Abstract
Protein chemical cross-linking and mass spectrometry enable the analysis of protein-protein interactions and protein topologies, however complicated cross-linked peptide spectra require specialized algorithms to identify interacting sites. The Kojak cross-linking software application is a new, efficient approach to identify cross-linked peptides, enabling large-scale analysis of protein-protein interactions by chemical cross-linking techniques. The algorithm integrates spectral processing and scoring schemes adopted from traditional database search algorithms, and can identify cross-linked peptides using many different chemical cross-linkers, with or without heavy isotope labels. Kojak was used to analyze both novel and existing datasets, and was compared with existing cross-linking algorithms. The algorithm provided increased cross-link identifications over existing algorithms, and equally importantly, the results in a fraction of computational time. The Kojak algorithm is open-source, cross-platform, and freely available. This software provides both existing and new cross-linking researchers alike an effective way to derive additional cross-link identifications from new or existing datasets. For new users, it provides a simple analytical resource resulting in more cross-link identifications than other methods.
Keywords: Proteomics, cross-linking, mass spectrometry, protein structure
Introduction
Analysis of protein-protein interactions and protein topologies via mass spectrometry is possible with the use of chemical cross-linking techniques (1, 2). Common to most of these techniques, proteins are cross-linked either in vitro or in vivo (3–5), enzymatically digested to peptides, and analyzed using reversed phase liquid chromatography and shotgun mass spectrometry (6). While conceptually simple, in practice the observation and identification of cross-linked peptides is very difficult: 1) the dynamic range of samples containing high abundances of non-cross-linked peptides hinders the ability to trigger acquisition of tandem mass spectra (MS/MS) of low abundance cross-linked peptides; 2) MS/MS spectra obtained from these cross-linked peptide precursors are difficult to interpret due to fragmentation along both peptide backbones; and 3) cross-linked peptide MS/MS spectra cannot be identified using standard database search programs (7). Several creative strategies have been developed to facilitate the identification of MS/MS spectra derived from cross-linked peptides. These include isotopic labeling (8–10), affinity purification tags (7, 11, 12), and cleavable cross-linker bonds (7). Concurrent with these strategies are specialized algorithms designed for data analysis within the bounds of the experimental method.
Data analysis is often seen as the largest hurdle in understanding chemical cross-links and successful identification of protein-protein interactions. Even when MS/MS spectra have been identified as originating from cross-linked peptides, deriving the sequences of the two peptides is problematic since standard database search algorithms are unsuitable for solving cross-linked peptide fragmentation spectra. Several cross-linking-specific algorithms exist, but all have room for improvement as, despite decades of development, the application of cross-linking methods in conjunction with mass spectrometry is still confined to a few specialist laboratories. Many current algorithms rely on methods that utilize isotope labels (13–15), require specialized cross-linkers (16, 17), or have limitations on the number of proteins that can be analyzed (18–23). The algorithm presented here is amenable to many experimental designs and overcomes limitations in the number of proteins analyzed.
An ideal cross-linking analysis pipeline would be experimentally simple, scalable, and label-free. It would contain standard techniques already applied in proteomics approaches for peptide identification. This pipeline would include a fully automatic, easy to use data analysis algorithm that produced statistically valid and accurate results. The algorithm source code would be publically available for scrutiny and further development by the community in a collaborative approach. With this approach, the capability of cross-linked protein analysis will now be available to almost all research laboratories, vastly increasing the rate of identification of protein-protein interactions and protein topologies. Recent developments in cross-linked peptide database searching algorithms have produced excellent tools that differ in features, performance, and results. Among them, xQuest (9) and xProphet (24) have been widely adopted, but perform best when using heavy isotope labeled cross-linker. Protein Prospector (25) is operational from its web interface or additionally by command line on a local installation after obtaining a licensed copy. pLink (26) has been shown to perform whole proteome analysis, but reports results at a fixed error rate limiting the extent of downstream analysis. Although free, pLink is not provided with source code to modify its functionality and improve its utility. These differences in design and deployment highlight the need to further develop tools that improve upon existing designs and extend the capabilities of cross-linking data analysis.
Here we present Kojak, a computationally efficient, easy to use, open source, and free algorithm for analyzing cross-linked peptide spectra. The algorithm is used to identify peptides cross-linked using various different cross-linking chemistries. Use of heavy isotope-labeled spectra is possible, but not required for analysis, simplifying the experimental approach. All components of the algorithm are supplied in a single application with a simple set of parameters that is executable from the command line for easy integration into analytical pipelines. The spectral processing and scoring algorithms were designed for high computational efficiency to process large-scale protein complexes, and can operate on single core desktops or large-scale multithreaded systems. Results are exported in easily filtered text-based spreadsheets and formatted for validation using Percolator (27) at a user-defined false discovery rate (FDR). Evaluation of Kojak is provided through the analysis of several protein complexes. Detailed technical discussion of the major algorithm components is provided along with the source code and technical information as a roadmap for the further evolution of algorithms for analysis of cross-linked proteins using mass spectrometry.
Methods
Sample Preparation
SCF(FBXL3) ubiquitin ligase complex was obtained as a kind gift from Ning Zheng and Weiman Xing. The complex was expressed and purified as described previously (28). The protein complex was first buffer exchanged into HB200 buffer (40 mM HEPES, pH 7.5, 200 mM NaCl) using protein desalting spin columns (Pierce) according to the manufacturer’s instructions. 10% glycerol was added to the desalted protein. 20 µg protein was diluted with HB200 buffer to a final volume of 46.5 µL. Cross-linker concentration was brought to 0.06 mM by adding 2.9 mM DSS (Pierce). The reaction was allowed to proceed for 30 mins at room temperature before quenching with 7 µL 500 mM NH4HCO3. After quenching all reactions were buffer exchanged to HB500 (40 mM HEPES, pH 7.5, 500 mM NaCl) using protein desalting spin columns (Pierce) according to the manufacturer’s instructions. The complex was reduced with 5 mM dithiothreitol for 30 minutes at 60°C and alkylated with 15 mM iodoacetamide for 30 minutes at room temperature, in the dark.
Mass Spectrometry
After digesting the complex with trypsin (1:100 ratio, 37°C overnight), the peptides were analyzed by reversed phase liquid chromatography and mass spectrometry at high-resolution using a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Chromatography was conducted using a Waters nanoAcquity binary UPLC pump and autosampler operated with a binary mobile phase gradient. Mobile phase A was water with 0.1% formic acid, and mobile phase B was acetonitrile with 0.1% formic acid. A 250nl/min gradient was operated that consisted of 2–35% mobile phase B for 120 minutes, 35–60% mobile phase B for 10 minutes with an additional 10 minutes at constant 60% mobile phase B. Flow was increased to 500 nl/min and the column was washed with 95% mobile phase B for 10 minutes with 30 minutes equilibration at 2% mobile phase B. Samples were eluted from a pulled fused-silica capillary column 75-µm i.d.) packed with 30 cm of Reprosil-Pur C18-AQ (3-µm bead diameter, Dr. Maisch). The mass analyzer duty cycle was set to acquire full scans at 70,000 resolution followed by 6 data dependent MS/MS spectra at 35,000 resolution with higher energy collisional dissociation (HCD), and normalized collision energy set to 25. The target AGC was set to 1e6 for both MS and MS/MS scan events. Dynamic exclusion was set to 10 seconds, and charge exclusion set for +1, +2, and unassigned precursor charge states. The precursor scan range was 400 to 1600 Thomsons, and all spectra were acquired in profile mode, although profile mode is not a requirement for analysis with the Kojak algorithm.
Analytical Software Overview
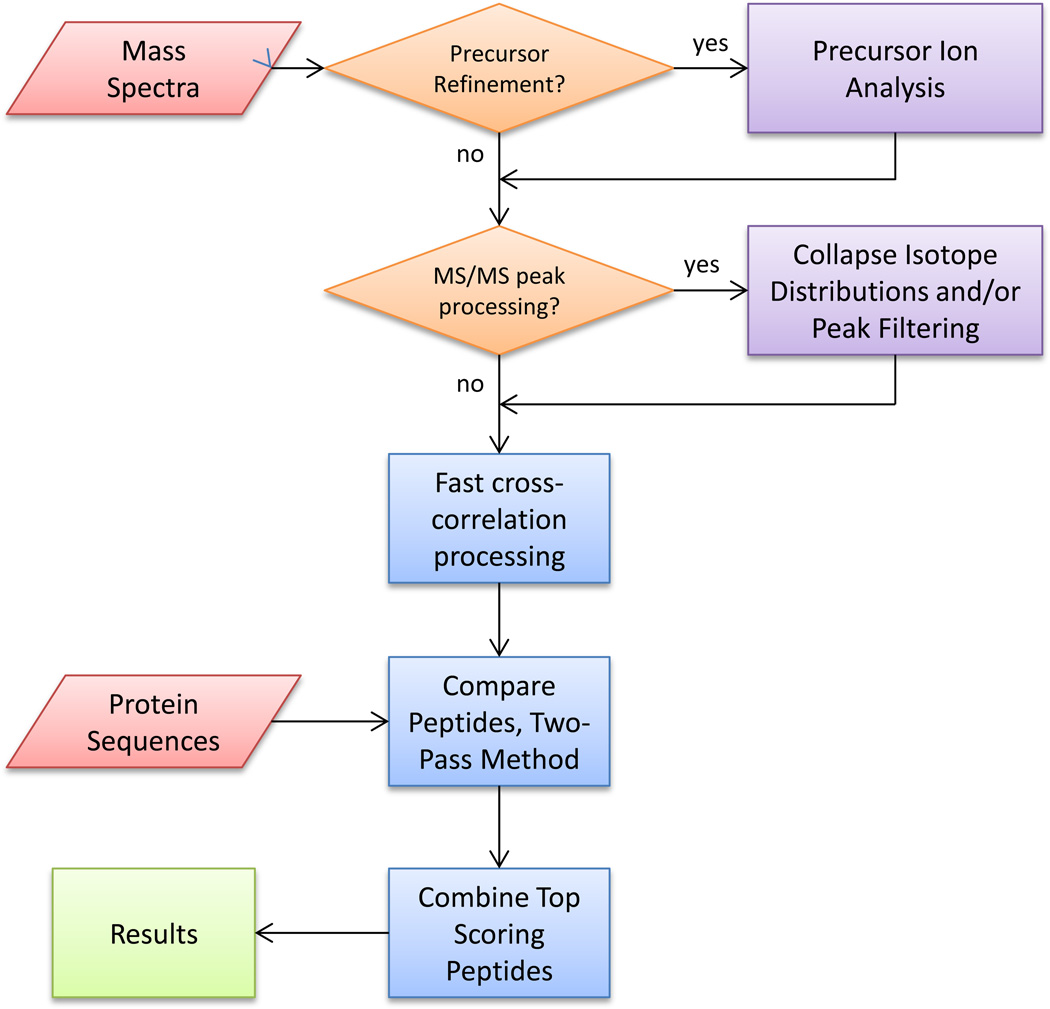
Kojak was designed for identification of peptides cross-linked using various chemistries, including, but not limited to, amine-reactive and carboxyl-reactive reagents. This stand-alone software application combines spectral processing and database searching (Figure 1). The software was written in C++, supports multi-threaded computation, and is open source and freely available at http://www.kojak-ms.org/. For convenience, pre-compiled binary formats are also provided for both Windows and Linux operating systems. Instructions and supported file formats (including mzXML and mzML) are provided on the website. All data analysis was performed on a desktop computer running Windows 7 32-bit), unless otherwise specified.
Figure 1.
Flow diagram of the Kojak algorithm.
Spectral Processing
MS/MS spectra are processed prior to analysis to improve algorithm speed and accuracy. The monoisotopic mass peak for a spectrum was optionally predicted using Kojak from the precursor ion scans. This prediction step narrows the list of target peptide sequences that must be searched to obtain the correct PSM (29) and improves algorithm speed. To obtain a clear representation of the precursor ion isotope envelope, precursor ion peaks were averaged across neighboring scan events at the apex of chromatographic elution. The monoisotopic precursor mass is then predicted from the composite using a model-based precursor fitting function (30) included in the source and compiled in Kojak (Supplementary Figure 1). A second function can be optionally activated for processing high-resolution MS/MS spectra with resolved product ion isotope envelopes. Isotope envelopes are identified by iterating through the peaks from most to least intense and observing sets of peaks equally spaced by the inverse relationship to charge state. Peaks within an isotope envelope are summed together and collapsed to the monoisotopic value, in Thomsons, leaving only a single peak for each isotope envelope. Peaks that are not grouped into an isotope envelope are unchanged (Supplementary Figure 2). This processing step improves scoring of candidate peptides, whose product ion masses are computed at monoisotopic values. Whether or not isotope peaks are reduced, the next step in MS/MS spectral processing optionally reduces the spectra to a user-defined number of most intense peaks. The last processing function implements a modified fast cross-correlation algorithm previously described (31, 32). This pre-processing step allows calculation of a Comet (32) algorithm-like cross-correlation score, described in detail below, by summation of matched theoretical product ion masses.
Database Searching and Scoring Algorithm
Database searching was performed by computing theoretical product ion masses for candidate peptide sequences and matching them to the observed peaks in the processed MS/MS spectra. Cross-linked peptides were identified using a two-pass algorithm. In the first pass all single peptides were searched with a differential modification mass on linkable residues equal to the difference between the precursor ion mass and the peptide mass, similar to the open-modification search strategy (33). The top 250 (user defined) scoring single peptides for each spectrum were stored in memory. In the second pass, for each spectrum, the top scoring peptides from the first pass were paired to search for cross-linked peptides. If two peptide masses from the top scoring list plus the cross-linker mass equaled the precursor ion mass, a match was assigned to the spectrum with a score equal to the sum of each individual peptide. Searches for loop-linked peptides - single peptides with a linker spanning two residues - and non-cross-linked peptides were also performed. These searches were also considered with differential modifications, including the situation where the cross-linker molecule attaches to only a single site (mono-link), modifying the peptide fragmentation masses without producing a cross-link or loop-link. These comprehensive peptide searches are important as few cross-linked peptides are expected to be observed amongst all the ions selected. The highest scoring peptide, whether cross-linked or otherwise, is kept after considering all possibilities. In the case of a tie for the top score, all such peptides are kept.
Candidate peptide sequences were scored using a modified form of the fast cross-correlation algorithm developed for the Comet (32) search tool. Briefly, the fast cross-correlation score is an efficient implementation of the SEQUEST (34) scoring algorithm that eliminates the Fourier transform calculations. Details of this change have been previously published (31), and involve preprocessing each spectrum so that the cross-correlation score is calculated from the sum of processed spectrum intensity values for each theoretical fragment ion mass. Theoretical b- and y-ions were computed from the peptide sequences plus any modification masses and matched to the observed peaks of the processed MS/MS spectra. The sum of matched processed peak intensities produced the peptide score for each peptide comparison. The Comet algorithm uses an array of bins to quickly index matched peaks, which scales with spectrum resolution. Using Comet, searching high-resolution spectra requires large amounts of RAM, multiple passes, or significant computational overhead if using its use_sparse_matrix parameter. To overcome these limitations, the Comet scoring algorithm was modified in Kojak to reduce data consumption with a lower-precision sparse array. The array is rapidly traversed with minimal overhead using an approach similar to a hash function. Because of these modifications, Kojak can be used to rapidly analyze large spectral datasets of either low- or high-resolution data (Supplementary Figure 3 and Supplementary Table 4).
Analysis of MS/MS spectra
Mass spec data were searched using Kojak with parameters specific to each data set (Table 1). All parameters are specified in a single, human-readable text file which is parsed when Kojak is executed from the command prompt. In addition to the SCF(FBXL3) ubiquitin ligase complex data generated for this study, additional datasets were analyzed. Raw spectral data for the Cop9 signalosome (35) and 26S proteasome (36) were kindly provided by the authors of their original studies. The SCF(FBXL3) ubiquitin ligase complex data were searched against the three target protein sequences, and numerous background and decoy sequences to produce a sizable database of 292 protein sequences (see Supplementary Information). The Cop9 signalosome and 26S proteasome search databases were smaller to match the published search sequences. Eight human Cop9 signalosome complex protein sequences and 33 Schizosaccharomyces pombe 26S proteasome protein sequences were used, plus trypsin and chymotrypsin (26S data only), and an equal number of decoys for totals of 18 and 70 protein sequences, respectively. The Cop9 signalosome data were searched with d0/d12 DSS cross-linker, and the 26S proteasome data were searched with d0/d8-ADH and DMTMM cross-linkers. Detailed descriptions of all Kojak parameters used are provided in the Supplementary Information.
Table 1.
Parameters used in each analysis with Kojak.
| Parameter1 | SCF(FBXL3) ubiquitin ligase complex |
Cop9 signalosome | 26S proteasome |
|---|---|---|---|
| cross-linked modification mass2 | 138.0680742 (a-a) | 138.0680742 (a-a) 150.1434042 (a-a) |
138.09055 (b-b) 146.14076 (b-b) −18.010595 (a-b) |
| differential amino acid modification mass (Da) | K=14.015894 K=28.031788 K=42.047682 M=15.9949 N-term=42.01055 |
K=14.015894 K=28.031788 K=42.047682 M=15.9949 |
K=14.015894 K=28.031788 K=42.047682 M=15.9949 |
| enzyme specificity | trypsin | trypsin | trypsin |
| maximum number of missed cleavages (includes linkage site) | 4 | 4 | 3 |
| maximum peptide mass (Da) | 8000 | 8000 | 8000 |
| minimum peptide mass (Da) | 500 | 500 | 500 |
| mono-linked modification mass2 | 155.0946 (a) 156.0786 (a) |
155.0946 (a) 156.0786 (a) 167.16993 (a) 168.15393 (a) |
156.10111 (b) 164.15132 (b) |
| precursor mass prediction | yes | yes | no |
| precursor mass tolerance (ppm) | 15 | 10 | 15 |
| scoring algorithm bin offset (Th)3 | 0 | 0.4 | 0.4 |
| scoring algorithm bin size (Th)3 | 0.03 | 1.0005 | 1.0005 |
| spectral processing | yes | no | no |
| static amino acid modification mass (Da) | C=57.02146 | C=57.02146 | C=57.02146 |
| top number of single peptides | 250 | 250 | 250 |
Parameters listed here are relevant to the differences for each analysis. Detailed description of these and other Kojak parameters are provided in Supplementary Table 5.
‘a’ designates linkage to primary amines (K or protein N-terminus). ‘b’ designates linkage to carboxyl groups (D, E, or C-terminus). For example ‘a–b’ indicates the cross-linker binds an amine to a carboxyl-terminating side chain.
bin offset and bin size are set to match the resolution of the data set. High resolution data analysis requires smaller bins and no offset.
Cross-linked Peptide Validation
Peptide-spectrum matches (PSMs) are automatically exported from Kojak in tab-delimited text formatted as input for Percolator (27) (version 2.07). Percolator is a semi-supervised algorithm that assigns false discovery rate (FDR) to each PSM through analysis of the target and decoy PSM distributions. Decoy PSMs derive from peptide sequences known to be false, such as from reversed or shuffled protein sequences. Cross-link PSMs were considered false if at least one of the peptides was from a decoy protein sequence. The top scoring PSM for each spectrum was used as input to Percolator. Multiple different scoring metrics were used as parameters for Percolator and are described in detail in the Supplementary Information. In particular, the lower score of the two peptides in a cross-link was used as a parameter to evaluate whether a PSM contained evidence for both peptides or is instead highly promoted by only a single member (37). The results were filtered for cross-linked PSMs prior to validation. The cross-linked PSMs were further separated into two sets, intraprotein and interprotein PSMs, and analyzed separately. This separation ensures proper FDR assessment as the error rates of each type of PSM vary from each other (24, 37). Non-linked and loop-linked PSMs were validated using the same filtering method.
Results and Discussion
Analysis of cross-linked CRY2-FBXL3-SKP1 complex using Kojak
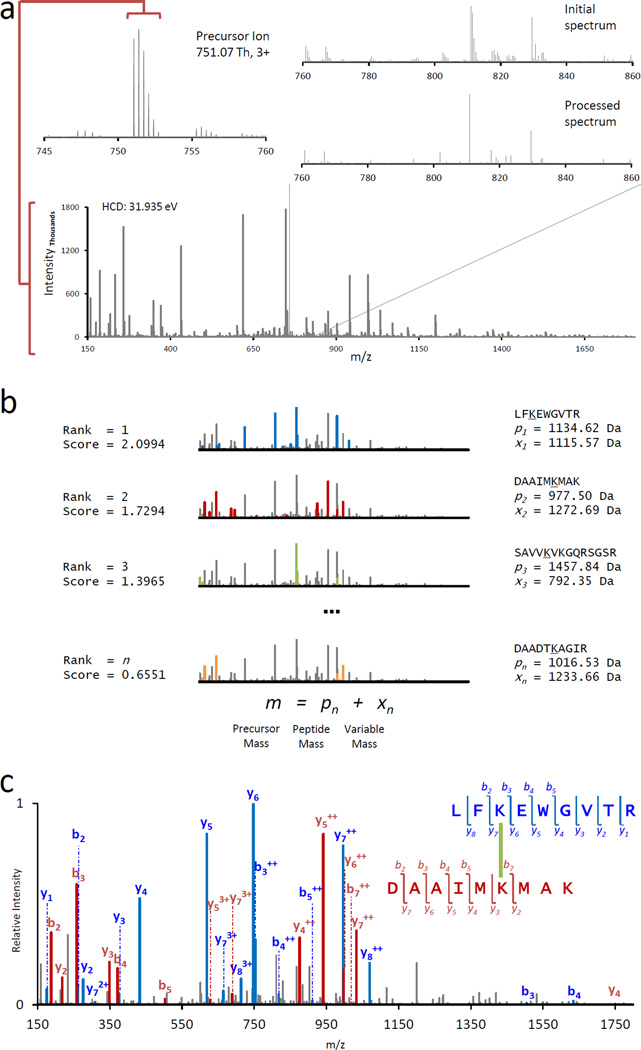
Purified CRY2-FBXL3-SKP1 was cross-linked, digested with trypsin, and analyzed by shotgun MS on a Q-Exactive mass spectrometer. The resulting 81,256 peptide MS/MS spectra were analyzed with Kojak to identify peptide spectrum matches (PSMs). The typical analysis performed by Kojak on all MS/MS spectra is illustrated in Figure 2, which shows an identified cross-linked PSM from our analysis of the CRY2-FBXL3-SKP1 complex. Prior to database searching, MS/MS spectra are processed to improve the precursor ion detection and reduce spectral complexity by collapsing isotope peaks to a single value (Figure 2A). Database searching was performed using a modified fast cross-correlation algorithm for single candidate peptides with a differential modification mass at the proposed site of linkage (Figure 2B). The mass difference for each candidate peptide equaled the remainder after subtracting the peptide mass from the precursor ion mass. All candidate peptides were ranked by cross-correlation score, keeping the top 250 (user-defined) peptides. This top set of peptides was then parsed to find pairs that sum by mass plus the cross-linker mass to equal the precursor ion mass. The sum of each peptide score in the pair equaled the score of the cross-linked PSM. Figure 2C illustrates a high-quality PSM. The two peptides were ranked #1 and #2 in the top set, and both peptides were typically observed in the upper echelon of the top set for high-quality PSMs in the CRY2-FBXL3-SKP1 analysis.
Figure 2.
Example analysis of a cross-linked peptide spectrum using Kojak. (a) The MS/MS spectrum of a triply-charged precursor ion of m/z 751.07. The spectrum is processed (inset) to collapse isotope envelope distributions and facilitate fast and accurate peptide identification. (b) Matches to product ions for individually searched peptides are scored and ranked. pn is the theoretical peptide mass and xn is the modification mass of the internal lysine, which sum to the precursor mass. The top 250 (user-defined) scoring peptide sequences are then pairwise compared. Matches are made when two peptides plus the cross-linker mass sum to the precursor ion mass. (c) The best matching pair of peptides was the first and second ranked peptides from panel (b).
The CRY2-FBXL3-SKP1 MS/MS PSMs were divided into four subsets: intraprotein cross-links, interprotein cross-links, loop-links, and single peptide PSMs. Each subset was analyzed with Percolator and 13,808 PSMs observed at approximately 1% FDR (Table 2). More than 80% of the PSMs were to non-linked peptide sequences, confirming that a significant portion of the data in analysis was not derived from cross-linked peptides. Eleven percent of the PSMs were intraprotein or interprotein cross-links, including 91 uniquely paired CRY2-FBXL3-SKP1 cross-linked residues (Supplementary Tables 1–3).
Table 2.
Summary of CRY2-FBXL3-SKP1 PSMs observed with Kojak at 1% FDR
| PSM-type | PSMs | Non-redundant peptide sequences |
Unique XL sites1 |
|---|---|---|---|
| Single | 11433 | 441 | n/a |
| Loop-Link | 886 | 35 | 22 |
| Interprotein XL | 169 | 33 | 23 |
| Intraprotein XL | 1320 | 105 | 68 |
Represents the novel linkage between two residues by absolute protein position, thus removing redundant associations derived from elongated peptide sequences that result from enzymatic missed cleavages.
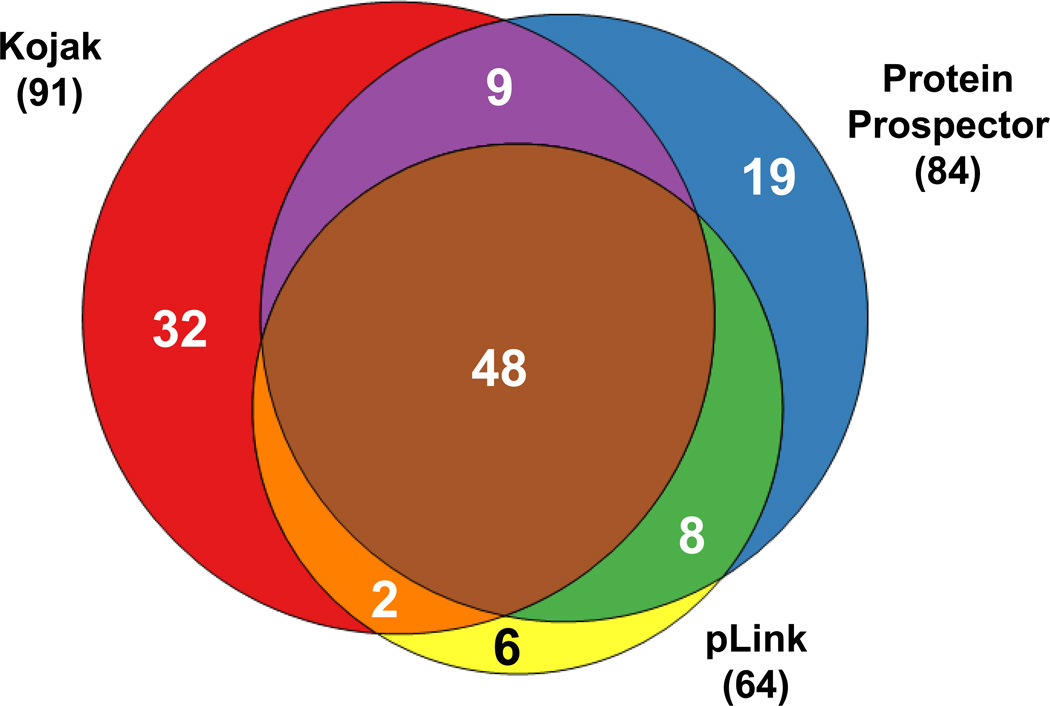
To assess the cross-links identified with Kojak, the CRY2-FBXL3-SKP1 data were analyzed in parallel with Protein Prospector (25) and pLink (26) and the results were compared (see Supplementary Information). Using Protein Prospector, 84 cross-linked sites were identified. pLink reported 64 linked sites. Beyond identifying more cross-linked sites, the results were further explored to illustrate the utility of the Kojak analysis. Of the 124 unique cross-linked sites identified by all three algorithms, only 48 were common to all three analyses (Figure 3). The PSMs relating to algorithm-specific cross-link sites were examined. Fifteen of the 19 sites specific to Protein Prospector contained a PSM with five or fewer amino acids. Ten of these had a peptide with three or fewer amino acids. Spectra from these cross-links must be interpreted carefully, as they contain little or no product ion evidence for the short peptides and high scoring PSMs often reflect the longer complementary peptide (37, 38). Though many were also identified with Kojak, PSMs from these spectra were not validated at the FDR threshold, due to the low score contribution of the smaller peptides. The Kojak specific PSMs, on the other hand, contained longer peptides, and included eight novel interprotein link sites missed with the other two algorithms. pLink specific PSMs likewise contained longer peptides, but only six novel cross-linked sites were identified with the algorithm. The additional PSMs identified with Kojak show the utility of the algorithm for cross-linked data analysis, whether used alone, or in combination with other algorithms to improve PSM identification, as is commonly done with traditional database searching (39). Additional performance comparisons are discussed in the Supplemental Information.
Figure 3.
Comparison of the uniquely cross-linked sites for the CRY2-FBXL3-SKP1 complex.
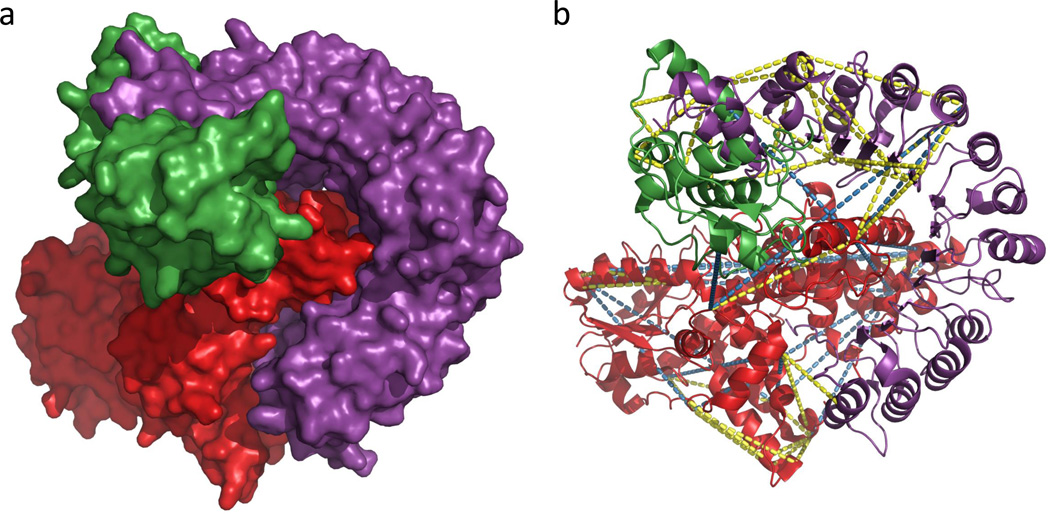
Cross-linked sites were compared to the published crystal structure (28) (PDB: 4I6J, Figure 4). Distances between linked residues were expected to be within 30 Å (Cα-Cα), given the DSS spacer arm length of 11.4 Å and structural flexibility of the protein complex. Of the 91 Kojak-identified cross-links, 23 fell in regions not present in the published structure. Of the 65 that were present in the published structure, 48 (74%) were within the 30 Å cutoff. The level of structural agreement was better with Kojak-produced results, than those observed using Protein Prospector (46 of 65, 71%) and pLink (34 of 49, 69%). The cross-links exceeding 30 Å were most likely a result of cross-complex linkage due to transient complex-complex interactions being trapped during the cross-linking reaction.
Figure 4.
Structure of the CRY2-FBXL3-SKP1 complex. CRY2 is shown in red, FBXL3 is shown in purple, and SKP1 is shown in green. (a) Surface representation of the overall complex structure (PDB code: 4I6J). (b) Ribbon diagram of the structure with cross-linked lysine residues superimposed. Cross-links were identified using Kojak with a 1% FDR cutoff. Yellow lines indicate Cα-Cα within 30 Å, and blue lines indicate Cα-Cα distances exceeding 30 Å.
The cross-linked sites identified using Kojak and Protein Prospector are the most interesting in that many of the sites were specific to either algorithm. The differences between these two subsets were described above, and suggest that each algorithm may be better suited towards identification of cross-linked peptides with particular attributes. Though continued development of each algorithm will likely improve the number of cross-links that either identifies, using a combination of algorithms when analyzing cross-linked data can possibly provide additional insight and validation over relying on a single algorithm. As Kojak performance shows many novel cross-linked sites, it has utility both on its own or supplemental to existing analyses.
Cop9 Signalsome
To demonstrate Kojak’s versatility and performance on diverse datasets, Kojak was used to analyze a previously published dataset of the Cop9 signalosome using xQuest (35). Kojak parameters were adjusted to match those used in the published analysis so that the results could be directly compared. PSM validation was performed separately for intraprotein and interprotein cross-links and comparisons were made at a 5% FDR cutoff. Overall, Kojak identified 94 unique cross-linked sites, compared to the 38 originally published (Supplementary Figure 4 and Supplementary Table 6). The majority (57 of 94) of the identifications was intraprotein cross-links, with all 15 published intraprotein cross-links observed and an additional 42 identified. 37 interprotein cross-links were identified, with 12 matching the published results, and an additional 25 previously unreported. Of the remaining 11 interprotein cross-links only found with xQuest, an additional 5 were within 10% FDR in the Kojak results. All of the xQuest cross-links were observed at a 50% FDR, indicating that although not validated at the desired FDR, none of the published cross-links were missed with Kojak. As could be expected, a greater number of intraprotein cross-links than interprotein cross-links were identified with Kojak, although, interestingly, a larger ratio of interprotein cross-links was reported using xQuest. Analysis of the Cop9 dataset also highlighted key capabilities of the Kojak algorithm. In particular, Kojak was capable of analyzing heavy isotope labeled data and supported low-resolution MS/MS spectral analysis.
S. pombe 26S ribosome
Kojak performance was also compared to another published analysis of significant complexity, the Schizosaccharomyces pombe 26S proteasome analyzed with xQuest (36). This data set offered several challenges in that the MS/MS spectra are low-resolution, both DMTMM and d0/d8-ADH cross-linkers were used simultaneously, and the cross-linkers have different specificities, with DMTMM linking carboxyl-terminating side chains, and ADH producing hybrid amine-to-carboxyl links. Parameters for Kojak were adjusted to match those used in the xQuest analysis. The parameters allowed for both cross-linking chemistries to be searched simultaneously in the analysis. PSM validation was performed separately for intraprotein and interprotein cross-links and comparisons were made at a 5% FDR cutoff to match the threshold used in the published results. PSMs were reduced to unique combinations of two linked sites. Several of the cross-links reported in the xQuest analysis had ambiguity where a second possible site on a peptide was suggested. In these cases, both sites were counted as independent cross-links, despite the likelihood that only one of the sites is correct.
Kojak obtained a considerable increase in the identification of cross-linked peptides. Of the intraprotein cross-links, Kojak identified 198 versus 125 with xQuest, but with a modest overlap of only 74 cross-links (Supplementary Figure 5a and Supplementary Table 6). Although cross-links found only with xQuest were also observed at higher FDR thresholds in the Kojak analysis, 33 were unique to the xQuest analysis. Inspection of these cross-links showed many to be the aforementioned ambiguous sites. For nearly all remaining xQuest-specific cross-links, one of the peptides scored highly using Kojak’s two-pass algorithm, with the second peptide as far as 2000th place amongst the top-scoring peptide sequences, and beyond the top 250 cutoff specified in the parameters. These spectra provide almost no product ion evidence for the second peptide and thus they were not observed using Kojak. 39 of the cross-links observed only with Kojak contained a peptide sequence of only four or five residues. Cross-links with a peptide fewer than six residues were filtered from the xQuest analysis prior to using xProphet, presumably to improve FDR thresholds. It is possible many of these cross-links would otherwise have been identified with xQuest, but such measures were not required with the Kojak results, significantly improving the number of cross-links identified. For interprotein analysis, 41 cross-links were found with Kojak compared to 56 with xQuest, with an overlap of 18 cross-links (Supplementary Figure 5b). Inspection of the xQuest-only cross-links showed that either different sites of linking within the same peptides were preferred, or that only one peptide was observed amongst the top scoring peptides in the two-pass method of Kojak. Consistent between the xQuest and Kojak results, a greater number of cross-links, both interprotein and intraprotein, were observed from DMTMM than from ADH.
The 26S proteasome data represent some of the biggest challenges in cross-linked peptide analysis. The data were obtained from a protein complex with more than 60 subunits, and the analysis of such large protein complexes is becoming more commonplace. The use of multiple cross-linkers with different cross-linking chemistries can potentially increase the structural information obtained from cross-linking experiments. As such, some of the most interesting cross-linking experiments may include or require cross-linkers that differ significantly from the routinely used amine-reactive reagents. In addition, isotope-labeled cross-linkers were used that increased the complexity of the data and the analysis. Despite these challenges, Kojak was capable of analyzing the data and identifying a significant number of additional cross-linked sites that were missed in the original analysis. The Kojak PSM scores included the individual contribution of both peptides in a cross-link, which aids in the validation of problematic PSMs where product ion peaks dominantly represent only one of the peptides. As most of the cross-links specific to the original analysis showed only a dominant product ion series to a single peptide, the analysis with Kojak adds useful information in the evaluation of these cross-links. Indeed, this artifact was a major reason for the fewer interprotein cross-links observed using Kojak; however, many novel cross-links were observed using Kojak, with product ion evidence for both peptides.
Conclusions
The Kojak algorithm is a robust software tool for the analysis of cross-linked proteins using mass spectrometry. The processing and scoring functions produced a greater number of cross-linked sites, with improved conformity to available published structures, than observed with other algorithms used for comparison. Furthermore, the algorithm is adaptable to the analysis of data with a variety of cross-linker chemistries and spectral resolutions, making it applicable to a wide range of experimental designs, and accessible to a large number of users. The source code is open and freely available for further exploration and development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Ning Zheng and Dr. Weiman Xing for providing SCF(FBXL3) ubiquitin ligase complex and helpful discussions. We also thank Drs. Alexander Leitner and Ruedi Aebersold for access to the Cop9 signalosome and 26S proteasome raw data used for software comparisons. This work was supported in part with federal funds from the National Science Foundation MRI grant No. 0923536, from the National Institutes of Health National Institute of General Medical Sciences under grant Nos. 2P50 GM076547/Center for Systems Biology, GM087221, S10RR027584, and P41 GM103533.
ABBREVIATIONS
- FDR
false discovery rate
- PSM
peptide-spectrum match
Footnotes
ASSOCIATED CONTENT
Supporting Information Available
Supplemental information is provided in separate Microsoft Word & Excel documents, and as zipped file archives, and contains additional text, figures, and tables as referenced in the manuscript.
Supplementary Figure 1: Precursor ion spectral processing.
Supplementary Figure 2: MS/MS spectral processing
Supplementary Figure 3: Computation times.
Supplementary Figure 4: Comparison of Cop9 signalosome intraprotein and interprotein cross-links.
Supplementary Figure 5: Comparison of 26S proteasome intraprotein and interprotein cross-links.
Supplementary Table 1: CRY2-FBXL3-SKP1 interprotein cross-links, 1% FDR
Supplementary Table 2: CRY2-FBXL3-SKP1 intraprotein cross-links, 1% FDR
Supplementary Table 3: CRY2-FBXL3-SKP1 cross-linked site evaluation compared between the different algorithms and the published PDB structure.
Supplementary Table 4: Comparison of database search speeds for 991 MS/MS spectra using different approaches to spectral searching.
Supplementary Table 5: Kojak parameter descriptions.
Supplementary Table 6: Kojak PSMs for Cop9 and 26S at 5% FDR.
This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Bruce JE. In vivo protein complex topologies: sights through a cross-linking lens. Proteomics. 2012;12(10):1565–1575. doi: 10.1002/pmic.201100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J Mass Spectrom. 2003;38(12):1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Tang X, Weisbrod CR, Munske GR, Eng JK, von Haller PD, Kaiser NK, Bruce JE. A photocleavable and mass spectrometry identifiable cross-linker for protein interaction studies. Anal Chem. 2010;82(9):3556–3566. doi: 10.1021/ac902615g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Tang X, Munske GR, Tolic N, Anderson GA, Bruce JE. Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol Cell Proteomics. 2009;8(3):409–420. doi: 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng C, Yang L, Hoopmann MR, Eng JK, Tang X, Weisbrod CR, Bruce JE. Cross-linking measurements of in vivo protein complex topologies. Mol Cell Proteomics. 2011;10(10) doi: 10.1074/mcp.M110.006841. M110 006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173(3):530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang X, Bruce JE. A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol Biosyst. 2010;6(6):939–947. doi: 10.1039/b920876c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zelter A, Hoopmann MR, Vernon R, Baker D, MacCoss MJ, Davis TN. Isotope signatures allow identification of chemically cross-linked peptides by mass spectrometry: a novel method to determine interresidue distances in protein structures through cross-linking. J Proteome Res. 2010;9(7):3583–3589. doi: 10.1021/pr1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5(4):315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrotchenko EV, Olkhovik VK, Borchers CH. Isotopically coded cleavable cross-linker for studying protein-protein interaction and protein complexes. Mol Cell Proteomics. 2005;4(8):1167–1179. doi: 10.1074/mcp.T400016-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Sohn CH, Agnew HD, Lee JE, Sweredoski MJ, Graham RL, Smith GT, Hess S, Czerwieniec G, Loo JA, Heath JR, Deshaies RJ, Beauchamp JL. Designer reagents for mass spectrometry-based proteomics: clickable cross-linkers for elucidation of protein structures and interactions. Anal Chem. 2012;84(6):2662–2669. doi: 10.1021/ac202637n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang S, Mou L, Lanman J, Velu S, Brouillette WJ, Prevelige PE., Jr Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun Mass Spectrom. 2009;23(11):1719–1726. doi: 10.1002/rcm.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q, Xue S, Doneanu CE, Shaffer SA, Goodlett DR, Nelson SD. Pro-CrossLink. Software tool for protein cross-linking and mass spectrometry. Anal Chem. 2006;78(7):2145–2149. doi: 10.1021/ac051339c. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Zhang Z, Zang T, Spahr C, Cheetham J, Ren D, Zhou ZS. Discovery of undefined protein cross-linking chemistry: a comprehensive methodology utilizing 18O-labeling and mass spectrometry. Anal Chem. 2013;85(12):5900–5908. doi: 10.1021/ac400666p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrotchenko EV, Borchers CH. ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinformatics. 2010;11:64. doi: 10.1186/1471-2105-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoopmann MR, Weisbrod CR, Bruce JE. Improved strategies for rapid identification of chemically cross-linked peptides using protein interaction reporter technology. J Proteome Res. 2010;9(12):6323–6333. doi: 10.1021/pr100572u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson GA, Tolic N, Tang X, Zheng C, Bruce JE. Informatics strategies for large-scale novel cross-linking analysis. J Proteome Res. 2007;6(9):3412–3421. doi: 10.1021/pr070035z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YJ, Lackner LL, Nunnari JM, Phinney BS. Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J Proteome Res. 2007;6(10):3908–3917. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 19.Tang Y, Chen Y, Lichti CF, Hall RA, Raney KD, Jennings SF. CLPM: a cross-linked peptide mapping algorithm for mass spectrometric analysis. BMC Bioinformatics. 2005;6(Suppl 2):S9. doi: 10.1186/1471-2105-6-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadeau OW, Wyckoff GJ, Paschall JE, Artigues A, Sage J, Villar MT, Carlson GM. CrossSearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins. Mol Cell Proteomics. 2008;7(4):739–749. doi: 10.1074/mcp.M800020-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du X, Chowdhury SM, Manes NP, Wu S, Mayer MU, Adkins JN, Anderson GA, Smith RD. Xlink-identifier: an automated data analysis platform for confident identifications of chemically cross-linked peptides using tandem mass spectrometry. J Proteome Res. 2011;10(3):923–931. doi: 10.1021/pr100848a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIlwain S, Draghicescu P, Singh P, Goodlett DR, Noble WS. Detecting cross-linked peptides by searching against a database of cross-linked peptide pairs. J Proteome Res. 2010;9(5):2488–2495. doi: 10.1021/pr901163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panchaud A, Singh P, Shaffer SA, Goodlett DR. xComb: a cross-linked peptide database approach to protein-protein interaction analysis. J Proteome Res. 2010;9(5):2508–2515. doi: 10.1021/pr9011816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walzthoeni T, Claassen M, Leitner A, Herzog F, Bohn S, Forster F, Beck M, Aebersold R. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9(9):901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 25.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol Cell Proteomics. 2008;7(12):2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9(9):904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 27.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 28.Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, Zheng N. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496(7443):64–68. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh EJ, Hoopmann MR, MacLean B, MacCoss MJ. Comparison of database search strategies for high precursor mass accuracy MS/MS data. J Proteome Res. 2010;9(2):1138–1143. doi: 10.1021/pr900816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoopmann MR, Finney GL, MacCoss MJ. High-speed data reduction, feature detection, and MS/MS spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Anal Chem. 2007;79(15):5620–5632. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eng JK, Fischer B, Grossmann J, Maccoss MJ. A fast SEQUEST cross correlation algorithm. J Proteome Res. 2008;7(10):4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- 32.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. PROTEOMICS. 2013;13(1):22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 33.Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzner RA, Larson Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal Chem. 2008;80(22):8799–8806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 35.Birol M, Enchev RI, Padilla A, Stengel F, Aebersold R, Betzi S, Yang Y, Hoh F, Peter M, Dumas C, Echalier A. Structural and biochemical characterization of the Cop9 signalosome CSN5/CSN6 heterodimer. PLoS One. 2014;9(8):e105688. doi: 10.1371/journal.pone.0105688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leitner A, Joachimiak LA, Unverdorben P, Walzthoeni T, Frydman J, Forster F, Aebersold R. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc Natl Acad Sci U S A. 2014;111(26):9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trnka MJ, Baker PR, Robinson PJ, Burlingame AL, Chalkley RJ. Matching cross-linked peptide spectra: only as good as the worse identification. Mol Cell Proteomics. 2014;13(2):420–434. doi: 10.1074/mcp.M113.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9(8):1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10(12) doi: 10.1074/mcp.M111.007690. M111 007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.