Abstract
Gallbladder cancer is relatively uncommon with high incidence in certain geographic locations, including Latin America, East and South Asia and Eastern Europe. Molecular characterization of this disease has been limited and targeted therapy options for advanced disease remain an open area of investigation. In the present study, surgical pathology obtained from resected gallbladder cancer cases (n=72) was examined for the presence of targetable, somatic mutations. All cases were formalin-fixed and paraffin-embedded (FFPE). Two approaches were used: a) mass spectroscopy-based profiling for 159 point (‘hot-spot’) mutations in 33 genes commonly involved in solid tumors and b) next-generation sequencing (NGS) platform that examined the complete coding sequence of in 182 cancer-related genes. Fifty-seven cases were analyzed for hotspot mutations and 15 for NGS. Fourteen hotspot mutations were identified in nine cases. Of these, KRAS mutation was significantly associated with poor survival on multivariate analysis. Other targetable mutations included PIK3CA (N=2) and ALK (N=1). On NGS, 26 mutations were noted in 15 cases. P53 and PI3 kinase pathway (STK11, RICTOR,TSC2) mutations were common. One case had FGF10 amplification while another had FGF3-TACC gene fusion, not previously described in gallbladder cancer. In conclusion, somatic mutation profiling using archival FFPE samples from gallbladder cancer is feasible. NGS, in particular may be a useful platform for identifying novel mutations for targeted therapy.
Keywords: gallbladder neoplasms, mutational analysis, DNA Sequencing
INTRODUCTION
Gallbladder cancer affects over 140,000 patients annually worldwide and over 100,000 will die each year from this disease.(1) Women are affected more than men and in the U.S.; Hispanic population and Alaskan natives have a disproportionately high incidence of gallbladder cancer.(2) There is a remarkable geographic variation with the highest incidence rates reported in India, Korea, Japan, Czech Republic, Slovakia, Spain, Columbia, Chile, Peru, Bolivia, and Ecuador. Etiologies include chronic cholelithiasias, Salmonella infections, toxin exposure, obesity and rarely due to genetic diseases like Hereditary Non-Polyposis Cancer Coli (HNPCC) and type 1 neurofibromatosis. Gallbladder cancer is thought to be at least partly the consequence of chronic inflammation-induced genetic changes.
The current molecular profiling data of gallbladder cancer are limited to small case series or case reports that include one or more oncogenes. High-throughput screening for targetable mutations in this disease is lacking. An understanding of the molecular characteristics and heterogeneity of gallbladder cancer is critical towards improving the treatment paradigm for this disease. An impetus for such characterization is the potential of targeted therapies directed against the products of these molecular aberrations including the tumor proteomic profile. Once the underlying molecular abnormalities of a cancer are identified, targeted inhibitors can be discovered and result in incremental benefit even in genetically heterogeneous malignancies. For instance, in lung cancer the identification of echinoderm microtubule associated protein like 4 - anaplastic lymphoma kinase (EML4-ALK) mutation has led to a targeted approach with crizotinib and tumors with epidermal growth factor receptor (EGFR) mutations to the development of erlotinib or gefitinib.(3) High-throughput technologies that can rapidly screen for somatic mutations in archival formalin-fixed, paraffin-embedded specimens are critical for this effort. The Sequenom Massarray™ system is ideally suited for the detection of low abundance mutations and can be customized towards targeted therapeutics.(4, 5) In the present study, we used the high-throughput Sequenom MassArray™ approach to investigate mutations in 33 genes in a cohort of gallbladder cancer cases to determine the frequency of genetic mutations in this population. We also explored next generation sequencing (NGS) to examine a wider panel of genetic aberrations in a limited number of gallbladder cancer cases.
MATERIAL AND METHODS
Tumor samples
Surgically resected, formalin fixed paraffin embedded (FFPE) specimens were obtained for 72 patients with gallbladder cancer. The paraffin embedded blocks were sectioned, and hematoxylin & eosin (H&E) stained slides were reviewed by surgical pathology to confirm the tumor content in each section. Ten serial sections (4µm) were cut from selected tissue blocks and areas with tumor tissue were micro dissected from those slides using the H&E slides as templates. Approval for the study was obtained from the institutional review board at MD Anderson Cancer Center.
DNA Extraction
The samples were deparaffinized using xylene washes followed by ethanol (100%) washes. DNA extraction was performed using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer protocol. DNA was quantitated using the NanoQuant system (Tecan Group, Männedorf, Switzerland).
Sequenom MassArray
Hotspot mutational analysis was performed using the Sequenom MassARRAY™ using the iPLEX™ technology (Sequenom, Inc, San Diego, CA). This technology allows for parallel high-throughput screening while using minimal DNA obtained from FFPE specimens (6). Mutations were screened by using amplification through polymerase chain reaction (PCR) and single-base primer extension where the wild type or mutated base was identified by mass spectrometry. Briefly, for each mutation site, PCR and extension primers were designed using Sequenom, Inc. Assay Design. PCR reactions were run following manufacture’s protocol. After PCR, amplicons were cleaned using EXO-SAP® kit (Sequenom) in a GeneAmp 9700 thermocycler (Applied Biosystems). Then the primer was then extended by IPLEX™ chemistry, desalted using Clean Resin (Sequenom), and spotted onto SpectroChip matrix chips (Sequenom) using a nanodispenser (Samsung). Chips were run in duplicate on a Sequenom MassArray Matrix-assisted laser desorption/ionization-Time of Flight (MALDI-TOF) MassArray system. We used Sequenom Typer Software for visual inspection and interpretation of mass spectra. Reactions where the mutant peak represented more than 10% of the wild type peak were scored as positive. The data analysis was performed using MassArray TYPER 4.0 genotyping software (Sequenom) where the SNP calls were divided in 3 groups: conservative, moderate and aggressive calls, depending on the level of confidence.
The Sequenom panel used here was previously designed by the Characterized Cell Line Core (Core Shared Resources – CCSG) at MD Anderson Cancer Center with the aim of detecting somatic DNA alterations in cancer samples. The Sequenom panel was designed based on data form the Catalogue of Somatic Mutations in Cancer (COSMIC) and the Cancer Genome Atlas (TCGA) that reported those alterations (and others in the panel) as somatic mutations previously. A total of 159 point mutations in 33 genes frequently mutated in solid tumors including were analyzed. The analytical sensitivity of the assay [limit of detection (LOD) 5%–10% of mutant DNA in total DNA] is higher than conventional Sanger sequencing (LOD: 10%–20%) and similar to pyrosequencing (LOD: 5%–10%). The advantages offered by the MassARRAY system include high-throughput screening for many hot-spot mutations in parallel, use of minimal DNA isolated from formalin-fixed paraffin-embedded tissues, ability to detect coexisting multiple mutations, and cost and time effectiveness. Appendix 1 lists the genes and mutations investigated in this study.
Next Generation Sequencing
The pathologic diagnosis of each case of gallbladder cancer was confirmed on routine hematoxylin- and eosin-stained slides. All samples sent for DNA extraction contained a minimum of 20% DNA derived from tumor cells. DNA was extracted from 40 mm of FFPE tissue using the Maxwell 16 FFPE Plus LEV DNA Purification kit (Promega™) and quantified using a standardized PicoGreen fluorescence assay (Invitrogen™). Library construction was performed as described previously, using 50–200 ng of DNA sheared by sonication to B100–400 bp before end-repair, dA addition and ligation of indexed, Illumina™ sequencing adaptors (7, 8). Enrichment of target sequences (3320 exons of 182 cancer-related genes and 37 introns from 14 genes recurrently rearranged in cancer representing approximately 1.1Mb of the human genome) was achieved by solution-based hybrid capture with a custom Agilent SureSelect™ biotinylated RNA baitset (8). The selected libraries were sequenced on an Illumina HiSeq 2000 platform using 49149 paired-end reads. Sequence data from genomic DNA was mapped to the reference human genome (hg19) using the Burrows-Wheeler Aligner™ and were processed using the publicly available Sequence Alignment/Map (SAMtools), Picard and Genome Analysis Toolkit (9, 10). Point mutations were identified by a Bayesian algorithm; short insertions and deletions determined by local assembly; gene copy number alterations (amplifications) by comparison to process matched normal controls; and gene fusions/rearrangements were detected by clustering chimeric reads mapped to targeted introns as described previously (11).
Statistical Analysis
Given the limited number of cases analyzed for NGS, only the cases analyzed for hotspot mutations (n=57) were analyzed for their association with survival. Overall survival (OS) was calculated as the number of months from surgery (or core biopsy) to death or last follow-up date. Patients who were alive at their last follow-up were censored on that date. Time to Progression (TTP) was calculated as the number of months from surgery (or core biopsy) to progression. Patients without tumor progression at their last follow-up were censored on that date. The Kaplan-Meier product limit method was used to estimate the median OS for each clinical/demographic factor.(12) Univariate Cox proportional hazards regression was used to model the association between potential predictors and OS. Multivariate Cox proportional hazards regression was used to model all the statistically significant variables in the univariate setting. Backwards selection method was used to remove variables that did not remain significant in the multivariate model.(13) For each factor, medians, hazard ratios (HR), their 95% confidence intervals (CI), and proportional hazards regression p-values are presented in tables. Similar analyses were performed for time to progression. Statistical significance was considered at P-values of <0.05.
Statistical analysis was performed using STATA/SE version 12.1 statistical software (Stata Corp. LP, College Station, TX).
RESULTS
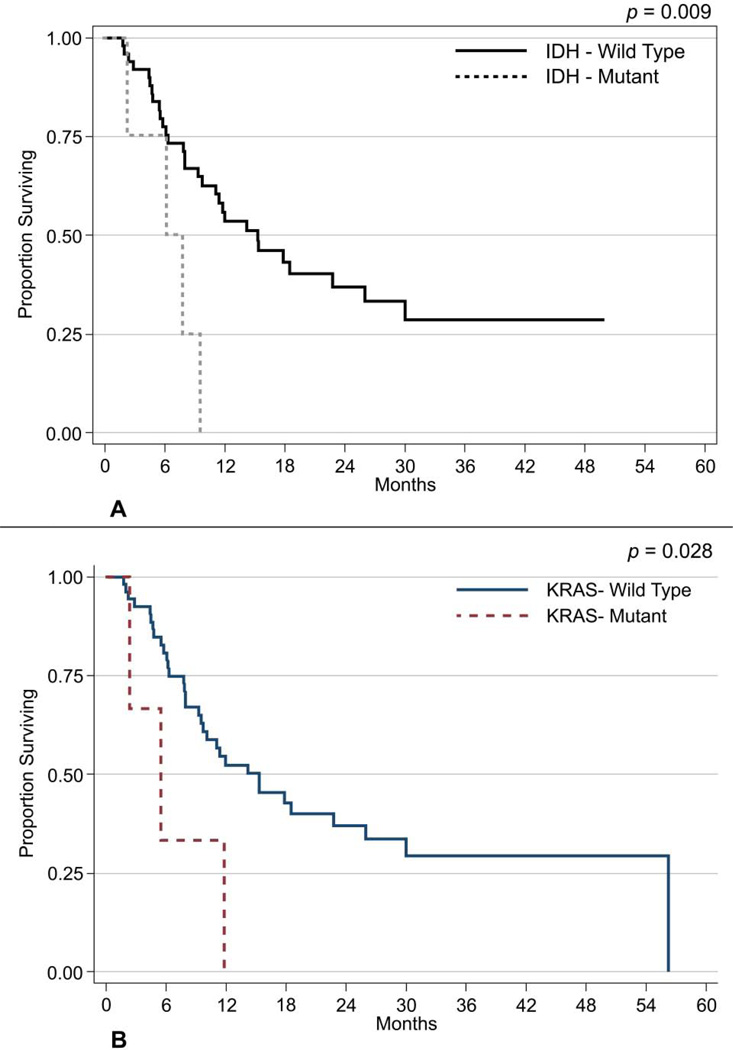
Fifty-seven cases of gallbladder cancer were analyzed for hotspot mutations and 15 for NGS. Patient demographics are described in Table 1. Fourteen hotspot mutations (Table 2) were identified from eleven different tumors within this sample set, with three cases demonstrating more than 1 mutation. IDH1 mutations were the most frequent (n=4). The others identified included mutations of KRAS (n=3), NRAS (n=3), PIK3CA (n=2) and MET (n=1). Of these, IDH1 and MET may represent germline polymorphisms rather than somatic mutations as discussed below. Figure 1 demonstrates the PIK3CA, IDH1 and KRAS mutations. Figures 2A–2D depict the histologies (H&E) of four gallbladder cancer cases along with their corresponding mutations. A total of 36/57 (63.2%) patients enrolled in the study have expired to date. A univariate survival analysis on these data demonstrated a significant relationship of overall survival with six factors. The overall risk of mortality was associated with treatment with chemotherapy (HR: 2.84; 95%CI: 1.23–6.53; p=0.014), lymphatic infiltration (HR: 2.72; 95%CI: 1.22–6.04; p=0.014), venous infiltration (HR: 2.27; 95%CI: 1.08–4.79; p=0.031), perineural infiltration (HR: 2.14; 95%CI: 1.06–4.33; p=0.033), positive KRAS mutation (HR: 3.56; 95%CI: 1.06–11.92; p=0.040), and with a positive IDH1 mutation (HR: 4.04; 95%CI: 1.35–12.13; p=0.013) (Fig.3a). In addition, patients who had chemotherapy were at greater risk of progressing than non-treated patients (HR: 13.82; 95%CI: 1.84–103.84; p=0.011).
Table 1.
Summary Statistics of Patient Demographics and Tumor Characteristics
| CHARACTERISTICS | Type of Analysis | |
|---|---|---|
| Hotspot Mutation Analysis (N = 57) |
Next Generation Sequencing (N=15) |
|
| Age (years) | MEDIAN (RANGE) 62 (30–84) |
MEDIAN (RANGE) 62 (48–78) |
| N (%) | N (%) | |
| Sex | ||
| Male | 25 (44%) | 5 (33%) |
| Female | 32 (56%) | 10 (67%) |
| Ethnicity | ||
| Asian | 1 (2%) | 1 (7%) |
| Hispanic | 8 (14%) | 1 (7%) |
| Black | 5 (9%) | 0 (0%) |
| White | 43 (75%) | 13 (86%) |
| Type of Surgery | ||
| None | 3 (5%) | 4 (27%) |
| Simple (Laparoscopic) | 31 (54%) | 6 (40%) |
| Radical | 23 (41%) | 5 (33%) |
| Adjuvant Therapy | ||
| Chemotherapy | 25 (44%) | 13 (87%) |
| Chemotherapy & Radiation | 11 (19%) | 2 (13%) |
| None | 21 (37%) | 0 (0%) |
| Histological Type | ||
| Adenocarcinoma | 51 (90%) | 15 (100%) |
| Adenosquamous | 4 (7%) | 0 (0%) |
| Carcinosarcoma | 2(3%) | 0 (0%) |
| Degree of Differentiation | ||
| Poor | 16 (28%) | 7 (47%) |
| Moderate | 38 (67%) | 6 (40%) |
| Well | 2 (4%) | 2 (13%) |
| N/A | 1 (2%) | 0 (0%) |
| Lymphatic Infiltration* | ||
| No | 21 (39%) | 1 (10%) |
| Yes | 33 (61%) | 9 (90%) |
| Venous Infiltration* | ||
| No | 22 (41%) | 1 (10%) |
| Yes | 32 (59%) | 9 (90%) |
| Perineural Infiltration* | ||
| No | 29 (54%) | 3 (30%) |
| Yes | 25 (46%) | 7 (70%) |
N=Patient numbers;
Surgical samples only (Hotspot N=54, NGS N=10)
Table 2.
Genetic Mutations identified through Hotspot Analysis
| Sample ID | Histology | Mutations |
|---|---|---|
| 13 | Adenocarcinoma | IDH1_V178I_G532A* PIK3CA_H1047RL_A3140GT |
| 26 | Adenosquamous | KRAS_G12DAV_G35ACT NRAS_Q61RPL_A182GCT |
| 32 | Adenocarcinoma | IDH1_V178I_G532A* |
| 34 | Adenocarcinoma | NRAS_G12DAV_G35ACT |
| 42 | Adenocarcinoma | IDH1_V178I_G532A* |
| 44 | Adenocarcinoma | KRAS_G12DAV_G35ACT MET_N375S_A1124G* |
| 46 | Adenocarcinoma | KRAS_G13DAV_G38ACT |
| 47 | Adenocarcinoma | IDH1_V178I_G532A* |
| 49 | Adenocarcinoma | PIK3CA_M1043I_G3129ATC |
| 56 | Adenocarcinoma | ALK_F1174L_C3522AG |
| 57 | Adenocarcinoma | NRAS_G12DAV_G35ACT |
Most likely to represent genomic variation (SNP)
Figure 1.
Peaks for PIK3CA, IDH1 and KRAS mutations (Sequenom Massarray)
Figure 2.
2a) IDH1 mutation and association with overall survival.
2b) KRAS mutation and association with overall survival
Figure 3.
Schematic of FGFR3-TACC3 Fusion Gene in Gallbladder Cancer
A multivariate analysis of overall survival was also performed using backward elimination methods. Overall survival was seen to be associated with patients age 62–79 (HR: 5.93; 95%CI: 1.76 – 20.00; p=0.004), and age ≥ 70 (HR: 3.84; 95%CI: 1.19 – 12.39; p=0.024), clinical stages 3a, 3b, 4a & 4b (HR: 2.60; 95%CI: 1.03–6.59; p=0.044), venous infiltration (HR: 3.42; 95%CI: 1.46–8.03; p=0.005) and KRAS (HR: 8.91; 95%CI: 1.99–39.94; p=0.004-Fig.3b).
On NGS, 26 mutations were noted in 15 cases (Tables 3). P53 was most common and there was relative preponderance of mutations involving the PI3 kinase pathway: STK11, RICTOR, TSC2. Two cases had FGF pathway aberrations: FGF10 amplification and one case of FGF3-TACC fusion gene (Fig 4). Two cases are illustrated wherein the mutational data were utilized for targeted therapeutics with success (Fig 5a; Fig 5b).
Table 3.
Genetic Alterations Identified Through NGS (N=15)
| GENE | Alterations (With allele frequency or copy number) |
|---|---|
| TP53 | V274F (10%) R282G (50%) R213* (29%) Y220C (2%) R342* (24%) C141* (21%) Splice site 559+1G>T (21%) F109V (46%) V272L |
| STK11 | R86* (11%) E120* (15%) K62fs*98 (6%) |
| CCNE1 | Amplification (copy no 11×) Amplification (copy no 13×) |
| MDM2 | Amplification (copy no 6×) Amplification (copy no 16×) |
| MYC | Amplification (copy no 12×) Amplification (copy no 7×) |
| RICTOR | Amplification (copy no 12×) Amplification (copy no 7×) |
| APC | S2113fs*25 (21%) |
| ARID1A | G284fs*78 (18%) |
| AURKA | S398L (48%) |
| CDKN2A | Truncation - exon 1 |
| CDKN2A/B | Loss Loss |
| CRKL | Amplification (copy no 12×) |
| FGF10 | Amplification (copy no 7×) |
| FGFR3-TACC | FGFR3-TACC3 fusion, Amplification (copy no 8×) |
| KRAS | G12C, 3% |
| MCL1 | Amplification (copy no 8×) Amplification (copy no 8×) |
| PRKAR1A | R97* (33%) |
| SMAD4 | Truncation |
| SMARCA4 | D558fs*6 (26%) |
| TSC2 | Loss |
| BAP1 | splice site 438-1delGTTTTTCCCC AG, 10% 1delGTTTTTCCCC AG, 10% |
| ERBB2 | Amplification (copy no 20×) Amplification (copy no 9×) |
| PIK3CA | Amplification (copy no 7×) |
| ZNF703 | Amplification (copy no 7×) |
Figure 4.
Illustrations of mutational data successfully utilized for targeted therapeutics.
4a) Erlotinib in combination with gemcitabine and oxaliplatin before therapy and 4 months post therapy.
4b) Trastuzumab in combination with 5-fluorouracil, leucovorin and oxaliplatin as second-line therapy before therapy and 3 months post-therapy.
Figure 5.
Representative histopathology of samples with corresponding mutations used for Sequenom analysis and NGS.
5A) KRAS
5B) TP53, ERBB2
5C) FGFR3-TACC3, CCNE1, MCL1, MYC, TP53
5D)ARID1A
DISCUSSION
Gallbladder cancer has been referred to as an ‘orphan’ cancer, given its relative infrequency in the Western population. Molecular research in this disease has lagged behind the commoner gastrointestinal cancers, such as colorectal and gastric cancer. The known genetic alterations include mutations of K-RAS (in 3–40%, more likely in East Asia), PI3KCA (12%), p53 (40%) and BRAF (33%) oncogenes, and amplification of Her-2/ Neu (15%).(14, 15) Other genetic alterations described include loss of expression fragile histidine triad (FHIT) gene, microsatellite instability, overexpression of P13-K/Akt, VEGF and p21.(16, 17) Key limitations of the above data include the small number of cases tested, geographic and ethnic variation.
The present study, to our knowledge represents the largest number of surgically resected gallbladder cancer cases that had somatic mutation profiling. All of our specimens were FFPE and therefore we chose a platform that had non-fastidious DNA requirements and could detect low-abundance mutations. Sequenom Massarray technique is ideal in this situation for profiling single nucleotide mutations and polymorphisms. A limitation of this retrospective study is that we did not have parallel blood or normal tissue to assess if the mutations we noted were germline or somatic. We have used preselected panels, which included targetable oncogenes from the COSMIC and TCGA database. While the plan was to include somatic mutations only, in these panels, subsequent studies have reported that at least two of the genetic alterations (IDH1 and met) were germline.
Sanger sequencing has been effectively used for somatic mutation discovery. However, when there is a heterogeneous mixture of cancerous and normal tissue, Sanger sequencing may be unable to detect low frequency mutations. In one published study, sequencing failed to detect EGFR (Epidermal Growth Factor Receptor) mutations in tumors with roughly 10% allele frequencies.(18) Clinical somatic mutation detection will require high degree of sensitivity than standard sequencing. The Massarray™ system combines PCR with matrix-assisted laser desorption/ ionization time of flight mass spectrometry for rapidly multiplexed nucleic acid analysis. Furthermore, this system can rapidly profile hundreds of mutations in FFPE samples with as little as 5% mutation abundance with a short turn-around time. However, the podisadvantages of this approach is that these multiplex genomic tests only detect the expression of pre-selected hotspot mutations and do not lead to the discovery of novel targets. This limitation is particularly relevant to the less common tumors, such as gallbladder cancer.
Our findings indicated that IDH1_V178I was the commonest DNA variation on Sequenom Massarray. It is estimated that another mutation on IDH1_R132 occurs in upto 20% of high grade glioma and this mutation is associated with a better prognosis and response to therapy.(19) On the other hand, the same somatic mutation in acute myeloid leukemia is associated with a poor prognosis and lack of complete response, particularly in otherwise cytogenetically normal cases.(20) A poor prognosis was noted in our study with IDH1_V178I mutation. In a prior study, IDH1 mutations (IDH1_R132) were noted in cholangiocarcinoma, but none were noted in the 25 cases of gallbladder cancer studied. (21) Isocitrate dehydrogenase (IDH) catalyzes the conversion of isocitrate to α-ketoglutarate and mutations in this pathway is a relevant target for therapy given the development of IDH inhibitors.(22) These mutations also conferred an enzymatic gain-of-function: the novel NADPH-dependent reduction of α-ketoglutarate to the normally trace metabolite R(−)-2-hydroxyglutarate (2-HG), which is oncogenic.(23) Measurement of intracellular 2-HG can therefore be used to assess the functional impact of the mutation. In case of IDH1_V178I, no elevation of 2-HG was noted, which raises the question of whether this mutation represents a non-functional polymorphism or if the functional oncogenic effect includes a non 2-HG metabolic pathway. Several SNPs related to the lipid metabolism, estrogen receptor and DNA repair have been associated with survival in gallbladder cancer (24–26). One case had ALK mutation (ALK_F1174L_C3522AG), which has not yet been described in this disease and offers effective targeted therapy options.
The next generation sequencing approach offers several advantages over the traditional methods, including the ability to simultaneously sequence hundreds of genes in a single test, have a higher depth of coverage and thereby heightened sensitivity for mutation detection, ideal for ‘precision medicine’.(27) In addition, these technologies can detect deletions, amplifications, translocations and base substitutions at a relatively rapid rate. The disadvantage includes cost, high computational requirements and high tissue requirement that make the technology unsuitable for smaller biopsies, circulating tumor cells and circulating plasma DNA. A notable finding in our study was the relatively common occurrence PI3-kinase pathway mutations (TCS2, STK11, RICTOR), which opens potential options for targeted therapies directed against these proteins. Deshpande et al, had noted the relative frequency of PI3KCA mutations in this population.(21) Other targetable mutations included AURKA and BAP, which may potentially be treated with aurora kinase inhibitors and DNA damaging agents [such as cisplatin and poly ADP ribose polymerase (PARP) inhibitors], respectively.
A novel finding in our study was the detection of fusion between Fibroblast Growth Factor Receptor (FGFR3) and Transforming Acidic Coiled-Coil (TACC) [in-frame fusion between exons 1–17 of FGFR3 (containing the kinase domain) and exons 11 to the C-terminus of TACC3 (containing the coiled coil TACC domain)]. The FGFR family plays an important role in cellular proliferation and angiogenesis and gain of function mutations in FGFRs have been reported in several malignancies.(28) FGFR3 mutation or amplification has not been reported in gallbladder cancer to our knowledge. Similar fusions between FGFR3 and TACC3 have recently been reported in a small percentage of glioblastomas.(29) These fusions have also recently been described in cholangiocarcinoma, are proven to be oncogenic and the resulting tumors may be susceptible to FGFR inhibitors.(30)
In conclusion, gallbladder cancer is amenable to precise interventions with targeted therapies and novel sequencing techniques may provide prognostic and therapeutic opportunities.
Acknowledgments
Funding Disclosures: Supported by a grant from Global Academic Programs, MD Anderson Cancer Center and Project Fondecyt 1120208.
Appendix 1
Appendix 1.
GENES AND MUTATIONS INVESTIGATED
| AKT1_E17K_G49A | FGFR1_S125L_C374T | MET_Y1248C_A3743G |
| AKT2_E17K_G49A | FGFR2_N549KK_T1647GA | MET_Y1248HD_T3742CG |
| AKT3_E17K_G49A | FGFR2_S252W_C755G | MET_Y1253D_T3757G |
| ALK_F1174CS_T3521GC | FGFR3_G370C_G1108T | MGA_T1747N_C5421A |
| ALK_F1174L_C3522AG | FGFR3_G380R_G1138A | NRAS_A146T_G436A |
| ALK_F1174LIV_T3520CAG | FGFR3_G697C_G2089T | NRAS_G12DAV_G35ACT |
| ALK_F1245C_T3734G | FGFR3_K650MT_A1949TC | NRAS_G12SRC_G34ACT |
| ALK_F1245L_C3735AG | FGFR3_R248C_C742T | NRAS_G13DAV_G38ACT |
| ALK_F1245VI_T3733GA | FGFR3_S249C_C746G | NRAS_G13SRC_G37ACT |
| ALK_I1171N_T3512A | FGFR3_Y373C_A1118G | NRAS_Q61EKX_C181GAT |
| ALK_R1275QL_G3824AT | FOXL2_C134W_C402G | NRAS_Q61HHQ_A183TCG |
| BCOR_N1407STI_A4220GCT | GNA11_Q209LP_A626TC | NRAS_Q61RPL_A182GCT |
| BRAF_D594GV_A1781GT | GNA11_R183C_C547T | PDGFRA_D842V_A2525T |
| BRAF_E586K_G1756A | GNAQ_Q209H_A627T | PDGFRA_D842YN_G2524TA |
| BRAF_G464EVA_G1391ATC | GNAQ_Q209LPR_A626TCG | PDGFRA_N659K_C1977A |
| BRAF_G466EVA_G1397ATC | GNAS_R201H_G602A | PDGFRA_N659Y_A1975T |
| BRAF_G466R_G1396CA | GNAS_R201SC_C601AT | PDGFRA_V561D_T1682A |
| BRAF_G469EVA_G1406ATC | GRM3_E870K_G2608A | PIK3CA_A1046V_C3137T |
| BRAF_G469R_G1405CA | IDH1_G70D_G209A | PIK3CA_C420R_T1258C |
| BRAF_K601E_A1801G | IDH1_R132CGS_C394TGA | PIK3CA_E110K_G328A |
| BRAF_L597RQ_T1790GA | IDH1_R132HL_G395AT | PIK3CA_E418K_G1252A |
| BRAF_V600_G1800 | IDH1_V178I_G532A | PIK3CA_E453K_G1357A |
| BRAF_V600EAG_T1799ACG_F | IDH2_R140LQ_G419TA | PIK3CA_E542KQ_G1624AC |
| BRAF_V600EAG_T1799ACG_R | IDH2_R140W_C418T | PIK3CA_E542VG_A1625TG |
| BRAF_V600LM_G1798TA | IDH2_R172GW_A514GT | PIK3CA_E545AGV_A1634CGT |
| CC2D1A_L913V_C3036G | IDH2_R172MK_G515TA | PIK3CA_E545D_G1635CT |
| CDK4_R24C_C70T | IDH2_R172S_G516T | PIK3CA_E545KQ_G1633AC |
| CDK4_R24H_G71A | JAK2_V617F_G1849T | PIK3CA_F909L_C2727G |
| CSMD1_A409S_G1225T | KIT_D816GVA_A2447GTC | PIK3CA_G118D_G353A |
| CSMD1_Q3005X_C9013T | KIT_D816HNY_G2446CAT | PIK3CA_H1047RL_A3140GT_F |
| CTNNB1_D32AGV_A95CGT | KIT_K642E_A1924G | PIK3CA_H1047RL_A3140GT_R |
| CTNNB1_D32HNY_G94CAT | KIT_L576P_T1727C | PIK3CA_H1047Y_C3139T |
| CTNNB1_G34EVA_G101ATC | KIT_N566D_A1696G | PIK3CA_H701P_A2102C |
| CTNNB1_H36PRY_A107CGT | KIT_N822KNK_T2466GCA | PIK3CA_K111N_G333C |
| CTNNB1_I35NST_T104AGC | KIT_N822YHD_A2464TCG | PIK3CA_M1043I_G3129ATC |
| CTNNB1_S33APT_T97GCA | KIT_R634W_C1900T | PIK3CA_M1043V_A3127G |
| CTNNB1_S37CFY_C110GTA | KIT_V559ADG_T1676CAG | PIK3CA_N345K_T1035A |
| CTNNB1_S45APT_T133GCA | KIT_V560DGA_T1679AGC | PIK3CA_P539R_C1616G |
| CTNNB1_S45CFY_C134GTA | KIT_V825A_T2474C | PIK3CA_Q060K_C178A |
| CTNNB1_T41APS_A121GCT | KIT_Y553N_T1657A | PIK3CA_Q546EK_C1636GA |
| CTNNB1_T41INS_C122TAG | KRAS_A146PT_G436CA | PIK3CA_Q546LPR_A1637TCG |
| EGFR_G719CS_G2155TA | KRAS_G10R_G28A | PIK3CA_R088Q_G263A |
| EGFR_K860I_A2579T | KRAS_G12DAV_G35ACT | PIK3CA_S405F_C1214T |
| EGFR_L858R_T2573G | KRAS_G12SRC_G34ACT | PIK3CA_T1025SA_A3073TG |
| EGFR_L861QR_T2582AG | KRAS_G13DAV_G38ACT | PIK3CA_Y1021C_A3062G |
| EGFR_S720P_T2158C | KRAS_G13SRC_G37ACT | PIK3CA_Y1021HN_T3061CA |
| EGFR_T790M_C2369T | KRAS_Q61EKX_C181GAT | PPP2R1A_W257G_T769G |
| EGFR_T854I_C2561T | KRAS_Q61HHQ_A183CTG | RAF1_A319S_G955T |
| EGFR_Y813C_A2438G | KRAS_Q61LPR_A182TCG | RAF1_L613V_C1837G |
| EPHA3_K761NN_G2283TC | MAP2K2_E207KQ_G619AC | RAF1_N115S_A344G |
| FBXO4_L23Q_T68A | MAP2K7_D290D_C870T | RAF1_Q335H_G1005C |
| FBXO4_P76T_C226A | MAP2K7_R162H_G485A | RAF1_S259A_T775G |
| FBXO4_S8R_C24AG | MAP2K7_S271T_T811A | RAF1_Y340D_T1018G |
| FBXW7_R465C_C1393T | MAP2K7_S311L_C932T | RET_M918T_T2753C |
| FBXW7_R465HL_G1394AT | MET_H1112RL_A3335GT | RPL22_K15TRM_A44CGT |
| FBXW7_R479QL_G1436AT | MET_H1112Y_C3334T | SFRS9_Y192X_C722A |
| FBXW7_R505CS_C1513TA | MET_M1268T_T3803C | SMO_A324T_G970A |
| FBXW7_R505HLP_G1514ATC | MET_N375S_A1124G | SRC_Q531X_C1591T |
| FBXW7_S582L_C1745T | MET_R988C_C2962T | TGM2_S212P_T734C |
| FGFR1_P252T_C754A | MET_T1010I_C3029T |
Appendix 2
APPENDIX 2.
GENETIC MUTATIONS SEQUENCED USING NGS
| 182 genes sequenced across entire coding sequence | ||||
| Gene | Gene | Gene | Gene | Gene |
| ABL1 | CDK6 | FLT4 | MEN1 | PTPN11 |
| ABL2 | CDK8 | FOXP4 | MET | PTPRD |
| AKT1 | CDKN2A | GATA1 | MITF | RAF1 |
| AKT2 | CDKN2B | GNA11 | MLH1 | RARA |
| AKT3 | CDKN2C | GNAQ | MLL | RB1 |
| ALK | CEBPA | GNAS | MPL | RET |
| APC | CHEK1 | GPR124 | MRE11A | RICTOR |
| AR | CHEK2 | GUCY1A2 | MSH2 | RPTOR |
| ARAF | CRKL | HQXA3 | MSH6 | RUNX1 |
| ARFRP1 | CRLF2 | HRAS | MTOR | SMAD2 |
| ARID1A | CTNNB1 | HSP9OAA1 | MUTYI-1 | SMAD3 |
| ATM | DDR2 | IDH1 | MYC | SMAD4 |
| ATR | DNMT3A | IDH2 | MYCL1 | SMARCA4 |
| AURKA | DOT1L | IGF1R | MYCN | SMARCB1 |
| AURKS | EGFR | IGF2R | NFl | SMO |
| BAP1 | EPI-1A3 | IKBKE | NF2 | SOX1O |
| BCL2 | EPF-1A5 | IKZF1 | NKX2-1 | SOX2 |
| BCL2A1 | EPHA6 | INHBA | NOTCH1 | SRC |
| BCL2L1 | EPHA7 | INSR | NPM1 | STAT3 |
| BCL2L2 | EPHB1 | IRS2 | NRAS | STK11 |
| BCL6 | EPHB4 | JAK1 | NTRK1 | SUFU |
| BRAF | EPHB6 | JAK2 | NTRK2 | T5X22 |
| BRCA1 | ERBB2 | JAK3 | NTRK3 | TET2 |
| BRCA2 | ERBB3 | JUN | PAK3 | TGFBR2 |
| CARD11 | ERBB4 | KDM6A | PAX5 | TNFAIP3 |
| CBL | ERCC2 | KDR | PDGFRA | TNKS |
| CCND1 | ERG | KIT | PDGFRB | TNKS2 |
| CCND2 | ESR1 | KRAS | PHLPP2 | TOP1 |
| CCND3 | EZH2 | LRP1B | PIK3CA | TP53 |
| CCNE1 | FANCA | LRP6 | PIK3CG | TSC1 |
| CD79A | FBXW7 | LTK | PIK3R1 | TSC2 |
| CD79B | FGFR1 | MAP2K1 | PKHD1 | USP9X |
| CDH1 | FGFR2 | MAP2K2 | PLCG1 | VHL |
| CDH2 | FGFR3 | MAP2K4 | PRKDC | WT1 |
| CDH2O | FGFR4 | MCL1 | PTCH1 | |
| CDH5 | FLT1 | MDM2 | PTCH2 | |
| CDK4 | FLT3 | MDM4 | PTEN | |
| 14 genes sequenced across selected iritrons | ||||
| Gene | ||||
| ALK | ||||
| BCR | ||||
| BRAF | ||||
| EGFR | ||||
| ETV1 | ||||
| ETV4 | ||||
| ETV5 | ||||
| ETV6 | ||||
| EWSR1 | ||||
| MLL | ||||
| RAF1 | ||||
| RARA | ||||
| RET | ||||
| TMPRSS2 | ||||
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None
References
- 1.Wistuba II, Gazdar AF. Gallbladder cancer: lessons from a rare tumour. Nature reviews Cancer. 2004;4:695–706. doi: 10.1038/nrc1429. [DOI] [PubMed] [Google Scholar]
- 2.Zatonski WA, Lowenfels AB, Boyle P, Maisonneuve P, Bueno de Mesquita HB, Ghadirian P, Jain M, Przewozniak K, Baghurst P, Moerman CJ, Simard A, Howe GR, McMichael AJ, Hsieh CC, Walker AM. Epidemiologic aspects of gallbladder cancer: a case-control study of the SEARCH Program of the International Agency for Research on Cancer. J Natl Cancer Inst. 1997;89:1132–1138. doi: 10.1093/jnci/89.15.1132. [DOI] [PubMed] [Google Scholar]
- 3.Trial watch: success for crizotinib in ALK-driven cancer. Nat Rev Drug Discov. 2010;9:908. doi: 10.1038/nrd3328. [DOI] [PubMed] [Google Scholar]
- 4.Stemke-Hale K, Shipman K, Kitsou-Mylona I, de Castro DG, Hird V, Brown R, Flanagan J, Gabra H, Mills GB, Agarwal R, El-Bahrawy M. Frequency of mutations and polymorphisms in borderline ovarian tumors of known cancer genes. Mod Pathol. 2013;26:544–552. doi: 10.1038/modpathol.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang LE, Ma H, Hale KS, Yin M, Meyer LA, Liu H, Li J, Lu KH, Hennessy BT, Li X, Spitz MR, Wei Q, Mills GB. Roles of genetic variants in the PI3K and RAS/RAF pathways in susceptibility to endometrial cancer and clinical outcomes. Journal of cancer research and clinical oncology. 2012;138:377–385. doi: 10.1007/s00432-011-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreou A, Kopetz S, Maru DM, Chen SS, Zimmitti G, Brouquet A, Shindoh J, Curley SA, Garrett C, Overman MJ, Aloia TA, Vauthey J-N. Adjuvant chemotherapy with FOLFOX for primary colorectal cancer is associated with increased somatic gene mutations and inferior survival in patients undergoing hepatectomy for metachronous liver metastases. Annals of Surgery. 2012;256:642–650. doi: 10.1097/SLA.0b013e31826b4dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol. 2011;136:527–539. doi: 10.1309/AJCPR1SVT1VHUGXW. [DOI] [PubMed] [Google Scholar]
- 8.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, Donahue A, Downing SR, Frampton GM, Garcia L, Juhn F, Mitchell KC, White E, White J, Zwirko Z, Peretz T, Nechushtan H, Soussan-Gutman L, Kim J, Sasaki H, Kim HR, Park SI, Ercan D, Sheehan CE, Ross JS, Cronin MT, Janne PA, Stephens PJ. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of the American Statistical Association. 1958;53:457–481. [Google Scholar]
- 13.Cox DR. Regression models and life tables (with discussion) Journal of the Royal Statistical Society B. 1972;34:187–220. [Google Scholar]
- 14.Zhu AX, Hezel AF. Development of molecularly targeted therapies in biliary tract cancers: reassessing the challenges and opportunities. Hepatology. 2011;53:695–704. doi: 10.1002/hep.24145. [DOI] [PubMed] [Google Scholar]
- 15.Rashid A. Cellular and molecular biology of biliary tract cancers. Surg Oncol Clin N Am. 2002;11:995–1009. doi: 10.1016/s1055-3207(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 16.Kawamoto T, Krishnamurthy S, Tarco E, Trivedi S, Wistuba II, Li D, Roa I, Roa JC, Thomas MB. HER Receptor Family: Novel Candidate for Targeted Therapy for Gallbladder and Extrahepatic Bile Duct Cancer. Gastrointest Cancer Res. 2007;1:221–227. [PMC free article] [PubMed] [Google Scholar]
- 17.Roa JC, Anabalon L, Roa I, Melo A, Araya JC, Tapia O, de Aretxabala X, Munoz S, Schneider B. Promoter methylation profile in gallbladder cancer. J Gastroenterol. 2006;41:269–275. doi: 10.1007/s00535-005-1752-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang HP, Ruan L, Zheng LM, Bai DY, Zhang HF, Liao YQ, Ding Y. Screening for EGFR mutations in lung cancer by a novel real-time PCR with double-loop probe and Sanger DNA sequencing. Zhonghua zhong liu za zhi [Chinese journal of oncology] 2013;35:28–32. doi: 10.3760/cma.j.issn.0253-3766.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Labussiere M, Sanson M, Idbaih A, Delattre JY. IDH1 gene mutations: a new paradigm in glioma prognosis and therapy? Oncologist. 2010;15:196–199. doi: 10.1634/theoncologist.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, Metzeler KH, Powell BL, Carter TH, Kolitz JE, Wetzler M, Carroll AJ, Baer MR, Caligiuri MA, Larson RA, Bloomfield CD. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, Kwak EL, Clark JW, Ryan DP, Deshpande V, Dias-Santagata D, Ellisen LW, Zhu AX, Iafrate AJ. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 23.Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, Thompson CB. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. The Journal of biological chemistry. 2013;288:3804–3815. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreotti G, Chen J, Gao YT, Rashid A, Chen BE, Rosenberg P, Sakoda LC, Deng J, Shen MC, Wang BS, Han TQ, Zhang BH, Yeager M, Welch R, Chanock S, Fraumeni JF, Jr, Hsing AW. Polymorphisms of genes in the lipid metabolism pathway and risk of biliary tract cancers and stones: a population-based case-control study in Shanghai, China. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:525–534. doi: 10.1158/1055-9965.EPI-07-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SK, Andreotti G, Sakoda LC, Gao YT, Rashid A, Chen J, Chen BE, Rosenberg PS, Shen MC, Wang BS, Han TQ, Zhang BH, Yeager M, Chanock S, Hsing AW. Variants in hormone-related genes and the risk of biliary tract cancers and stones: a population-based study in China. Carcinogenesis. 2009;30:606–614. doi: 10.1093/carcin/bgp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava K, Srivastava A, Mittal B. Polymorphisms in ERCC2, MSH2, and OGG1 DNA repair genes and gallbladder cancer risk in a population of Northern India. Cancer. 2010;116:3160–3169. doi: 10.1002/cncr.25063. [DOI] [PubMed] [Google Scholar]
- 27.Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:1803–1805. doi: 10.1200/JCO.2013.49.4799. [DOI] [PubMed] [Google Scholar]
- 28.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, Qiu K, Gao Z, Ceccarelli M, Riccardi R, Brat DJ, Guha A, Aldape K, Golfinos JG, Zagzag D, Mikkelsen T, Finocchiaro G, Lasorella A, Rabadan R, Iavarone A. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, Cheng AJ, Kunju LP, Siddiqui J, Tomlins SA, Wyngaard P, Sadis S, Roychowdhury S, Hussain MH, Feng FY, Zalupski MM, Talpaz M, Pienta KJ, Rhodes DR, Robinson DR, Chinnaiyan AM. Identification of Targetable FGFR Gene Fusions in Diverse Cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]