Abstract
BRCAPRO is a widely used model for genetic risk prediction of breast cancer. It is a function within the R package BayesMendel and is used to calculate the probabilities of being a carrier of a deleterious mutation in one or both of the BRCA genes, as well as the probability of being affected with breast and ovarian cancer within a defined time window. Both predictions are based on information contained in the counselee’s family history of cancer. During the last decade, BRCAPRO has undergone several rounds of successive refinements: the current version is part of release 2.1 of BayesMendel. In this review, we showcase some of the most notable features of the software resulting from these recent changes. We provide examples highlighting each feature, using artificial pedigrees motivated by complex clinical examples. We illustrate how BRCAPRO is a comprehensive software for genetic risk prediction with many useful features that allow users the flexibility to incorporate varying amounts of available information.
Keywords: BRCAPRO, BRCA1, BRCA2, mutation carrier, breast cancer, ovarian cancer
Introduction: Background
The BRCAPRO genetic risk prediction model1 is widely used in genetic counseling and is freely available through the open source R package BayesMendel2 and through a web based risk service3 interface. Genetic counseling packages Cancer-Gene4 and HughesRiskApps (HRA)5 also embed BRCAPRO calculations.
BayesMendel is a statistical package designed to calculate Mendelian risk prediction of different types of inherited disease, according to the family history provided as an input. It includes several modules: BRCAPRO [breast and ovarian cancer (BC and OC)], MMRpro (colorectal and endometrial cancer), PancPRO (pancreatic cancer), and MelaPRO (melanoma) models, as well as the functionality to adapt these models to specific populations and to develop new Mendelian risk prediction models for other syndromes.
The BayesMendel R package has been consistently updated during the last decade: 11 versions were released from 2004 to 2007 (versions 1.1–15 to 1.4–3). These versions allowed input of family history up through relatives of first and second degree to the counselee. Starting with version 2.0 in 2008, relatives of any degree could be included in the family history, and 11 versions of BayesMendel were released from 2008 to 2014 (versions 2.0–1 to the currently available 2.1–1). The different risk modules of the BayesMendel package, in general, return as output
the probabilities of carrying a germline mutation in genes relevant to the specific cancer (eg, BRCA1 or BRCA2 or both for BRCAPRO);
- the future risk of cancer within a user-specified time window: in particular
- the future risk of BC for an unaffected counselee,
- the future risk of contralateral BC (CBC) for a counselee already diagnosed with unilateral BC, and
- the future risk of OC for an unaffected counselee.
As it will be shown in one example, the standard time window used for the risk prediction is 5 years, but this can be modified by the user to any value.
Here we review some of the most useful features that have been incorporated in the recent releases of BayesMendel, focusing only on BRCAPRO, and illustrating each feature with examples. Only in one case we will refer directly to the output of the R version of BRCAPRO for listing the results that the software returns under different scenarios. The same results are provided by CancerGene, HRA, and the online risk service. In fact, CancerGene and HRA are directly used for counseling, while the role of the R version is at the back-end of these tools, to supply the numerical results only. Thus, these clinical tools are better equipped with counseling-friendly features such as graphs and tables showing various diagnosis and risks to help make clinical decisions. Here we focus on illustrating various features of the R package directly, rather than specific outputs of CancerGene and HRA.
Methods
For the purpose of illustration, we use artificially generated families. In these examples, we consider “large” (29 individuals or more) as well as “small” (10 individuals or less) families. We have chosen families with different types and numbers of members to illustrate variations in the distributions of the diseases and age of onset across the family members. Specifically, we include families with occurrence of BC only, families with BC and OC diagnoses, a family with no cancer diagnoses, a family with one BC within a pair of twin sisters, and a family including rare occurrence of male BC. These choices allow us to give a more complete description of the software capabilities, and show how its applicability is not restricted to family with a specific size or structure. From a computational standpoint, we note that BRCAPRO results are not affected by the size or type of the pedigree, but it is reasonable to expect that the accuracy of the risk estimates will increase with more information (including size) on the pedigree if that information is correct.
In a few examples, we modify some of the information in a pedigree to demonstrate specific features of BRCAPRO. Specifically, the covered modifications will include removal of affection ages, to mimic the situation in which such information is missing; addition of information about tumor markers for the counselee (if affected) or for a different affected member of the family; addition of information on oophorectomy for an unaffected proband or relative; modification of ethnicity information on the paternal and/or maternal side, and modification of the information on race.
Families
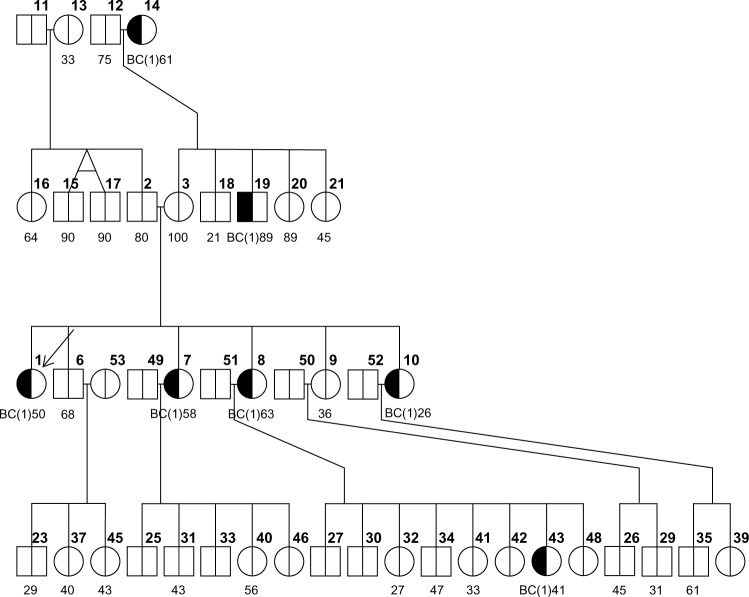
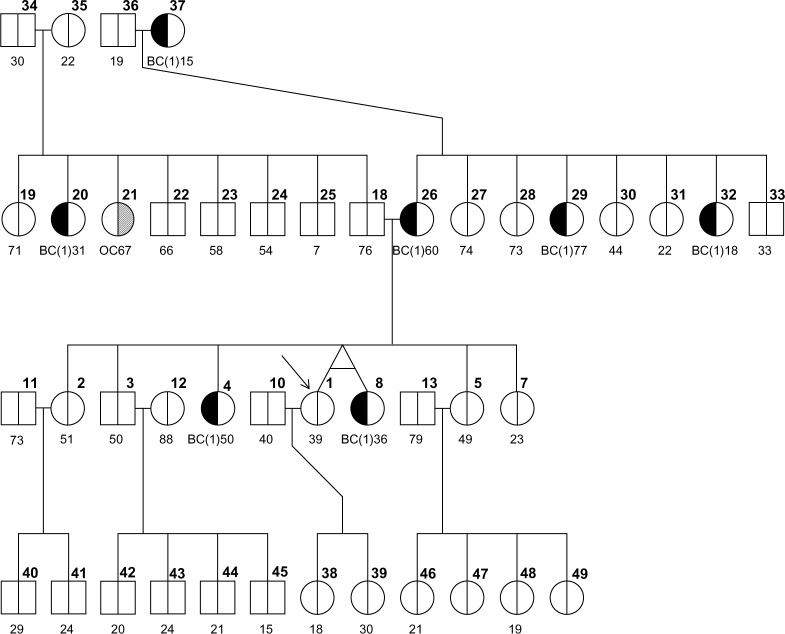
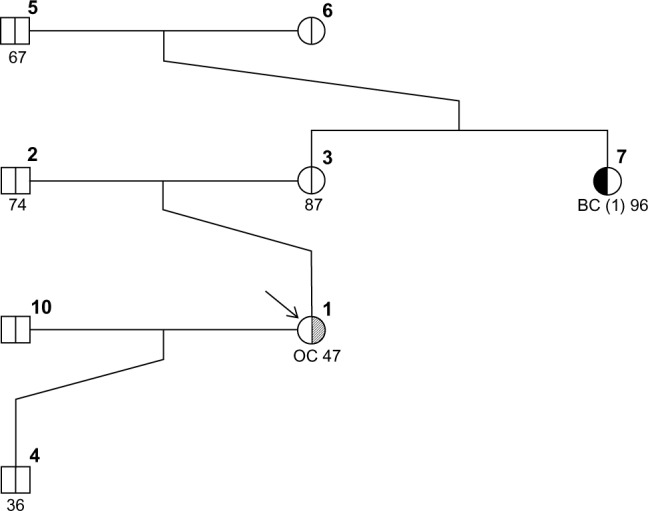
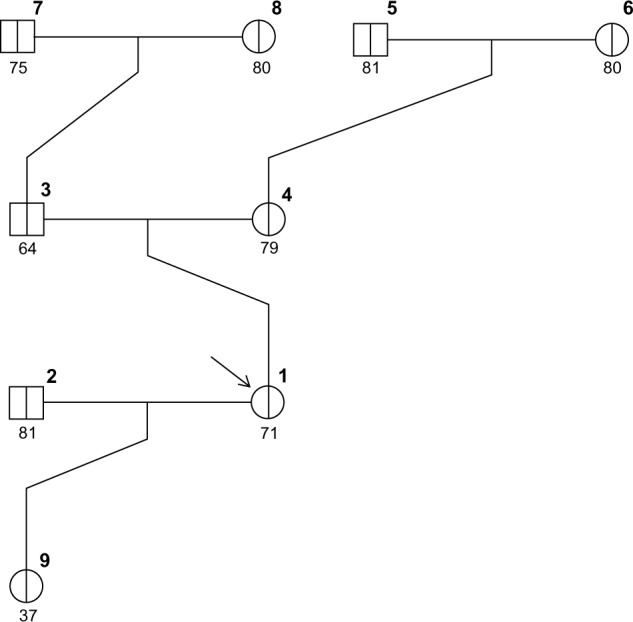
Figures 1–3 show three randomly generated “small” pedigrees with different family structures and different combinations of affected (with either BC or OC) and unaffected probands and relatives. In each figure, the boldface number above each member indicates his/her identifying number (ID) within the pedigree. The number below each member is either the affection age (if affected) or the current age/age at death (if unaffected), whenever available. The member indicated by an arrow is the proband or counselee, for which the results are calculated by the model. Diagnoses of BC and OC are indicated accordingly, and the diagnosis of unilateral (as opposed to bilateral) BC is indicated by the symbol “(1)” next to BC. The pedigree plotting package that BRCAPRO internally calls (R package kinship2) automatically assigns a number to each individual in the pedigree; the numbers are not returned when the pedigree is plotted, but in this case they have been added in bold in the upper-right corner of each symbol for the sake of easy reference.
Figure 1.

Family 1s.
Figure 2.
Family 2s.
Figure 3.
Family 3s.
To account for more complex family structures, we have also randomly generated three “big” pedigrees with different structures and different combinations of affected (with either BC or OC) and unaffected probands and relatives. These are represented in Figures 4–6 using the same notation as in Figures 1–3. We also observe that the plotting package kinship2 does not show whether a person is alive or deceased. For example, in family 2b, individual 12 (with age listed as 88) has a child with ID 45 of age 15. However, both of these ages denote the age of death (and not the current age) and hence it does not lead us to conclude that an 88-year-old mother has a 15-year-old son or even that the 15-year-old son died at the same time when his mother was 88.
Figure 4.
Family 1b.
Figure 5.
Family 2b.
Figure 6.
Family 3b.
BRCA prevalences and penetrances
A dataset of penetrance information by sex, mutation status on BRCA1/2, and age is used by BRCAPRO as one of the main parameter sources. Entries in this dataset are the net probability of developing cancer in a 1-year interval, in the absence of death or censoring. The female penetrances are obtained by combining the best available published estimates with a large set of tested families assembled through the National Cancer Institute’s Cancer Genetics Network. More specifically, they are based on a meta-analysis of nine studies using the DerSimonian and Laird random effect modeling approach.6 The male penetrance estimates are based on one of the largest US-based cohort collected retrospectively through the Cancer Genetics Network.7
The default prevalence values8 of BRCA1 and BRCA2 for non-AJ (Ashkenazi Jewish) are 0.000583 and 0.000676, and those for AJ are 0.006098 and 0.006797, respectively. The users can provide their values of prevalences (as well as penetrances) by simply specifying an optional parameter in the software.
Main Features in the Recent Versions of BRCAPRO
Risk of contralateral breast cancer
Following recently published results9, BRCAPRO has been expanded with the possibility of calculating the age-specific risk of contralateral BC. The risk values are automatically returned for the counselees who are affected with BC. For example, consider family 1s in which the counselee (ID #1) has been diagnosed with a BC at age 37, and her current age is 38. The default BRCAPRO outputs are the probability of being a BRCA mutation carrier and the marginal probabilities of being a BRCA1 or a BRCA2 mutation carrier. In addition to these, as the counselee is affected with BC, BRCAPRO returns her risk of developing CBC and OC separately and stratified by age. The standard output shows risk values up to eight decimal points; in this case, for reading purposes, only four decimal points are retained. In particular, we show here an example of the R output of a BRCAPRO session, listing the risk by 5-year age intervals by default as follows:
The probability of being a carrier is 0.0911
an BRCA1 carrier 0.0400
an BRCA2 carrier 0.0500
The risks of developing cancers are
| By age | CBC risk | OC risk | |
|---|---|---|---|
| 1 | 43 | 0.0367 | 0.0015 |
| 2 | 48 | 0.0717 | 0.0042 |
| 3 | 53 | 0.1050 | 0.0084 |
| 4 | 58 | 0.1363 | 0.0137 |
| 5 | 63 | 0.1651 | 0.0203 |
| 6 | 68 | 0.1903 | 0.0276 |
| 7 | 73 | 0.2118 | 0.0350 |
| 8 | 78 | 0.2277 | 0.0420 |
| 9 | 83 | 0.2383 | 0.0480 |
If a counselee is only diagnosed with OC, as, for instance, the proband (ID #1) in family 2s, BRCAPRO returns only the risk of being diagnosed with the first BC, as shown below in the output returned by the R version of the software:
The probability of being a carrier is 0.0197
an BRCA1 carrier 0.0145
an BRCA2 carrier 0.0052
The risks of developing cancers are
| By age | BC risk | OC risk | |
|---|---|---|---|
| 1 | 69 | 0.0198 | NA |
| 2 | 74 | 0.0393 | NA |
| 3 | 79 | 0.0578 | NA |
| 4 | 84 | 0.0733 | NA |
If, on the contrary, a counselee is unaffected, the risks of being diagnosed with both BC and OC are provided. For example, for the unaffected counselee in family 3s, the 2-year risks of both BC and OC are calculated as follows:
The probability of being a carrier is 0.0020
an BRCA1 carrier 0.0007
an BRCA2 carrier 0.0013
The risks of developing cancers are
| By age | BC risk | OC risk | |
|---|---|---|---|
| 1 | 73 | 0.0082 | 0.0010 |
| 2 | 75 | 0.0163 | 0.0020 |
| 3 | 77 | 0.0242 | 0.0029 |
| 4 | 79 | 0.0318 | 0.0039 |
| 5 | 81 | 0.0389 | 0.0048 |
| 6 | 83 | 0.0453 | 0.0057 |
| 7 | 85 | 0.0510 | 0.0065 |
Note that in this latter case we have used a simple option to display risk evaluations in a 2-year time window as opposed to the default 5-year time window provided in the first two examples.
Note also that the mutation probabilities and cancer risks are the only outputs returned by the software: here, the general problem is estimating the risk of developing either BC or OC rather than testing of a hypothesis. Thus, no significance is attached to the risk values. The use of BRCAPRO in a clinical setting involves dealing with several problems of communication and understanding the results; the developers’ team, in consultation with clinicians, felt that even the presence of a 95% confidence interval (CI) for each risk value could raise an excessive difficulty on the patients’ side to understand the statistical meaning of the results: this is the main reason why no CIs are shown with the risk values, even though they can be calculated in a relatively straightforward manner.10
Imputation of current and affection ages
One of the main features of a recent release of BRCAPRO is the ability to deal with missing current ages or ages at diagnosis. For instance, in both families 1s and 2s we observe that there are some individuals with missing information on current age (the brother, niece, and sister-in-law of the counselee, respectively, with ID #4, 10, and 11 in family 1s; and the grandmother and the brother of the counselee with ID #6 and 10 in family 2s). BRCAPRO 2.1 automatically imputes the values of the current ages of unaffected relatives based on the algorithm BRCAPROLYTE-Plus11, and displays the following warning message:
“Warning: Unknown ages of some unaffected and affected family members have been imputed. You may want to get more information about family member ages and re-run the calculation”, and then proceeds with the calculation of the results.
Let us consider family 1s and summarize into Table 1 the results obtained for different scenarios, with the increments and decrements in probabilities from the initial status (first row) indicated with ↑ and ↓, respectively:
Table 1.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 1s for varying age status of her relatives.
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| Missing current ages for individuals #4, #10, and #11 | 0.0911 | – | 0.0410 | – | 0.0500 | – |
| As above and affection age missing for the aunt (#8) | 0.1154 | ↑ | 0.0589 | ↑ | 0.0563 | ↑ |
| As above, and affection age missing for individuals #6, #8 | 0.1204 | ↑ | 0.0670 | ↑ | 0.0532 | ↑ |
We observe an increase in the probability of a deleterious mutation in the BRCA genes of the proband when the affection ages of the aunt and the grandmother are imputed by the model, together with imputation of the already missing ages of the nonaffected brother, sister-in-law, and niece. BRCAPRO imputes the missing affection ages of family members using a multiple imputation approach. In particular, a large number of affection ages (by default 100) are sampled using the Surveillance, Epidemiology, and End Results (SEER) program incidence rates of BC and OC; for each sampled age, the probabilities are calculated and then averaged, which is what is finally returned in the output.
To get further insight into the functioning of the multiple imputation algorithm, we consider a variation of family 1s whose results were shown in Table 1. In the original configuration, wherein only the missing current ages for the nonaffected brother, sister-in-law, and niece of the proband are imputed, we artificially lower the affection ages of the aunt (#8) and grandmother (#6) of the proband, respectively, to 35 and 32 (in the original dataset they were 63 and 58).
Table 2 shows the results with the increments and decrements in probabilities from the original status indicated with ↑ and ↓, as usual. Also, we have added a row for the scenario where the affection ages of these two relatives are assumed to be missing and are imputed (same as the last row of Table 1) for ease of comparison.
Table 2.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 1s for varying age status of her aunt and grandmother.
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| Missing current ages for #4, #10, and #11, real affection ages for #8 and #6 resp. 63 and 58 | 0.0911 | – | 0.0410 | – | 0.0500 | – |
| Missing current ages for #4, #10, and #11, artificial affection ages for #8 and #6 resp. 35 and 32 | 0.4792 | ↑ | 0.2680 | ↑ | 0.2103 | ↑ |
| Missing current ages for #4, #10, and #11, missing affection ages for #8 and #6 | 0.1204 | ↑ | 0.0670 | ↑ | 0.0532 | ↑ |
Note that the risk is much larger when artificial ages of 35 and 32 are used compared to when those affection ages are imputed. This is because the imputed age, which is an average over multiple imputations, is in this case higher than these artificial ages but is less than the real ages of 63 and 58, and younger affection ages are stronger indication of mutation. The imputation is consistent with the fact that SEER’s average affection age of BC is between 60 and 80.
Tumor marker information
The information on tumor markers – estrogen receptor (ER) status, expression of two of the basal cytokeratins (CK14, CK5/6), progesteron receptor (PR) status, and HER2 status – can be incorporated whenever available for the counselee or any affected relative. The marker status can be positive, negative, or unknown.
We illustrate this by considering family 1s. With no information on tumor markers, the proband (ID # 1) has carrier probabilities as listed in the first row of Table 3. Further, the table shows how carrier probabilities change when changing tumor status for the proband; in particular, the differences are indicated by either ↑ or ↓ from the baseline probability, where no tumor marker information is used:
Table 3.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 1s for different tumor marker status for the proband.
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| No information (baseline) | 0.0911 | – | 0.0409 | – | 0.0500 | – |
| Proband ER+ | 0.0625 | ↓ | 0.0081 | ↓ | 0.0544 | ↑ |
| Proband ER− | 0.1482 | ↑ | 0.1068 | ↑ | 0.0411 | ↓ |
| Proband ER− and HER2+ | 0.0244 | ↓ | 0.0152 | ↓ | 0.0091 | ↓ |
| Proband ER−, CK14+, CK5/6+ | 0.5677 | ↑ | 0.5454 | ↑ | 0.0206 | ↓ |
| Proband ER−, CK14−, CK5/6− | 0.0808 | ↓ | 0.0363 | ↓ | 0.0443 | ↓ |
The results shown in Table 3 are consistent with what is known about the relations between molecular subtypes of breast cancers and BRCA mutations as we discuss in the following. It is widely acknowledged that triple-negative BCs (presenting with ER−, PR–, and HER2–) are an overlapping category with basal-like BCs (featuring cells with similar characteristics to the outer cells surrounding the mammary ducts). Most triple-negative tumors are basal-like and most basal-like tumors are triple negative; however, not all triple-negative tumors are basal-like, and not all basal-like tumors are triple negative. In particular, most BCs for patients carrying BRCA1 mutation are both triple negative and basal-like: this is the reason why a proband with a negative ER status (row 2 of Table 3) is shown to have an increased probability of a BRCA1 mutation (and a decreased probability of a BRCA2 mutation).
On the other hand, most BRCA2-related BCs tend to be ER+, which justifies the increased probability of BRCA2 mutation (and a decrease in BRCA1 probability) in row 1 of Table 3 compared to row 2. Moreover, basal cytokines CK5/6 and CK14 are expressed in basal-like tumors, resulting in an even more increased probability of BRCA1 mutation and an even more decreased probability of BRCA2 mutation, as seen in row 4 of Table 3 (representing, in fact, basal-like and triple-negative BCs).
Usually, if a person is ER−, she is more likely to be a carrier; however, if a woman is both ER− and HER2+, she is more likely to be a noncarrier. HER2+ BCs are in general ER− and present, as a distinctive feature, with a mutation in genes other than BRCA1 or BRCA2: this is why the probability of a mutation in both BRCA genes is decreased in row 3 of the table. These differences illustrate the importance of using joint marker status information, whenever available.
From a computational standpoint, when the testing result for ER is negative, and the results for both CK14 and CK5/6 are also available, these three markers are treated as a group, and the calculations of carrier probabilities will incorporate their joint probabilities. If ER is positive, the testing results for CK14 or CK5/6 are not considered. If the result for either CK14 or CK5/6 is not available, the calculations of carrier probabilities will involve either the marginal conditional probability of ER or, if HER2 result is available, the joint probabilities of ER and HER2. If all information on ER, HER2, and CK markers is available, then the joint probabilities of ER and CKs are utilized.
For any family member, if the testing result for ER is available, the testing result for PR will be ignored even if it is also available, due to strong association between ER and PR results. That is, PR will not be included in carrier probability calculation when ER is available. PR will be used only when either PR only or PR and HER2 testing are available, because of their ability to indicate the possible presence of a triple-negative BC, which is often aggressive and usually has a poor prognosis.
The same trend is seen if tumor marker information is used for relatives other than the counselee. Consider, for instance, a series of tumor marker information on the counselee’s aunt (individual # 8) for her BC. The results for the proband as returned by BRCAPRO, and the differences from the baseline probabilities, indicated by either ↑ or ↓, are summarized in Table 4. A similar trend was found if the grandmother (individual # 6) instead of the aunt has the same tumor marker information (results not shown). Finally, in Table 5 we show same set of results when tumor marker information is available on both the aunt (ID # 8) and the grandmother (ID # 6).
Table 4.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 1s for different tumor marker status for proband’s aunt (ID #8).
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| No information (baseline) | 0.0911 | – | 0.0409 | – | 0.0500 | – |
| ID #8 ER+ | 0.0782 | ↓ | 0.0255 | ↓ | 0.0525 | ↑ |
| ID #8 ER− | 0.1181 | ↑ | 0.0733 | ↑ | 0.0446 | ↓ |
| ID #8 ER− and HER2+ | 0.0540 | ↓ | 0.0294 | ↓ | 0.0248 | ↓ |
| ID #8 ER−, CK14+, CK56+ | 0.3835 | ↑ | 0.3534 | ↑ | 0.0294 | ↓ |
| ID #8 ER−, CK14−, CK56− | 0.0854 | ↓ | 0.0388 | ↓ | 0.0464 | ↓ |
Table 5.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 1s for different tumor marker status for proband’s aunt and grandmother (IDs #6 and #8).
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| No information (baseline) | 0.0911 | – | 0.0409 | – | 0.0500 | – |
| IDs #6 and 8 ER+ | 0.0720 | ↓ | 0.0170 | ↓ | 0.0550 | ↑ |
| IDs #6 and 8 ER− | 0.1834 | ↑ | 0.1456 | ↑ | 0.0376 | ↓ |
| IDs #6 and 8 ER− and HER2+ | 0.1122 | ↑ | 0.0634 | ↑ | 0.0487 | ↓ |
| IDs #6 and 8 ER−, CK14+, CK56+ | 0.8090 | ↑ | 0.8027 | ↑ | 0.0049 | ↓ |
| IDs #6 and 8 ER−, CK14−, CK56− | 0.0795 | ↓ | 0.0365 | ↓ | 0.0429 | ↓ |
Note that ER−/Her2+ status of just the aunt’s BC on the proband’s risk of a mutation is reflected in a reduction of the carrier probability compared to baseline, due to the molecular subtype classification explained above; however, if we combine the information of the tumor markers of both the BC-affected aunt and grandmother, the evidence for the presence of a deleterious mutation overwhelms the effect seen above. This is reflected in Table 5 as an increase of the probability of a BRCA1 mutation for ER−/Her+ compared to baseline, which drives the increase of the overall probability of BRCA mutation. Nonetheless, we note that this probability decreases compared to when both relatives are just ER− (same trend as in Tables 3 and 4).
Oophorectomy information
If a family member has undergone oophorectomy, this information can be accounted for in BRCAPRO. Let us consider family 3s as an example. The fact that the proband herself as well as her mother and grandmother (IDs #1,4, and 6, respectively) have survived up to relatively older ages without getting BC or OC in the absence of oophorectomy makes a very strong case in favor of the absence of BRCA mutations, and thus the proband has a low carrier probability of ∼0.2%.
Now let us assume that both mother and grandmother (IDs #4 and #6) had undergone oophorectomy intervention at ages 47 and 61, respectively: the carrier probabilities of the proband would be slightly increased. These changes, compared to when oophorectomy had not been performed, are explained by oophorectomy’s protective effect against developing cancer. The fact that these women are cancer-free at older ages is now partly attributable to their oophorectomy intervention, which makes the relatives less likely to experience a cancer even if they carry a BRCA mutation. As a result, the counselee’s chances of carrying a mutation are slightly increased.
Table 6 summarizes the scenarios described above, with rows describing incremental steps in the acquisition of information about oophorectomy for the examined family members, and the last row adding also a scenario in which the counselee herself has undergone the intervention at age 65. The changes in probability with respect to the situation in which no oophorectomy information is known are, as usual, indicated with ↑ for increments, and ↓ for decrements.
Table 6.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 3s in different oophorectomy scenarios.
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| No information (baseline) | 0.0020 | – | 0.00066 | – | 0.0013 | – |
| Oophorectomy for ID #4 (mother) at age 47 | 0.0022 | ↑ | 0.00074 | ↑ | 0.00148 | ↑ |
| Oophorectomy for ID #4 and 7 at ages 47 and 61 | 0.0023 | ↑ | 0.00076 | ↑ | 0.00154 | ↑ |
| As above, and oophorectomy for proband at age 65 | 0.0026 | ↑ | 0.00090 | ↑ | 0.00170 | ↑ |
We would like to note in this case that, although we see a very slight effect of oophorectomy on the mutation probability for the proband, it is widely acknowledged that oophorectomy has a larger protective effect on the risk of BC when it is performed at younger ages (generally ≤60).12 Following this logic, BRCAPRO is specifically designed not to modify the hazard ratio for oophorectomy on the risk of BC for ages 60 and higher; that is, the risk will increase if oophorectomy is performed at age more than 60.
Mastectomy (including male mastectomy) information
Following the same logic as oophorectomy, BRCAPRO can account for bilateral mastectomy in carrier probability calculation. In particular, starting from version 2.1, BRCAPRO can allow for female and male mastectomies as possible interventions, as we show using family 1b.
We now assume to have information about a bilateral mastectomy for the counselee (ID #1) at age 53 and the uncle of the proband (ID #19), affected by male BC, at age 89. Incorporating these interventions into calculations, we obtain an increase in the carrier probabilities. As in the previous section, these increased probabilities, compared to when the mastectomy is not present, account for the fact that the intervention makes the individuals less likely to experience a cancer even if they carry a BRCA mutation. As a result, the chances of carrying a mutation are increased.
Table 7 displays the incremental changes in the probabilities returned by BRCAPRO when we start from a scenario in which no information about possible mastectomies is known, and we add subsequent information about the proband and the uncle of the proband.
Table 7.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (any BRCA), BRCA1, and BRCA2 for the proband in family 1b in different mastectomy scenarios.
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| No information (baseline) | 0.3827 | – | 0.0424 | – | 0.3402 | – |
| Mastectomy for ID #1 (proband) at age 53 | 0.4131 | ↑ | 0.0493 | ↑ | 0.3637 | ↑ |
| Mastectomy for ID #1 and 19 (uncle) at ages 53 and 89 | 0.4446 | ↑ | 0.0481 | ↑ | 0.3964 | ↑ |
When information about interventions is included in BRCAPRO, the age-specific penetrance curves for BRCA1 and BRCA2 get modified. The presence of a male member affected with breast cancer is a strong indicator of BRCA2 mutation and, consequently, BRCA2 mutation risk increases when the mastectomy for the uncle of the proband is considered (as with a mastectomy, the uncle is less likely to experience bilateral BC even if he is actually a BRCA2 carrier). The slight decrease (0.001) in BRCA1 risk compared to the situation in which only the proband undergoes mastectomy may be due to the way the penetrance of BRCA1 for male individuals at that age is modified by a mastectomy.
Specification of family race
The recent releases of BRCAPRO have included a set of race/ethnicity-specific baseline penetrances for noncarriers. The user can specify one of five different inputs: Asian, Black, Hispanic, Native American and White, while the current default assumes that the race/ethnicity of the input family is unknown, ie, none of the five possible groups, and is representative of the general population.
Race/ethnicity categories and estimates were derived using the DevCan software provided by NCI.13 The baseline penetrance values need to be loaded into BRCAPRO from the database named BRCAbaseline.race.2008, so that a particular race among the ones indicated above can be specified. Let us consider family 2b and calculate carrier probabilities by specifying different races (Table 8). We see that knowledge of race makes a difference in the probabilities.
Table 8.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 2b for different race values.
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| Unknown (baseline) | 0.9251 | – | 0.5022 | – | 0.3863 | – |
| Asian | 0.9480 | ↑ | 0.4809 | ↓ | 0.4040 | ↑ |
| Black | 0.8810 | ↓ | 0.5007 | ↓ | 0.3498 | ↓ |
| Hispanic | 0.9557 | ↑ | 0.4927 | ↓ | 0.4097 | ↑ |
| Native American | 0.9202 | ↓ | 0.6329 | ↑ | 0.2695 | ↓ |
| White | 0.9304 | ↑ | 0.5035 | ↑ | 0.3910 | ↑ |
Specification of ethnicity
Five different choices are available to specify the ethnicity of each member of a family: Ashkenazi Jewish (“AJ”),“non-AJ”, “Italian”, “Other”, or NA. If at least one family member is “AJ”, the default is to use the prevalence associated with the “AJ” for family members with unknown ethnicity. Otherwise, the prevalence associated with “non-AJ” is used for family members with unknown ethnicity. Such care in handling the “AJ” ethnicity is justified by the fact, as is well known from literature14, that in the US, BC risk is slightly higher among Jewish women compared to other women. This is likely due to a high prevalence of BRCA1 and BRCA2 mutations in Jewish women of Eastern European descent (Ashkenazi Jews) (about 1/40 versus between 1/400 and 1/800 in the general population).
With this feature, it is straightforward to incorporate different ethnicities for different lineages of the same family. Let us consider family 3b; by default, the ethnicity “non-AJ” is used, so in the “baseline” situation we will have all “non-AJ” individuals. Running BRCAPRO for the BC-affected proband (ID #1) in this family results in the output shown in the first row of Table 9.
Table 9.
Probabilities of carrying mutation in either BRCA1 or BRCA2 (Any BRCA), BRCA1, and BRCA2 for the proband in family 3b for different ethnicity scenarios.
| CARRIER PROBABILITIES FOR THE PROBAND | ||||||
|---|---|---|---|---|---|---|
| ANY BRCA | BRCA1 | BRCA2 | ||||
| Original ethnicity: unspecified/non-AJ (baseline) | 0.0010 | – | 0.0006 | – | 0.0005 | – |
| Mother’s (ID #3) lineage: Italian, Others: unspecified/non-AJ. | 0.0013 | ↑ | 0.0007 | ↑ | 0.0006 | ↑ |
| Mother’s lineage: Italian, Father’s (ID #2) lineage: AJ, Others: unspecified/AJ. | 0.0092 | ↑ | 0.0054 | ↑ | 0.0037 | ↑ |
| Same as above but paternal grandmother (ID #17) non-AJ | 0.0074 | ↑ | 0.0046 | ↑ | 0.0028 | ↑ |
| Same as above but also maternal grandfather (ID #16) non-AJ | 0.0073 | ↑ | 0.0045 | ↑ | 0.0027 | ↑ |
Now let us suppose that the mother of the proband (ID #3) comes from an Italian family, and all the other individuals remain unspecified (NA option, and in this case these other members will be treated as “non-AJ” by default). The resulting risks for the proband for this scenario are now modified, as shown in the second row of Table 9, and they are very close to the baseline scenario.
Now let us further modify the previous situation by assuming the father’s (ID #2) lineage, including the proband, as “AJ”: observe how there is an increment of almost an order of magnitude in the risk for the proband with respect to the baseline situation. This happens because, if at least one family member is “AJ”, BRCAPRO by default uses the prevalence associated with “AJ” for family members with unspecified ethnicity also. Note that this may lead to overestimation of the risk if, in fact, some of the unspecified relatives are not “AJ”. So, this risk estimate is a more conservative estimate (erring on the side of caution).
We note, in the last two rows of the table, how the risk of the proband decreases from the previous scenario when sequentially only one grandparent (say, the paternal grandmother, ID #17) and then also the maternal grandfather (ID #16) are assumed to be “non-AJ”.
Discussion
In this paper, we reviewed and showcased some of the recent additions to the widely used BRCAPRO software. BRCAPRO is part of a more comprehensive R package called BayesMendel, which includes prediction models for colorectal, endometrial, and pancreatic cancer, and melanoma.
These models are employed by CancerGene, which distributes the BayesMendel package to more than 4,000 users in more than 75 countries. They are also available through a risk service, which provided, in 2014, around 15,000 risk evaluations per month, primarily through the HughesRiskApps software. The two most recent releases of the R package BayesMendel (2.0–9, released in March 2014 and 2.1, released in October 2014) have been downloaded 99 times by users in academia and 79 times by organizations outside the academic world.
BRCAPRO has allowed its users to more specifically tailor calculations for families through its incorporation of clinical intervention information (such as mastectomy, both for male and female individuals, and oophorectomy) and tumor markers for breast cancer. In version 2.0–8, released in January 2013, BRCAPRO improved the risk assessment of a recurring contralateral malignancy in patients already diagnosed with BC by including separate penetrance functions for contralateral BC. Notably, the most recent release of BRCAPRO accommodates the real-life challenges of family history data collection, specifically with its ability to impute missing ages both for affected and nonaffected individuals, and the possibility of modifying the predicted risk values according to race and ethnicity. In this last case, the software is capable of accepting multiple combinations of ethnicities for different members or lineages of the family.
For most of these new features, BRCAPRO has been evaluated in independent data and is calibrated; the results of calibration are described for the inclusion of Asian ethnicity15 in version 2.0-2, released in March 2009; for the unclusion of joint tumor marker information16 in version 2.0-6, released in October 2011; for the addition of the penetrance functions of CBC9 in version 2.0-8, released in January 2013, and for imputation of current ages11 in version 2.1, released in October 2014.
Nonetheless, even with the improvements highlighted here, some limitations still exist in the model. For example, it is widely acknowledged that BRCA-mutation carriers in different ethnic groups present with different susceptibility to BC; moreover, within a broadly defined ethnicity such as “white”, there exist significant differences in the annual incidence of BC, for instance, in BRCA1 mutation carriers.17 One direction for future work in expanding the BRCAPRO model is to incorporate specific prevalences for different and more finely defined ethnic groups as relevant data become available. This process was undertaken, for instance, for including a separate “Italian” ethnicity in the model because a separate set of prevalence values was available for Italians. Another direction of possible improvement would be the inclusion of different malignancies in family history and a more populated family of genes modifying the susceptibility of the individuals to specific cancers.
A limitation of working with clinical data is that, in general, family histories are not always clean and detailed, as data misreporting happens frequently: BRCAPRO is designed in such a way as to make specific assumptions in presence of missing data, and these assumption may lead, in some cases, to imprecise estimates.
Overall, we conclude that with the newly added features BRCAPRO now encompasses a wide variety of situations wherein different amounts and types of information may be available for different counselees. In many cases, BRCAPRO is expected to make more accurate predictions compared to its earlier versions, as it can now utilize added information. Thus, BRCAPRO can now reach a wider population with better prediction capabilities, and thereby have an even more widespread impact in familial BC counseling.
Acknowledgments
We thank the three anonymous reviewers for their constructive comments and suggestions.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
FUNDING: This work was supported in part by National Cancer Institute grants 1R03CA173834–02 and 2P30CA006516–47, and the Dana-Farber Cancer Institute. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: GP discloses SAB membership and stock options in HughesRisk Apps. GP and EM disclose that their employer, Dana-Farber Cancer Institute, provides risk calculations for BRCAPRO. Other authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: EM, GP, SB. Analyzed the data: EM, AB, SB. Wrote the first draft of the manuscript: EM. Contributed to the writing of the manuscript: EM, AB, SB. Agree with manuscript results and conclusions: EM, AB, GP, SB. Jointly developed the structure and arguments for the paper: EM, AB, GP, SB. Made critical revisions and approved final version: EM, AB, GP, SB. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–58. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. BayesMendel: an R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1063. Article21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chipman J, Drohan B, Blackford A, Parmigiani G, Hughes K, Bosinoff P. Providing aggess to risk prediction tools via the HL7 XML-formatted risk web service. Breast Cancer Res Treat. 2013;140(1):187–93. doi: 10.1007/s10549-013-2605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Available at: https://www4.utsouthwestern.edu/breasthealth/cagene.
- 5. Available at: http://www.hughesriskapps.com/
- 6.Chen S, Iversen ES, Friebel T, et al. Characterization of BRCA1 and BRCA2 mutations in a large US sample. J Clin Oncol. 2006;24(6):863–71. doi: 10.1200/JCO.2005.03.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai YC, Domchek S, Parmigiani G, Chen S. Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99(23):1811–14. doi: 10.1093/jnci/djm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antoniou AC, Pharoah PD, McMullan G, et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86(1):76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazzola E, Chipman J, Cheng SC, Parmigiani G. Recent BRCAPRO upgrades significantly improve calibration. Cancer Epidemiol Biomarkers Prev. 2014;23(8):1689–95. doi: 10.1158/1055-9965.EPI-13-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas S, Berry DA. Determining joint carrier probabilities of cancer-causing genes using Markov chain Monte Carlo methods. Genet Epidemiol. 2005;29:141–54. doi: 10.1002/gepi.20082. [DOI] [PubMed] [Google Scholar]
- 11.Biswas S, Atienza P, Chipman J, et al. Simplifying clinical use of the genetic risk prediction model BRCAPRO. Breast Cancer Res Treat. 2013;139(2):571–9. doi: 10.1007/s10549-013-2564-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rebbeck TR, Kauff ND, Domchek SM. Meta-analysis of risk reduction estimates associated with risk-reducing salpingo-oophorectomy in BRCA1 or BRCA2 mutation carriers. J Natl Cancer Inst. 2009;101(2):80–7. doi: 10.1093/jnci/djn442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Available at: http://surveillance.cancer.gov/devcan.
- 14.Egan KM, Newcomb PA, Longnecker MP, et al. Jewish religion and risk of breast cancer. Lancet. 1996;346(9016):1645–6. doi: 10.1016/s0140-6736(96)91485-3. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Blackford A, Parmigiani G. Tailoring BRCAPRO to Asian-Americans. J Clin Oncol. 2009;27:642–3. doi: 10.1200/JCO.2008.20.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biswas S, Tankhiwale N, Blackford A, et al. Assessing the added value of breast tumor markers in genetic risk prediction model BRCAPRO. Breast Cancer Res Treat. 2012;133:347–55. doi: 10.1007/s10549-012-1958-z. [DOI] [PubMed] [Google Scholar]
- 17.Lubinski J, Huzarski T, Byrski T, et al. Hereditary Breast Cancer Clinical Study Group The risk of breast cancer in women with a BRCA1 mutation in North America and Poland. Int J Cancer. 2012;131:229–34. doi: 10.1002/ijc.26369. [DOI] [PMC free article] [PubMed] [Google Scholar]