Abstract
Here we examined the impact of a commonly employed method used to measure nitrogen fixation, the acetylene reduction assay (ARA), on a marine sediment community. Historically, the ARA technique has been broadly employed for its ease of use, in spite of numerous known artifacts. To gauge the severity of these effects in a natural environment, we employed high-throughput 16S rRNA gene sequencing to detect differences in acetylene-treated sediments vs. non-treated control sediments after a 7 h incubation. Within this short time period, significant differences were seen across all activity of microbes identified in the sediment, implying that the changes induced by acetylene occur quickly. The results have important implications for our understanding of marine nitrogen budgets. Moreover, because the ARA technique has been widely used in terrestrial and freshwater habitats, these results may be applicable to other ecosystems.
Keywords: heterotrophic nitrogen fixation, acetylene reduction assay, sediments, sulfate-reducing bacteria, high throughput sequencing
Introduction
Every sampling effort and each experimental design impacts the environment and the processes we wish to measure. Quantifying the nature and extent of that effect is a necessary step in our broader understanding of microbial processes and for the interpretation of the data we collect. High-throughput 16S rRNA gene-based bacterial community surveys provide a powerful mechanism for us to examine the impact of commonly employed geochemical methods on the microbial population.
Of particular interest to many aquatic biogeochemists is the microbially-mediated process of nitrogen fixation (N-fixation), which converts inert dinitrogen (N2) gas to biologically reactive nitrogen (N). While Earth’s atmosphere is comprised primarily of N2 gas, most organisms cannot tap into this reservoir. Thus, N-fixation provides critical links between biological organisms, this unreactive N pool, and N concentrations in the environment. Indeed, the ability to fix N2 provides a significant advantage to organisms with this capability. Once N is fixed, various processes transform reactive N until it is returned to the atmosphere through denitrification. Because N is an essential and often limiting nutrient for primary productivity, its availability, at least in part, constrains global primary productivity (Tyrrell, 1999; Elser et al., 2007).
Direct measurements of N-fixation rates from decreases in N2 concentration are difficult, as they require detection of small changes in a large reservoir of N2 gas. Equally challenging is the measurement of in situ total N increases to quantify N fixation, because this method also requires detection of a small amount of N against a much larger background of environmental organic nitrogen (Seitzinger and Garber, 1987). In recent years, our ability to measure such small changes has increased substantially, but they are time consuming and expensive. As such, for the last five decades we have used the methodologically more simple acetylene reduction assay (ARA) to measure N-fixation (Dilworth, 1966; Hardy et al., 1968; Stewart et al., 1968). The ARA relies on the flexibility of nitrogenase, the enzyme responsible for N2 reduction, as it can reduce acetylene (C2H2) to ethylene (C2H4), which can then be directly quantified. Theoretically, every three moles of ethylene produced corresponds to the reduction of 1 mole of N2 to ammonia.
However, the ARA is known to have numerous experimental artifacts. Some of these are newly discovered and related to the addition of acetylene and subsequent incubation of the sample (Mohr et al., 2010; Wilson et al., 2012). Other factors compromising this technique have been known for much longer and are related to shifts in the activity and ability of the microbial community to process nutrients and carbon in the presence of acetylene. For example, acetylene irreversibly inhibits methanogenesis in cultures and sediments (Oremland and Taylor, 1975). Acetylene also reversibly blocks aerobic nitrification (Hynes and Knowles, 1982) and nitrous oxide reduction in anaerobic denitrifiers (Balderston et al., 1976). Almost thirty years ago, acetylene was shown to partially or completely inhibit carbon dioxide production and growth of two sulfate reducing bacteria, Desulfovibrio desulfuricans and Desulfovibrio gigas although another species, Desulfotomaculum ruminis, was not effected at all (Payne and Grant, 1982). This is particularly relevant for marine ecosystems where sulfur- and sulfate-reducing bacteria are often significant contributors to the N-fixing community (Nielsen et al., 2001; Bertics et al., 2013; Brown and Jenkins, 2014).
To examine the broader applicability of the foundational efforts of Payne and Grant (1982), we used sediments from a temperate New England estuary to investigate the influence of the ARA on the total active microbial community of marine sediment. We directly measured net N2 fluxes and then measured N-fixation using ARA. Following the ARA, we conducted a 16S rRNA bacterial gene community survey to examine the change in active community activity after exposure to acetylene. The results are provided in the context of how the ARA technique alters processes within the community it is intended to unobtrusively query.
Materials and Methods
Study Site and Sample Collection
Samples were collected in July 2011 from a station in Narragansett Bay, RI, USA (41°35.3′, 071°22.3′) where the water column depth was 8 m. The in situ temperature was 17°C and the salinity was 31.3 psu. Intact triplicate sediment cores were hand collected by SCUBA divers with the vertical architecture of the sediment maintained during core collection and handling (Fulweiler and Heiss, 2014). Capped cores were transported with in situ water headspace in coolers filled with site water to an environmental chamber set to ambient temperature at the Graduate School of Oceanography at the University of Rhode Island. The sediment cores were placed in a 17°C water bath in the dark with air gently bubbling through the surface water until the incubation began (∼8 h).
Net N2 Measurements
We first completed an N2/Ar incubation to measure the net flux of N2 across the sediment-water interface. These methods have been documented previously (Fulweiler and Nixon, 2009, 2012). Briefly, before the incubation we carefully replaced the overlying water with filtered (0.2-μm) site water, the cores were sealed with a gas tight lid (no air headspace), gently stirred (∼40 rpm), and replicate samples taken for N2/Ar analysis at five points over the course of an incubation. Each sample was preserved with 20 μl of saturated ZnCl solution. All incubations took place in the dark and lasted ∼ 11 h until we observed an overlying water oxygen concentration drop of at least 2 mg L-1 (62.5 μM), but at no point did the cores approach hypoxia. Dissolved N2 and Ar gas concentrations were analyzed on a quadrupole membrane inlet mass spectrometer with a precision of ± 0.03% (Sisler and ZoBell, 1951; Heip, 1995; Kana et al., 1998). N2 change for each of the triplicate cores was determined from a five-point linear regression. Rates were then prorated for the volume of water overlying the core and the sediment area of the core. The rates calculated from the N2/Ar technique actually present a measure of net N2 flux (gross denitrification – gross N fixation).
ARA Measurements
After net N2 measurements were made, we quantified the reduction of acetylene (C2H2) to ethylene (C2H4) as a proxy for N-fixation (Hardy et al., 1968). Duplicate sub-cores (∼2.5 cm i.d.) were collected from each core and sectioned into 0–2m and 2–4 cm increments, halved, and placed in 40 mL glass vials. For each depth, each core duplicate sample halves were separated into the following treatments: acetylene plus filtered seawater, acetylene plus 40 mM sodium molybdate (Na2MoO4), seawater alone, and 40 mM Na2MoO4 alone. Molybdate is an established inhibitor of sulfate-reducing bacteria and used as an indicator of N-fixation by sulfate reducing bacteria (Postgate, 1949; Oremland and Taylor, 1978). Samples were placed in sealed vials, the air headspace was evacuated, and replaced with argon gas after which 10 mL of C2H2 was injected into the vials (Hamilton et al., 2011; Cole and McGlathery, 2012). The ARA incubation lasted 7 h. Gas samples were run on a Shimadzu gas chromatograph equipped with a flame ionization detector using a Porapak N column, mesh size 80/100 at the Graduate School of Oceanography at the University of Rhode Island. The halved sub-core samples were then extrapolated to area units and rates of sediment acetylene reduction were converted to N-fixed using the common 3:1 molar conversion (Seitzinger and Garber, 1987). All samples were then frozen at -80°C and shipped overnight to the University of Tennessee for molecular analysis.
Molecular Analysis
RNA was extracted from all 24 sediment samples using the MoBio PowerSoilTM RNA isolation kit (MoBio, Carlsbad, CA, USA) according to manufacturer’s protocols and checked for contaminating DNA by PCR (below). cDNA was produced from extracted RNA using 16S rRNA gene-specific primers (9F and 1522R, E. coli numbering) using the Invitrogen Superscript III First-Strand Synthesis SuperMix kit according the manufacturer’s protocols. We PCR amplified bacterial 16S rRNA using primers targeting bases 338–926 of the 16S rRNA gene (E. coli numbering), which contains the V3–V5 region, with the following PCR protocol: 95°C for 5 min, followed by 30 rounds of (95°C for 30 s, 55°C for 30 s, 72°C for 30 s) and then a final extension step at 72°C for 10 min. Product amplification was verified on a 1% agarose gel stained with ethidium bromide and viewed on a UV transilluminator. Individual samples were processed to remove unincorporated primers and nucleotides using the Qiaquick PCR cleanup kit (Qiagen, Valencia, CA, USA). Amplicon concentrations were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Individual sample amplicons were barcoded (six additional PCR cycles : 95°C for 30 s, 55°C for 30 s, 72°C for 30 s) with primers that contained a unique 8-bp barcode attached to the 454 fusion primers (LeCleir et al., 2014; Wilhelm et al., 2014). The barcoding primers were designed for unidirectional sequencing on a 454 GSFLX sequencer (454 Life Sciences, Branford, CT, USA). This strategy required the use of the Lib-L kit (see Roche application brief 001-2009). We opted for unidirectional sequencing as our PCR product was larger than the average read length of the available 454 Titanium sequencing chemistry. This approach ensured sequences would overlap for the longest length possible. All barcoding reactions were prepared to have 0.5 ng/μl of amplicon DNA per reaction. After the barcoding reaction, we again verified our amplicons on an agarose gel before pooling all barcoded amplicons. Barcoded amplicons were processed to remove unincorporated primers and nucleotides using a single Qiagen Qiaquick column. Sequencing was completed at the University of Tennessee/Oak Ridge National Laboratory Joint institute of Biological Sciences. Sequence information has been deposited in the NCBI short-read archive in bioproject PRJNA271790.
We used the Mothur software package (version 1.27.0; Schloss et al., 2009) to process our sequences for sufficient length and quality. We processed our sequences similar to the Schloss SOP1 with some modifications of the shhh.flows command; we changed the number of flows value in the shhh.flows command to 360–720 from 450 (Quince et al., 2009). Mothur was also used to cluster sequences into operational taxonomic units (OTUs) and for phylogenetic classification. A 0.03 cutoff (97% identity) was chosen for OTU determination. To visualize the influence of acetylene on OTU abundance, proportional abundances for each OTU were compared for samples collected in the presence vs. absence of acetylene. Outputs were visualized after clustering (using the Euclidean distance method) with Cluster 3.02 using Java TreeView (Saldanha, 2004).
The Primer-E software package (Version 6.0; Clarke and Gorley, 2006) was used to more deeply interrogate relationships between OTUs across samples and to also look for correlations between OTU presence/abundance and environmental parameters. The “shared” file (a matrix file containing OTU abundances for each sample) created by Mothur was imported directly into the Primer-E software package. All OTUs with an abundance (sum of counts across all libraries) >25 sequences were used for further analyses. OTUs meeting these criteria were standardized to the total number of sequences per barcoded library (proportional abundances). These standardized abundances were then square-root transformed to partially deemphasize more highly abundant OTUs. A Bray-Curtis similarity matrix was constructed and used to perform non-metric multidimensional scaling analysis (NMDS) for visualization of community structure relationships between the different samples. The ANOSIM and PERMANOVA programs within Primer-E were used to document the statistical significance of our findings. Both tests were performed on our Bray-Curtis similarity matrix of OTU abundances. A principal components analysis (PCA) was performed on the square-root transformed data to visualize community structure differences and identify OTUs driving the differences in the bacterial communities. Phylogenetic identities of OTUs were determined using the Ribosomal Database Project (RDP) classification within Mothur. We also employed SIMPER to define discriminating species between treatment and control (Clarke, 1993) as well as the software package LEfSe (Segata et al., 2011) to statistically compare differentially abundant OTUs.
Results
Net N2 Fluxes and N-Fixation Estimates
The net N2 flux across the sediment water-interface was quantified to determine if the sediments were net denitrifying (positive N2-flux dominated) or net nitrogen fixing (negative N2-flux dominated). Net N2 fluxes across the sediment-water interface for the individuals cores ranged from -10 to 28.6 μmol N2-N m-2 h-1, suggesting that both N-fixation and denitrification were occurring (Table 1). N-fixation rates measured with the ARA, similarly, revealed high variability among the cores and, in some cases rates differed by depth (Table 1). After the addition of molybdate, N-fixation rates were reduced on average by almost 70%, indicating that sulfate reducers were responsible for a significant component of the observed N-fixation (Gandy and Yoch, 1988).
Table 1.
Geochemical measurements of the sediments used in this study.
| Core ID | Net N2–N flux, μmol m-2 h-1 | Depth, cm | Acetylene reduction assay (ARA) | Treated with Molybdate (M) or Seawater (S) | ARA N2 Flux, μmol m-2 h-1 | % change between SW and Mo |
|---|---|---|---|---|---|---|
| Core 1 | 7.5 | 0–2 | Y | S | 22.7 | -77 |
| 0–2 | Y | M | 5.1 | |||
| 2–4 | Y | S | 19.6 | -55 | ||
| 2–4 | Y | M | 8.9 | |||
| 0–2 | N | S | n.a. | |||
| 0–2 | N | M | n.a. | |||
| 2–4 | N | S | n.a. | |||
| 2–4 | N | M | n.a. | |||
| Core 2 | 28.6 | 0–2 | Y | S | 27.1 | -57 |
| 0–2 | Y | M | 11.6 | |||
| 2–4 | Y | S | 30.5 | -67 | ||
| 2–4 | Y | M | 10.0 | |||
| 0–2 | N | S | n.a. | |||
| 0–2 | N | M | n.a. | |||
| 2–4 | N | S | n.a. | |||
| 2–4 | N | M | n.a. | |||
| Core 3 | -10.3 | 0–2 | Y | S | 44.3 | -60 |
| 0–2 | Y | M | 17.6 | |||
| 2–4 | Y | S | 62.4 | -100 | ||
| 2–4 | Y | M | 0.0 | |||
| 0–2 | N | S | n.a. | |||
| 0–2 | N | M | n.a. | |||
| 2–4 | N | S | n.a. | |||
| 2–4 | N | M | n.a. |
Whole core net N2–N fluxes were measured by the N2/Ar technique for three cores at the onset of this experiment. These cores were then sub-cored in duplicate, separated by depth (0–2 cm or 2–4 cm), and divided into two groups: a control group not measured by the acetylene reduction assay (ARA) and a treatment group measured with the ARA. In each of these groups, the sub-cores were further divided into two groups: molybdate and seawater treated or seawater treated only. In the sub-cores measured by the ARA we used the commonly employed 3:1 ratio to convert ethylene production to N fixation rate. When geochemical measurements were finished all 24 samples were processed for molecular analysis. (n.a. = not applicable).
Activity of the Microbial Community
After screening sequences to ensure sufficient quality and length as well as removal of chimeric sequences, 115,319 16S rRNA gene sequences remained across the 24 different samples. Total sequence abundances (per library) ranged from a minimum of 1,379 sequences to a maximum of 10,569 sequences, with an average of 4,805 sequences per library. After clustering, 9,979 OTUs at 0.03 cut-off (97% similarity) remained.
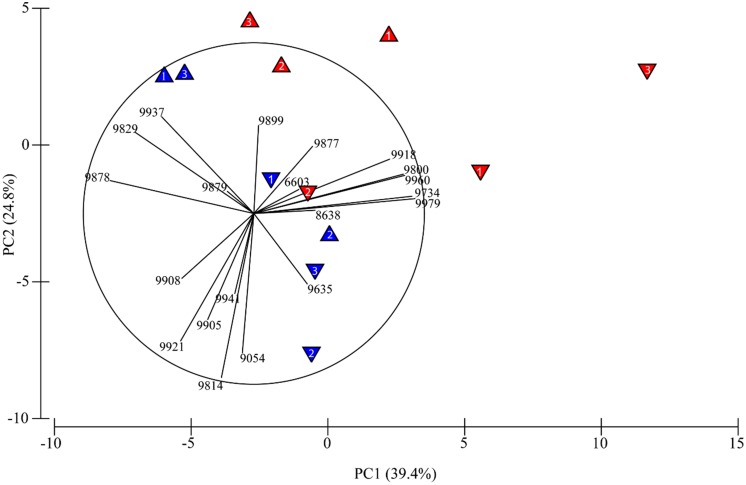
To determine how acetylene influenced total community structure, we compared total community 16S rRNA expression by principle component analyses (Figure 1; Table 2). Importantly, we performed PCA analysis only for those samples treated with acetylene and the control groups. Samples treated with molybdate (with or without acetylene) were excluded, as we found no impact of molybdate on the active microbial community (Table 3). The PCA highlights two clear patterns. First, there is distinct separation between those samples treated with acetylene and the control along principal component one (PC1). And secondly, depth appears to also be a key driver separating samples along principal component two (PC2). Similarly, the ANOSIM and PERMANOVA analyses show the significant impact of acetylene and depth with the latter being the most significant driver (Table 3).
FIGURE 1.
Results of principal components analysis (PCA) of operational taxonomic units (OTUs) and how they separate between acetylene treated (red triangles) and untreated (blue triangles) and by depth (0–2 cm: upward facing triangles and 2–4 cm: downward facing triangles). The number in each triangle indicates which core the sample came from. No molybdate treated samples are included here, as molybdate did not significantly alter the active sediment microbial community. Principal component (PC1 and PC2) together, account for 64.2% of the variance in these data.
Table 2.
Dominant operational taxonomic units (OTUs) that drive the separation of the acetylene treated sediments versus the control sediments.
| OTU | OTU count | Phylum | Genus (best hit) |
|---|---|---|---|
| Over-represented in acetylene treatment | |||
| 9877∗ | 3738 | Proteobacteria | Thioprofundum sp. |
| 9979∗†‡ | 3313 | Proteobacteria | Thiohalomonas sp. |
| 9899∗† | 1660 | Bacteroidetes | Unclassified |
| 9800∗†‡ | 1489 | Proteobacteria | Pelobacter sp. |
| 9635∗† | 1044 | Bacteroidetes | Prolixibacter sp. |
| 9960∗†‡ | 1033 | Proteobacteria | Desulfobulbus sp. |
| 9734∗† | 1005 | Proteobacteria | Thiohalomonas sp. |
| 9918∗†‡ | 912 | Bacteroidetes | Arcicella sp. |
| 9879∗ | 497 | Spirochaetes | Spirochaeta sp. |
| 8638∗†‡ | 128 | Proteobacteria | Desulfobacula sp. |
| 9780†‡ | 165 | Proteobacteria | Pelobacter sp. |
| 9819†‡ | 98 | Proteobacteria | Thioprofundum sp. |
| Over-represented in control treatment | |||
| 9878∗†‡ | 4110 | Proteobacteria | Methylomicrobium sp. |
| 9814∗† | 3642 | Proteobacteria | Desulfosalsimonas sp. |
| 9829∗† | 2373 | Cyanobacteria | Bacillariophyta sp. |
| 9905∗‡ | 2328 | Bacteroidetes | Meniscus sp. |
| 9921∗†‡ | 1912 | Bacteroidetes | Prolixibacter sp. |
| 9941∗† | 1436 | Bacteroidetes | Fulvivirga sp. |
| 9937∗† | 1347 | Bacteroidetes | Roseivirga sp. |
| 9054∗† | 607 | Bacteroidetes | Owenweeksia sp. |
| 9908∗†‡ | 560 | Proteobacteria | Desulfocapsa sp. |
| 6603∗† | 508 | Proteobacteria | Neptuniibacter sp. |
| 9633†‡ | 516 | Bacteroidetes | Persicobacter sp. |
| 9608†‡ | 265 | Proteobacteria | Sulfurovum sp. |
| 5645†‡ | 160 | Proteobacteria | Sedimenticola sp. |
| 7286‡ | 30 | Lentisphaerae | Lentisphaerae sp. |
OTUs that were in the top 20 of significance in the in the principal components analysis (PCA; ∗) and LEfSe analyses (‡), along with those deemed significant by SIMPER(†) analyses are noted for discriminating species.
Table 3.
Results from the ANOSIM and PERMANOVA analyses describing the impact acetylene, depth, or molybdate has on the active sediment microbial community.
| Treatment | ANOSIM |
PERMANOVA |
||
|---|---|---|---|---|
| Sample statistic (R) | Significance level | Pseudo-F | p-value | |
| ARA | 0.15 | 1.1 | 2.461 | 0.003 |
| Depth | 0.347 | 0.1 | 3.739 | 0.001 |
| Molybdate | -0.016 | 57.8 | 0.974 | 0.495 |
Together, the first and second components of the PCA explained just over 64% of the variation in the samples (Figure 1). The treatments separated primarily along the first component which showed a positive relationship with OTU 9979 (Thiohalomonas) and a negative relationship with OTU 9878 (Methylomicrobium). The samples also appear to separate by depth along PC2 which is dominated by a negative relationship with OTU 9814 (Desulfosalsimonas) and a positive relationship with OTU 9829 (Bacillariophyta). In some cases, treatment with acetylene led to increased representation of a number of OTUs within the active community (Table 2). Concomitantly, we also observed a number of OTUs that appear to be underrepresented in the acetylene treated versus control groups suggesting that the acetylene may inhibit or alter their activity (Table 2). Overall, nine OTUs were identified by all three approaches (PCA, SIMPER, and LEfSe) as significant drivers of difference in the communities while other combinations of approaches (different by SIMPER and LEfSe, 4; different by SIMPER and PCA 9; different by LEfSe and PCA, 1) revealed another 14 OTUs of interest.
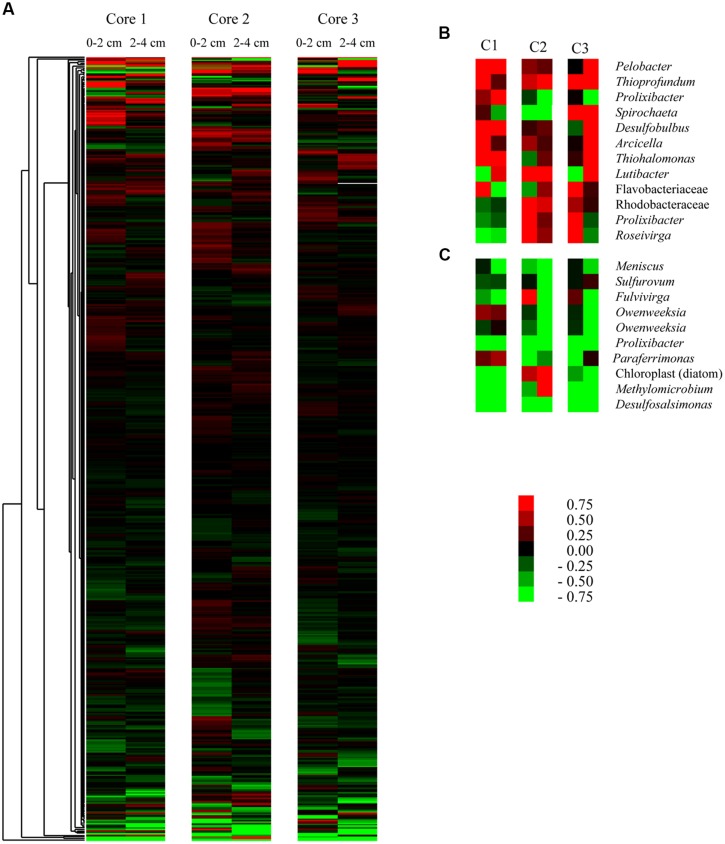
To visualize which OTUs were driving observed relationships, we clustered OTU expression based on differences between control and acetylene-treated replicates (Figure 2). We then identified the dominant OTUs that were over-represented under acetylene (Figure 2B) and those that were under-represented (Figure 2C). Within these groups, changes were subtle: the most upregulated groups (OTUs most closely identified as Thiohalamonas, Thioprofundum, and Pelobacter) demonstrated increases in 16S rRNA representation of up to 9.5%. In parallel, the most down-regulated OTUs in any sample showed decreases in representation of ∼5.0% (Paraferrimonas and Desulfosalimonas).
FIGURE 2.
Influence of acetylene on the distribution of individual (OTUs) from the sediment cores used in this study. Heat map (A) of all OTUs with greater than 25 representative signatures in the complete data set. Data within libraries were normalized to percent of total reads and then expressed as differences between treatment and control [values above zero (red) signify over-representation in the treatment where values below zero (green) represent underrepresentation in the treatment]. Data are shown for three replicate cores and for 0–2 cm and 2–4 cm depth fractions and for seawater treated only (no molybdate). Insets show the 10 most over-represented (B) and underrepresented (C) groups with cores (C1–C3) and depths (0–2 or 2–4 cm depth) presented in the same order as in (A).
Discussion
Geochemistry of the Samples
We directly quantified the flux of N2 gas across the sediment-water interface using the N2/Ar technique. We chose this technique because it is the least intrusive and yet direct measurement of N2 flux. Unfortunately, it only provides a net measurement and thus only reports the balance between sediment N-fixation and its opposite process, denitrification (the microbial conversion of nitrate to N2). At the time of this experiment, the sediments exhibited net positive N2 fluxes (or net denitrification) as well as net negative N2 fluxes (or N-fixation; Table 1). It is not unusual for sediments at this site to show such variability. In fact, the sediments at this site routinely alternate between net denitrification and net N-fixation and the month following these measurements the sediments were dominated by net N-fixation (Fulweiler and Heiss, 2014). Additionally, we did observe the conversion of acetylene to ethylene for each core and at every depth. The conversion of acetylene to ethylene was considerably reduced when exposed to molybdate suggesting sulfate-reducing bacteria are driving the N-fixation in these sediments. These results are consistent with previous research at this site where N-fixation was observed in core incubations and in large experimental mesocosm studies and, in all cases, the active N-fixing community was dominated by anaerobic sulfate reducers and sulfur/iron reducers (Brown, 2013; Fulweiler et al., 2013; Brown and Jenkins, 2014).
ARA Impacts on Sulfate Reducing Bacteria and N-Fixation
Several dominant OTUs related to sulfur (S) cycling were observed in this study (Table 2). Specifically, we observed OTUs closely related to both sulfur and sulfate reducers (e.g., Desulfosalsimonas, Desulfobulbus, etc.) and those related to sulfur oxidizers (e.g., Thioprofundum and Thiohalomonas). Sulfur and sulfate reducing bacteria, specifically, Desulfovibrio spp. and Desulfobacter sp. have long been known to fix N in culture (Sisler and ZoBell, 1951; Widdel, 1987). A rich literature in sea grass beds (Welsh et al., 1996; McGlathery et al., 1998) and salt marshes (Gandy and Yoch, 1988; Piceno and Lovell, 2000) have credited sulfate reducers as the primary N-fixers. More recently, in Baltic Sea sediments, acetylene reduction, and sulfate reduction rates showed similar seasonal patterns and molecular analysis revealed the presence of two sulfate reducing bacteria, Desulfovibrio vulgaris, and Desulfonema limicola (Bertics and Ziebis, 2010). As the work that motivated this research highlighted, over three decades ago Payne and Grant (1982) reported that acetylene partially or completely inhibited the growth of two sulfate reducing bacteria, Desulfovibrio desulfuricans, and Desulfovibrio gigas while a third species, Desulfotomaculum ruminis, was completely unaffected. In this study, we find a similar species or closely related groups of bacteria responding in different ways to the acetylene (Figure 2). The different responses may have important consequences for our understanding of heterotrophic N cycling in marine sediments. Sulfur- and sulfate-reducers are important N-fixers in a range of marine environments, and they experience up or down regulation in the presence of acetylene, one of the most commonly used techniques to measure N-fixation. Thus, we have likely been under- or over-estimating N-fixation rates in these environments. Complicating the picture further is that response appears to be species specific, thus we cannot apply a correction across measurements leaving us with a methodologically introduced unknown amount of error.
These findings are significant because ARA has been and is still widely used in both terrestrial and aquatic ecosystem studies of N-fixation. This may be particularly important for marine systems, where debate exists concerning whether the modern N budget is balanced. Although some estimates have suggested that the oceanic fixed N budget is balanced, they are plagued with gross uncertainties (Gruber, 2004). More recent estimates indicate that the budget is unbalanced with a substantial N deficit, although these budgets also contain considerable uncertainty (Codispoti et al., 1992; Brandes and Devol, 2002; Deutsch et al., 2007). This deficit is driven mainly by larger denitrification rates, as denitrification is thought to be a dominant process in the N cycle, while sediment N fixation is traditionally considered to be inconsequential (Howarth et al., 1988). Thus, to balance the modern marine N budget, rates of denitrification need to be decreased and/or rates of N-fixation increased. Several recent lines of evidence argue for the latter as it appears N-fixation may be more important than originally anticipated (open ocean: Zehr et al., 2001; Davis and McGillicuddy, 2006; shallow coastal systems: Gardner et al., 2006; Fulweiler et al., 2007; Mortazavi et al., 2012). Importantly, these more recent studies have used other techniques besides ARA to measure rates of N-fixation (e.g., N2/Ar technique, nifH expression, etc.). We propose one important driver delaying our thorough understanding of the role of N-fixation in marine environments is the widespread historic and current use of ARA. Furthermore, because ARA is also used in terrestrial and freshwater ecosystems rates of N-fixation rates may also be inaccurate by varying degrees across these environments.
ARA Impacts on the Active Microbial Community
We expected to see significant changes in the active microbial community and this was observed in our 16S rRNA bacterial community survey data. We found a significant difference in sediments treated with acetylene versus those in the control samples (Table 3). However, the impact of acetylene was not uniform across phylum or genera (Table 2). Instead, we observed the up or down regulation of different OTUs associated with Proteobacteria, Spirochaetes, and Bacteroidetes. OTUs dominant under the acetylene treatment most closely identified with a range of bacteria, including Pelobacter and Arcicella. In contrast, other OTUs were more active in the control samples such as Methylomicrobium, Roseiviriga, and Bacillariophyta.
The data also highlight the heterogenic nature of the sediment microbial community and provide hints about how this heterogeneity alters the community response to disturbance. In this experiment, sediment cores were hand collected by divers within close distance to each other. Despite the careful nature of the collection and the proximity of the sediments, sediment core 2 appears dramatically different to sediments cores 1 and 3 (Table 1; Figure 2). In core 2 we measured the highest rate of net denitrification (29 μmol N2-N m-2 h-1) and a different microbial response to acetylene as compared to the other cores. The difference in active community response is seen most clearly in the PCA where the 2–4 cm depth sample in core 2 did not change across PC 1. In addition, depth was the most significant driver of differences in the microbial community (Table 3). This makes sense as the sediment microbial community was exposed to acetylene for only 7 h and presumably they had much more time to acclimate to there location within the sediments.
Our observations are not surprising as the ARA technique has long been known to inhibit various organisms (Oremland and Taylor, 1975; Schink, 1985; Knowles, 1990). In fact, it is this inhibitory power that has been harnessed to measure denitrification rates in the acetylene block technique (Joye et al., 1996; Welsh et al., 2001) and the importance of methane production and consumption processes (Prior and Dalton, 1985). Thus, as expected we observed repression in the activity of an OTU identifying as Methylomicrobium, a strictly aerobic group of methanotrophs (Hanson and Hanson, 1996; Vuilleumier et al., 2012). However, to our knowledge this is the first study to show total active community changes driven by acetylene using 16S rRNA data. These data also allowed us to observe a rapid change in active community metabolism as incubations only lasted 7 h. Overall, our observations concerning the rapid effects of acetylene on microbial community activity give pause concerning the robustness of the technique, adding yet another cautionary note when interpreting ARA data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This material is based on work funded through a development grant (2011-R/RC-124-PD) to RWF from the MIT Sea Grant College Program (NOAA). Additionally, this work was supported through grants from the Rhode Island Sea Grant College Program (NOAA) and NSF (OCE 0926859) to RWF, and OCE-1030518 and OCE-1061352 to SWW. We thank the Sloan Foundation for additional support to RWF. We thank Sarah Foster, Lindsey Fields, Matt Horn, Jason Krumholz, Connor McManus, and Jeff Mercer for field help. We thank Luke Cole and Melanie Hayn for method development help long ago. Additionally, we thank Leanna Heffner and Dennis Graham for GC instruction and troubleshooting and David Smith, Art Spivack, and Steve D’Hondt for allowing us to use the GC at the Graduate School of Oceanography at the University of Rhode Island.
Footnotes
References
- Balderston W. L., Sherr B., Payne W. (1976). Blockage by acetylene of nitrous oxide reduction in Pseudomonas perfectomarinus. Appl. Environ. Microbiol. 31 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertics V. J., Loscher C. R., Salonen I., Dale A. W., Gier J., Schmitz R. A., et al. (2013). Occurrence of benthic microbial nitrogen fixation coupled to sulfate reduction in the seasonally hypoxic Eckernforde Bay, Baltic Sea. Biogeosciences 10 1243–1258 10.5194/bg-10-1243-2013 [DOI] [Google Scholar]
- Bertics V. J., Ziebis W. (2010). Bioturbation and the role of microniches for sulfate reduction in coastal marine sediments. Environ. Microbiol. 12 3022–3034 10.1111/j.1462-2920.2010.02279.x [DOI] [PubMed] [Google Scholar]
- Brandes J. A., Devol A. H. (2002). A global marine-fixed nitrogen isotopic budget: implications for Holocene nitrogen cycling. Global Biogeochem. Cycles 146 67-61–67-14. 10.1029/2001GB001856 [DOI] [Google Scholar]
- Brown S. M. (2013). Using Molecular Tools to Elucidate Controls on Microbes Driving the Nitrogen Cycle in Marine Sediments. Kingston, RI: University of Rhode Island. [Google Scholar]
- Brown S., Jenkins B. (2014). Profiling gene expression to distinguish the likely active diazotrophs from a sea of genetic potential in marine sediments. Environ. Microbiol. 16 3128–3142 10.1111/1462-2920.12403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18 17–143 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- Clarke K., Gorley R. (2006). PRIMER v6: User Manual/Tutorial. Plymouth: Plymouth Marine Laboratory. [Google Scholar]
- Codispoti L. A., Elkins J. W., Yoshinari T., Friederich G. E., Sakamoto C. M., Packard T. T. (1992). “On the nitrous-oxide flux from productive regions that contain low oxygen waters,” in Oceanography of the Indian Ocean ed. Desai B. N. (Rotterdam: A. A. Balkema; ) 271–284. [Google Scholar]
- Cole L. W., McGlathery K. J. (2012). Nitrogen fixation in restored eelgrass meadows. Mar. Ecol. Progr. Ser. 448 235–246 10.3354/Meps09512 [DOI] [Google Scholar]
- Davis C. S., McGillicuddy D. J. (2006). Transatlantic abundance of the N2-fixing colonial cyanobacterium Trichodesmium. Science 312 1517–1520 10.1126/science.1123570 [DOI] [PubMed] [Google Scholar]
- Deutsch C., Sarmiento J. L., Sigman D. M., Gruber N., Dunne J. P. (2007). Spatial coupling of nitrogen inputs and losses in the ocean. Nature 445 163–167 10.1038/nature05392 [DOI] [PubMed] [Google Scholar]
- Dilworth M. (1966). Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim. Biophys. Acta Gen. Subj. 127 285–294 10.1016/0304-4165(66)90383-7 [DOI] [PubMed] [Google Scholar]
- Elser J. J., Bracken M. E. S., Cleland E. E., Gruner D. S., Harpole W. S., Hillebrand H., et al. (2007). Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 10 1135–1142 10.1111/j.1461-0248.2007.01113.x [DOI] [PubMed] [Google Scholar]
- Fulweiler R. W., Brown S. M., Nixon S. W., Jenkins B. D. (2013). Evidence and a conceptual model for the co-occurrence of nitrogen fixation and denitrification in heterotrophic marine sediments. Mar. Ecol. Progr. Ser. 482 57–68 10.3354/Meps10240 [DOI] [Google Scholar]
- Fulweiler R., Heiss E. (2014). (Nearly) a decade of directly measured sediment N2 fluxes: what can Narragansett Bay tell us about the global ocean nitrogen budget. Oceanography 27 184–195 10.5670/oceanog.2014.22 [DOI] [Google Scholar]
- Fulweiler R. W., Nixon S. W. (2009). Responses of benthic-pelagic coupling to climate change in a temperate estuary. Hydrobiologia 629 147–156 10.1007/s10750-009-9766-0 [DOI] [Google Scholar]
- Fulweiler R. W., Nixon S. W. (2012). Net Sediment N2 fluxes in a southern new england estuary - variations in space and time. Biogeochemistry 111 111–124 10.1007/s10533-011-9660-5 [DOI] [Google Scholar]
- Fulweiler R. W., Nixon S. W., Buckley B. A., Granger S. L. (2007). Reversal of the net dinitrogen gas flux in coastal marine sediments. Nature 448 180–182 10.1038/Nature05963 [DOI] [PubMed] [Google Scholar]
- Gandy E. L., Yoch D. C. (1988). Relationship between nitrogen-fixing sulfate reducers and fermenters in salt-marsh sediments and roots of Spartina-Alterniflora. Appl. Environ. Microbiol. 54 2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner W. S., Mccarthy M. J., An S. M., Sobolev D., Sell K. S., Brock D. (2006). Nitrogen fixation and dissimilatory nitrate reduction to ammonium (DNRA) support nitrogen dynamics in Texas estuaries. Limnol. Oceanogr. 51 558–568 10.4319/lo.2006.51.1_part_2.0558 [DOI] [Google Scholar]
- Gruber N. (2004). “The dynamics of the marine nitrogen cycle and its influence on atmospheric CO2 variations,” in The Ocean Carbon Cycle and Climate eds Follows M., Oguz T. (Berlin: Springer; ) 97–148. [Google Scholar]
- Hamilton T. L., Lange R. K., Boyd E. S., Peters J. W. (2011). Biological nitrogen fixation in acidic high-temperature geothermal springs in Yellowstone National Park, Wyoming. Environ. Microbiol. 13 2204–2215 10.1111/j.1462-2920.2011.02475.x [DOI] [PubMed] [Google Scholar]
- Hanson R. S., Hanson T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R. W., Holsten R., Jackson E., Burns R. (1968). The acetylene-ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiol. 43 1185–1207 10.1104/pp.43.8.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heip C. (1995). Eutrophication and zoobenthos dynamics. Ophelia 41 113–136 10.1080/00785236.1995.10422040 [DOI] [Google Scholar]
- Howarth R. W., Marino R., Lane J., Cole J. J. (1988). Nitrogen-fixation in fresh-water, estuarine, and marine ecosystems.1. Rates and importance. Limnol. Oceanogr. 33 669–687 10.4319/lo.1988.33.4_part_2.0669 [DOI] [Google Scholar]
- Hynes R., Knowles R. (1982). Effect of acetylene on autotrophic and heterotrophic nitrification. Can. J. Microbiol. 28 334–340 10.1139/m82-049 [DOI] [Google Scholar]
- Joye S. B., Smith S. V., Hollibaugh J. T., Paerl H. W. (1996). Estimating denitrification rates in estuarine sediments: a comparison of stoichiometric and acetylene based methods. Biogeochemistry 33 197–215 10.1007/BF02181072 [DOI] [Google Scholar]
- Kana T. M., Sullivan M. B., Cornwell J. C., Groszkowski K. M. (1998). Denitrification in estuarine sediments determined by membrane inlet mass spectrometry. Limnol. Oceanogr. 43 334–339 10.4319/lo.1998.43.2.0334 [DOI] [Google Scholar]
- Knowles R. (1990). “Acetylene inhibition technique: development, advantages, and potential problems,” in Denitrification in Soil and Sediment eds Revsbech N. P., Sørensen J. (Berlin: Springer; ) 151–166. [Google Scholar]
- LeCleir G. R., Debruyn J. M., Maas E. W., Boyd P. W., Wilhelm S. W. (2014). Temporal changes in particle-associated microbial communities after interception by non-lethal sediment traps. FEMS Microbiol. Ecol. 87 153–163 10.1111/1574-6941.12213 [DOI] [PubMed] [Google Scholar]
- McGlathery K. J., Risgaard-Petersen N., Christensen P. B. (1998). Temporal and spatial variation in nitrogen fixation activity in the eelgrass Zostera marina rhizosphere. Mar. Ecol. Progr. Ser. 168 245–258 10.3354/Meps168245 [DOI] [Google Scholar]
- Mohr W., Grosskopf T., Wallace D. W. R., Laroche J. (2010). Methodological underestimation of oceanic nitrogen fixation rates. PLoS ONE 5:e12583 10.1371/journal.pone.0012583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi B., Riggs A. A., Caffrey J. M., Genet H., Phipps S. W. (2012). The contribution of benthic nutrient regeneration to primary production in a shallow eutrophic estuary, Weeks Bay, Alabama. Estuaries Coasts 35 862–877 10.1007/s12237-012-9478-y [DOI] [Google Scholar]
- Nielsen L. B., Finster K., Welsh D. T., Donelly A., Herbert R. A., De Wit R., et al. (2001). Sulphate reduction and nitrogen fixation rates associated with roots, rhizomes and sediments from Zostera noltii and Spartina maritima meadows. Environ. Microbiol. 3 63–71 10.1046/j.1462-2920.2001.00160.x [DOI] [PubMed] [Google Scholar]
- Oremland R. S., Taylor B. F. (1975). Inhibition of methanogenesis in marine sediments by acetylene and ethylene: validity of the acetylene reduction assay for anaerobic microcosms. Appl. Microbiol. 30 707–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland R. S., Taylor B. F. (1978). Sulfate reduction and methanogenesis in marine sediments. Geochim. Cosmochim. Acta 42 209–214 10.1016/0016-7037(78)90133-3 [DOI] [Google Scholar]
- Payne W. J., Grant M. A. (1982). Influence of acetylene on growth of sulfate-respiring bacteria. Appl. Environ. Microbiol. 43 727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piceno Y. M., Lovell C. R. (2000). Stability in natural bacterial communities: II. Plant resource allocation effects on rhizosphere diazotroph assemblage composition. Microbial. Ecol. 39 41–48 10.1007/s002489900191 [DOI] [PubMed] [Google Scholar]
- Postgate J. (1949). Competitive inhibition of sulphate reduction by selenate. Nature 164 670–671 10.1038/164670b0 [DOI] [Google Scholar]
- Prior S., Dalton H. (1985). Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 29 105–109 10.1111/j.1574-6968.1985.tb00843.x [DOI] [Google Scholar]
- Quince C., Lanzen A., Curtis T. P., Davenport R. J., Hall N., Head I. M., et al. (2009). Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6 639–627 10.1038/nmeth.1361 [DOI] [PubMed] [Google Scholar]
- Saldanha A. J. (2004). Java Treeview – extensible visualization of microarray data. Bioinformatics 20 3246–3248 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Schink B. (1985). Inhibition of methanogenesis by ethylene and other unsaturated hydrocarbons. FEMS Microbiol. Lett. 31 63–68 10.1111/j.1574-6968.1985.tb01132.x [DOI] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541 10.1128/aem.01541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12 R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitzinger S. P., Garber J. H. (1987). Nitrogen-fixation and N-15(2) calibration of the acetylene-reduction assay in coastal marine-sediments. Mar. Ecol. Progr. Ser. 37 65–73 10.3354/Meps037065 [DOI] [Google Scholar]
- Sisler F. D., ZoBell C. E. (1951). Nitrogen fixation by sulfate reducing bacteria indicated by Nitrogen/Argon ratios. Science 113 511 10.1126/science.113.2940.511 [DOI] [PubMed] [Google Scholar]
- Stewart W., Fitzgerald G., Burris R. (1968). Acetylene reduction by nitrogen-fixing blue-green algae. Archiv für Mikrobiol. 62 336–348 10.1007/BF00425639 [DOI] [PubMed] [Google Scholar]
- Tyrrell T. (1999). The relative influences of nitrogen and phosphorus on oceanic primary production. Nature 400 525–531 10.1038/22941 [DOI] [Google Scholar]
- Vuilleumier S., Khmelenina V. N., Bringel F., Reshetnikov A. S., Lajus A., Mangenot S., et al. (2012). Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J. Bacteriol. 194 551–552 10.1128/JB.06392-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh D. T., Bourgues S., Dewit R., Herbert R. A. (1996). Seasonal variation in rates of heterotrophic nitrogen fixation (acetylene reduction) in Zostera noltii meadows and uncolonised sediments of the Bassin d’Arcachon, south-west France. Hydrobiologia 329 161–174 10.1007/Bf00034555 [DOI] [Google Scholar]
- Welsh D., Castadelli G., Bartoli M., Poli D., Careri M., De Wit R., et al. (2001). Denitrification in an intertidal seagrass meadow, a comparison of 15N-isotope and acetylene-block techniques: dissimilatory nitrate reduction to ammonia as a source of N2O? Mar. Biol. 139 1029–1036 10.1007/s002270100672 [DOI] [Google Scholar]
- Widdel F. (1987). New types of acetate-oxidizing, sulfate-reducing desulfobacter species, D-hydrogenophilus Sp-Nov, D-latus Sp-Nov, and D-curvatus Sp-Nov. Arch. Microbiol. 148 286–291 10.1007/BF00456706 [DOI] [Google Scholar]
- Wilhelm S. W., Lecleir G. R., Bullerjahn G. S., Mckay R. M., Saxton M. A., Twiss M. R., et al. (2014). Seasonal changes in microbial community structure and activity imply winter production is linked to summer hypoxia in a large lake. FEMS Microbiol. Ecol. 87 475–485 10.1111/1574-6941.12238 [DOI] [PubMed] [Google Scholar]
- Wilson S. T., Bottjer D., Church M. J., Karl D. M. (2012). Comparative assessment of nitrogen fixation methodologies, conducted in the Oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 78 6516–6523 10.1128/Aem.01146-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr J. P., Waterbury J. B., Turner P. J., Montoya J. P., Omoregie E., Steward G. F., et al. (2001). Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412 635–638 10.1038/35088063 [DOI] [PubMed] [Google Scholar]