Abstract
Background
MiR-137 dysregulation has been implicated in the etiology of schizophrenia, but its functional role remains to be determined.
Methods
Functional magnetic resonance imaging scans were acquired on 48 schizophrenia patients and 63 healthy volunteers (total sample size n=111 subjects), with similar mean age and sex distribution, while subjects performed a Sternberg Item Response Paradigm with memory loads of 1, 3, and 5 numbers. Dorsolateral prefrontal cortex (DLPFC) retrieval activation for the working memory load of 3 numbers, for which hyperactivation had been shown in schizophrenia patients compared with controls, was extracted. The genome-wide association study confirmed schizophrenia risk SNP rs1625579 (miR-137 locus) was genotyped (schizophrenia: GG n=0, GT n=9, TT n=39; healthy volunteers: GG=2, GT n=15, and TT n=46). Fisher's Exact Test examined the effect of diagnosis on rs1625579 allele frequency distribution (p=ns). Mixed model regression analyses examined the effects of diagnosis and genotype on working memory performance measures and DLPFC activation.
Results
Patients showed significantly higher left DLPFC retrieval activation on working memory load 3, lower working memory performance and longer response times compared with controls. There was no effect of genotype on working memory performance or response times in either group. However, individuals with the rs1625579 TT genotype had significantly higher left DLPFC activation than those with the GG/GT genotypes.
Conclusion
Our study suggests that the rs1625579 TT (miR-137 locus) schizophrenia risk genotype is associated with the schizophrenia risk phenotype DLPFC hyperactivation commonly considered a measure of brain inefficiency.
Keywords: schizophrenia, rs1625579, mir137, imaging, working memory, genes
Introduction
Aberrant dorsolateral prefrontal cortex (DLPFC) activation, as measured using functional magnetic resonance imaging (fMRI) during working memory processing, is a robust correlate of schizophrenia (1, 2). Consistent with DLPFC inefficiency - more activation for a similar level of performance - in schizophrenia (3), we have shown DLPFC hyper-activation during a working memory load of 3 items on the Sternberg Item Recognition Paradigm (SIRP) in schizophrenia patients compared with controls even when task performance is equivalent (4). We have identified several genetic polymorphisms that are associated with DLPFC activation in schizophrenia (5, 6) and subsequently identified several other putative schizophrenia risk genes as well as microRNAs based on a gene set enrichment analysis (7). MicroRNAs are short (<∼27 nucleotides) non-coding RNAs that regulate the expression levels of other genes (8) via mRNA cleavage or translation inhibition (9) and are hypothesized to play a role in the genetic liability for schizophrenia (10, 11). The microRNAs identified in our prior study included miR-137 (7), which was recently suggested to play an etiological role in schizophrenia based on a very large genome-wide case-control association study of 51,695 individuals that found a genome-wide significance of p=1.6×10-11 for the rs1625579 SNP in linkage disequilibrium with the miR-137 primary transcript as well as significant associations with predicted miR-137 targets (12).
Apart from its observed role in cancer (13-15), miR-137 is shown to regulate neural stem cell proliferation and differentiation in mouse embryonic neural stem cells (NSC) (16), neurogenesis in mouse adult NSC (17) and neuronal maturation, including regulation of dendrite length, branch points, end points and spine density in mouse adult hippocampal neuroprogenitor-derived and mouse fetal hippocampus neurons (18). Decreased spine density has also been observed in the dorsolateral cortex of patients with schizophrenia compared with controls consistent with poor memory performance and suggests altered synaptic connectivity (19). Combined, these studies implicate miR-137-dysregulation in the etiology of schizophrenia and suggest that miR-137 may be involved in regulating DLPFC activation by affecting neural connectivity within the DLPFC resulting in abnormal (inefficient) neural transmission. The putative miR-137 rs1625579 schizophrenia risk SNP was not part of the Illumina Infinium HumanHap300 Bead Array used for genotyping in our prior work (5). In this study we genotyped the rs1625579 SNP in order to directly examine the association between the rs1625579 (miR-137 locus) schizophrenia risk alleles (genotypes) and SIRP 3 item probe DLPFC activation on the Sternberg Item Recognition Test; the SIRP contrast shown to best separate schizophrenia versus control DLPFC activation (4).
To date, four human studies have examined the functional role of the rs1625579 (miR-137) SNP in schizophrenia (20-23). Cummings and colleagues (2012) (23) recently found, in a sample of 399 schizophrenia, bipolar I disorder, and schizoaffective disorder patients and 171 controls, that rs1625579 TT genotype was associated with poor working memory, episodic memory and attention. In contrast, Green and colleagues (2011) (12), who grouped schizophrenia patients into those with and without cognitive deficits, found that the rs1625579 SNP alone was not predictive of cognitive dysfunction severity or group status, but that the G-allele predicted cognitive dysfunction group status when negative symptoms were included in a multinomial regression analysis. The latter finding is hard to interpret given that the common rs1625579 T allele is associated with schizophrenia risk status.
An EEG study by Decoster and colleagues (21) found that the rs1625579 SNP T allele was associated with lower P300 amplitude. This finding is in the hypothesized direction in that P300 amplitude for common rs1625579 risk allele (T) appeared to be associated with lower P300 amplitude, a well-known quantitative endophenotype for schizophrenia (24). While the finding did not survive stringent Bonferroni correction, it nevertheless suggests a contribution of the rs1625579 (miR-137) SNP to the P300 deficit observed in schizophrenia that warrants further study.
A fMRI study by Whalley and colleagues (22) examined the effects of rs1625579 genotype on brain activation during a sentence completion task in individuals at genetic high-risk for schizophrenia or bipolar disorder compared with healthy controls. They found a significant group by genotype interaction on left amygdala and pre/post central gyrus activation. Decomposition of the interaction effect showed that the interaction was due to higher activation in schizophrenia T allele homozygotes and lower activation in control T allele homozygotes compared with their respective G allele carriers during sentence completion versus baseline condition in the left amygdala and pre/post central gyrus. They also found lower right posterior medial frontal gryus activation in rs1625579 T allele homozygotes compared with G allele carriers for a contrast sensitive to the difficulty of sentence completion irrespective of group status. These findings suggest that miR-137 gene regulation may affect brain physiology as measured with fMRI.
In this first working memory study examining the effects of rs1625579 genotypes on BOLD activation in schizophrenia, we hypothesize (1) lower SIRP load 3 working memory performance and longer response times in schizophrenia patients compared with healthy volunteers and (2) higher left DLPFC SIRP probe 3 activation in schizophrenia patients compared with healthy volunteers, consistent with findings from our previous report (4). Looking at specific effects of the rs1625579 (miR-137) SNP, our explicit directional hypothesis is (3) higher left DLPFC SIRP probe 3 activation for the rs1625579 T (schizophrenia risk) allele compared with G (non-risk) allele carriers (TT > GT > GG genotypes) consistent with higher neural inefficiency in the schizophrenia rs1625579 risk allele carriers. Given the low frequency of the rare GG genotype we collapsed across the GG and GT genotypes in the analyses and compared GG/GT with TT genotypes. We specifically focus on DLPFC retrieval activation on SIRP load 3 because it showed the strongest hyperactivation in patients with schizophrenia compared with controls even when behavioral performance was matched between the groups (4). We also explored whether the hypothesized genetic effect added to or interacted with diagnosis.
Methods and Materials
Participants
Forty-eight patients with schizophrenia (mean age±standard deviation (SD) = 37.0±10.7, 35 males) and 63 healthy volunteers (mean age±SD = 37.6±12.4, 39 males) with similar mean age, sex, handedness and race distributions, recruited from 9 sites part of the Function Biomedical Informatics Network Phase 2 study on multi-center functional imaging, participated in the study (Table 1). Study participants were between the ages of 18-70, had regular hearing levels (<25-db loss in either ear), had sufficient eyesight to see the task stimuli, were fluent in English and able to perform the cognitive tasks in the study. Inclusion criteria for the patients were a schizophrenia diagnosis based on the Structured Clinical Interview for DSM-IV-TR Axis I Disorders (SCID-I/P) (25). All patients were clinically stable on antipsychotic medication for at least two months. Schizophrenia patients and healthy volunteers with a history of major medical illness, a previous head injury or prolonged unconsciousness, substance and/or alcohol dependence, current migraine treatments, contraindications for MRI, and/or an IQ less than 75 were excluded. Patients with schizophreniform disorder or significant extrapyramidal symptoms / tardive dyskinesia and healthy volunteers with a current or past history of major neurological or psychiatric illness (SCID-I/NP) (26) or with a first-degree relative with an Axis-I psychotic disorder diagnosis were also excluded.
Table 1. Sample Demographics.
| Schizophrenia Patients (n=48) | Healthy Volunteers (n=63) | Statistic | p-value | |
|---|---|---|---|---|
| Mean Age (SD) | 37.0 (10.7) | 37.6 (12.4) | t109=0.28 | 0.78 |
| Gender (M/F) | 35/13 | 39/24 | χ21=1.49 | 0.22 |
| Handedness (bilateral/left/right) | 1/6/41 | 1/5/57 | FET5 | 0.77 |
| Mean premorbid FSIQ1 estimate (SD) | 105.1 (9.5) | 113.3 (7.2) | t105= -5.05 | <0.0001 |
| Subject's Mean Years of Education (SD) | 13.3 (1.5) | 16.0 (1.9) | t100=-7.86 | <0.0001 |
| Mother's Mean Years of Education (SD) | 13.3 (3.0) | 13.8 (3.0) | t91=-0.83 | 0.41 |
| Father's Mean Years of Education (SD) | 14.6 (3.4) | 14.9 (3.6) | t91=-0.40 | 0.69 |
| Race (% Caucasian) | 66% | 75.8 | ||
| Race6 | FET | 0.20 | ||
| American Indian or Alaskan Native | 0 | 1 | ||
| Asian | 3 | 3 | ||
| Black or African American | 10 | 5 | ||
| White | 34 | 52 | ||
| SANS (global measures sum) | 7.49 (3.71) | |||
| SAPS (global measures sum) | 6.28 (2.68) | |||
| InterSePT suicidality scale (sum of 12 items) | 2.26 (3.18) | |||
| Calgary Depression Scale (total score) | 5.36 (5.04) | |||
| rs1625579 genotype (GG/GT/TT) | 0/9/39 | 2/15/46 | FET | 0.47 |
| SIRP Load 3 % Correct (SD) / Mean RT (SD)2,3 | ||||
| GG | 100 (0) / 6774 | |||
| GT | 89.6 (12.2) / 967 (235) | 97.4 (3.8) / 736 (110) | ||
| TT | 94.2 (8.5) / 784 (130) | 96.5 (4.8) / 696 (91) | ||
| Absolute Mean % (SD) left DLPFC Activation2 | ||||
| GG | … | -0.023 (0.242) | ||
| GT | 0.195 (0.380) | 0.075 (0.340) | ||
| TT | 0.298 (0.323) | 0.216 (0.250) |
FSIQ = Full Scale Intelligence Quotient; derived from the NAART (29)
Absolute means are provided for all three rs1625579 genotypes (GG, GT, and TT) though the analyses combine the less frequent GG and GT genotypes.
Response times were collected at 8 of the 9 sites (90 of 111 subjects).
No SD, only a single subject with response time.
FET = Fisher's Exact Test.
Race is missing for one patient and 2 controls.
Both patients and controls were assessed with demographic and other socioeconomic questionnaires, the Edinburgh Handedness Questionnaire (27), the Fagerstrom Test for Nicotine Dependence (28) and the North American Adult Reading Test (29). Patients were assessed with the Scales for the Assessment of Positive (30) and Negative (31) Symptoms, Schedule for the Deficit Syndrome (32), Calgary Depression Scale (33), InterSePT Scale for Suicidal Thinking (34), Abnormal Involuntary Movement Scale (35), Barnes Akathisia Rating Scale (36), and the Simpson-Angus Scale (37). Ratings were standardized across sites through cross-site group training sessions with experienced clinical raters and by rating videotapes from several patients for comparisons with expert assessments.
Written informed consent, including permission to share de-identified data between the centers and with the wider research community, approved by University of California Irvine, California Los Angeles, New Mexico, Iowa, Minnesota, North Carolina as well as Brigham and Women's Hospital, Massachusetts General Hospital, Yale and Duke University Institutional Review Boards, was obtained from all study participants.
Genotyping
We genotyped the rs1625579 SNP reported in the recent GWAS study (12). Three intergenic Infinium HumanHap300 Bead Array SNPs [rs1938570 (LD=0.828), rs1702292 (LD=0.806), and rs4378243 (LD=0.8)] are in > 80% LD with rs1625579, but are at greater distance to miR-137. Genotyping of rs1625579 was performed by real-time PCR using TaqMan® probes and primers (Applied Biosystems). We performed PCRs on 10ng of genomic DNA in a 5μL final volume using the TaqMan® Genotyping Master Mix and TaqMan® SNP genotyping assay on a 7900 HT real time sequence detector (Applied Biosystems). The TaqMan assay was previously validated by Sanger sequencing with 100% concordance on 10 random samples.
Sternberg Item Recognition Paradigm
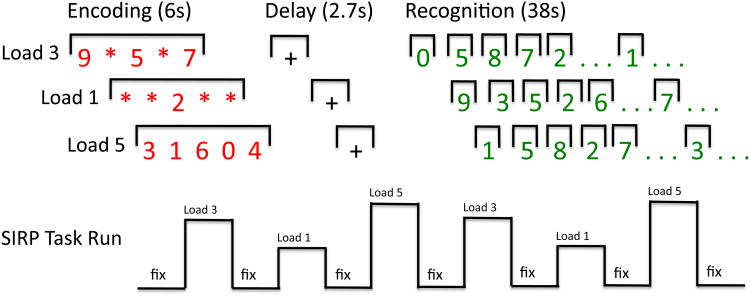
Participants performed the Sternberg Item Recognition Paradigm (SIRP), a working memory paradigm comprised of encoding, delay, and recognition phases (Figure 1). During encoding participants memorized 1, 3, or 5 red digits that were visually presented for 6 seconds. Following a 2.7s delay, 14 green single probe digits were presented serially (50% targets) and participants indicated, via button-box presses, whether a probe was a target or foil (member or non-member of the memorized set, respectively). Probe digits were presented for 1.1s followed by a jittered delay (0.6s-2.486s). After each task block (Load 1, 3, or 5) a jittered duration (mean=12s, range: 4-20s) screen with a flickering cross was presented. Each of the 3 working memory loads was presented twice in pseudo-random order within a run and each participant performed 3 runs. Stimulus presentation and response collection was performed with E-Prime (Psychology Software Tools, Inc.) and participants performed at least one practice run independently prior to the scan to ensure that that they were able to achieve > 75% accuracy on the task. To sustain subject motivation throughout the imaging session, participants were able to earn 5 cents for each correct task response. Mean accuracy and mean response times were calculated for each of the working memory loads (for additional task paradigm details see (4)).
Figure 1. Sternberg Item Response Paradigm.
The Sternberg Item Response Paradigm (SIRP) is a working memory paradigm in which participants study 1, 3, or 5 digits. After a delay they are presented with 14 probe digits. Participants are asked to indicate via button press whether or not the probe digits were part of the study set.
Image Acquisition
Imaging data were acquired at nine imaging centers (4) and included: a localizer for identification of the AC-PC axis, a high-resolution sagittal T1-weighted scan (GE: FSPGR; Siemens: MP-RAGE, with FOV=24cm, 1.2-1.5mm slice thickness, 160-170 slices), a T2-weighted scan to set slice prescription for the functional imaging scans (FOV=22cm, matrix=256×192, 27 slices [thickness/gap=4mm/1mm]), b0 field maps on the Siemens scanners, and T2*-weighted gradient echo EPI scans (FOV=22cm, TR/TE=2s/30ms, flip angle=90°, matrix=64×64, and 27 AC-PC aligned slices [thickness/gap=4mm/1mm]). Participants were required to have a normal night of sleep and no more than one alcoholic drink the night prior to the scan. Subjects refrained from caffeinated beverages two hours and smoking 40 minutes before the scan.
Image Analysis
We obtained DLPFC activation data from one of our previous studies (4). Detailed image analysis procedures are provided in Potkin et al. (2009) (4) and detailed multi-center fMRI quality assurance procedures have been reported on in Glover et al. (2012). Briefly, blind to group errors in the imaging data were identified via image meta-data files, NIfTI image headers, by visual inspection of the raw EPI data (e.g., poor fat suppression, poor slice prescription), and by AFNI's 3dToutcout (38) tool (runs with more than 34 out of 177 volumes with outlier spikes were removed from the analyses) (4). EPI data were corrected for head movement using FMRIB Software Library (FSL(39))'s MCFLIRT by aligning all EPI volumes to the middle volume of each run, FSL's PRELUDE and FUGE were used to B0 correct images, slice-time correction was performed and images were smoothed to 8mm FWHM (40).
First-level statistical analyses were carried out for each run using the general linear model, predicting time series functional imaging data with encoding and recognition epochs convolved with a double gamma hemodynamic response function. Temporal derivatives were included in the model to allow for a better fit of the data by accounting for temporal variation. Statistical contrasts included encoding and recognition epochs versus fixation baseline as well as between-load contrasts. First-level statistical images were registered to the MNI-152 standard space using a 12-parameter affine transformation (41) and second-level fixed effects analyses combined the three runs from each subject. Blind to group base images with obvious structural flaws or a Jaccard (overlap) index of the base image to the MNI-152 atlas larger than 1.5 interquartile range above the 75th percentile of the remaining values, and second-level statistical maps with clear evidence of excessive noise (n=6) were excluded from further analyses. Third-level random effects analyses were performed to create contrast maps for patients and controls separately (p<0.05, FDR correction).
The left DLPFC region of interest (ROI) used in this study included the union of all gray matter voxels that showed significant probe-associated increases with working memory load (p<0.05, based on FDR correction) in either the SZ or HV group analysis within Brodmann areas 9 and 46 of the WFU PickAtlas (42, 43). This combined structural-functional ROI selection method was chosen to confine the extraction of percent signal change to the DLPFC while allowing for group differences in activation patterns across subregions of the DLPFC and reducing the likelihood of washing out our hypothesized group differences due to signal averaging over a large region. DLPFC percent signal change was extracted from the second level contrast of parameter estimates (copes) images.
Statistical Analyses
Given the low frequency of the rs16255579 GG genotype, the distribution of rs1625579 genotypes across diagnoses was examined using the Fisher's Exact Test (Statistical Analysis Systems [SAS 9.2], Proc Freq). The frequency distribution of rs1625579 genotypes did not differ between patients and controls (Table 1). The rs1625579 genotype frequency distributions for both patients and controls were in Hardy-Weinberg Equilibrium. Given that only two controls had the rare GG genotype, GG and GT genotypes were collapsed in all subsequent analyses. Group (patient/control) and genotype (GG+GT/TT) effects on left DLPFC SIRP load 3 probe activation were examined using four univariate mixed model regression analyses (SAS 9.2, Proc Mixed) in which left DLPFC probe 3 activation was predicted either with: 1) diagnosis (patient/control), to confirm DLPFC hyper-activation in patients compared with controls; 2) genotype (GG+GT/TT), to test the hypothesis that the rs1625579 T allele is associated with DLPFC hyper-activation, 3) diagnosis and genotype, to examine whether group or genotype add in predicting DLPFC activation, and 4) diagnosis, genotype, and the diagnosis × genotype interaction, to examine whether group and genotype interact in predicting DLPFC activation ([SZ-TT versus HV-TT] > [SZ-GG/GT versus HV-GG/GT]). All analyses included site, sex, and age as covariates and used planned one-tailed contrast analyses to test the specified directional hypotheses for diagnosis, genotype, and diagnosis × genotype effects. Analyses using similar models were performed to examine diagnosis and possible rs1625579 genotype effects on SIRP load 3 working memory performance and response times. Even though a prior analyses showed no evidence of sample stratification (5), in the current analysis we controlled for the possible confound of rs1625579 allele frequency differences between ethnicities by re-analysis of the data in a CEU-only sample, identified based on a HAPMAP3-based multi-dimensional scaling and cluster analysis (http://pngu.mgh.harvard.edu/purcell/plink; (44)). In addition to our hypothesis-driven DLPFC region of interest analysis, we performed an exploratory analysis in image space examining the effect of rs1625579 genotype on 3 item SIRP probe activation (z>2.3, p<0.05) with a mixed effects analysis using FSL(39)'s FLAME controlling for age, sex, site, and diagnosis.
Results
Mixed model regression analyses confirmed left DLPFC hyper-activation for SIRP load 3 probes in SZ patients compared with health volunteers (t100=1.91, p=0.03). Individuals carrying the common miR-137 schizophrenia risk genotype (rs1625579 TT) showed hyper-activation compared with those carrying the less frequent GG/GT genotype (t100=2.17, p=0.02) (Figure 2). The analysis of additive effects of genotype and diagnoses showed significant effects of genotype (TT > GG/GT; t99=1.99, p=0.03) as well as diagnosis (SZ > HV; t99=1.77, p<0.05) on left DLPFC activation. The analysis of interaction effects of genotype and diagnosis did not show a significant interaction effect (t98=0.35, p=0.36) on left DLPFC activation, while the main effects for diagnosis and genotype remained similar. Analysis of the CEU-only sample showed similar, but more highly significant findings for diagnosis (t65=3.89, p=0.02) and genotype (t65=4.20, p=0.01) and no significant interaction. Behavioral data analysis confirmed the significantly lower working memory performance (t96=-3.5, p=0.0007) and longer response times (t76=40.95, p=0.0005) on SIRP load 3 in schizophrenia patients (LSMean±StdErr=89%±1.3; 951ms±27) compared with healthy volunteers (94%±1.0; 837ms±20) previously observed (4), but working memory performance and response times were similar for rs1625579 genotypes: 92%±0.8 versus 92%±1.4 and 869ms±17 versus 918ms±29 for TT and GG/GT genotypes, respectively. Exploratory whole brain comparisons found two significant clusters in which individuals with rs1625579 TT genotypes showed higher activations compared with rs1625579 GG/TG genotypes (Figure 3). There were no significant rs1625579 GG/TG > TT clusters. While the DLPFC activations were not part of the significant clusters, there was a peak activation in the left DLPFC (z=2.67, P<0.004, uncorrected; see Figure 3).
Figure 2. miR-137 rs1625579 TT Genotype associated with DLPFC Hyper-activation during Working Memory.
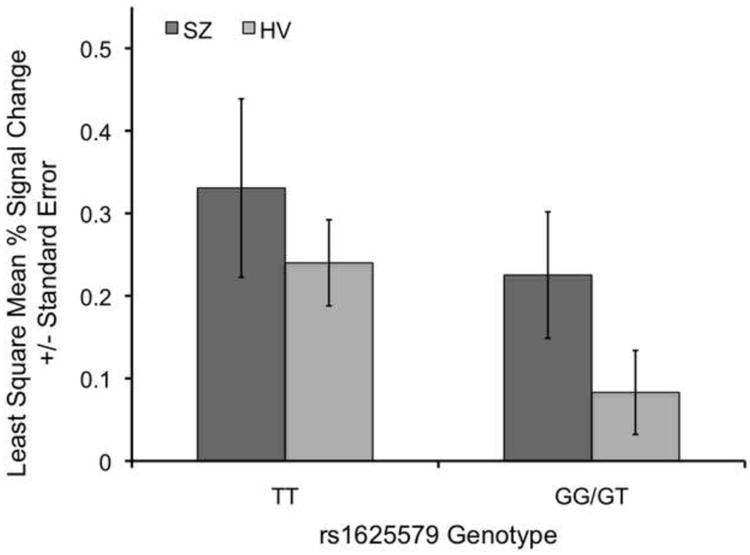
Individuals with rs1625579 TT genotypes show higher left dorsolateral prefrontal cortex probe activation on SIRP working memory load 3 than individuals with the rs1625579 GT or GG genotypes. Schizophrenia patients (SZ) show higher dorsolateral prefrontal cortex probe activation on working memory load 3 than healthy volunteers (HV).
Figure 3.
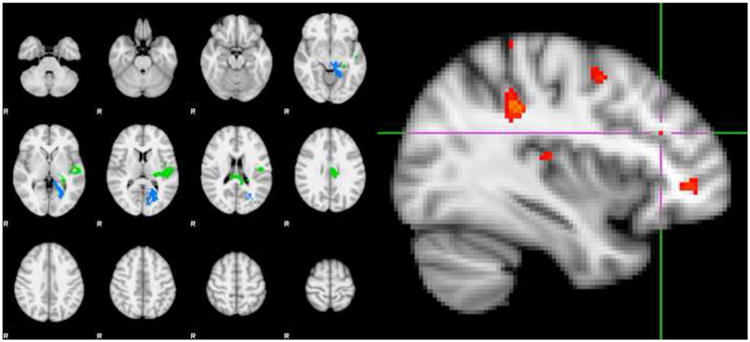
Exploratory Whole Brain Analysis.
|
| |||||
| Cluster Index | Z | x | y | z | FSL Harvard-Oxford Cortical Atlas Label |
|
| |||||
| Blue | 3.8 | -22 | -68 | 8 | Left Intracalcarine Cortex |
| Blue | 3.84 | -18 | -48 | -2 | Left Lingual Gyrus |
| Blue | 3.98 | 20 | 72 | 10 | Left Intracalcarine Cortex |
| Blue | 4.25 | -12 | -40 | -4 | Left Parahippocampal Gyrus, posterior division; extending into the Left Lingual Gyrus |
| Green | 3.45 | -46 | -14 | 18 | Left Central Opercular Cortex |
| Green | 3.46 | -50 | -24 | 2 | Left Planum Temporale |
| Green | 3.72 | -48 | -22 | 6 | Left Heschl's Gyrus |
| Green | 4.11 | -6 | -22 | 28 | Left Cingulate Gyrus, posterior division |
|
| |||||
| Peak DLPFC Voxel | 2.67 | -36 | 30 | 24 | Left Middle Frontal Gyrus (p<0.004, uncorrected) |
|
| |||||
Exploratory whole brain comparison of rs1625579 TT versus GG/TG genotypes showed two significant clusters (z=2.3, p<0.05), which included peak activations in the Left Cingulate Gyrus - posterior division, Left Heschl's Gyrus extending into the Left Planum Temporale, the left Central Opercular Cortex, the Left Parahippocampal Gyrus – posterior division, extending in the Left Lingual Gyrus and Left Intracalcarine Cortex; Left Figure). Exploratory examination beyond the significant clusters, for the rs1625579 TT > rs1625579 GG/TG genotype contrast at z=2.67, p<0.004 (uncorrected) showed significant voxels in the Left Middle Frontal Gyrus activation (Right Figure).
Discussion
The main finding of our study is that the rs1625579 TT (miR-137) schizophrenia risk genotype is associated with left DLPFC hyperactivation compared with the combined GG/GT genotype irrespective of diagnosis. Given equivalent working memory performance between rs1625579 TT and GG/GT carriers, we interpret the observed DLPFC hyperactivation in individuals with the rs1625579 TT genotype compared with those with the GG/GT genotypes as being consistent with neural inefficiency in rs1625579 schizophrenia risk allele carriers; similar to DLPFC inefficiency observed in patients with schizophrenia (3, 4). To our knowledge, this is the first report of a link between the rs1625579 SNP (miR-137) and working memory associated brain activation as measured with fMRI.
The biology underlying the observed miR-137 TT genotype associated neural inefficiency is as of yet unknown though studies of miR-137 expression in mice have shown that miR-137 regulates neural stem cell proliferation and differentiation (16), neurogenesis (17), and neuronal maturation (18). Together with evidence that miR-137 is highly expressed in the rat neuron synaptosome compared to the cytosome (45) and evidence of reduced spine density in DLPFC cortical pyramidal neurons in schizophrenia, these findings suggest that miR-137 may be involved in the development of dysfunctional (neural, synaptic) connectivity within the DLPFC (19) as well as possibly between brain regions (46, 47).
To date, no human post-mortem studies have shown aberrant expression of miR-137 in brains of adult patients with schizophrenia compared with controls; for review see (48). However, recent in-vitro data that 5 schizophrenia candidate genes are regulated by miR-137 (49, 50) strongly suggests a role for miR-137 in schizophrenia. Moreover, it is possible that other mechanisms like differential expression miR-137 across the lifespan or epigenetic mechanisms regulating miR-137 target genes may account for the lack of observed effects in published postmortem work. In fact, miR-137 does not function in isolation but is indirectly regulated by miR-132 (17) whose expression has been found to differ between patients and controls in several studies (51) and may also be influenced by more general perturbations in the microRNA biogenesis pathway (48).
With regard to the functional phenotype associated with the miR-137 risk SNP in living humans, there is some convincing evidence of an impact of miR-137 on cognition (23, 45), some evidence of an impact on the P300 ERP component (21) and on the fMRI-based BOLD signal (22), including that reported in the current study. Of interest is also that some cases diagnosed with autism spectrum disorder with hemizygous deletions on chromosome 1p21.3 have been reported (52). Autism has established clinical and genetic overlap with schizophrenia (53-55) and the possible involvement of miR-137 in chromosome 1p21.3 deletion syndrome was recently investigated (45).
In sum, there are several lines of converging evidence that miR-137 may play a role in schizophrenia. The GWAS finding by Ripke and colleagues (12) in conjunction with the GSEA finding by Potkin and colleagues (7), miR-137 involvement in developmental brain processes (16-18, 45), the confirmed regulation of schizophrenia candidate genes by miR-137 (49, 50), some recent observations of rs1625579 associations with cognition (23, 45), EEG (21), and fMRI (22) measures as well as the therapeutic promise of microRNA related treatments provide impetus for future studies of the role of miR-137 in schizophrenia. The findings from this paper suggest that miR-137 may regulate neural efficiency shown to be abnormal in schizophrenia.
Our findings lend further support to the use of fMRI-based physiological activations to understand the brain effects of schizophrenia liability genes (56) including those that regulate other genes (e.g, miRNAs). They encourage further investigation of miR-137 microstructure, expression of miRNA in different cell types and compartments, its interactions with other non-coding elements [e.g., miR-132] and its target genes (49, 50). It must be noted that DLPFC inefficiency is influenced by other gene variations, including ones that affect neurotransmitter levels such as DAOA and COMT (57) and spine density such as DISC1 (58), corroborating the interpretation that genes contributing to altered synaptic connectivity may influence DLPFC inefficiency.
Strengths of the study are 1) that it used a well-established Sternberg item response functional imaging paradigm that shows robust hyperactivation in schizophrenia patients compared with controls (for retrieval at load 3) when using data collected from multiple scanners even when behavioral performance is matched between the groups (4), 2) that it examined a gene identified by a very large GWAS study with independent sample replication (12) as well as implicated by an independent group (7), 3) that activation differences were observed between genotypes despite similar SIRP load 3 working memory performance and response times, 4) that the rs1625579 genotypes were based on a validated genotyping assay, which is considered less ambiguous than based on imputation (59), 5) that the strength of the findings increased in a more homogeneous, CEU-only sample which controls for possible stratification confounds, and 6) that a voxelwise comparison, while not cluster-level significant, showed a peak activation (rs1625579 TT > GG/TG) in the left DLPFC. Some limitations must also be noted. The sample size is relatively small for a genetic study without a replication cohort. However, concerns about type I error are minimized given that 1) the study examines the functional role of the rs1625579 risk SNP which was identified as a schizophrenia risk allele based on a GWA study with a cohort of 51,695 individuals that included a replication sample, 2) the significant rs1625579 risk SNP effects on DLPFC activation were in the a priori predicted direction (DLPFC hyperactivation in patients with schizophrenia as well as in individuals with the schizophrenia risk allele (T)), and 3) the effects were in the same direction for both patients and controls. The G allele is rare and our sample included only two controls with the GG genotype and therefore did not allow for a more parametric analysis of the genotype effect (TT>GT>GG). The rs1625579 SNP maps about 9kb from the pre-miR137 sequence within an intron of the MIR137HG locus telomeric to the pre-miR137 sequence that maps within the 3rd exon of the MIR137HG. The high LD of the region extends to include the gene DPYD (60). A clump-LD analysis cannot exclude that rs1625579 may map to DPYD which is also a predicted miR-137 target (61).
MiR-137 was previously implicated in a gene set enrichment analysis (7), a SNP (rs1625579) recently showed the strongest effect in a GWA study with a discovery sample of 21,856 and a replication sample of 29,839 independent individuals (12), and shows an association with a schizophrenia phenotype that has been well-characterized in a large functional imaging study (4). This phenotype that has been shown to be highly heritable (62) in a region shown to be highly correlated with genetic liability for schizophrenia (63, 64) lending significant weight to the finding and encouraging further exploration of the functional role of miR-137 in schizophrenia etiology.
Our study suggests that the putative miR-137 locus rs1627759 TT schizophrenia risk genotype (12) is associated with DLPFC hyper-activation; a known schizophrenia risk phenotype and a measure of brain inefficiency. These findings suggest that the functional implications of miR-137 gene-regulation are measurable with functional magnetic resonance imaging even at relatively modest sample sizes and that the miR-137 gene regulatory network merits further investigation with regard to its etiological role in schizophrenia.
In This Issue Statement.
The SNP rs1625579 (miR-137 locus) confers risk for schizophrenia based on a recent genome wide association study including 51,695 individuals, but its functional role remains to be determined. This study examined the association between rs1625579 genotypes and dorsolateral prefrontal cortex activation during working memory. The study found that the schizophrenia risk genotype rs1625579 TT is associated with the schizophrenia risk phenotype DLPFC hyperactivation commonly considered a measure of brain inefficiency.
Acknowledgments
We are thankful to Liv McMillan for overall study coordination, Linda Morgan for genotype assay validation, and to the research subjects for their participation.
Funding: This work was supported by the National Center for Research Resources at the National Institutes of Health (grant numbers: NIH 1 U24 U24 RR021992 (Function Biomedical Informatics Research Network) and NIH 1 U24 RR025736-01 (Biomedical Informatics Research Network Coordinating Center; http://www.birncommunity.org), the William Lion Penzner Foundation (University of California, Irvine, Department of Psychiatry and Human Behavior), and the National Institutes of Mental Health (grant number R01MH085801 to MVP).
Footnotes
Financial Disclosures: Dr. Van Erp is funded by NIH. Dr. Potkin's is funded by the NIH. Dr. Potkin has financial interests in Bristol-Myers Squibb, Eisai, Inc., Eli Lilly, Forest Laboratories, Genentech, Janssen Pharmaceutical, Lundbeck, Merck, Novartis, Organon, Pfizer, Roche, Sunovion, Takeda Pharmaceutical, Vanda Pharmaceutical, Novartis, Lundbeck, Merck, Sunovion and has received grant funding from Amgen, Baxter, Bristol-Myers Squibb, Cephalon, Inc., Eli Lilly, Forest Laboratories, Genentech, Janssen Pharmaceutical, Merck, Otsuka, Pfizer, Roche, Sunovion, Takeda Pharmaceutical, Vanda Pharmaceutical, NIAAA, NIBIB, NIH/NCRR, University of Southern California, UCSF, UCSD, Baylor College of Medicine. Dr. Bustillo has financial interests in Novartis Pharmaceuticals. Dr. Mathalon consults for Bristol-Myers Squibb. The remaining authors declare no potential conflict of interest.
References
- 1.Van Snellenberg JX, Torres IJ, Thornton AE. Functional neuroimaging of working memory in schizophrenia: task performance as a moderating variable. Neuropsychology. 2006;20:497–510. doi: 10.1037/0894-4105.20.5.497. [DOI] [PubMed] [Google Scholar]
- 2.Barch DM, Moore H, Nee DE, Manoach DS, Luck SJ. CNTRICS imaging biomarkers selection: Working memory. Schizophr Bull. 2011;38:43–52. doi: 10.1093/schbul/sbr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callicott JH, Mattay VS, Verchinski BA, Marenco S, Egan MF, Weinberger DR. Complexity of prefrontal cortical dysfunction in schizophrenia: more than up or down. Am J Psychiatry. 2003;160:2209–2215. doi: 10.1176/appi.ajp.160.12.2209. [DOI] [PubMed] [Google Scholar]
- 4.Potkin SG, Turner JA, Brown GG, McCarthy G, Greve DN, Glover GH, et al. Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr Bull. 2009;35:19–31. doi: 10.1093/schbul/sbn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, et al. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potkin SG, Turner JA, Fallon JA, Lakatos A, Keator DB, Guffanti G, et al. Gene discovery through imaging genetics: identification of two novel genes associated with schizophrenia. Mol Psychiatry. 2009;14:416–428. doi: 10.1038/mp.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potkin SG, Macciardi F, Guffanti G, Fallon JH, Wang Q, Turner JA, et al. Identifying gene regulatory networks in schizophrenia. Neuroimage. 2010;53:839–847. doi: 10.1016/j.neuroimage.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins DO, Jeffries C, Sullivan P. Expanding the ‘central dogma’: the regulatory role of nonprotein coding genes and implications for the genetic liability to schizophrenia. Mol Psychiatry. 2005;10:69–78. doi: 10.1038/sj.mp.4001577. [DOI] [PubMed] [Google Scholar]
- 12.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 14.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Y, Deng H, Bi F, Liu J, Bemis LT, Norris D, et al. MicroRNA-137 targets carboxyl-terminal binding protein 1 in melanoma cell lines. Int J Biol Sci. 2011;7:133–137. doi: 10.7150/ijbs.7.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 20.Green MJ, Cairns MJ, Wu J, Dragovic M, Jablensky A, Tooney PA, et al. Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.84. [DOI] [PubMed] [Google Scholar]
- 21.Decoster J, De Hert M, Viechtbauer W, Nagels G, Myin-Germeys I, Peuskens J, et al. Genetic association study of the P300 endophenotype in schizophrenia. Schizophr Res. 2012 doi: 10.1016/j.schres.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 22.Whalley HC, Papmeyer M, Romaniuk L, Sprooten E, Johnstone EC, Hall J, et al. Impact of a microRNA MIR137 Susceptibility Variant on Brain Function in People at High Genetic Risk of Schizophrenia or Bipolar Disorder. Neuropsychopharmacology. 2012 doi: 10.1038/npp.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummings E, Donohoe G, Hargreaves A, Moore S, Fahey C, Dinan TG, et al. Mood congruent psychotic symptoms and specific cognitive deficits in carriers of the novel schizophrenia risk variant at MIR-137. Neurosci Lett. 2012 doi: 10.1016/j.neulet.2012.08.065. [DOI] [PubMed] [Google Scholar]
- 24.Turetsky BI, Calkins ME, Light GA, Olincy A, Radant AD, Swerdlow NR. Neurophysiological endophenotypes of schizophrenia: the viability of selected candidate measures. Schizophr Bull. 2007;33:69–94. doi: 10.1093/schbul/sbl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.First MB, Spitzer RL, Gibbon MG, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders - Patient Edition (SCID-I/P, 11/2002 revision) New York: 2002. [Google Scholar]
- 26.First MB, Spitzer RL, Gibbon MG, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders - Patient Edition (SCID-I/NP, 11/2002 revision) New York: 2002. [Google Scholar]
- 27.Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9:97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- 28.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 29.Uttl B. North American Adult Reading Test: age norms, reliability, and validity. J Clin Exp Neuropsychol. 2002;24:1123–1137. doi: 10.1076/jcen.24.8.1123.8375. [DOI] [PubMed] [Google Scholar]
- 30.Andreasen N. The scale for the assessment of positive symp-toms (SAPS) Iowa City: University of Iowa; 1984. [Google Scholar]
- 31.Andreasen N. The scale for the assessment of negative symptoms (SANS) Iowa City: University of Iowa; 1983. [Google Scholar]
- 32.Kirkpatrick B, Buchanan RW, McKenney PD, Alphs LD, Carpenter WT., Jr The Schedule for the Deficit syndrome: an instrument for research in schizophrenia. Psychiatry Res. 1989;30:119–123. doi: 10.1016/0165-1781(89)90153-4. [DOI] [PubMed] [Google Scholar]
- 33.Addington D, Addington J, Schissel B. A depression rating scale for schizophrenics. Schizophr Res. 1990;3:247–251. doi: 10.1016/0920-9964(90)90005-r. [DOI] [PubMed] [Google Scholar]
- 34.Lindenmayer JP, Czobor P, Alphs L, Nathan AM, Anand R, Islam Z, et al. The InterSePT scale for suicidal thinking reliability and validity. Schizophr Res. 2003;63:161–170. doi: 10.1016/s0920-9964(02)00335-3. [DOI] [PubMed] [Google Scholar]
- 35.Abnormal Involuntary Movement Scale (AIMS) Psychopharmacol Bull. 1988;24:781–783. [PubMed] [Google Scholar]
- 36.Barnes TR. The Barnes Akathisia Rating Scale--revisited. J Psychopharmacol. 2003;17:365–370. doi: 10.1177/0269881103174013. [DOI] [PubMed] [Google Scholar]
- 37.Simpson GM, Angus JW. A rating scale for extrapyramidal side effects. Acta Psychiatr Scand Suppl. 1970;212:11–19. doi: 10.1111/j.1600-0447.1970.tb02066.x. [DOI] [PubMed] [Google Scholar]
- 38.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 39.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 40.Friedman L, Glover GH. Reducing interscanner variability of activation in a multicenter fMRI study: controlling for signal-to-fluctuation-noise-ratio (SFNR) differences. Neuroimage. 2006;33:471–481. doi: 10.1016/j.neuroimage.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 42.Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- 43.Maldjian JA, Laurienti PJ, Burdette JH. Precentral gyrus discrepancy in electronic versions of the Talairach atlas. Neuroimage. 2004;21:450–455. doi: 10.1016/j.neuroimage.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 44.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willemsen MH, Valles A, Kirkels LA, Mastebroek M, Olde Loohuis N, Kos A, et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- 46.Weinberger DR, Berman KF, Suddath R, Torrey EF. Evidence of dysfunction of a prefrontal-limbic network in schizophrenia: a magnetic resonance imaging and regional cerebral blood flow study of discordant monozygotic twins. Am J Psychiatry. 1992;149:890–897. doi: 10.1176/ajp.149.7.890. [DOI] [PubMed] [Google Scholar]
- 47.Wolf DH, Gur RC, Valdez JN, Loughead J, Elliott MA, Gur RE, et al. Alterations of fronto-temporal connectivity during word encoding in schizophrenia. Psychiatry Res. 2007;154:221–232. doi: 10.1016/j.pscychresns.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 49.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- 50.Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr Res. 2012 doi: 10.1016/j.schres.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter MT, Nikkel SM, Fernandez BA, Marshall CR, Noor A, Lionel AC, et al. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin Genet. 2010;80:435–443. doi: 10.1111/j.1399-0004.2010.01578.x. [DOI] [PubMed] [Google Scholar]
- 53.Rapoport J, Chavez A, Greenstein D, Addington A, Gogtay N. Autism spectrum disorders and childhood-onset schizophrenia: clinical and biological contributions to a relation revisited. J Am Acad Child Adolesc Psychiatry. 2009;48:10–18. doi: 10.1097/CHI.0b013e31818b1c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King BH, Lord C. Is schizophrenia on the autism spectrum? Brain Res. 2011;1380:34–41. doi: 10.1016/j.brainres.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 55.Bevan Jones R, Thapar A, Lewis G, Zammit S. The association between early autistic traits and psychotic experiences in adolescence. Schizophr Res. 2012;135:164–169. doi: 10.1016/j.schres.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 56.Walton E, Turner J, Gollub RL, Manoach DS, Yendiki A, Ho BC, et al. Cumulative Genetic Risk and Prefrontal Activity in Patients With Schizophrenia. Schizophr Bull. 2012 doi: 10.1093/schbul/sbr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nixon DC, Prust MJ, Sambataro F, Tan HY, Mattay VS, Weinberger DR, et al. Interactive effects of DAOA (G72) and catechol-O-methyltransferase on neurophysiology in prefrontal cortex. Biol Psychiatry. 2011;69:1006–1008. doi: 10.1016/j.biopsych.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 58.Brauns S, Gollub RL, Roffman JL, Yendiki A, Ho BC, Wassink TH, et al. DISC1 is associated with cortical thickness and neural efficiency. Neuroimage. 2011;57:1591–1600. doi: 10.1016/j.neuroimage.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A. A comprehensive evaluation of SNP genotype imputation. Hum Genet. 2009;125:163–171. doi: 10.1007/s00439-008-0606-5. [DOI] [PubMed] [Google Scholar]
- 60.Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012 doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blokland GA, McMahon KL, Hoffman J, Zhu G, Meredith M, Martin NG, et al. Quantifying the heritability of task-related brain activation and performance during the N-back working memory task: a twin fMRI study. Biol Psychol. 2008;79:70–79. doi: 10.1016/j.biopsycho.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cannon TD, Thompson PM, van Erp TG, Toga AW, Poutanen VP, Huttunen M, et al. Cortex mapping reveals regionally specific patterns of genetic and disease-specific gray-matter deficits in twins discordant for schizophrenia. Proc Natl Acad Sci U S A. 2002;99:3228–3233. doi: 10.1073/pnas.052023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlsgodt KH, Glahn DC, van Erp TG, Therman S, Huttunen M, Manninen M, et al. The relationship between performance and fMRI signal during working memory in patients with schizophrenia, unaffected co-twins, and control subjects. Schizophr Res. 2007;89:191–197. doi: 10.1016/j.schres.2006.08.016. [DOI] [PubMed] [Google Scholar]