coats. Nature. 1981;289:420–422. doi: 10.1038/289420a0. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Unanue ER, Branton D. Functional and structural studies on clathrin triskelions and baskets. Cold Spring Harbor Symp Quant Biol. 1982;46(2):723–731. doi: 10.1101/sqb.1982.046.01.069. [DOI] [PubMed] [Google Scholar]
- Vigers GP, Crowther RA, Pearse BM. Three-dimensional structure of clathrin cages in ice. EMBO J. 1986;5:529–534. doi: 10.1002/j.1460-2075.1986.tb04242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Wilde A, Brodsky FM. In vivo phosphorylation of adaptors regulates their interaction with clathrin. J Cell Biol. 1996;135:635–645. doi: 10.1083/jcb.135.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler FK, Stanley KK. Clathrin heavy chain, light chain interactions. EMBO J. 1983;2:1393–1400. doi: 10.1002/j.1460-2075.1983.tb01597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba S, Keen JH. Assembly polypeptides from coated vesicles mediate reassembly of unique clathrin coats. J Cell Biol. 1983;97:1339–1347. doi: 10.1083/jcb.97.5.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.