Summary
The acquisition and metabolism of iron (Fe) by the human pathogen Staphylococcus aureus is critical for disease progression. S. aureus requires Fe to synthesize inorganic cofactors called iron-sulfur (Fe-S) clusters, which are required for functional Fe-S proteins. In this study we investigated the mechanisms utilized by S. aureus to metabolize Fe-S clusters. We identified that S. aureus utilizes the Suf biosynthetic system to synthesize Fe-S clusters and we provide genetic evidence suggesting that the sufU and sufB gene products are essential. Additional biochemical and genetic analyses identified Nfu as a Fe-S cluster carrier, which aids in the maturation of Fe-S proteins. We find that deletion of the nfu gene negatively impacts staphylococcal physiology and pathogenicity. A nfu mutant accumulates both increased intracellular non-incorporated Fe and endogenous reactive oxygen species (ROS) resulting in DNA damage. In addition, a strain lacking Nfu is sensitive to exogenously supplied ROS and reactive nitrogen species. Congruous with ex vivo findings, a nfu mutant strain is more susceptible to oxidative killing by human polymorphonuclear leukocytes and displays decreased tissue colonization in a murine model of infection. We conclude that Nfu is necessary for staphylococcal pathogenesis and establish Fe-S cluster metabolism as an attractive antimicrobial target.
Keywords: Iron, Sulfur, Cluster, Neutrophil, Staphylococcus
Introduction
Iron is the most abundant transition metal in the human body and vertebrates go to great lengths to prevent microbes from acquiring it. Iron chelating molecules compete with microbes for Fe resulting in low concentrations of free Fe in body fluids. The process of actively limiting Fe from invading microorganisms has been called a form of nutritional immunity and it plays a major role in preventing infection (Weinberg, 1978). Staphylococcus aureus is ahuman commensal and a leading cause of morbidity and mortality worldwide. For successful infection S. aureus must circumvent the nutritional immunity of the host to acquire and metabolize iron (Graves et al., 2010, Hammer & Skaar, 2011).
The acquisition of Fe is critical for S. aureus to colonize host tissues and for subsequent disease progression (Skaar et al., 2004). S. aureus has numerous systems to acquire iron including siderophore-mediated systems and one heme-mediated system (Skaar et al., 2004, Cheung et al., 2009, Beasley et al., 2009, Sebulsky et al., 2000). Upon heme uptake Fe is removed from the heme porphyrin using a heme monooxygenase (Reniere et al., 2010). Iron captured by siderophores is reduced to the ferrous state by ferric reductases upon internalization (Schroder et al., 2003).
Although much is known about the mechanisms of Fe acquisition, little is known about how internalized Fe is metabolized in S. aureus. Iron is incorporated into proteins as Fe-S clusters, heme, or as a mononuclear cofactor. Only a small pool of non-incorporated Fe exists inside of bacterial cells (Keyer & Imlay, 1996). Iron metabolism is a double-edged sword; Fe is essential for S. aureus survival, but intracellular Fe not incorporated into macromolecules (non-incorporated Fe) can be toxic. Non-incorporated Fe can react with hydrogen peroxide to form the highly reactive hydroxyl radical (Fenton chemistry) (Haber & Weiss, 1932), which damages cellular macromolecules resulting in cell death (Aruoma et al., 1989, Imlay & Linn, 1988). Internalized Fe must be shielded from endogenous peroxides by chelation or trafficked to the desired apo-protein or storage molecule (Imlay et al., 1988, Morrissey et al., 2004). Therefore, for successful infection, S. aureus must acquire Fe and make it available for cellular processes, yet prevent it from participating in detrimental chemistry. How S. aureus accomplishes this vital task is not known.
Polymorphonuclear leukocytes (PMNs) and macrophages are a part of the first line of defense against bacterial infections. These cells phagocytose invading bacteria and assault them with reactive-oxygen species (ROS) produced by the enzymes NADPH oxidase and myeloperoxidase (Nauseef, 2008). Individuals that have a genetic disposition rendering their phagocytes unable to form ROS often have chronic and reoccurring S. aureus infections emphasizing the importance of ROS in preventing and combating S. aureus disease (Song et al., 2011). Macrophages also possess the ability to produce reactive nitrogen species (RNS) such as nitric oxide via an inducible nitric oxide synthase (iNOS) (Vouldoukis et al., 1995, Annane et al., 2000). Mice that have a mutation in the NOS2 gene are incapable of producing high levels of nitric oxide and are more susceptible to S. aureus infections (Richardson et al., 2006).
Among the cellular targets of ROS and RNS toxicity are inorganic cofactors composed of Fe and S called iron-sulfur (Fe-S) clusters (Soum & Drapier, 2003, Hurst et al., 1991, Duan et al., 2009, Flint et al., 1993b, Keyer & Imlay, 1997, Jang & Imlay, 2007). Solvent exposed Fe-S clusters can be oxidized by ROS resulting in cluster disintegration and the release of Fe (Djaman et al., 2004). Most organisms are reliant upon Fe-S cluster chemistry for metabolism, and therefore, disintegration of the cluster can result in enzyme inactivation and metabolic standstill. S. aureus uses proteins requiring Fe-S clusters for diverse cellular functions including: redox reactions (Orme-Johnson et al., 1968), DNA repair (Yeeles et al., 2009), environmental sensing (Sun et al., 2011), cofactor biosynthesis (Layer et al., 2005) and antibiotic resistance (Yan et al., 2010).
Three biosynthetic machineries (Nif, Suf, and Isc) that use Fe2+, S2-, and electrons to build [Fe2-S2] and [Fe4-S4] clusters have been discovered in bacteria (Takahashi & Tokumoto, 2002, Zheng et al., 1998, Zheng et al., 1993). In all three systems S2- is typically generated from cysteine (Zheng et al., 1993). The Nif and Isc systems use the “U-type” proteins NifU and IscU as molecular scaffolds whereon the Fe-S clusters are synthesized (Smith et al., 2001, Smith et al., 2005). The Suf system uses the SufBCD proteins to synthesize Fe-S clusters while SufB acts as the scaffold (Wollers et al., 2010, Saini et al., 2010). Biochemical analysis found that holo-scaffolds can transfer Fe-S clusters to apo-proteins (Chahal & Outten, 2012, Chandramouli & Johnson, 2006), but the frequency at which this happens in vivo is unknown. Alternatively, Fe-S clusters can be transferred to carrier molecules that traffic and insert Fe-S clusters into target apo-proteins. Combined genetic and biochemical studies have described six potential Fe-S cluster carrier/trafficking proteins in bacteria: A-type carriers (SufA) (Vinella et al., 2009), NfuA proteins (Py et al., 2012, Bandyopadhyay et al., 2008), NifU proteins (Smith et al., 2005), ApbC (Mrp) proteins (Boyd et al., 2008b, Boyd et al., 2009a), monothiol glutaredoxins (Iwema et al., 2009), and Nfu proteins (Nishio & Nakai, 2000, Jin et al., 2008).
The characterized bacterial Nfu proteins are approximately 80 amino acids in length and contain two conserved cysteine (Cys) residues separated by two amino acids (C-X-X-C), which is suggestive of Fe-S cluster binding. Biophysical analyses found that the Synechococcus sp. Nfu bound a [Fe4-S4] cluster that could be transferred to apo-proteins in vitro (Nishio & Nakai, 2000, Jin et al., 2008). Genetic analysis suggests that the nfu gene is essential in Synechococcus sp. strain PCC 7002 (Balasubramanian et al., 2006).
The NifU proteins are modular and contain a central ferredoxin [Fe2-S2] cluster binding domain, a N-terminal U-type scaffold domain and a C-terminal Nfu domain (Dos Santos et al., 2004, Fu et al., 1994). Biochemical and biophysical analyses have found that NifU is capable of ligating an [Fe4-S4] cluster in both the U-type and Nfu domains and each domain is independently capable of transferring the cluster to an apo-protein target (Smith et al., 2005).
The NfuA proteins are also modular with a N-terminal domain that resembles an A-type carrier and a C-terminal Nfu domain (Bandyopadhyay et al., 2008, Angelini et al., 2008). A majority of the NfuA proteins are missing the conserved Cys residues in the A-type domain that are necessary for Fe-S cluster ligation and evidence suggests that this “degenerate” A-type domain promotes interaction between NfuA and target apo-proteins (Py et al., 2012). The C-terminal Nfu domain can ligate a [Fe4-S4] cluster and transfer it to apo-proteins (Py et al., 2012, Bandyopadhyay et al., 2008). NfuA is required for growth under oxidative stress conditions and has a degree of functional overlap with NifU (Bandyopadhyay et al., 2008, Angelini et al., 2008).
The study herein is the first to test the hypothesis that effective Fe-S cluster metabolism is necessary for S. aureus pathogenesis. To initiate testing this hypothesis we engineered a S. aureus strain defective in Fe-S cluster metabolism by deleting the nfu gene, which is predicted to encode for a Nfu-type protein. We provide in vivo and in vitro evidence suggesting that Nfu is an Fe-S cluster carrier. Over the course of our studies we characterized how the absence of Nfu impacts a) cellular physiology, and b) staphylococcal pathogenesis. Our results show that a S. aureus nfu mutant strain has increased cellular pools of non-incorporated Fe and ROS, which result in DNA damage. We also find that the presence of Nfu is necessary for S. aureus to survive the antimicrobial action of the respiratory burst of PMNs and to successfully colonize host tissues using a murine model of systemic infection.
Results
Genomic and genetic analyses of S. aureus Fe-S cluster biosynthetic machinery
Unless otherwise stated the studies presented herein were conducted in the USA300_LAC strain (wild-type or WT) of S. aureus, which differs from the genome of USA300_FPR3757 by a few single nucleotide polymorphisms (Li et al., 2009). We analyzed the S. aureus USA300_FPR3757 genome for Fe-S cluster metabolic components. We identified a set of genes, arranged in a putative operon, that are homologues to Escherichia coli genes encoding for the SufBCDS iron-sulfur biosynthetic system. A schematic of the putative S. aureus suf operon is illustrated in Figure 1A. The components include a cysteine desulfurase (SufS; USA300_0820 (Zheng et al., 1993)), a U-type Fe-S cluster biogenesis molecular scaffold protein and/or sulfur trafficking molecule (SufU; SAUSA300_0821 (Albrecht et al., 2010, Selbach et al., 2014)), and the SufBCD Fe-S cluster biosynthetic scaffold machinery (SAUSA300_0822, 0818, 0819, respectively (Takahashi & Tokumoto, 2002)). We were unable to identify operons in S. aureus genomes with similarity to operons encoding for the E. coli Isc or the Azotobacter vinelandii Nif Fe-S cluster assembly systems. Downstream of the proposed suf operon we identified a gene encoding for a potential A-type Fe-S cluster trafficking protein (SufA; SAUSA300_0843 (Vinellaet al., 2009)). An additional gene was identified (nfu; SAUSA300_0839) that displayed homology to the nfuA and nifU genes from A. vinelandii, but the shared homology is confined to the C-terminal “Nfu” domains of these proteins (Dos Santos et al., 2004, Bandyopadhyay et al., 2008).
Figure 1. The chromosomal locations of Fe-S cluster biosynthesis genes and a working model for Fe-S cluster biogenesis in S. aureus.
Panel A: Chromosomal locations of potential Fe-S cluster biosynthetic genes in S. aureus. Examining the genome of the S. aureus strain USA300_FPR3757 revealed loci potentially involved in Fe-S cluster metabolism. The genes for the proposed biosynthetic machinery are organized in an apparent operon (sufCDSUB) and the genes encoding the proposed Fe-S cluster carrier molecules nfu and sufA have a proximal chromosomal location. Panel B: A working model for Fe-S cluster biogenesis and trafficking in S. aureus. SufS provides S0 for Fe-S cluster biosynthesis. Fe-S clusters are synthesized on the SufBCD or SufU scaffolds followed by transfer of the cluster to either 1) an apo-protein, or 2) an Fe-S cluster carrier molecule (Nfu, SufA or uncharacterized) for delivery to a target apo-protein.
Based on our bioinformatic analyses and studies conducted by alternate groups (reviewed in (Py & Barras, 2010)) we generated a working model for Fe-S cluster biogenesis and trafficking in S. aureus (Figure 1B). In the model, S2- is supplied by the SufS cysteine desulfurase. SufS transfers the S0 atoms to either the SufU or the SufBCD scaffolding proteins for Fe-S cluster formation. The Fe-S cluster can then be transferred directly from SufBCD or SufU to an apo-protein or to an Fe-S cluster carrier protein that subsequently traffics the Fe-S cluster to a apo-protein target. We propose that Nfu, SufA or alternate unidentified molecules act as Fe-S cluster carriers in S. aureus.
We aimed to test our model using a reverse genetics approach. We attempted to construct mutant strains in the USA300 community-acquired methicillin resistant S. aureus strain LAC that contained individual chromosomal deletions in the sufU, sufB, sufA, or nfu genes. We were unable to construct homozygous strains with mutations in either sufU or sufB under either aerobic or anaerobic growth conditions. Attempted mutant construction resulted in diploid or merodiploid cells, which retained a wild-type copy of the allele of interest (data not shown). Once the selective pressure to maintain the sufB::tet or sufU::tet alleles was removed the cells became haploid and retained only the wild-type copy of the respective allele.
We were able to construct mutant strains that contained chromosomal deletions in the nfu and the sufA genes (Figure S1 and Table 1). The doubling times for the nfu and sufA mutant strains during exponential growth phase were similar to the WT strain when cultured in complex or defined media, but the Δnfu strain displayed a decreased growth rate in the post-exponential growth phases when cultured in tryptic soy broth (TSB) (Figure S2A).
Table 1.
Strains and plasmids used in this study.
| Strains used in this study. | |||
|---|---|---|---|
| S. aureus Strains | Genotype/Description | Genetic Background | Source/Reference |
| JMB1100 | USA300_LAC | LAC | (Boles et al., 2010) |
| RN4220 | Restriction minus | NCTC8325 | (Kreiswirth et al., 1983) |
| JMB1165 | SAUSA300_0839(nfu)Δ | LAC | |
| JMB1580 | nfu::kanR | LAC | |
| JMB2102 | nfu::tetM | RN4220 | |
| JMB2316 | nfu::tetM | LAC | |
| JMB1144 | SAUSA300_0843(sufA)Δ | LAC | |
| JMB2220 | sufA::tetM | RN4220 | |
| JMB2223 | sufA::tetM | LAC | |
| JMB1836 | attP::pLL39 | RN4220 | |
| JMB2501 | nfu::kanR, sufA::tetM | LAC | |
| JMB1886 | attP::pLL39 | LAC | |
| JMB1888 | nfuΔ, attP::pLL39 | LAC | |
| JMB1889 | nfuΔ, attP::pLL39_nfu | LAC | |
| JMB1432 | fur::tetM (SAUSA300_1514) | LAC | (Horsburgh et al., 2001b) |
| JMB2715 | SAUSA300_1874(ftn)Δ | LAC | |
| JMB2957 | nfu::Tn(ermB) | LAC | |
| JMB2078 | kat::Tn(ermB) (SAUSA300_1232) | LAC | V. Torres |
| JMB2961 | sodA::Tn(ermB) | LAC | |
| JMB2080 | ahpC::Tn(ermB | LAC | |
| JMB2081 | aphF::Tn(ermB) nfuΔ | LAC | |
| JMB2151 | perR::kan | LAC | (Horsburgh et al., 2001a) |
| JMB1163 | acnA::tet | LAC | (Sadykov et al., 2010) |
| NE861 | SAUSA300_1246(acnA)::TN(ermB) | NARSA(Fey et al., 2013) | |
| NE892 | SAUSA300_2012(leuC)::TN(ermB) | NARSA(Fey et al., 2013) | |
| NE805 | SAUSA300_1178 (recA)::TN(ermB) | NARSA(Fey et al., 2013) | |
| NE911 | SAUSA300_0380(ahpC)::TN(ermB) | NARSA(Fey et al., 2013) | |
| NE718 | SAUSA300_2006(ilvD)::TN(ermB) | NARSA(Fey et al., 2013) | |
| NE1175 | SAUSA300_0839(nfu)::TN(ermB) | NARSA(Fey et al., 2013) | |
| NE1932 | sodA::TN(ermB) (SAUSA300_1513) | NARSA(Fey et al., 2013) | |
| NE1224 | sodM::TN(ermB) (SAUSA300_0135) | NARSA(Fey et al., 2013) | |
| NE1543 | qoxC::TN(ermB) (SAUSA300_0961) | NARSA(Fey et al., 2013) | |
| JMB2496 | recA::TN(ermB) | LAC | |
| JMB6068 | qoxC::TN(ermB) | LAC | |
| JMB2500 | recA::TN(ermB) nfuΔ | LAC | |
| JMB3076 | attP::pLL39_acnA_FLAG, acnA::TN(ermB) | LAC | |
| JMB4007 | nfuΔ, attP::pLL39_acnA_FLAG, acnA::TN(ermB) | LAC | |
| JMB4365 | attP::pLL39, acnA::TN(ermB) | LAC | |
| JMB3967 | nfuΔ, attP::pLL39, acnA::TN(ermB) | LAC | |
| JMB3954 | nfuΔ, attP::pLL39_nfu, acnA::TN(ermB) | LAC | |
| JMB3965 | attP::pLL39, leuC::TN(ermB) | LAC | |
| JMB3971 | nfuΔ, attP::pLL39, leuC::TN(ermB) | LAC | |
| JMB3952 | nfuΔ, attP::pLL39_nfu, leuC::TN(ermB) | LAC | |
| JMB3966 | attP::pLL39, ilvD::TN(ermB) | LAC | |
| JMB3970 | nfuΔ, attP::pLL39, ilvD::TN(ermB) | LAC | |
| JMB3955 | nfuΔ, attP::pLL39_nfu, ilvD::TN(ermB) | LAC | |
| UAMS-1 | parent | UAMS-1 | (Gillaspy et al., 1995) |
| JMB2399 | nfu::tetM | UAMS-1 | |
| MW2 | parent | MW2 | (Baba et al., 2002) |
| JMB2511 | nfu::tetM | MW2 | |
| SH1000 | parent | SH1000 | (Herbert et al., 2010) |
| JMB2426 | nfu::tetM | SH1000 | |
| Other Strains | Genotype/Description | Source/Reference | |
| Escherichia coli PX5 | Cloning strain | Protein Express | |
| S. epidermidis | ATCC12228 | ATCC | |
| S, cerevisiae FY2 | ura3-52 | W. Beiden (Winston et al., 1995) | |
| Plasmids used in this study | |||
|---|---|---|---|
| Plasmid name | Insert Locus/function | Source/Reference | |
| pJB38 | Mutant construction | (Bose et al., 2013) | |
| pJB38_Δnfu | SAUSA300_0839 (nfu) chromosomal deletion | ||
| pJB38_Δftn | SAUSA300_1874(ftn) chromosomal deletion | ||
| pJB38_ΔsufA | SAUSA300_0843 (sufA) chromosomal deletion | ||
| pJB38_ΔsufU | SAUSA300_0821 (sufU) chromosomal deletion | ||
| pJB38_ΔsufB | SAUSA300_0822 (sufB) chromosomal deletion | ||
| pJB38_Δdps | SAUSA300_2092 (dps) chromosomal deletion | ||
| pJB38_nfu::tetM | Allelic replacement | ||
| pJB38_nfu::kanR | Allelic replacement | ||
| pJB39_sufA::tetM | Allelic replacement | ||
| pJB38_sufU::tetM | Allelic replacement | ||
| pJB38_sufB::tetM | Allelic replacement | ||
| pLL39 | Chromosomal genetic complementation | (Luong & Lee, 2007) | |
| pLL39_nfu | SAUSA300_0839 | ||
| pLL2787 | φ11 int | (Luong & Lee, 2007) | |
| pDG783 | kanR | (Guerout-Fleury et al., 1995) | |
| pCM28 | Genetic complementation | ||
| pCM28_nfu | SAUSA300_0839 | ||
| pEPSA5 | Genetic complementation | (Forsyth et al., 2002) | |
| pEPSA5_nfu | SAUSA300_0839 | ||
| pEPSA5_acnA | SAUSA300_1246 | ||
| pEPSA5_FLAG_leuCD | SAUSA300_2012-3 | ||
| pEPSA5_IlvD | SAUSA300_2006 | ||
| pEPSA5_ahpCF | SAUSA300_0379-80 | ||
| pCM11 | Transcriptional fusion containing promoterless gfp | (Malone et al., 2009) | |
| pCM11_acnAp | SAUSA300_1246 promoter | ||
| pCM11_dpsp | |||
| pCM11_recAp | SAUSA300_1178 promoter | ||
| pET20b | Protein production | ||
| pET20b_nfu | SAUSA300_0839 | ||
| pET24a | Protein production | EMD Millipore | |
| pET24a_acnA | SAUSA300_1246 | ||
| pXEN-1_isdBp | SAUSA300_1028 promoter | V. torres | |
Aconitase protein has lower enzymatic activity in a nfu mutant strain
The TCA cycle protein aconitase (AcnA) requires an [Fe4-S4] cluster for enzymatic function (Kennedy et al., 1983). We assessed the activity of the AcnA enzyme in cell-free lysates harvested from the WT, Δnfu and ΔsufA strains. The activity of AcnA in the ΔsufA and WT strains was similar. However, AcnA activity in the Δnfu mutant strain was ~50% that of WT (Figure 2A). We constructed a nfu sufA double mutant strain and found that AcnA activity was not significantly different from the nfu single mutant strain. AcnA activity was also lower in other clinical isolates lacking Nfu (Figure S4A) suggesting that decreased AcnA activity is a phenotype that is common for S. aureus nfu mutant strains. The sufA mutant strains were not further examined in this study.
Figure 2. Aconitase activity is decreased in a S. aureus nfu mutant strain.
Panel A: Aconitase (AcnA) activity is decreased in a strain lacking Nfu. The activity of the AcnA enzyme was assessed in cell-free lysates harvested from the WT (JMB1100), ΔsufA (JMB 1144), Δnfu (JMB1165) and the sufA nfu double mutant (JMB2501) strains. Panel B: AcnA activity is decreased in a nfu mutant independent of AcnA expression levels. AcnA activity was assessed in the parent (JMB4365), Δnfu mutant (nfu−; JMB3967), and the genetically complemented Δnfu mutant (nfu+; JMB3954) strains. All strains contained a null allele of the acnA gene and the pEPSA5_acnA plasmid (acnA under the transcriptional control of a xylose inducible promoter). Strains were cultured in the presence (induced) or absence (not induced) of 1% xylose before assessing AcnA activity in cell-free lysates. Panel C: AcnA activity is decreased in a nfu mutant despite AcnA protein accumulation. AcnA activity was assessed in cell extracts from strains containing a null allele of the native acnA gene and a second FLAG-tagged allele of acnA integrated at a secondary location on the chromosome. Activity was assessed in the parent (JMB3076) and the Δnfu mutant (JMB4007) strains containing either the pCM28 plasmid (WT and nfu−) or the pCM28_nfu (nfu+) plasmid. Inset: Western blot analyses of the AcnA_FLAG protein confirming that AcnA protein accumulated in all three strains. FLAG_AcnA protein abundance was determined in duplicate. Panel D: AcnA activity is decreased in a nfu mutant strain when cultured in the absence of oxygen. The strains used in panel D are identical to those used in panel B. The strains were cultured anaerobically in the presence (induced) or absence (not induced) of 1% xylose prior to assessing AcnA activity in cell-free lysates. Data in Figure 2 represent the average of three independent experiments with standard deviations shown. Enzymatic activity was standardized with respect to the total protein concentration and subsequently to that of the WT/parent (parent induced in panels 2B and 2D). Paired t-tests were performed on the data and * denotes p< 0.05.
Five potential scenarios could result in the lower AcnA activity witnessed in the nfu mutant strain. The lower activity could be the result of decreased acnA expression, decreased AcnA protein abundance, diminished ability to interconvert between the inactive [Fe3-S4] cluster form of AcnA and the active [Fe4-S4] cluster form of AcnA, oxidative damage to the Fe-S cluster, or decreased occupancy of the Fe-S cluster. We conducted a series of experiments to discern between these scenarios.
We first examined whether the decreased AcnA activity was due to decreased acnA expression. The acnA gene was placed on a plasmid under the control of a xylose inducible promoter and the plasmid (pAcnA) was transduced into the parent, nfu+, and nfu− strains, which had the native acnA gene disrupted. Gene expression was induced and AcnA activity was assessed in cellular lysates. As illustrated in Figure 2B, the Δnfu mutant strain had ~4-fold lower AcnA activity than the parent strain and the enzymatic activity was partially restored when the nfu gene was returned to the chromosome at a secondary location (nfu+). Integration of the nfu gene onto the chromosome at an alternate location resulted in partial genetic complementation of the nfu mutant (Figures 2B, 2D and 10C). In contrast, returning the gene under its native promoter via an episome resulted in full genetic complementation (Figure 6A, 10A, 10B and S4). The reason for this discrepancy is currently unknown, but similar problems have been reported for the pLL chromosomal integration system (Mainiero et al., 2010, Yepes et al., 2014).
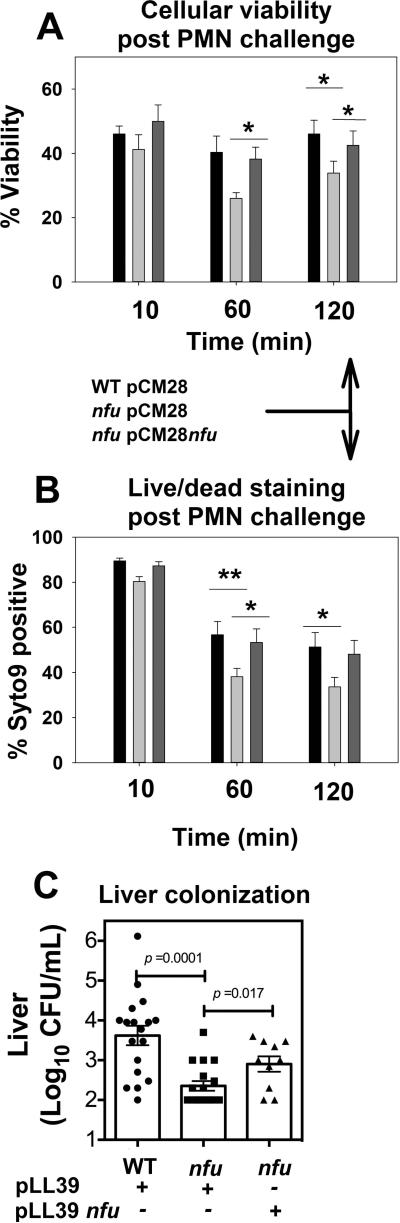
Figure 10. A S. aureus nfu mutant strain has decreased survival upon challenge with human neutrophils and attenuated liver colonization.
The WT with pCM28 (JMB1100; black), Δnfu with pCM28 (JMB1165; light gray) and Δnfu with pCM28_nfu (JMB1165; dark gray) were opsonized in pooled human serum and fed to normal or diphenyleneiodonium (DPI)-treated polymorphonuclear neutrophils (PMN) at a MOI of 5:1. Ingested bacteria were recovered at the indicated time points and viability was assessed by either enumerating CFUs by colony plating (Panel A) or live/dead staining with SYTO9 and propidium iodide (PI) (Panel B). For colony plating, at each time point, the viability of a strain is expressed as bacteria recovered from normal-PMN as a percent of the same strain recovered from DPI-treated PMN at time = 10. For live/dead stained cells, cells staining for Syto9 only were scored as live and those staining for PI only were scored as dead. For experiments shown in Panel A, a paired t-test was used to compare the bacterial strains at the indicated time points ((n=8 * denotes p< 0.05) and the data represent the mean ± standard error of the mean (SEM). For experiments shown in Panel B, strains were compared using 2-way ANOVA with a Bonferroni post test, (n=7) * denotes p< 0.05 and ** p denotes < 0.01. Panel C: A S. aureus nfu mutant is defective in colonizing liver in a systematic model of infection. ND4 Swiss Webster mice were infected systemically with the parent (JMB1886; n=18), Δnfu mutant (JMB1888;nfu−; n=18) and the complemented strain (JMB1889; nfu+; n=10) before the bacterial burden (CFUs) from livers was determined 96 hours post-infection. For the experiments in Panel C, a paired t-test was used to compare the bacterial strains (n=10 or 18) and bars represent the average CFUs with standard deviation shown.
Figure 6. A S. aureus nfu mutant has an increased cellular pool of non-incorporated Fe.
Panel A: A nfu mutant is sensitive to the antimicrobial compound streptonigrin. The WT (JMB1100), Δnfu (JMB1165), or Δftn (JMB2715) strains containing either pCM28 (nfu−) or pCM28_nfu (nfu+) were plated as top agar overlays on TSB agar plates with or without 1 mM of the cell permeable divalent metal chelator 2,2-dipyridyl. Four μg of streptonigrin was spotted and the diameter of the zone of growth inhibition was determined after 24 hours. Panel B: A nfu mutant strain has increased Fur accessible Fe. The WT (circle; JMB1100), Δnfu (triangle; JMB1165) and Δftn (square; JMB2715) strains containing the luxABCDE genes under the transcriptional control of the isdB promoter (pXEN-1_isdBp) were cultured aerobically and luminescence was monitored over time. The luminescence data were standardized to culture optical density. Panel C: The total cellular Fe load in a nfu mutant is the same as the WT strain. The total Fe in the cells (parts per billion (ppb)) was determined in the WT (JMB1100), Δnfu (JMB1165) and Δftn (JMB2715) strains using high resolution inductively coupled plasma mass spectrometry. The data presented in Figure 6 represent the average of three biological replicates with the standard deviations shown. Paired t-tests were performed on the data and * denotes p< 0.05 and N.S. denotes not significant. For the data shown in Panel B, paired t-tests were only performed on the data generated using the Δnfu and WT strains.
We next examined whether decreased AcnA accumulation resulted in decreased AcnA activity. We integrated a C-terminal FLAG affinity tagged allele of acnA under the transcriptional control of the native promoter at a secondary site on the chromosome and disrupted the native copy of the acnA gene. When compared to the parent strain the activity of AcnA_FLAG in cell-free lysates was ~50% in the nfu mutant strain and this phenotype could be genetically complemented (Figure 2C). Western blot analyses of the lysates indicated that AcnA_FLAG accumulated in all three strains.
The Fe-S cluster of AcnA can interconvert between [Fe3-S4] and [Fe4-S4] cluster forms (Kent et al., 1982, Kennedy et al., 1983). The [Fe3-S4] cluster form of AcnA is inactive and the addition of ferrous salts to cellular lysates aids in conversion to the [Fe4-S4] cluster resulting in restored enzymatic activity. It is possible that the lower AcnA activity witnessed in the Δnfu strain was as a result of enrichment of the inactive [Fe3-S4] AcnA. We assessed the activity of AcnA in cellular lysates from the Δnfu and WT strains after incubation in the presence and absence of ferrous salts. The addition of Fe2+ led to a slight increase in AcnA activity in lysates, but the increase was equal in both of the strains suggesting that the [Fe3-S4] form of AcnA was not enriched in lysates from the Δnfu strain (data not shown).
Solvent exposed Fe-S clusters can be damaged by oxygen or ROS (Imlay, 2006). We examined whether the decreased AcnA activity observed in the Δnfu strain could be the result of Fe-S cluster damage occurring under aerobic growth. We assessed AcnA activity in strains cultured anaerobically. As seen in Figure 2D, the activity of AcnA, in cell free lysates, was ~5-fold lower in the nfu mutant strain than the parent strain and this phenotype could be genetically complemented (nfu+). The results in Figure 2 suggest that the lower AcnA activity in a nfu mutant is a result of a defect in AcnA enzymatic activity and not the result of decreased acnA expression, decreased AcnA accumulation, decreased cluster interconversion or Fe-S cluster damage by ROS.
A nfu mutant strain has a general defect in Fe-S cluster metabolism
Biosynthesis of the branch chain amino acids (BCAA) Leu and Ile requires the enzymes isopropylmalate isomerase (LeuCD) and dihydroxy-acid dehydratase(IlvD), respectively. Both of these proteins require a [Fe4-S4] cluster for enzymatic function (Flint et al., 1993a, Hentze & Argos, 1991). We created strains that had mutations in the native copies of either the ilvD or leuC genes and returned a functional copy of the respective gene to the strains via a plasmid. The ilvD or leuCD genes were placed under the transciptional control of a xylose inducible promoter, which allowed us to grow our strains in a rich medium containing BCAA and induce leuCD or ilvD expression. We measured LeuCD and IlvD activity in cell-free lysates derived from the parent, nfu−, or nfu+ strains containing pleuCD and pilvD, repectively. As shown in Figures 3A and 3B, activities of LeuCD and IlvD were ~2-3-fold lower in cell-free lysates generated from the nfu− strains than the parent strains, and the defects could be genetically complemented. Importantly, the FLAG_LeuC protein accumulated to nearly equivalent levels in all three strains (Figure 3A, inset) indicating that the LeuCD protein is being produced, but is defective for enzymatic function in the nfu mutant strain.
Figure 3. A nfu mutant strain is defective in Fe-S cluster metabolism.
Panel A: LeuCD enzyme activity is decreased in a nfu mutant despite LeuC protein accumulation. LeuCD activity was assessed in the cell free lysates of the parent (JMB3965), Δnfu mutant (nfu−; JMB3971) and the genetically complemented Δnfu (nfu+; JMB3952) strains. The strains also contained a null allele of the native leuC gene and a second FLAG-tagged allele of leuC with leuD on a plasmid under the transcriptional control of a xylose inducible promoter (pEPSA5_leuCD). Inset: Western blot analyses of the FLAG_LeuC protein showing that LeuC protein accumulates in all strains. LeuC abundance was determined in duplicate. Panel B: IlvD activity is decreased in a nfu mutant strain. IlvD activity was assessed in cell-free lysates from the parent (JMB3966), Δnfu mutant (nfu+; JMB3955) and the genetically complemented Δnfu (nfu−; JMB3970) strains. The strains contained a null allele of ilvD and a second functional copy of ilvD on a plasmid under the transcriptional control of a xylose inducible promoter (pEPSA5_ilvD). The data are presented as the average of three independent experiments with standard deviations shown. Strains were cultured in the presence (induced) or absence (not induced) of 1% xylose. Enzymatic activities were standardized with respect to the total protein concentration and subsequently to the activity of the induced parent. Paired t-tests were performed on the data shown and * denotes p< 0.05.
Cellular respiration is reliant upon Fe-S cluster containing enymes. These enzymes are invoved in both the movement of electrons through respiratory pathways and the ability of the cell to produce sufficient reductant to drive respiration. When cultured in a complex medium (TSB) the growth of the Δnfu strain was similar to the WT in early growth phase, which is principally fermentative (Figure S2A). The nfu mutant displayed a growth defect in the latter phases of growth, which are principally respiratory (Ledala et al., 2014). As a positive control we examined the growth of a strain lacking a cytochrome oxidase (qox), which is defective in respiring dioxygen (Hammer et al., 2013). We found that the Δnfu strain displayed a growth pattern that was similar to the qox mutant in the latter growth phases suggesting that the nfu mutant strain was defective in cellular respiration (Figure S2A).
We next cultured the WT and Δnfu strains in complex media in the presence and absence of the respiratory inhibitor sodium azide. As illustrated in Figure S2B the Δnfu strain displayed a more severe growth defect than the parent when grown in the presence of sodium azide.
The chemical XTT can be used to monitor respiratory activity and the rate of reduction of XTT is proportional to the rate of electron flux though respiratory pathways (Berridge et al., 2005). We monitored the rate of XTT reduction in cell suspensions of the Δnfu and WT strains and found that the Δnfu strain had a reduced rate of XTT reduction consistent with this strain being deficient in respiration (Figure S2C). Collectively, the data presented suggest that a nfu mutant strain is defective in Fe-S metabolism and that this defect is manifested in the decreased activity of Fe-S cluster enzymes, as well as, an impaired ability to respire dioxygen.
Expression of S. aureus genes involved in Fe-S cluster metabolism are altered when cells are starved for iron or exposed to O2 or H2O2
Determining the factors that influence gene expression can aid in deciphering gene function. Oxygen and ROS can damage Fe-S clusters requiring de novo synthesis or repair of the Fe-S cluster (Imlay, 2006). We predicted that the physiological response to alterations in iron availability, exposure to oxygen or ROS insult would result in the altered expression of genes required to metabolize Fe-S clusters (nfu and sufC).
To examine the effect of oxygen we cultured WT S. aureus anaerobically and at mid-exponental growth phase switched one half of the cultures to an aerobic atomosphere. As shown in Figure 4A, the mRNA transcript levels from the nfu and sufC genes, as well as, the positive control cytochrome oxidase (cydB) were significantly increased upon the introduction of oxygen.
Figure 4. Fe starvation or exposure to Oxygen or ROS alters the mRNA abundance from genes necessary for Fe-S cluster metabolism.
Relative mRNA abundance was determined using quantitative real-time PCR (qPCR) using the WT strain (JMB1100). All data were normalized to 16s rRNA transcript abundance. Panel A: mRNA abundance for the cydB (cytochrome oxidase), sufC and the nfu genes under anaerobic growth conditions (black bars) and 25 minutes post introduction of oxygen (gray bars). Data are presented as fold-change relative to the mRNA abundance from cells cultured anaerobically. Panel B: mRNA abundance for the ahpC, nfu and the sufC genes 25 minutes after the addition of 10 mM H2O2 (gray bars) to aerobically grown cultures. Data are presented as fold change in mRNA abundance relative to non-treated samples (black bars). Panel C: mRNA abundance of the isdI, isdB, nfu and the sufC, genes 25 minutes after the addition of the cell permeable divalent metal chelator 2,2-dypyridyl (gray bars) to aerobically grown cultures. Data are presented as fold change in mRNA abundance relative to non-treated samples (black bars). Samples were prepared in biological triplicates yielding three cDNA libraries and each library was analyzed two times. Data represent averages with standard deviations shown. Paired t-tests were performed on the data and * denotes p< 0.05; ** denotes p < 0.01; *** denotes p < 0.001.
To examine the effect of ROS insult we cultured the WT strain aerobically and challenged one-half of the cultures with a bolus of H2O2. As shown in Figure 4B, the mRNA transcripts from the nfu and sufC genes, as well as, the positive control ahpC increased in response to H2O2 challenge.
To examine the effect of Fe deprivation we cultured WT S. aureus in the presence and absence of the cell permeable divalent metal chelator 2,2-dipyridyl (DIP), which has a high affinity for Fe2+ (Rauen et al., 2007). As shown in Figure 4C, the mRNA transcript levels from the nfu and sufC genes were downregulated in the presence of DIP. A similar decrease in SufC protein abundance upon DIP treatment was previously noted (Friedman et al., 2006). The isdB and isdI genes are under the regulation of Fur and their expression is derepressed under iron starvation conditions (Torres et al., 2010). As previously shown, the transcript levels of isdB and isdI were increased upon DIP treatment (Torres et al., 2010). Although DIP treatment did result in the increased transcription of the isdB and isdI genes it should be noted that DIP can chelate other divalent metals in addition to Fe, and therefore, we cannot rule out the possiblity that chelation of a divalent metal other than Fe2+ is resulting in the altered abundance of the nfu transcript.
The data in Figure 4 show that 1) the transcription of the nfu gene is altered upon Fe starvation, H2O2 exposure, and O2 introduction, and 2) the transcriptional pattern of the nfu gene under the tested physiological challenges is similar to those observed for sufC, which encodes for a core component of the the Fe-S cluster biosynthetic apparatus.
Nfu can bind and effectively transfer an Fe-S cluster to apo-aconitase
The results presented in conjunction with the proposed functions of ortholog proteins (Nfu/NifU/NfuA) led us to hypothesize that Nfu could bind and transfer Fe-S clusters in vitro. We found that purified Nfu was slightly brown and had the visible absorption characteristics of an Fe-S cluster-containing protein. The color dissipated after aerobic dialysis suggesting that the protein co-purified with a labile or dissociable cofactor. Post dialysis we were not able to detect significant quantities of iron or acid-labile sulfide associated with the protein using either ferene or N,N-dimethyl-p-phenylene-diamide to measure iron and sulfide, respectively.
We transferred the Nfu protein to an anaerobic chamber and treated it with DTT, followed by the addition of Fe2+ and S2- to facilitate the chemical reconstitution of an Fe-S cluster on Nfu. After removing excess Fe and S, the protein retained a dark-brown color and exhibited UV-Visible absorption and circular dichroism spectra characteristic of Fe-S binding proteins (Figure 5A and 5B) (Gao et al., 2013). Chemically reconstituted Nfu bound 2.2 ± 0.3 iron and 1.9 ± 0.3 acid labile sulfide (n=4) atoms per monomer.
Figure 5. Nfu can bind and effectively transfer Fe-S clusters.
Panel A: Representative UV-visible absorption spectrum of the reconstituted Nfu protein (55 μM). Panel B: Representative circular dichroism spectra of the as isolated (dashed; 350 μM) and the chemically reconstituted (solid; 350 μM) Nfu proteins. Panel C: Holo-Nfu protein can activate the enzymatic activity of apo-AcnA protein. Apo-AcnA (4 μM) was incubated with holo-Nfu (8 μM) (closed circles) or 16 μM Fe2+ and 16 μM S2- (open circles). Aliquots of the samples were removed periodically and assayed for AcnA activity. Panel D: Approximately two holo-Nfu are required to activate one AcnA protein. AcnA activation assays contained 4 μM apo-AcnA protein and 0-16 μM holo-Nfu. AcnA activity was assessed after a two-hour anaerobic incubation. For cluster transfer assays, data are represented as the average of three experiments with standard deviations shown. Standard deviations are shown for all data presented in panels C and D; however, in some cases they are smaller than the symbols shown.
We next examined whether holo-Nfu could transfer an Fe-S cluster to an apo-protein. Recombinantly produced S. aureus AcnA was purified from E. coli. We removed the Fe-S cluster and adventitiously bound Fe from AcnA resulting in apo-AcnA that was enzymatically inactive. Apo-AcnA was transferred to an anaerobic chamber, incubated with DTT and subsequently combined with either 1) holo-Nfu, 2) apo-Nfu, or 3) Fe2+ and S2-. At periodic intervals the assay mixtures were examined for AcnA activity. As illustrated in Figure 5C the activity of AcnA increased as a variable of time in the sample containing holo-Nfu. Importantly, over the course of the assay no significant AcnA activation was observed in the samples containing either Fe2+ and S2-, or apo-Nfu (Figure 5C and data not shown).
We next determined the amount of holo-Nfu necessary to activate apo-AcnA. We titrated a fixed concentration of apo-AcnA with holo-Nfu and the protein mixtures were subsequently incubated for two hours before measuring AcnA activity. As illustrated in Figure 5D, activation of AcnA required approximately two holo-Nfu. The biochemical and genetic data presented, in combination with the similarity of S. aureus Nfu protein to previously described Nfu proteins, support the hypothesis that S. aureus Nfu is an Fe-S cluster carrier.
A S. aureus nfu mutant strain has an increased intracellular non-incorporated iron pool
Studies using the bacterium Erwinia chrysanthemi have shown that strains lacking Fe-S cluster biosynthetic components have an increased cellular pool of iron not incorporated into macromolecules (non-incorporated Fe) (Nachinet al., 2001). In our model, Nfu serves as an Fe-S cluster carrier and we hypothesized that non-incorporated Fe would accumulate in the absence of Nfu.
We examined the sensitivity of the nfu mutant strain to the aminoquinone antibiotic streptonigrin. In combination with iron and an intracellular reducing agent, streptonigrin causes DNA and RNA damage, which ultimately results in cell death (Bolzan & Bianchi, 2001). The WT and Δnfu strains were plated as top-agar overlays and streptonigrin was spotted. The ftn gene encodes for ferritin (Ftn), which is an iron storage protein. Previous work found that the absence of Ftn results in Fe accumulation in the cytosol, and therefore, we included a ftn mutant in our assays as a positive control (Velayudhan et al., 2007). As shown in Figure 6A, the Δnfu and Δftn strains had a larger zone of growth inhibition than the WT strain. Importantly, the phenotype of the Δnfu strain could be genetically complemented. To ensure that the inhibition of growth was a result of intracellular iron concurrent experiments were conducted in the presence of divalent metal chelator DIP. The presence of DIP reduced the zone of growth inhibition in all the assayed strains. Fe-S cluster oxidation by ROS can cause cluster disintegration (Imlay, 2003) and Fe-S cluster damage may contribute to the increase in non-chelated Fe in the Δnfu strain cultured aerobically. We examined the streptonigrin sensitivity of the Δnfu and WT strains cultured anaerobically. We found that a nfu mutant strain is more sensitive to streptonigrin toxicity than the WT when cultured in the absence of oxygen suggesting that ROS alone was not causing increased streptonigrin sensitivity (Figure S3).
The ferric uptake regulator (Fur) is a DNA binding transcriptional regulator that alters gene transcription when bound to Fe (Xiong et al., 2000). The isdB gene is under the transcriptional control of the Fur protein (Torres et al., 2006). To qualitatively monitor Fur transcriptional activity we constructed a transcriptional reporter by placing the luxABCDE genes, which encode for luciferase and the enzymes that produce its substrate, under the transcriptional control of the isdB promoter. We found that luciferase activity was increased in a fur mutant strain consistent with isdB being under the transcriptional control of the Fur protein (data not shown). The Δnfu and Δftn strains had lower isdB transcriptional activity than the WT strain throughout growth (Figure 6B). These data suggest that the Δnfu and Δftn mutant strains have increased Fur accessible Fe resulting in the Fur mediated transcriptional repression of the isdB promoter.
The results from the isdB promoter activity experiments corroborated our streptonigrin sensitivity findings. However, it was possible that the repression of the Fur regulated genes and the streptonigrin sensitivity of a nfu mutant were a consequence of an increase in the total amount of cellular Fe, rather than an increase in the Fur associated Fe pool. To this end we investigated the cellular iron load in a nfu mutant strain. We used high-resolution inductively coupled plasma mass spectrometry (HR ICP-MS) to determine the overall Fe load in the WT and Δnfu strains. Previous analysis found that cells lacking Ftn had a decreased Fe load (Abdul-Tehrani et al., 1999), and therefore, we included a Δftn mutant in the assay as a negative control. As illustrated in Figure 6C, the total amount of cellular 56Fe was indistinguishable between the WT and Δnfu strains. As previously noted the total Fe was decreased in the Δftn mutant. Collectively, the data presented in Figure 6 are consistent with the hypothesis that a nfu mutant strain has an increased intracellular non-incorporated iron pool.
A nfu mutant strain has increased DNA damage, which is abrogated by Fe chelation or anaerobiosis
Non-chelated Fe can react with H2O2 resulting in hydroxyl radicals, which can damage DNA (Keyer & Imlay, 1996, Maringanti & Imlay, 1999). Results presented in Figure 6 lead us to hypothesize that a nfu mutant strain would have increased DNA damage when cultured aerobically. We monitored the rate of spontaneous mutagenesis in the WT and Δnfu strains by determining the frequency that the strains acquire a mutation conferring resistance to the antibiotic rifampicin (Ezekiel & Hutchins, 1968). As shown in Figure 7A, the Δnfu strain had a mutagenesis frequency that was ~3-fold higher than the WT when grown aerobically. We hypothesized that the DNA damage would be abrogated upon either oxygen deprivation or iron chelation. Consistent with this hypothesis growth in the presence of DIP or in the absence of oxygen abrogated the increased mutagenesis frequency. These data suggest that both oxygen and non-incorporated Fe are responsible for the increased mutagenesis frequency seen in a nfu mutant.
Figure 7. A nfu mutant strain has increased DNA damage.
Panel A: A nfu mutant strain has an increased mutagenesis frequency, which is abrogated by chelating Fe or culturing in the absence of oxygen. The mutagenesis frequency of the WT (JMB1100; black bars) and Δnfu (JMB1165; white bars) strains was determined in the presence or absence of the divalent metal chelator 2,2-dipyridyl (DIP; 200 μM) or oxygen. Data were plotted as fold-changes in the mutagenesis frequency relative to the mutagenesis frequency of the WT under each individual growth condition. The data represent the average of 10 biological replicates and experimental variation is shown as standard deviation. Panel B: The transcriptional activity of the recA gene is increased in a nfu mutant strain. The transcriptional activity of the recA promoter was assessed by monitoring GFP fluorescence over time in the WT (JMB1100; black bars) and Δnfu (JMB1165; white bars) strains containing a construct encoding for gfp under the transcriptional control of the recA promoter (pCM11_recAp). Fluorescence data were standardized to culture optical density (A600). The data are presented as the averages of biological triplicates with standard deviations shown. Paired t-tests were performed on the data in Figure 7 and * denotes p< 0.05 and *** denotes p < 0.001 and N.S. denotes not significant.
The SOS response is induced upon the detection of DNA damage (van der Veen & Abee, 2011). The RecA protein has a role in the repair of damaged DNA and transcription of the recA gene is induced as a part of the SOS response (Schlacher et al., 2006). We used a recA transcriptional reporter to qualitatively monitor recA transcriptional activity in the WT and Δnfu strains. As shown in Figure 7B, recA promoter activity was increased in the Δnfu strain during aerobic growth. Collectively, the data in Figure 7 led to the conclusion that a nfu mutant strain has increased DNA damage and that this phenotype requires the presence of both oxygen and chelatable Fe.
A nfu mutant strain has increased intracellular ROS. Studies have shown that aerobic bacteria generate sufficient ROS to damage their DNA (Keyer & Imlay, 1996). However, the cellular concentrations of ROS typically do not accumulate to levels that cause an impairment of growth due to the titers of scavenging enzymes (reviewed in (Imlay, 2008)). The Δnfu strain displayed an oxygen dependent DNA damage phenotype, which was mitigated by Fe chelation. Therefore, we hypothesized that a nfu mutant has increased intracellular ROS that interacts with the non-incorporated iron resulting in increased DNA damage.
We qualitatively measured intracellular ROS in the WT and Δnfu strains using the cell permeable compound 2’,7’-dichlorofluorescein diacetate (DHCF-DA). DHCF-DA is converted to 2,7 dichlorofluorescein (DHCF) upon oxidation in the presence of ROS and/or a cellular peroxidase (Myhre et al., 2003, Arenas et al., 2011). An alkyl hydroperoxidase (ahpC) mutant was assayed as a positive control. As shown in Figure 8A, the nfu and ahpC mutant strains had a greater rate of DHCF formation than the WT, consistent with a nfu mutant having increased intracellular ROS.
Figure 8. A strain lacking Nfu has increased endogenous reactive oxygen species.
Panel A: A nfu mutant strain has an increased rate of 2’,7’-dichlorofluorescein diacetate (DHCF-DA) oxidation. The relative concentrations of endogenous ROS were measured in the WT (JMB1100; closed circles), Δnfu (JMB1165; closed triangles) and ahpC (JMB2080; open circles) strains using the cell permeable fluorophore DHCF-DA. Panel B: The absence of Nfu results in the increased transcriptional activity of a gene repressed by PerR. The activity of the dps promoter was assessed in late-exponential growth phase cultures of the WT (JMB1100), Δnfu (JMB1165) and perR (JMB2151) strains containing a construct encoding for gfp under the transcriptional control of the dps promoter. Panel C: The transcriptional activity of the alkyl hydroperoxidase (ahpC) gene is decreased in a nfu mutant strain. The activity of the ahpC promoter was assessed by monitoring GFP fluorescence over time in the WT (JMB1100; filled circles) and the Δnfu (JMB1165; open circles) strains containing a construct encoding for gfp under the transcriptional control of the ahpC promoter (pCM11_ahpC). Fluorescence data in panels B and C were standardized to culture optical density (A600). The data shown in Figure 8 represent the averages of biological triplicates with standard deviations shown. Standard deviations are shown for all data presented; however, in some cases they are smaller than the symbols shown. Paired t-tests were performed on the data and * denotes p< 0.05. For the data shown in Panel A, paired t-tests were only performed on the data generated using the Δnfu and WT strains.
S. aureus uses the PerR transcriptional repressor to sense and respond to H2O2 (Lee & Helmann, 2006). PerR regulates transcription of the dps gene and dps expression can be used as a proxy for PerR dependent alterations in transcription (Horsburgh et al., 2001a). As illustrated in Figure 8B the transcriptional activity of the dps gene was increased in the nfu and perR mutant strains. These data confirm that PerR is a repressor of dps transcription and suggest that a nfu mutant has elevated levels of endogenous H2O2 resulting in decreased binding of PerR to the dps promoter.
We hypothesized that the increased ROS in a nfu mutant strain was the result of decreased transcription of genes encoding ROS scavenging proteins. The alkyl hydroperoxidase system (Ahp) has been proposed to be the major intracellular H2O2 scavenger in bacteria (Seaver & Imlay, 2001). We monitored the transcriptional activity of the ahp promoter in the WT and Δnfu strains. As illustrated in Figure 8C the transcriptional activity of the ahp promoter was decreased in a Δnfu strain. Collectively, the data presented in Figure 8 suggest that a strain lacking Nfu 1) has increased intracellular titers of ROS when cultured aerobically, and 2) has decreased transcriptional activity of at least one ROS metabolism gene.
A S. aureus nfu mutant strain is sensitive to reactive oxygen and reactive nitrogen species
Macrophages and PMNs kill S. aureus using ROS. In addition, macrophages kill using RNS. A primary mechanism of action of these ROS and RNS is to oxidize and modify Fe-S clusters, which can result in cluster damage (Imlay, 2006, Hurst et al., 1991, Fang, 2004). We hypothesized that a nfu mutant would have increased sensitivity to ROS and RNS. We cultured the WT and Δnfu strains in the presence of the redox cycling molecule methyl viologen or the nitrosative stress inducing molecule sodium nitroprusside. As shown in Figures S5B and S5C, the Δnfu strain had a growth defect when grown in the presence of these chemicals. Importantly, these phenotypes were corrected by genetic complementation.
PMNs generate ROS through the action of the enzyme NADPH oxidase, a multicomponent electron transferase that generates superoxide anion, which dismutates to H2O2 (Nauseef, 2008). In the presence of the PMN granule protein myeloperoxidase, released into phagosomes concomitantly with oxidase activation, H2O2 is consumed in the production of HOCl (Klebanoff et al., 2013). We examined the susceptibility of a nfu mutant strain to H2O2 and hypochlorite (-OCl) by culturing the WT and Δnfu mutant strains in the presence or absence of H2O2 or −OCl. As shown in Figures S5D and S5E the nfu mutant displayed heightened sensitivities to H2O2 and −OCl. The data in Figure S5 indicate that efficient metabolism of Fe-S clusters is a prerequisite for cellular survival when faced with ROS and RNS insult.
H2O2 toxifies a nfu mutant strain by interacting with Fe and damaging Fe-S clusters
The role of H2O2 in catalyzing endogenous Fenton chemistry is well established and previous studies found that strains with increased pools of intracellular non-incorporated iron display greater susceptibility towards H2O2 toxicity (Imlay et al., 1988, Keyer & Imlay, 1996).
We hypothesized that the increased non-incorporated Fe in the nfu mutant was reacting with H2O2 and resulting in cell death. We cultured the WT and Δnfu strains to different stages of growth and challenged with a bolus of H2O2 and the number of surviving bacteria were quantified. At every growth stage examined the Δnfu strain displayed decreased survival when challenged with H2O2 when compared to the WT strain and the phenotype could be genetically complemented (Figure 9A and data not shown). A number of alternate S. aureus clinical isolates lacking Nfu also displayed decreased survival (when compared to the respective parent strain) when challenged with H2O2 (Figure S4B)
Figure 9. H2O2 toxifies a nfu mutant strain by interacting with Fe and damaging Fe-S clusters.
Panel A: A nfu mutant strain is sensitive to H2O2 killing. Cultures were grown in TSB for 18 hours (optical density A600 ~10) before diluting, standardizing and challenging with H2O2. The data represent bacterial survival of the WT with pCM28 (black bars; JMB1100) or the Δnfu mutant (JMB1165) with pCM28 (light gray bars) or pCM28_nfu (dark gray bars). Panel B: Iron chelation protects a nfu mutant strain cultured to early-exponential growth phase from H2O2 killing. Cultures of the Δnfu strain (JMB1165) were grown to an optical density (A600) of 1 in TSB before challenging with H2O2. The cell permeable divalent metal chelator 2,2-dipyridyl (1mM) was added to one-half of the samples 20 minutes prior to H2O2 challenge. The WT strain did not display killing with 75 mM H2O2, and therefore, these data were not included in panel B. Panel C: A strain that accumulates endogenous H2O2 and lacks Nfu phenocopies the growth of a strain lacking aconitase. Representative growth traces of the ahpC (JMB4573; closed squares), Δnfu (JMB1165; open circles), acnA (JMB1163; closed triangles) and ahpC nfu double mutant (JMB2081; open squares) strains in TSB liquid are shown. Panel D: Intracellular H2O2 accumulation results in decreased activity of AcnA in a strain lacking Nfu. Strains were identical to those in Panel C. AcnA activity was monitored in cell-free lysates of cultures grown to an optical density of 5 (A600). Data are presented as the percent AcnA activity relative to the activity of the WT strain. The data in Panels A, B and D represent the averages of biological triplicates with standard deviations shown. Paired t-tests were performed on the data in Panels A, B and D and * denotes p< 0.05.
To examine whether Fe had a role in the killing of cells lacking Nfu, we repeated the H2O2 challenge assays in the presence and absence of the cell permeable divalent metal chelator DIP. We found that the inclusion of DIP increased the survival of Δnfu cells that had been grown to early exponential growth phase (<1 O.D. in TSB) (prior to challenge) (Figure 9B), but DIP did not provide protection to cells cultured to late-exponential or stationary phase (>1 O.D.; data not shown). The WT strain grown to early-exponential-phase had the same survival when challenged with or without 75 mM H2O2, and therefore, these data were not included in Figure 9B.
The inability of DIP to protect cells cultured to mid/late-exponential growth phase against H2O2 killing led to the hypothesis that H2O2 was toxifying the nfu mutant strain by more than one mechanism. Studies using E. coli have found that H2O2 has at least two modes of toxifying the cell and that H2O2 damages protein bound Fe-S clusters (Jang & Imlay, 2007, Imlay et al., 1988). We hypothesized that H2O2 was damaging Fe-S clusters and that the damage was more severe in a strain lacking Nfu. We examined the effect of intracellular H2O2 accumulation on a strain lacking Nfu. We monitored the growth of the nfu and ahpC single mutant strains, as well as, a nfu ahpC double mutant strain in TSB. We found that the nfu ahpC double mutant had a growth defect that was more severe than the growth defect of the nfu single mutant.
The growth defect of the nfu ahpC double mutant strain was manifested in the latter phases of growth, corresponding to the state of growth during which S. aureus switches from fermenting carbon to respiring fermentation byproducts (Ledala et al., 2014). The growth data suggested that the accumulation of intracellular ROS in the absence of AhpC toxifies the nfu mutant strain and is manifested as a decreased ability of the cell to respire. Growth on fermentative byproducts requires an optimally functioning TCA cycle and AcnA plays a pivotal role in TCA cycle function (Somerville et al., 2002). We hypothesized that the impaired growth of a nfu ahpC double mutant was, in part, due to damage to the Fe-S cofactor of the AcnA protein. We found that an acnA mutant strain grew similar to the nfu ahpC double mutant strain (Figure 9C).
We assessed AcnA activity in the cell-free lysates of the parent, nfu, ahpC, and nfu ahpC mutant strains. The activity of AcnA was only slightly decreased in lysates from an ahpC mutant strain, but the AcnA activity in the nfu ahpC double mutant was significantly less than that of the Δnfu strain (Figure 9D).
Collectively the data presented in Figure 9 show that 1) a nfu mutant strain has heightened sensitivity to H2O2, 2) DIP protects a nfu mutant strain from H2O2 toxicity, but only cells that were cultured to early-exponential growth phase, and 3) Fe-S clusters are a target of ROS toxicity in S. aureus.
Nfu contributes to virulence in S. aureus
The nfu mutant strain was more susceptible, under standard laboratory growth conditions, to H2O2 or −OCl, which are responsible for a majority of the ROS-mediated killing of staphylococci by PMNs (reviewed in (Nauseef, 2007a)). These data led us to hypothesize that a nfu mutant would have decreased survival when fed to PMNs. The WT and Δnfu mutant strains were fed to primary human PMNs and the fate of the ingested bacteria was determined. Prior to the addition of S. aureus the PMN were incubated in the presence or absence of diphenyleneiodonium chloride (DPI), which results in the pharmacological inhibition of the enzyme NADPH oxidase.
The fate of S. aureus was assessed using two methods. First, colony plating was used to determine the number of ingested bacteria that remained viable in PMNs over time. Second, ingested bacteria were recovered from PMNs and stained with the LIVE/DEAD stains SYTO9 and PI.
Within the first 10 minutes after ingestion by primary human PMNs, the viability of all three strains was similar (Figures 10A and 10B). Over 60 minutes the number of surviving bacteria in all strains decreased as they succumbed to the antimicrobial effects of the phagosomal milieu. The nfu mutant strain exhibited a significant decrease in survival relative to the WT strain as confirmed by both colony plating and LIVE/DEAD staining and this phenotype could be genetically complemented. A similar trend was also observed at 120 minutes. These data suggest that the presence of Nfu is necessary for optimal defense against the oxidative burst of host PMN and that the survival of S. aureus was compromised in its absence.
The physiological defects of the nfu mutant and the decreased survival in PMNs led to the hypothesis that a nfu mutant would be defective during in vivo infection. To test this hypothesis, we used a murine model of systemic infection. Mice were infected with the WT, Δnfu (nfu−) and the Δnfu complemented (nfu+) strains. Ninety-six hours post infection organs were dissected, homogenized, serially diluted and then plated to determine bacterial burden. The nfu mutant strain exhibited ~1.5 log reduction in bacterial burden in the liver compared to the WT strain and this phenotype was partially genetically complemented (Figure 10C). No difference in bacterial burden was observed in the kidneys or spleen (data not shown). Taken together, our data from the in vitro studies using human PMNs and in vivo data using a murine model of infection demonstrate that the presence of a functional Nfu in S. aureus is necessary for optimal defense against host PMN and replication in vivo.
Discussion
The goals of this study were to examine the effects of defective Fe-S cluster metabolism on S. aureus cellular physiology and pathogenesis. To our knowledge, the work presented here is the first to 1) examine the process of intracellular Fe metabolism in the staphylococci, 2) genetically and biochemically characterize the nfu locus in the staphylococci, and 3) examine Fe-S metabolism within the context of staphylococcal cellular physiology and pathogenesis.
The genome of S. aureus encodes for two potential Fe-S cluster-scaffolding systems (SufBCD and SufU) that are located in an apparent operon. Despite repeated attempts we were unable to obtain chromosomal deletions in genes encoding for SufB or SufU. Our findings are consistent with high-density transposon screens that were unable to isolate strains with a transposon in the sufCDSUB genes in S. aureus (Chaudhuri et al., 2009, Bae et al., 2004, Fey et al., 2013, Valentino et al., 2014). In E. coli, the Isc Fe-S cluster biosynthetic system predominates under non-stress growth conditions and the Suf system predominates under Fe limitation and ROS stress, but only one system is required for viability (Outten et al., 2004). The absence of a similar backup system in S. aureus likely places more emphasis on the Suf system making it indispensible.
We created a S. aureus strain crippled in Fe-S cluster metabolism by deleting the nfu gene. Data suggest that the nfu gene is essential for the viability of the bacterium Synechococcus sp. 7002 (Jin et al., 2008), but it is unknown why the nfu gene is not essential for S. aureus. In alternative organisms, a number of Fe-S cluster carrier molecules can interact with the biosynthetic components and functional overlap exists between the carrier molecules (Boyd et al., 2008a, Angelini et al., 2008, Vinella et al., 2009, Dos Santos et al., 2007, Bandyopadhyay et al., 2008). It is possible that a factor other than Nfu or SufA facilitates cluster trafficking in the absence of these carriers. It is also possible that SufBCD or SufU transfer Fe-S clusters directly to apo-proteins bypassing the necessity of trafficking molecules (Figure 1B). It should be noted that S. aureus does not produce glutathione nor does it possess homologues of Grx or ApbC, which are thought to serve trafficking functions (Shakamuri et al., 2012, Boyd et al., 2009a). Therefore, if an alternative trafficking mechanism exists in S. aureus it would be novel.
Our in vivo and in vitro data are consistent with the hypothesis that Nfu is an Fe-S cluster carrier protein. Chemically reconstituted Nfu binds ~2 Fe and ~2 S per monomer and holo-Nfu has a UV-Visible absorption spectrum similar to characterized Fe-S cluster binding proteins with ε280 = 12.7 mM−1 cm−1 and ε400 = 6.9 mM−1 cm−1, which are values similar to the ε280 and ε400 values reported for Nfu proteins (Bandyopadhyay et al., 2008, Gao et al., 2013, Smith et al., 2005). Chemically reconstituted S. aureus Nfu has a CD spectrum with positive bands at 280 and 450 nm and one negative band at 390 nm, which is comparable to the CD spectra of Fe-S cluster binding proteins, but different than the CD spectra of the Arabidopsis thaliana [Fe4-S4] Nfu2 that has peaks at 290 nm and 384 nm and a negative band at 440 nm or the [Fe2-S2] Nfu2 that has a peak at 360 nm and a valley at 400 nm.
Holo-Nfu can activate apo-aconitase, presumably through direct Fe-S cluster transfer, but the rate for holo-Nfu mediated activation of apo-AcnA is ~4 to 5-fold lower than the reported rates with which other characterized holo-Nfu proteins (or domains) activate apo-proteins (Gao et al., 2013, Py et al., 2012). Studies in our lab have also found that the S. aureus SufA protein activates apo-AcnA at a rate that is ~4 to 5-fold lower than the rates reported for the activation of apo-proteins by A-type carriers (Rosario-Cruz et al., under review). Importantly, during the duration of our assays we did not see activation of apo-AcnA when it was incubated with Fe2+, S2- and DTT in the absence of SufA or Nfu. The reason for the decreased rate of S. aureus AcnA activation by the proposed Fe-S cluster carriers is unknown and is currently under investigation.
Phenotypic analyses shed light on the importance of effective Fe-S cluster trafficking on staphylococcal physiology. The nfu mutant strain was defective in cellular respiration. S. aureus fermentation byproducts such as acetate are oxidized in the TCA cycle resulting in NADH, succinate, and malate, which serve as electron donors for respiratory pathways. A nfu mutant had decreased AcnA activity and would be expected to have decreased carbon flux through the TCA cycle. Thus, it is possible that the defect in respiration of a nfu mutant is not the result of impaired function of respiratory proteins, but rather a consequence of an impaired ability of the cell to produce sufficient reductant to drive respiration.
A nfu mutant strain had increased accumulation of ROS when cultured aerobically. Respiratory molecules are a source of intracellular ROS (Messner & Imlay, 1999, Seaver & Imlay, 2004, Messner & Imlay, 2002). In E. coli, an increase in O2 tension or allowing electrons to build on NADH oxidase results in an increased formation of H2O2 (Messner & Imlay, 1999). Proteins containing Fe-S clusters and/or heme mediate the movement of electrons through respiratory pathways. The biosynthesis of heme, a cofactor required for respiration, requires the Fe-S cluster dependent enzyme HemN (Layer et al., 2002). It is possible that the absence of heme and/or Fe-S clusters in a respiratory protein, such as succinate dehydrogenase, results in the buildup of electrons on a flavin bound to a respiratory enzyme allowing for dioxygen reduction and superoxide formation.
The transcriptional activity of the ahpC gene was decreased in a strain lacking Nfu providing an alternative mechanism for ROS accumulation. We also noted that transcription of the PerR regulated gene dps was derepressed in a nfu mutant. These data suggest that the PerR protein is sensing ROS in a nfu mutant and derepressing its regulon, which would be expected if the nfu mutant were accumulating H2O2. The ahpC gene is also under the transcriptional control of PerR (Horsburgh et al., 2001a). One explanation for these conflicting findings is that one or more alternative regulatory systems are negating the derepression of ahpC by PerR resulting in lower ahpC transcriptional activity in a nfu mutant.
Bacteria that live in aerobic environments have evolved mechanisms to tightly control the size of their non-incorporated intracellular pool of Fe (reviewed in (Hantke, 2001, Masse et al., 2007)). In light of this, it is interesting that a S. aureus strain lacking Nfu has an increased non-incorporated Fe pool. Studies by Nachin et al. found that E. chrysanthemi strains lacking the individual suf genes are sensitive to streptonigrin leading to the hypothesis that these strains have increased non-incorporated Fe (Nachin et al., 2001). Subsequent studies found that Fe incorporation into bacterioferritin was decreased in the suf mutants and demonstrated a functional link between Fe storage proteins and the Fe-S cluster metabolism machinery (Expert et al., 2008).
The nfu mutant strain had increased Fur accessible Fe despite having the same overall Fe load as the WT. These results suggest that increased Fur occupancy by Fe does not result in an overall decrease in Fe uptake in a nfu mutant. Previous studies in S. aureus found that accumulation of the Suf proteins was dependent upon Fe concentration, but independent of the Fur protein (Friedman et al., 2006). The findings by Friedman et al, in conjunction with the findings herein, suggest that S. aureus has a regulatory protein(s) or system(s), in addition to Fur, that responds to defects in Fe or Fe-S cluster status by increasing Fe uptake.
A nfu mutant had an increased DNA damage phenotype that was abrogated upon growth in the presence of either a cell permeable Fe chelator or by removing oxygen suggesting that Fenton chemistry was leading to the phenotype. Fe-S cluster oxidation by ROS can cause cluster disintegration (Imlay, 2003) and Fe-S cluster damage may contribute to the increase in non-chelated Fe in a nfu mutant strain cultured aerobically. A nfu mutant strain is also more sensitive to streptonigrin toxicity than the WT when cultured in the absence of oxygen suggesting that ROS alone was not causing increased non-chelated Fe pools.
Does the mammalian immune system target Fe-S cluster metabolism to kill or inhibit growth of S. aureus? Among the various bactericidal molecules generated and used by human PMNs to clear bacterial infections, ROS, such as H2O2 and HOCl, are critical for preventing and combating infection (Nauseef, 2007a). Solvent exposed Fe-S clusters are one of the primary targets of ROS and RNS toxicity and cluster oxidation can result in cluster destruction, inactivation, or modification (Soum et al., 2003, Hurst et al., 1991, Duan et al., 2009, Flint et al., 1993b, Keyer & Imlay, 1997, Jang & Imlay, 2007). Disintegration of Fe-S clusters results in the release of iron and subsequently an increase in the amount of non-incorporated intracellular Fe (Djaman et al., 2004). Damaged Fe-S clusters must be repaired or rebuilt. Consistent with these facts, in S. aureus the mRNA abundance from the suf and nfu genes increases upon oxidative stress and neutrophil phagocytosis (data herein and (Voyich et al., 2005)). Upregulation of these genes suggested that these cellular processes might be required to mitigate ROS stress and the environment encountered within the phagosomal milieu. We found that a S. aureus nfu mutant strain had a growth defect in the presence of ROS or RNS and was sensitive to killing with H2O2 and −OCl. Consistent with in vitro data we found that a nfu mutant had lower survival when challenged with PMNs and importantly this phenotype was dependent on the presence of a functional NADPH oxidase.
Our results using PMNs prompted us to investigate the importance of Fe-S cluster biogenesis within the context of a murine model of systemic infection. The nfu mutant displayed a defect in colonizing the liver. Intriguingly we found this to be a tissue specific defect in colonization since the nfu mutant colonized the kidneys and the spleen to levels observed with the WT strain. The role of the liver in facilitating the resolution of systemic bacterial infections is well documented (reviewed in (Mackaness, 1962)). Most bacteria that enter the bloodstream are sequestered within and rapidly cleared by the liver (Benacerrafet al., 1959). This function of the liver is mediated by a coordinated defense mechanism on the part of the body that results in the recruitment of PMNs to the liver (McDonald et al., 2012, Gregory et al., 1996). The PMNs in turn synergize with the Kupffer cells (fixed tissue macrophages) present in the liver sinusoids resulting in clearance of the bacterial infections (reviewed in (Gregory & Wing, 2002)). Indeed, studies found that neutrophil deficient mice are impaired in their ability to clear S. aureus infections via the liver (Gregory et al., 1996).
Our data strongly suggest that the sensitivity of a S. aureus nfu mutant to ROS results in decreased virulence. The decreased pathogenesis could be the result of defective re-metalation of apo-proteins or an inability to repair oxidized or modified clusters when faced with ROS insult. It is worth noting that during our in vitro experiments we found that chelating intracellular Fe protected S. aureus cells in early exponential growth-phase from H2O2-dependent killing. Our experiments using human PMNs were conducted using bacteria grown to early exponential growth-phase. It is tempting to speculate that Fenton chemistry is contributing to cell death of the nfu mutant in PMN.
In summary, in this study we used in vivo and in vitro experimentation to define a role for the Nfu protein. We then used a nfu mutant strain as a model system to examine the consequences of defective Fe-S metabolism on staphylococcal physiology and pathogenesis. The studies presented provide a framework for future studies examining the links between Fe-S cluster metabolism and staphylococcal pathogenesis.
Materials and Methods
Materials
Restriction enzymes, quick DNA ligase kit, deoxynucleoside triphosphates and Phusion DNA polymerase were purchased from New England Biolabs. The plasmid mini-prep kit, gel extraction kit and RNA protect were purchased from Qiagen. DNase I was purchased from Ambion. Lysostaphin was purchased from Ambi products. Oligonucleotides were purchased from Integrated DNA Technologies and sequences are listed in Table S1. Trizol, High-Capacity cDNA Reverse Transcription Kits and XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide) were purchased from Life Technologies. DL -Threo-3-isopropylmalic acid was purchased from Wako Pure Chemical Co. Tryptic Soy broth (TSB) was purchased from MP biomedical. Difco BiTek agar was added (15 g l−1) for solid medium. Unless specified all chemicals were purchased from Sigma-Aldrich and were of the highest purity available.
Bacterial strains, media, and growth conditions
Unless otherwise stated, the S. aureus strains used in this study (Table 1) were constructed in the community-associated S. aureus USA300_LAC strain that was cured of the native plasmid pUSA03 that confers erythromycin resistance (Boles et al., 2010). For aerobic growth S. aureus were cultured in 25 ml culture tubes containing 5 ml TSB at 37°C with shaking at 200 rpm unless otherwise indicated. For anaerobic growth cells were grown as described previously (Fuchs et al., 2007). A defined minimal medium was used for phenotypic analyses. The staphylococcal defined medium contained: 10 mg ml−1 (NH4)2SO4; 45 mg ml−1 KH2PO4, 105 mg ml−1 K2HPO4, 6.42 mg ml−1 NaCl, 2.23 mg ml−1 KCl, 0.5 μg ml−1 nicotinic acid, 0.5 μg ml−1 thiamine, 0.5 μg ml−1 pantothenic acid, 3 ng ml−1 biotin and 0.25 ng ml−1 of each individual amino acid. The 11 amino acid medium contained the following amino acids: PRMCHVYTFGL. Defined media contained 11 mM glucose as a carbon source. Top-agar overlays were created by diluting overnight TSB cultures 1:100 and adding 100 μl to 3.5 ml of 3.5% TSB-agar before laying over the top of TSB agar plates. Two μl of 2 mg ml−1 of streptonigrin, prepared in DMSO was spotted at the center of the plates. The minimal inhibitory concentration (MIC) for rifampicin was determined by following the protocols outlined by the Clinical and Laboratory Standards Institute (2009). When selecting for plasmids, antibiotics where added to the following concentrations: 150 μg ml−1 ampicillin; 30 μg ml−1 chloramphenicol (Cm); 10 μg ml−1 erythromycin (Erm); 3 μg ml−1 tetracycline (Tet); kanamycin, 125 μg ml−1 (Kan); anhydrotetracycline 150 ng ml−1; rifampicin 1.25 μg ml−1 (Rif). To maintain plasmids, the media was supplemented with 15 μg ml−1 or 5 μg ml−1 of chloramphenicol or erythromycin, respectively.
Recombinant DNA and genetic techniques
Escherichia coli PX5 was used as a cloning host for plasmid constructions. All clones were passaged through RN4220 (Kreiswirth et al., 1983) and subsequently transduced into the appropriate strains using bacteriophage 80α (Novick, 1991). All S. aureus mutant strains and plasmids were verified using PCR or by sequencing PCR products or plasmids. DNA sequencing was performed at Genewiz, South Plainfield, NJ.
Creation of plasmids and mutant strains
Unless otherwise specified chromosomal DNA from JMB1100 was used as the template for PCR reactions used in the construction of plasmids. Approximately 500 base pairs upstream and downstream of the nfu gene (SAUSA300_0839) was amplified using PCR and the following primer pairs; nfuup5EcoRI and nfuup3NheI; nfudwn3BamHI and nfudwn5MluI. Amplicons were joined by PCR using the nfuup5EcoRI and nfudwn3BamHI primer pair. The PCR product was, digested with EcoRI and SalI, and ligated into similarly digested pJB38 (Bose et al., 2013). Mutant strains were created as previously described (White et al., 2014). The primers nfuveri5 and nfuveri3 were used to verify the Δnfu mutation.
The ΔsufA (SAUSA300_0843) mutant was created using the same protocol as outlined above with the following exceptions: the upstream and downstream portions of sufA were amplified using the following primer pairs: 1) sufAup5EcoRI and sufAup3nheI; sufAdwn5MluI and sufAdwn3BamHI; and 2) the sufAup5EcoRI and sufAdwn3BamHI primers were used for joining the two PCR products.
The ftn (SAUSA300_1874) mutant was created using the same protocol as outlined above with the following exceptions: 1) the upstream and downstream portions of ftn were amplified using the following primer pairs: ftnup5SalI and ftnupfuse; ftndwnfuse and ftndownEcoRI; 2) the ftnup5SalI and ftndownEcoRI primers were used for joining the two PCR products.
The nfu::tet, sufA::tet, and nfu::kan strains were created by digesting the corresponding pJB38 plasmid with MluI and NheI and inserting the kan or tet genes between the up- and down-stream regions prior to mutant construction. The kan and tet alleles were a amplified using pDG783 or JMB1432 as template, respectively. The nfu::kan sufA::tet strain was created by transducing strain JMB1580 (nfu::kan) with the sufA::tet allele and scoring for Tet and Kan resistance. The nfu::kan, nfu::tet and Δnfu strains displayed equivalent decreases in AcnA activity and survival upon H2O2 challenge.
Additional plasmids were constructed by subcloning digested PCR products into similarly digested vectors or by using yeast homologous recombination cloning (YRC). Genes cloned into pEPSA5 (Forsyth et al., 2002) contained an engineered sodA ribosomal binding site. Genes cloned into pCM28 (Pang et al., 2010) or pLL39 (Luong & Lee, 2007) contained the native promoters. The pET20b_nfu and pET24a_acnA were constructed to encode a C-terminal polyhistidine affinity tag.
The pLL39_acnA_FLAG and pEPSA5_FLAG_leuCD plasmids were created using YRC as previously described (Joska et al., 2014). The pLL39_FLAG_srrAB plasmid was used as a template for the yeast cloning cassette (YCC) and the FLAG affinity tag (Joska et al., 2014). The pEPSA5 vector was linearized with SalI and the pLL39 vector was linearized with BamHI and SalI. The amplicons necessary for construction of pEPSA5_FLAG_leuCD were created using the following primer pairs: LeuCDFLAGfwd and LeuCDFLAGrev; LeuCDfwd and LeuCDrev; LeuCDyccfwd and LeuCDYccrev. The amplicons necessary for the construction of pLL39_acnA_FLAG were created using the following primer pairs: pLLYCC5 and yccAcnA3; YccAcnA5 and AcnAFLAG3; AcnAFLAG5 and FLAGpLL3.
Phenotypic analyses
Nutritional requirements were assessed in 200 μl cultures grown in defined media or TSB in 96-well plates using a BioTek 808E Visible absorption spectrophotometer with medium shake speed at 37 °C. Wells were inoculated using a 1:100 dilution from an overnight culture (>18 hours growth) that had been pelleted and suspended in 4x volume of phosphate buffered saline (PBS) (1:400 overall dilution). If present, sodium azide was included at 50 μM.
Growth analyses in the presence of ROS/RNS
Strains were grown overnight in TSB (~18-20 hours) to stationary phase. Optical density on all strains was standardized to 2.5 (A600) in a final volume of 1 ml of 1x PBS. 2 μl of the resuspended culture were used to inoculate 198 μl of the chemically defined medium in a 96-well microtiter plate. Growth was analyzed for each strain in the absence and the presence of oxidative stressors. A multichannel pipettor was used to amend the media with the indicated reactive oxygen/nitrogen species (hydrogen peroxide, sodium hypochlorite, methyl viologen, sodium nitroprusside) and mixed rapidly. Subsequently, strains were cultured with constant shaking at 37 °C in a microplate reader.
Transcriptional reporter fusion assays
Transcriptional reporter plasmids were constructed using either the pCM11 plasmid (Malone et al., 2009) or pXEN-1 (Xenogen Bioware). Strains containing pCM11 or pXEN-1 derivatives were grown overnight in TSB-Erm or TSB-Cm, respectively. The cultures were diluted 100-fold into fresh TSB (~0.1 A600) in a total volume of 5 ml and at periodic intervals 200 μl of cells were removed and assayed for culture optical density (A600) and fluorescence or luminescence using a Perkin Elmer Spectrometer HTS 7000 plus bioassay plate reader. GFP was excited at 485 nm and emission was read at 535 nm. For data analyses fluorescence or luminescence data were normalized with respect to culture optical density.
Cell free extract enzyme assays
Auxanographic analyses found that the pacnA, pleuCD, and pilvD genetically complemented acnA leuD, and ilvD mutant strains, respectively. Aconitase (AcnA) assays: Three ml of TSB was inoculated (1:50 dilution) with overnight cultures grown in TSB. When acnA was under the transcriptional control of the native promoter cultures were grown to an optical density A600 of 5 (Figure 9D) or 8 (Figure 2A and 2C) before the cultures were harvested by centrifugation. When acnA was under the transcriptional control of xylO, cultures were grown to an optical density of 2 (A600), 1% xylose was added, and the cultures were grown for an additional 3 hours before harvesting. Cellular pellets were placed in an anaerobic chamber and allowed to equilibrate for 10 min before suspension in 400 μl anaerobic lysis buffer (25 mM Tris-citrate, 150 mM NaCl, pH 7.4). Cells were lysed by the addition of 4 μg lysostaphin and 8 μg DNAse and incubated at 37 °C until confluent lysis was observed (~30 min). The cellular lysates were clarified using a 2 min high-speed spin. Twenty μl of extract was added to 680 μl of lysis buffer containing 20 mM DL-isocitrate. Aconitase activity was determined by monitoring the conversion of isocitrate to cis-aconitate spectrophotometrically using a Beckman Coulter DU530 UV-Vis absorption spectrophotometer (cis-aconitate ε240 nm = 3.6 mM−1cm−1 (Kennedy et al., 1983)).
For AcnA assays using anaerobically cultured S. aureus, strains were grown in 15 ml conical tubes containing 15 ml of TSB with and without 1% xylose. Strains were inoculated by diluting overnight cultures to a final optical density (A600) of 0.1 and strains were grown for 8 hours before harvesting by centrifugation. After centrifugation cells were processed as outlined above. For cluster conversion AcnA assays cells containing pAcnA were cultured as outlined above. Cells were lysed anaerobically and 100 μM Fe was added to the cell free supernatant. Samples were incubated anaerobically for 25 min at room temperature before AcnA activity assessed.
Isopropylmalate isomerase (LeuCD) assays
Three ml of TSB was inoculated (1:50 dilution) with overnight cultures grown in TSB to an optical density (A600) of 2, 1% xylose was added and the cultures were grown for an additional 3 hours before harvesting. Cell pellets were handled and lysed as described above. LeuCD was assayed by the addition of 20 μl of extract to 680 μl of assay buffer A (50 mM Tris, pH 8.0, 10 mM MgCl2, and 10 mM DL -Threo-3-isopropylmalic acid). LeuCD activity was assayed for the ability to convert 3-isopropylmalate to dimethylcitraconate acid spectrophotometrically (dimethylcitraconate ε235 nm = 4.35 mM−1cm−1 (Gross et al., 1963)) as described in (Boyd et al., 2008b).
Dihydroxy-acid dehydratase (IlvD) assays
Cultures were grown and lysed under conditions identical to those used to assess LeuCD activity. IlvD activity was assessed by adding 20 μl of cell free extract to 680 μl assay buffer B (50 mM Tris, pH 8.0, 10 mM MgCl2, and 10 mM D,L-2,3-dihydroxy-isovalerate). The ability of IlvD to produce keto acids from D,L-2,3-dihydroxy-isovalerate was monitored spectrophotometrically (keto acids ε240nm = 0.19 mM−1cm−1) (Flint et al., 1993a).
mRNA isolation and RT-PCR
Overnight cultures grown in TSB were diluted 100-fold into fresh TSB (~0.1 A600). For induction experiments strains were grown to an optical density of 6.5 A600 and one set of cultures was treated with either 10 mM hydrogen peroxide or 800 μM 2,2-dipyridyl. Cells were grown for an additional 25 minutes and harvested. For experiments in the presence and absence of an electron acceptor, cells were grown in capped microcentrifuge tubes until anaerobiosis was achieved (~1 A600). Anaerobic conditions were verified by addition of 0.001% resazurin to control tubes and the medium color was monitored over time. Cultures were grown for another 30 minutes to equilibrate completely to fermentative growth. Oxygen was reintroduced to one set of cultures by placing the cultures into glass tubes in an aerobic environment with a liquid to headspace ratio of 1:15. Cells were grown for an additional 25 minutes before 0.5 ml aliquots of cells were harvested and treated with RNAProtect reagent for 10 minutes at room temperature and pelleted by centrifugation. The pellets were washed once with 1 ml of lysis buffer (50 mM Tris, pH 8; RNase free). Cells were resuspended in 150 μl lysis buffer with 15 μg of lysostaphin and incubated at 37 °C until confluent lysis was observed (~30 min). RNA was extracted using Trizol reagent as per manufacturer instructions. The RNA extract was subsequently treated with DNase. cDNA libraries were constructed using isolated RNA as template. An Applied Biosystems StepOnePlus thermocycler was used to quantify DNA abundance. Primers for quantitative real-time PCR (qPCR) were designed using the Primer Express 3.0 software from Applied Biosystems.
Hydrogen peroxide killing assays
Cultures of S. aureus strains were grown to various stages of growth in TSB before harvesting. Cells were pelleted, supernatant decanted, pellets resuspended in 4x volume of PBS and the optical density recorded. Cells were diluted to optical density 0.25 (A600 ) with PBS in a final volume of 1 ml. Hydrogen peroxide (30% solution) or was added to a concentration of 30-1500 mM and cultures were incubated for one hour. The reaction was quenched by adding 50 μl of cell suspension to 950 μl of a 1300 U ml−1 catalase solution prepared in PBS. Bacterial viability was determined by serial dilution and plating on solid TSB.
Immunoblot analyses
A total of 40 or 80 μg of total protein (construct with a inducible or native promoter, respectively) was separated using a 12% SDS-PAGE gel. Proteins were then transferred to a PVDF membrane and incubated with mouse monoclonal anti-FLAG primary antibody (Sigma-Aldrich) (1:4000 dilution) and subsequently HRP conjugated secondary antibody (Bio-Rad) (1:12000 dilution). The blots were developed using chemiluminescent detection (ECL kit, Pierce). The blots were scanned as high quality TIFF images.
Quantification of endogenous ROS
DHCF-DA oxidation was measured as previously described (Nobre et al., 2010) with the following changes. Overnight TSB cultures were the diluted 100-fold into fresh TSB (~0.1 A600) and grown with shaking until all strains reached an optical density (A600) of 7. Cells were harvested by centrifugation and washed with PBS. DHCF-DA was added to cell suspensions to a final concentration of 10 μM. Fluorescence intensities were measured at 535 nm after exciting at 485 nm. Data was normalized with respect to a) a buffer blank containing 10 μM DHCF-DA to compensate for its autofluorescence, and b) optical density (A595) for each culture. Finally, data were plotted as percent increase in fluorescence, which negates differences in dye permeabilization in the diverse strain backgrounds.
Spontaneous mutagenesis frequency determination
Cultures propagated overnight in TSB were diluted 100-fold into fresh TSB (~0.1 A600) and grown at 37 °C with shaking for 24 hours before plating. For aerobic growth experiments, 1 ml cultures were grown with or without 200 μM 2,2-dipyridyl in 7 ml culture tubes. For anaerobic growth, 625 μl cultures were grown in sealed microcentrifuge tubes. Anaerobic conditions were verified by addition of 0.001% resazurin to control tubes and the medium color was monitored over time. Anaerobiosis was achieved at ~2 hours post inoculation. After 24 hours, 100 μl of each replicate was plated on TSB agar plates containing 1.25 μg ml−1 of Rif. Plates were incubated at 37 °C for 24 hours and CFUs determined. Aliquots of each replicate was also serially diluted and plated on TSB agar plates to determine the total colony forming units plated on the rifampicin plates. Mutational frequency was defined as the ratio of rifampicin resistant colonies to total CFUs.
Determination of whole cell iron concentrations
Overnight cultures grown in TSB in 15 ml metal free conical tubes were sub-cultured into 5 ml of TSB in a 15 ml metal free conical tube at a ratio of 1 to 100. Cultures were grown at 37°C 180 rpm until an optical density (A600) of 5.5-6.0 was observed. Cells were pelleted by centrifugation, supernatants removed, and the pellets washed with 10 ml of 0.5 M EDTA to remove extracellular metals. Cells were washed two more times with 1 ml of deionized water before suspending in 1 ml of deionized water. An aliquot of 0.5 ml was transferred into a metal free tube and 1 ml of 50% nitric acid was added (Fischer Optima Ultra-pure). Samples were incubated at 50 °C overnight and then diluted with 9 ml of deionized water.
The quantitative analysis of Fe56 in digested samples was performed on a Thermal Element II double-focusing sector field high-resolution inductively coupled plasma mass spectrometer (HR-ICPMS, Thermo Fisher Scientific, Bremen, Germany) equipped with ESI auto sampler (Elemental Scientific, Omaha, NE). The sample uptake was achieved through self-aspiration via a 0.50 mm ID sample probe and sample capillary. The Fe concentration was determined using the following operation parameters: RF power, 1250W; Cool gas, 16.00 L min−1; Auxiliary gas, 0.83 L min−1; Sample gas 1.03 L min−1; Resolution mode medium; 10 runs with 1 pass; 20 samples per peak; samples time 0.01 sec.
Recombinant Protein Over-Production and purification
Escherichia coli strains BL21(AI*) containing a protein production vector were grown at 37 °C in a 3 L Fernbach flasks containing 1 L of 2X standard LB medium. Cultures that had been grown to an optical density (A600) of 0.6 were cooled to 25 °C and arabinose (1 mM) and IPTG (0.1 mM) were added. Cultures were grown for an additional 12 hours before cells were harvested by centrifugation. Cell paste was flash frozen with liquid nitrogen and stored at -80 °C.
Frozen cell paste was suspended in two volumes of buffer A (50 mM Tris-HCl, pH 8.0, 150 mM NaCl) containing DNase (0.03 mg ml−1). Cell suspensions were passed three times through a chilled French pressure cell at 4 °C. Cell lysates were clarified by centrifugation (39,000 × g for 40 min at 4 °C). The cell extract was loaded onto a 1.6 × 10 cm pre-equilibrated column containing Ni2+-loaded Chelating Sepharose Fast Flow resin (Qiagen) and washed with 20 column volumes of 50 mM Tris, pH 8.0, 1 M NaCl. The column was equilibrated with buffer A and recombinant protein was eluted during a 30-column volume linear gradient from 0 to 100% elution buffer (50 mM Tris, pH 8.0, 250 mM imidazole). Fractions that contained the protein of interest at >95% purity by SDS-PAGE analysis were pooled and dialyzed overnight in 50 mM Tris-HCl, pH 8.0, 10% (v/v) glycerol and 150 mM NaCl. The Nfu and AcnA proteins were concentrated over a 3,000 or a 30,000 Da molecular mass cutoff membranes (Amicon), respectively. Finally, the proteins were pelleted into liquid nitrogen and stored at -80 °C until needed. All steps were performed at 4 °C and buffers used for dialysis had the pH adjusted at 4 °C.
Iron and sulfur determination was conducted as previously described (Lovenberg et al., 1963, Smith et al., 1984, Boyd et al., 2008b). The AcnA Fe-S cluster was destroyed and iron was removed from the sample as previously described (Kennedy & Beinert, 1988). EDTA was removed from the AcnA solution by dialysis against buffer A and three successive buffer exchanges. Protein concentration was determined using a copper/bicinchonic acid based colorimetric assay modified for a 96-well plate (Olson & Markwell, 2007). Bovine serum albumin (2 mg ml−1) was used as a standard.
Anaerobic Work
Anaerobic work was performed using a Coy anaerobic glove-box (Grass Lake, MI) or vacuum manifold. Bacterial growth on solid agar was conducted in an air incubator within the Coy chamber. Solutions, plastic-ware, and liquid and solid growth medium was allowed to equilibrate for >6 h inside the glove-box before use.
Fe-S cluster reconstitution
All the steps were performed under strictly anaerobic conditions (<1 ppm oxygen). Purified Nfu was adjusted to 60 μM in reconstitution buffer (50 mM Tris, 150 mM NaCl, 5 mM DTT, pH 7.5) and incubated anaerobically for one hour. Cluster reconstitution was initiated by adding a 5-fold excess of ferrous ammonium sulfate and lithium sulfide (300 μM) as previously described (Boyd et al., 2009b). The reaction mixture was allowed to proceed for one hour before excess Fe, S, and DTT were removed by desalting using a PD-10 column (GE Healthcare) that had been pre-equilibrated with reconstitution buffer. Reconstituted protein was concentrated using YM-3 Centriplus Centrifugal Concentrators (Millipore), prior to use in activity assays. UV-Visible absorption spectra were recorded using a Beckman-Coulter DU800 spectrophotometer. Circular dichroism spectra were recorded using an AVIV model 62A (Aviv Associates, Lakewood, NJ) spectropolarimeter.
Fe-S transfer assay
All steps were performed under anaerobic conditions (<1 ppm oxygen), in a Coy Anaerobic chamber. Purified AcnA, which had been stripped of Fe, was incubated in reconstitution buffer for 30 minutes. Cluster transfer mixtures were obtained by mixing together 4 μM AcnA and between 0-16 μM reconstituted Nfu, to a final volume of 20 μl. As a control, AcnA was mixed with 16 μM Fe2+ and 16 μM S2-. Mixtures were incubated at room temperature (23 °C) for the indicated amounts of time at which point 16 μl aliquots were removed and assayed for AcnA activity. Activity assays were performed in final volume of 1 ml and contained reconstitution buffer with 7.5 mM isocitrate. AcnA activity was determined by monitoring the conversion of cis-aconitate to isocitrate spectrophotometrically (cis-aconitate ε240 nm = 3.6 mM−1 cm−1 (Kennedy et al., 1983)).
Ethics Statement
Written informed consent was obtained from each individual following a protocol approved by the Institutional Review Board for human subjects at the University of Iowa. Peripheral blood was drawn from normal healthy volunteers and PMN were purified as previously described (Nauseef, 2007b). Animal work in this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the Animal Welfare Act and US federal law. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of New York University School of Medicine (protocol 120401).
PMN Killing Assay
Mid-log phase bacteria were pelleted for 5 minutes and resuspended in 20 mM HEPES buffered Hank's balanced salt solution (HBSS) containing Ca2+ and Mg2+. The measured absorbance at 550 nm of the suspension was converted to CFU ml−1; an A550 reading of 1.0 being equivalent to 3x108 CFU ml−1. Bacteria were used immediately or held on ice. Bacteria were opsonized by incubating the bacteria at 37°C for 20 minutes in the presence of 10% pooled human serum. Opsonized bacteria were fed to PMN or PMN pre-treated with 10 μM of diphenyleneiodonium chloride (DPI) at the desired MOI and tumbled end over end at 37°C to allow phagocytosis to proceed over 10 minutes. The PMN were then pelleted at 380x g for 5 minutes and the extracellular bacteria were aspirated away. The cell pellets were then resuspended to the original volume with 20 mM HEPES buffered HBSS. A 50 μl sample of this suspension was removed at the time = 10 time point. The remaining samples were tumbled at 37°C and 50 μL aliquots were removed at specified time points. For viability testing, the 50 μL samples were diluted and incubated with 2.5 ml of pH 11 water for 5 minutes to lyse the PMN (Decleva et al., 2006).
The released bacteria were then assessed for viability by colony plating or live/dead staining. For colony plating, these samples were vortexed and serial diluted into saline. Ten μL of each diluted sample were spotted onto TSA in at least triplicates. After 16 hours of growth at 37 °C the CFU were enumerated. From these the CFUs in the total PMN suspension for each sample was calculated. For assessing viability at each time point, the total calculated CFUs for each strain was expressed as a percentage of the starting CFUs for that strain. Because killing of ingested staphylococci by human PMN is completely inhibited by DPI (Decleva et al., 2006), the starting CFUs for each strain was taken as the CFUs recovered from bacteria fed to DPI-treated PMN at time = 0.
For live and dead staining, the bacterial samples were first supplemented with 4 mM HEPES buffer, pH 7-7.6, containing 150 mM sodium chloride. These were then stained with 0.3 nM of SYTO9 and 0.6 μM of propidium (PI) from the LIVE/DEAD BacLight Bacterial Viability Kit (Invitrogen). Both the membrane permeable dye SYTO9 and the impermeable dye PI bind to nucleic acids, although PI can quench SYTO9 fluorescence and the stains may also compete for access to nucleic acids (Stocks, 2004). Consequently, living bacteria with intact cell walls stain with SYTO9, whereas bacteria with compromised integrity will exhibit PI fluorescence. Staining of the bacteria recovered from PMN was assessed using the Accuri C6 flow cytometer. The total number of scorable events was defined as events that stained with only PI or Syto9. Events scoring double negative or double positive were not included due to their uncharacterized nature.
Murine model of infection
For in vivo experiments, 5-6-week old female ND4 Swiss Webster (Harlan) were used (Alonzo et al., 2012) and they were anesthetized with 250 μl of Avertin (2,2,2-tribromoethanol dissolved in tert-amyl-alcohol and diluted to a final concentration of 2.5% v/v in sterile saline), followed by retro-orbital injection of 1 × 107 CFU of isogenic S. aureus LAC strains (JMB1886, JMB1888, and JMB1889). 96-hours post infection, mice were sacrificed and organs were harvested and homogenized to evaluate the bacterial burden (CFUs).
Supplementary Material
Acknowledgements
The Boyd lab is supported by Rutgers University, the Charles and Johanna Busch foundation and USDA MRF project NE−1028. A.M. is supported by the Douglas Eveleigh fellowship from the Microbial Biology Graduate Program and Rutgers University. The Nauseef lab is supported by grants AI070958 AI044642. The Nauseef lab is also supported by a Merit Review award and use of facilities at the Iowa City Department of Veterans Affairs (VA) Medical Center, Iowa City, IA 52246. The Skaar lab is supported by the NIH grant AI069233. The Torres lab was supported by New York University School of Medicine development funds. M.A.B. was supported by an American Heart Association predoctoral fellowship (10PRE3420022). The authors would like to thank William Belden and Peter Kahn for the use of their real-time thermocycler and CD-spectrophotometer, respectively. We would also like to thank Kerrie May for editing the manuscript and providing valuable suggestions.
References
- Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht AG, Netz DJ, Miethke M, Pierik AJ, Burghaus O, Peuckert F, Lill R, Marahiel MA. SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J Bacteriol. 2010;192:1643–1651. doi: 10.1128/JB.01536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F, 3rd, Benson MA, Chen J, Novick RP, Shopsin B, Torres VJ. Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol Microbiol. 2012;83:423–435. doi: 10.1111/j.1365-2958.2011.07942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S, Gerez C, Ollagnier-de Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283:14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- Annane D, Sanquer S, Sebille V, Faye A, Djuranovic D, Raphael JC, Gajdos P, Bellissant E. Compartmentalised inducible nitric-oxide synthase activity in septic shock. Lancet. 2000;355:1143–1148. doi: 10.1016/S0140-6736(00)02063-8. [DOI] [PubMed] [Google Scholar]
- Arenas FA, Covarrubias PC, Sandoval JM, Perez-Donoso JM, Imlay JA, Vasquez CC. The Escherichia coli BtuE protein functions as a resistance determinant against reactive oxygen species. PLoS One. 2011;6:e15979. doi: 10.1371/journal.pone.0015979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Damage to the bases in DNA induced by hydrogen peroxide and ferric ion chelates. Journal of Biological Chemistry. 1989;264:20509–20512. [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U S A. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Shen G, Bryant DA, Golbeck JH. Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 2006;188:3182–3191. doi: 10.1128/JB.188.9.3182-3191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Naik SG, O'Carroll IP, Huynh BH, Dean DR, Johnson MK, Dos Santos PC. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J Biol Chem. 2008;283:14092–14099. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley FC, Vines ED, Grigg JC, Zheng Q, Liu S, Lajoie GA, Murphy ME, Heinrichs DE. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol Microbiol. 2009;72:947–963. doi: 10.1111/j.1365-2958.2009.06698.x. [DOI] [PubMed] [Google Scholar]
- Benacerraf B, Sebestyen MM, Schlossman S. A quantitative study of the kinetics of blood clearance of P32-labelled Escherichia coli and Staphylococci by the reticuloendothelial system. J Exp Med. 1959;110:27–48. doi: 10.1084/jem.110.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MV, Herst PM, Tan AS. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnology annual review. 2005;11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzan AD, Bianchi MS. Genotoxicity of streptonigrin: a review. Mutation research. 2001;488:25–37. doi: 10.1016/s1383-5742(00)00062-4. [DOI] [PubMed] [Google Scholar]
- Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Drevland RM, Downs DM, Graham DE. Archaeal ApbC/Nbp35 homologs function as iron-sulfur cluster carrier proteins. J Bacteriol. 2009a;191:1490–1497. doi: 10.1128/JB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Lewis JA, Escalante-Semerena JC, Downs DM. Salmonella enterica requires ApbC function for growth on tricarballylate: evidence of functional redundancy between ApbC and IscU. J Bacteriol. 2008a;190:4596–4602. doi: 10.1128/JB.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Pierik AJ, Netz DJ, Lill R, Downs DM. Bacterial ApbC can bind and effectively transfer iron-sulfur clusters. Biochemistry. 2008b;47:8195–8202. doi: 10.1021/bi800551y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Sondelski JL, Downs DM. Bacterial ApbC protein has two biochemical activities that are required for in vivo function. J Biol Chem. 2009b;284:110–118. doi: 10.1074/jbc.M807003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahal HK, Outten FW. Separate FeS scaffold and carrier functions for SufB(2)C(2) and SufA during in vitro maturation of [2Fe2S] Fdx. Journal of inorganic biochemistry. 2012;116:126–134. doi: 10.1016/j.jinorgbio.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC genomics. 2009;10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Beasley FC, Liu S, Lajoie GA, Heinrichs DE. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol Microbiol. 2009;74:594–608. doi: 10.1111/j.1365-2958.2009.06880.x. [DOI] [PubMed] [Google Scholar]
- Decleva E, Menegazzi R, Busetto S, Patriarca P, Dri P. Common methodology is inadequate for studies on the microbicidal activity of neutrophils. Journal of leukocyte biology. 2006;79:87–94. doi: 10.1189/jlb.0605338. [DOI] [PubMed] [Google Scholar]
- Djaman O, Outten FW, Imlay JA. Repair of oxidized iron-sulfur clusters in Escherichia coli. Journal of Biological Chemistry. 2004 doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- Dos Santos PC, Johnson DC, Ragle BE, Unciuleac MC, Dean DR. Controlled expression of nif and isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J Bacteriol. 2007;189:2854–2862. doi: 10.1128/JB.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos PC, Smith AD, Frazzon J, Cash VL, Johnson MK, Dean DR. Iron-sulfur cluster assembly: NifU-directed activation of the nitrogenase Fe protein. J Biol Chem. 2004;279:19705–19711. doi: 10.1074/jbc.M400278200. [DOI] [PubMed] [Google Scholar]
- Duan X, Yang J, Ren B, Tan G, Ding H. Reactivity of nitric oxide with the [4Fe-4S] cluster of dihydroxyacid dehydratase from Escherichia coli. Biochem J. 2009;417:783–789. doi: 10.1042/BJ20081423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert D, Boughammoura A, Franza T. Siderophore-controlled iron assimilation in the enterobacterium Erwinia chrysanthemi: evidence for the involvement of bacterioferritin and the Suf iron-sulfur cluster assembly machinery. J Biol Chem. 2008;283:36564–36572. doi: 10.1074/jbc.M807749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezekiel DH, Hutchins JE. Mutations affecting RNA polymerase associated with rifampicin resistance in Escherichia coli. Nature. 1968;220:276–277. doi: 10.1038/220276a0. [DOI] [PubMed] [Google Scholar]
- Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4:e00537–00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DH, Emptage MH, Finnegan MG, Fu W, Johnson MK. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. Journal of Biological Chemistry. 1993a;268:14732–14742. [PubMed] [Google Scholar]
- Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993b;268:22369–22376. [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, King KGC,P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Zy ZY, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2006;2:e87. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Jack RF, Morgan TV, Dean DR, Johnson MK. nifU gene product from Azotobacter vinelandii is a homodimer that contains two identical [2Fe- 2S] clusters. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol. 2007;189:4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Subramanian S, Couturier J, Naik SG, Kim SK, Leustek T, Knaff DB, Wu HC, Vignols F, Huynh BH, Rouhier N, Johnson MK. Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster1338 containing proteins. Biochemistry. 2013;52:6633–6645. doi: 10.1021/bi4007622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves SF, Kobayashi SD, DeLeo FR. Community-associated methicillin1343 resistant Staphylococcus aureus immune evasion and virulence. J Mol Med (Berl) 2010;88:109–114. doi: 10.1007/s00109-009-0573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SH, Sagnimeni AJ, Wing EJ. Bacteria in the bloodstream are trapped in the liver and killed by immigrating neutrophils. J Immunol. 1996;157:2514–2520. [PubMed] [Google Scholar]
- Gregory SH, Wing EJ. Neutrophil-Kupffer cell interaction: a critical component of host defenses to systemic bacterial infections. Journal of leukocyte biology. 2002;72:239–248. [PubMed] [Google Scholar]
- Gross SR, Burns RO, Umbarger HE. The Biosynthesis of Leucine. II. the Enzymic Isomerization of Beta-Carboxy-Beta-Hydroxyisocaproate and Alpha- Hydroxy-Beta-Carboxyisocaproate. Biochemistry. 1963;2:1046–1052. doi: 10.1021/bi00905a023. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic1355 resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- Haber F, Weiss J. On the catalysis of hydroperoxide. Naturwissenschaften. 1932;20:948–950. [Google Scholar]
- Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio. 2013;4 doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annual review of microbiology. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Argos P. Homology between IRE-BP, a regulatory RNA1366 binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res. 1991;19:1739–1740. doi: 10.1093/nar/19.8.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun. 2001a;69:3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol. 2001b;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst JK, Barrette WC, Jr., Michel BR, Rosen H. Hypochlorous acid and myeloperoxidase-catalyzed oxidation of iron-sulfur clusters in bacterial respiratory dehydrogenases. Eur J Biochem. 1991;202:1275–1282. doi: 10.1111/j.1432-1033.1991.tb16500.x. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Pathways of oxidative damage. Annual review of microbiology. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- Institute, C.a.L.S. CLSA document M07-A8. Clinical and Laboratory Standards Institute; Wayne, PA: 2009. CLSI. Methods for dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approvied Standard-Eighth Edition. [Google Scholar]
- Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Heinnickel M, Krebs C, Shen G, Golbeck JH, Bryant DA. Biogenesis of iron-sulfur clusters in photosystem I: Holo-NfuA from the cyanobacterium Synechococcus sp. PCC 7002 rapidly and efficiently transfers [4Fe-4S] clusters to apo-PsaC in vitro. J Biol Chem. 2008 doi: 10.1074/jbc.M803395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joska TM, Mashruwala A, Boyd JM, Belden WJ. A universal cloning method based on yeast homologous recombination that is simple, efficient, and versatile. J Microbiol Methods. 2014 doi: 10.1016/j.mimet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MC, Beinert H. The state of cluster SH and S2- of aconitase during cluster interconversions and removal. A convenient preparation of apoenzyme. J Biol Chem. 1988;263:8194–8198. [PubMed] [Google Scholar]
- Kennedy MC, Emptage MH, Dreyer JL, Beinert H. The role of iron in the activation-inactivation of aconitase. Journal of Biological Chemistry. 1983;258:11098–11105. [PubMed] [Google Scholar]
- Kent TA, Dreyer JL, Kennedy MC, Huynh BH, Emptage MH, Beinert H, Munck E. Mossbauer studies of beef heart aconitase: evidence for facile interconversions of iron-sulfur clusters. Proc Natl Acad Sci U S A. 1982;79:1096–1100. doi: 10.1073/pnas.79.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free1419 iron levels. Proc. Natl. Acad. Sci. USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Inactivation of dehydratase [4Fe-4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J Biol Chem. 1997;272:27652–27659. doi: 10.1074/jbc.272.44.27652. [DOI] [PubMed] [Google Scholar]
- Klebanoff SJ, Kettle AJ, Rosen H, Winterbourn CC, Nauseef WM. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. Journal of leukocyte biology. 2013;93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Layer G, Grage K, Teschner T, Schunemann V, Breckau D, Masoumi A, Jahn M, Heathcote P, Trautwein AX, Jahn D. Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines. J Biol Chem. 2005;280:29038–29046. doi: 10.1074/jbc.M501275200. [DOI] [PubMed] [Google Scholar]
- Layer G, Verfurth K, Mahlitz E, Jahn D. Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J Biol Chem. 2002;277:34136–34142. doi: 10.1074/jbc.M205247200. [DOI] [PubMed] [Google Scholar]
- Ledala N, Zhang B, Seravalli J, Powers R, Somerville GA. Influence of iron and aeration on Staphylococcus aureus growth, metabolism, and transcription. J Bacteriol. 2014;196:2178–2189. doi: 10.1128/JB.01475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community1444 associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovenberg W, Buchanan BB, Rabinowitz JC. Studies on the Chemical Nature of Clostridial Ferredoxin. J Biol Chem. 1963;238:3899–3913. [PubMed] [Google Scholar]
- Luong TT, Lee CY. Improved single-copy integration vectors for Staphylococcus aureus. J Microbiol Methods. 2007;70:186–190. doi: 10.1016/j.mimet.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackaness GB. Cellular resistance to infection. J Exp Med. 1962;116:381–406. [PubMed] [Google Scholar]
- Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol. 2010;192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringanti S, Imlay JA. An intracellular iron chelator pleiotropically suppresses enzymatic and growth defects of superoxide dismutase-deficient Escherichia coli. J Bacteriol. 1999;181:3792–3802. doi: 10.1128/jb.181.12.3792-3802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- McDonald B, Urrutia R, Yipp BG, Jenne CN, Kubes P. Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell Host Microbe. 2012;12:324–333. doi: 10.1016/j.chom.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. Journal of Biological Chemistry. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- Morrissey JA, Cockayne A, Brummell K, Williams P. The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect Immun. 2004;72:972–979. doi: 10.1128/IAI.72.2.972-979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhre O, Andersen JM, Aarnes H, Fonnum F. Evaluation of the probes 2',7'-dichlorofluorescin diacetate, luminol, and lucigenin as indicators of reactive species formation. Biochem Pharmacol. 2003;65:1575–1582. doi: 10.1016/s0006-2952(03)00083-2. [DOI] [PubMed] [Google Scholar]
- Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol Microbiol. 2001;39:960–972. doi: 10.1046/j.1365-2958.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunological reviews. 2007a;219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- Nauseef WM. Isolation of human neutrophils from venous blood. Methods Mol Biol. 2007b;412:15–20. doi: 10.1007/978-1-59745-467-4_2. [DOI] [PubMed] [Google Scholar]
- Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283:16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio K, Nakai M. Transfer of iron-sulfur cluster from NifU to apoferredoxin. J. Biol. Chem. 2000;275:22615–22618. doi: 10.1074/jbc.C000279200. [DOI] [PubMed] [Google Scholar]
- Nobre LS, Todorovic S, Tavares AF, Oldfield E, Hildebrandt P, Teixeira M, Saraiva LM. Binding of azole antibiotics to Staphylococcus aureus flavohemoglobin increases intracellular oxidative stress. J Bacteriol. 2010;192:1527–1533. doi: 10.1128/JB.01378-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- Olson BJ, Markwell J. Assays for determination of protein concentration. Current protocols in protein science / editorial board, John E. Coligan ... [et al.] 2007 doi: 10.1002/0471140864.ps0304s48. Chapter 3: Unit 3 4. [DOI] [PubMed] [Google Scholar]
- Orme-Johnson WH, Hansen RE, Beinert H, Tsibris JC, Bartholomaus RC, Gunsalus IC. On the sulfur components of iron-sulfur proteins. I. The number of acid-labile sulfur groups sharing an unpaired electron with iron. Proc Natl Acad Sci U S A. 1968;60:368–372. doi: 10.1073/pnas.60.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B, Barras F. Building Fe-S proteins: bacterial strategies. Nat Rev Microbiol. 2010;8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- Py B, Gerez C, Angelini S, Planel R, Vinella D, Loiseau L, Talla E, Brochier-Armanet C, Garcia Serres R, Latour JM, Ollagnier-de Choudens S, Fontecave M, Barras F. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol Microbiol. 2012;86:155–171. doi: 10.1111/j.1365-2958.2012.08181.x. [DOI] [PubMed] [Google Scholar]
- Rauen U, Springer A, Weisheit D, Petrat F, Korth HG, de Groot H, Sustmann R. Assessment of chelatable mitochondrial iron by using mitochondrion1517 selective fluorescent iron indicators with different iron-binding affinities. Chembiochem : a European journal of chemical biology. 2007;8:341–352. doi: 10.1002/cbic.200600311. [DOI] [PubMed] [Google Scholar]
- Reniere ML, Ukpabi GN, Harry SR, Stec DF, Krull R, Wright DW, Bachmann BO, Murphy ME, Skaar EP. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75:1529–1538. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- Sadykov MR, Mattes TA, Luong TT, Zhu Y, Day SR, Sifri CD, Lee CY, Somerville GA. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. J Bacteriol. 2010;192:1459–1462. doi: 10.1128/JB.01377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini A, Mapolelo DT, Chahal HK, Johnson MK, Outten FW. SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry. 2010;49:9402–9412. doi: 10.1021/bi1011546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Pham P, Cox MM, Goodman MF. Roles of DNA polymerase V and RecA protein in SOS damage-induced mutation. Chemical reviews. 2006;106:406–419. doi: 10.1021/cr0404951. [DOI] [PubMed] [Google Scholar]
- Schroder I, Johnson E, de Vries S. Microbial ferric iron reductases. FEMS microbiology reviews. 2003;27:427–447. doi: 10.1016/S0168-6445(03)00043-3. [DOI] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in. Escherichia coli. J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 2004;279:48742–48750. doi: 10.1074/jbc.M408754200. [DOI] [PubMed] [Google Scholar]
- Sebulsky MT, Hohnstein D, Hunter MD, Heinrichs DE. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J Bacteriol. 2000;182:4394–4400. doi: 10.1128/jb.182.16.4394-4400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach BP, Chung AH, Scott AD, George SJ, Cramer SP, Dos Santos PC. Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc1547 dependent sulfur transfer protein. Biochemistry. 2014;53:152–160. doi: 10.1021/bi4011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakamuri P, Zhang B, Johnson MK. Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins. J Am Chem Soc. 2012;134:15213–15216. doi: 10.1021/ja306061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. Journal of the American Chemical Society. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- Smith AD, Jameson GN, Dos Santos PC, Agar JN, Naik S, Krebs C, Frazzon J, Dean DR, Huynh BH, Johnson MK. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry. 2005;44:12955–12969. doi: 10.1021/bi051257i. [DOI] [PubMed] [Google Scholar]
- Smith FE, Herbert J, Gaudin J, Hennessy DJ, Reid GR. Serum iron determination using ferene triazine. Clinical biochemistry. 1984;17:306–310. doi: 10.1016/s0009-9120(84)90613-1. [DOI] [PubMed] [Google Scholar]
- Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun. 2002;70:6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Jaishankar GB, Saleh H, Jithpratuck W, Sahni R, Krishnaswamy G. Chronic granulomatous disease: a review of the infectious and inflammatory complications. Clinical and molecular allergy : CMA. 2011;9:10. doi: 10.1186/1476-7961-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soum E, Brazzolotto X, Goussias C, Bouton C, Moulis JM, Mattioli TA, Drapier JC. Peroxynitrite and nitric oxide differently target the iron1571 sulfur cluster and amino acid residues of human iron regulatory protein 1. Biochemistry. 2003;42:7648–7654. doi: 10.1021/bi030041i. [DOI] [PubMed] [Google Scholar]
- Soum E, Drapier JC. Nitric oxide and peroxynitrite promote complete disruption of the [4Fe-4S] cluster of recombinant human iron regulatory protein 1. J Biol Inorg Chem. 2003;8:226–232. doi: 10.1007/s00775-002-0412-9. [DOI] [PubMed] [Google Scholar]
- Stocks SM. Mechanism and use of the commercially available viability stain, BacLight. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2004;61:189–195. doi: 10.1002/cyto.a.20069. [DOI] [PubMed] [Google Scholar]
- Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C. AirSR, a [2Fe-2S] Cluster-Containing Two-Component System, Mediates Global Oxygen Sensing and Redox Signaling in Staphylococcus aureus. J Am Chem Soc. 2011 doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28393. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, Anderson KL, Stauff DL, McDonald WH, Zimmerman LJ, Friedman DB, Heinrichs DE, Dunman PM, Skaar EP. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun. 2010;78:1618–1628. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentino MD, Foulston L, Sadaka A, Kos VN, Villet RA, Santa Maria J, Jr., Lazinski DW, Camilli A, Walker S, Hooper DC, Gilmore MS. Genes Contributing to Staphylococcus aureus Fitness in Abscess- and Infection- Related Ecologies. mBio. 2014;5 doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen S, Abee T. Generation of variants in Listeria monocytogenes continuous-flow biofilms is dependent on radical-induced DNA damage and RecA-mediated repair. PLoS One. 2011;6:e28590. doi: 10.1371/journal.pone.0028590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J, Castor M, Richardson A, Main-Hester KL, Fang FC. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol Microbiol. 2007;63:1495–1507. doi: 10.1111/j.1365-2958.2007.05600.x. [DOI] [PubMed] [Google Scholar]
- Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009;5:e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouldoukis I, Riveros-Moreno V, Dugas B, Ouaaz F, Becherel P, Debre P, Moncada S, Mossalayi MD. The killing of Leishmania major by human macrophages is mediated by nitric oxide induced after ligation of the Fc epsilon RII/CD23 surface antigen. Proc Natl Acad Sci U S A. 1995;92:7804–7808. doi: 10.1073/pnas.92.17.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- Weinberg ED. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Boyd JM, Horswill AR, Nauseef WM. Phosphatidylinositol1619 specific phospholipase C contributes to survival of Staphylococcus aureus USA300 in human blood and neutrophils. Infect Immun. 2014 doi: 10.1128/IAI.01168-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wollers S, Layer G, Garcia-Serres R, Signor L, Clemancey M, Latour JM, Fontecave M, Ollagnier de Choudens S. Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J Biol Chem. 2010 doi: 10.1074/jbc.M110.127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong A, Singh VK, Cabrera G, Jayaswal RK. Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology. 2000;146(Pt 3):659–668. doi: 10.1099/00221287-146-3-659. [DOI] [PubMed] [Google Scholar]
- Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Cammack R, Dillingham MS. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J Biol Chem. 2009;284:7746–7755. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepes A, Koch G, Waldvogel A, Garcia-Betancur JC, Lopez D. Reconstruction of mreB expression in Staphylococcus aureus via a collection of new integrative plasmids. Appl Environ Microbiol. 2014;80:3868–3878. doi: 10.1128/AEM.00759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- Zheng L, White RH, Cash VL, Jack RF, Dean DR. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.