Abstract
MicroRNA research in humans and mammalian model organisms is in a crucial stage of development. Diagnostic and therapeutic values of microRNAs appear promising, but remain to be established. The physiological and pathophysiological significance of microRNAs is generally recognized, but much better understood in some organ systems and disease areas than others. In the present paper, we review several translational studies of microRNAs, including those showing the potential value of therapeutic agents targeting microRNAs and diagnostic or prognostic microRNA markers detectable in body fluids. We discuss the lessons learned and the experience gained from these studies. Several recent studies have begun to explore translational microRNA research in kidney disease and hypertension. Translational research of microRNAs in the kidney faces unique challenges, but provides many opportunities to develop and apply new methods, and to merge complementary basic and clinical approaches.
Keywords: diagnosis, hypertension, kidney, microRNA (miRNA), therapy, translational research
INTRODUCTION
miRNAs (microRNAs) are endogenously produced non-coding RNA molecules approximately 22 nucleotides long that play a ubiquitous and important role in regulating protein expression (Figure 1A). miRNAs, and non-coding RNA in general, are importantly involved in numerous disease processes as indicated by targeted studies, as well as exploratory studies such as genome-wide association studies [1]. The human genome contains genes encoding more than 1000 miRNAs [2]. With some exceptions, miRNAs generally reduce the abundance of target proteins through translational repression or mRNA degradation. Readers interested in the specific and detailed mechanisms mediating the biogenesis and molecular action of miRNAs are referred to numerous extensive reviews published elsewhere [3–8].
Figure 1. Translational research of miRNAs.
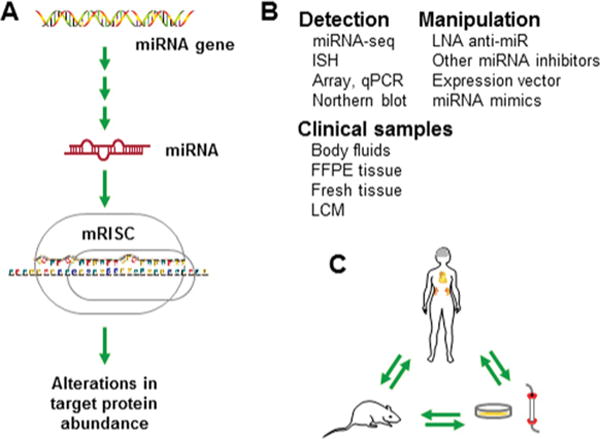
(A) miRNAs are encoded by endogenous genes and regulate the abundance of target proteins. (B) Methods and materials for translational study of miRNAs. (C) Translational research by merging studies of humans, animal models and in vitro systems. mRISC, miRNA-induced silencing complex; miRNA-seq, miRNA sequencing; ISH, in situ hybridization; FFPE, formalin-fixed paraffin-embedded; LCM, laser-capture microdissection.
An easy way for clinicians to get an initial understanding of miRNAs would be to compare miRNAs with mRNAs in terms of their biogenesis and transcriptional factors in terms of their action. Similar to mRNAs, miRNAs are transcribed from the genome by RNA polymerase II, processed until reaching their mature forms and degraded over time. miRNAs, however, are much shorter than mRNAs and are not translated into proteins.
Similar to transcriptional factors, miRNAs bind to their targets on the basis of nucleotide sequence, have a one-to-many and many-to-one relationship with their targets, and can potentially influence the expression of nearly all protein-coding genes. miRNAs, however, primarily bind to the 3′-UTR (untranslated region) of mRNA, although they could in some cases bind to other regions of mRNA as well as genomic DNA, including promoter regions. Several miRNA target prediction databases have been developed to aid in sequence-based target identification; however, all of the factors that contribute to miRNA targeting are not yet understood. miRNA target prediction tools provides a good starting point for functional studies. The long lists of predicted targets must still be sifted through with experimental validation.
miRNA research has undoubtedly been one of the most exciting and rapidly developing areas of biomedical research over the last decade. It appears, however, that miRNA research in humans and mammalian model organisms is entering a crucial stage of development. Diagnostic and therapeutic values of miRNAs appear promising, but remain to be established. The physiological and pathophysiological significance of miRNAs is generally recognized, but much better understood in some organ systems and disease areas than others.
Translational research is the critical testing ground for finding out whether any new directions of biomedical research, miRNAs included, are going to be ‘make or break’. In the present paper, we review several translational studies of miRNAs and discuss the lessons learned and the opportunities and challenges ahead for miRNA study, focussing on the area of hypertension and kidney disease research.
miRNAs AS THERAPEUTIC TARGETS
In many diseases, either excessive or insufficient miRNA expression levels have been found to alter gene expression in a way that may contribute to the pathology of the condition. Despite the fact that an miRNA can have multiple target genes, several studies have suggested that an miRNA can in some cases target genes that are functionally related and therefore have a great impact on the functional pathway. For example, we [9] and other investigators [9a–9d] have found that the miR-29 family targets nearly 20 genes related to extracellular matrix formation, providing one of the most dramatic examples of a single miRNA targeting a large number of functionally related genes. Recently published studies also suggest that a single miRNA or co-expressed miRNAs could act to co-ordinate a cellular change by regulating the expression of genes in multiple pathways [10,11]. With such powerful regulatory capability, the potential for miRNA therapies in treating disease are very exciting [12]. However, as with all clinical treatments, a therapeutic approach aimed at restoring physiological miRNA levels must be efficient and safe. In this section, we will review some of the progress made in this area in recent years.
The largest portion of progress in this area has been in reducing the expression of an miRNA that has been up-regulated with disease (Figure 1B). Chemically synthesized anti-miRNA oligonucleotides (anti-miRs) have a complementary sequence to the miRNA of interest and, when complexed, physically prevent mature miRNAs from interacting with their targets. Additionally, the duplexing of the anti-miR can target the miRNA for degradation [13,14]. The actionofanmiRNA can also be attenuated by excess pseudo-targets that can be provided experimentally or, interestingly, generated naturally [15–17].
Despite initial potent suppression of miRNA, preserving the long-term effectiveness of oligonucleotide therapy in vivo has been challenging due to the rapid degradation of miRNAs by endogenous nucleases and renal excretion. To overcome this, many chemical modifications have been introduced to synthesized oligonucleotides to provide increased protection and stability in vivo, such as phosphorothioate linkages, 2′-O-methyl RNA, 2′-fluoro modifications and LNA (locked-nucleic acid) modifications (Figure 1B) [9c,13,14,18–20]. In particular, LNA-modified anti-miR oligonuleotides have been reported to provide long-lasting, efficient and apparently safe suppression of microRNA in vitro and in vivo [18].
In 2008, Elmen et al. [18] demonstrated that this technology could effectively suppress miR-122 expression in both mice and African green monkeys. To test the efficacy of various modifications, they focused on an miRNA-regulated pathway involved with cholesterol production in the liver where miR-122 acts to suppress Aldoa (aldolase A) mRNA, resulting in the production of cholesterol. Intraperitoneal injection of LNA-modified anti-miR-122 in mice successfully suppressed miR-122 expression in the liver with a relatively low dosage when compared with other modifications attempted on anti-miRs. With just two weekly injections over a 6-week period, total cholesterol levels fell by 30 % in a mouse model of dietary obesity [18]. The effectiveness of the LNA-modified anti-miR was also tested in African green monkeys given three intravenous injections of the anti-miR on alternating days (1–10mg/kg of body weight) and followed over time. Remarkably, the dose-dependent reduction in total plasma cholesterol that resulted from the treatment was still evident 7 weeks after the initial treatment with the highest dosage. Lower dosages were also found to be efficacious.
That study [18] is significant for several reasons. The LNA modification appeared to confer long-term efficacy to an antagonizing oligonucleotide. The extended in vivo efficacy eliminates the need for chronic infusion. The efficient suppression with lower dosages of oligonucleotide compared with other anti-miR formulations was also an important advancement in this area. Of comparable translational significance was the demonstration that therapeutic treatment in both mice and non-human primates did not have any measureable adverse effects. LNA-modified anti-miRs have been reported to be effective in several recent studies, including our laboratory’s study in rats [9]. The effectiveness of LNA-modified anti-miR, however, appears to vary depending on specific miRNAs and the accessibility of target tissues.
Another approach to altering miRNA expression is increasing an miRNA that has been suppressed during the development of disease (Figure 1B) [22,23]. The loss of miRNA regulation may facilitate an increase in the expression of genes that contribute to the disease processes. In these instances, the restoration of normal expression of these regulatory miRNAs would be expected to be beneficial. In 2009, Kota et al. [24] described the intravenous delivery of a miR-26a AVV (adeno-associated virus) that successfully restored miR-26a levels that had been reduced in a murine model of liver cancer. They observed apoptosis of tumour cells without death of normal cells and no traces of toxicity [24].
Although the studies described above indicate that therapies targeting miRNA expression changes show promise, there are many issues that must be resolved before applying these types of treatments in humans. The first relates simply to the significance of the change in miRNA expression to the development of the disease. Determining whether the change is causal or secondary will be an important initial consideration when deciding if a particular disease would be most effectively treated by miRNA therapy. Additionally, many of the same challenges with siRNA (small interfering RNA)-based therapies and gene therapies also apply to miRNA-based therapies. Particularly important issues include target specificity, tissue specificity, cellular uptake and adverse reactions to delivery vehicle (Figure 2).
Figure 2. Challenges in the development of miRNA therapies.
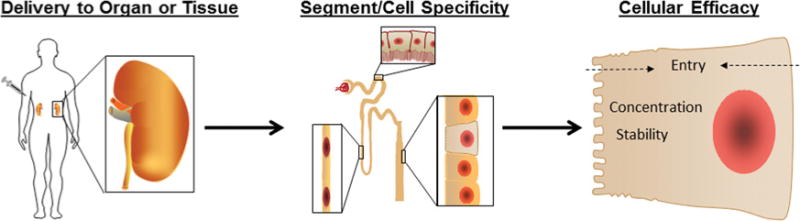
As undesired effects of miRNA therapies are of great concern, the route of delivery must be carefully considered. Once the therapeutic agent has reached the tissue, it will be important that it targets cell types involved with the disease process. This is especially important in the kidney where there are many cell types present, all with a unique function. Upon reaching the appropriate cell population, the therapeutic agent must gain entry into the cell in a sufficient concentration to be effective for the clinical treatment of disease.
The distribution of therapeutic agents within different organs and tissues is a prevailing concern in treatments which would alter gene expression. It is clear that miRNA profiles can vary greatly between tissue and cell types and within a single cell type during disease progression. It is possible that altering miRNA expression could be therapeutic in one tissue or cell type, while detrimental in another (Figure 2). On the other hand, cell-specific expression or action of miRNAs could be an advantage for miRNA therapies. For example, an anti-miR administered systemically would knock down an miRNA only in cells that express the miRNA while sparing cells that do not express the miRNA.
Intravenous delivery of therapeutics might be more effective in organs that receive the greatest portion of blood flow. The liver is a likely place for circulating oligonucleotides to accumulate, as demonstrated by Elmen et al. [18]. In our laboratory, we have observed effective suppression of miRNA by LNA-modified anti-miRNAs in the kidney [9,24a]. There is also growing evidence that LNA-modified anti-miRs are effective in other organs or tissues including tumours [25].
Once delivered to the tissue, the therapeutic agent needs to efficiently enter the cell. The route of uptake of oligonucleotides appears to vary between organs. In an isolated perfusion system, it has been shown that oligonucleotide uptake occurs non-specifically in both parenchymal and non-parenchymal cells of the liver [26]. Sawai et al. [27] have shown that the uptake of modified oligonucleotides in a perfused rat kidney resulted from tubular reabsorption and uptake from the capillaries. A greater understanding of organ-specific differences in uptake may allow us to improve the targeting of tissues with modifications to avoid off-target effects.
Although delivery of miRNA-based therapeutics directly to the diseased tissue may be more safe and effective, this would probably limit the clinical applications. The study by Kota et al. [24] restored miRNA levels using an AAV under a constitutively active promoter. If the safety of miRNA AVV delivery can be confirmed, it may be possible to drive expression of the miRNA using a tissue- or cell-specific promoter to diminish unintended effects.
BODY FLUID miRNAs AS DIAGNOSTIC OR PROGNOSTIC MARKERS
Stringent statistical criteria must be satisfied and clinical values beyond currently available markers must be demonstrated in order to establish any miRNA expression signatures as clinically useful diagnostic or prognostic markers. Numerous studies have reported the identification of miRNA expression patterns in cells or organ tissues that are associated with the diagnosis or prognosis of human disease. Few, if any, have come close to establishing new clinically relevant diagnostic or prognostic markers. However, analysis of miRNA expression patterns remains a promising possibility for discovering disease markers, particularly expression patterns that can be discerned in body fluids collected in non-invasive or minimally invasive manners (Figure 1B).
Over the past few years, several studies have shown that miRNAs are detectable in circulating blood; however, the mechanisms facilitating their entry into the bloodstream have not been fully characterized [28–30]. Early observations that endogenous plasma miRNAs exist in a form that is resistant to plasma RNase activity suggested that these blood miRNAs were not cell-associated, but rather located in protective microvesicles [28,30]. Several studies have indicated that miRNAs are both actively secreted into the blood within microvesicles [29–32] and released from apoptotic bodies originating from cells lining blood vessels [32]. miRNAs detectable in extracellular compartments have also been proposed to play a role as intercellular messengers [33].
Measurements obtained from plasma or serum indicated that both fluids may be suitable for miRNA biomarker analysis. Serum levels of various miRNAs remain stable for several hours at room temperature [28,30]. These are important characteristics that would permit expression analysis of circulating miRNAs in a clinical setting.
The potential use of circulating miRNAs in diagnosis has been investigated in various cardiovascular diseases, such as CAD (coronary artery disease), myocardial infarction and heart failure [34–36]. A study by Fichtlscherer et al. [34] used hybridization-based array to compare the expression profiles of circulating miRNAs in patients with stable CAD and healthy volunteers. This analysis revealed alterations in miRNA enriched in vascular endothelial cells, including miR-126, members of the miR-17~92 cluster (miR-17, miR-20a and miR-92a), miR-130a, miR-221, let-7d and miR-21. When expression levels of several cardiac and smooth muscle miRNAs were measured by TaqMan qPCR (quantitative PCR) in larger cohorts (n = 53), smooth-muscle-enriched miR-145 was significantly reduced and cardiac-muscle-enriched miRNAs miR-133 and miR-208a were significantly increased in CAD patients. The study indicated that alterations in circulating miRNAs are not restricted to endothelial or vascular miRNAs in CAD. The sensitivity of the detection assay may be an important consideration when deciding which techniques to use for detecting disease-related biomarkers. In that regard, recently developed technologies such as RNA-seq (RNA sequencing) [37] could provide unprecedented views of miRNA expression profiles (Figure 1B).
Another study by Zampetaki et al. [38] has described the plasma miRNA signature for DM (diabetes mellitus) in subjects from the prospective Bruneck population-based study. It was shown that 91 out of 99 (92%) controls and 56 out of 80 (70%) DM cases were correctly classified using expression profiles of the five most significant miRNAs. Interestingly, more than 50% of normoglycaemic subjects that developed DM within the 10-year follow-up period were already determined to be diabetics by this analysis, highlighting the potential for miRNA biomarkers to act as predictors of disease. Roderburg et al. [39] have reported that human cirrhotic livers had lower miR-29 expression compared with the control liver samples. Members of the miR-29 family have been shown to regulate the expression of extracellular matrix proteins which are involved in fibrosis [9,9c]. Patients with advanced liver cirrhosis had lower miR-29 levels than patients with early stage fibrosis, indicating its potential use as not only a marker of disease, but also as an indicator of disease progression and disease severity. miR-29 expression may be an indicator of the aetiology of fibrosis, as patients with alcoholic cirrhosis had a lower miR-29 levels compared with patients with viral hepatitis.
Detection of miRNAs in body fluids other than the plasma, such as urine, pleural fluid, peritoneal fluid, cerebrospinal fluid, breast milk, colostrum, saliva, seminal fluid, tears and amniotic fluid, have been reported [30,40]. In a study by Weber et al. [40], a variety of miRNAs were shown to be present at high levels in most body fluids, whereas others were specific to a type of body fluid. The amount of total RNA ranged from approximately100 μg/l to almost 50 mg/l. Breast milk and seminal fluid were richest in RNA, whereas urine, plasma and cerebrospinal fluid contained the least amount. The largest number of miRNAs was detected in saliva, seminal fluid and breast milk (~430 or more), whereas the number of detectable miRNAs in urine, pleural fluid and cerebrospinal fluid was approximately 50% less. Compared with blood, cerebrospinal fluid or other body fluids, saliva may have increased variability in the miRNA profile because of variations in the number of desquamated oral epithelial cells in the sample. The variability could be reduced by removing desquamated cells with centrifugation [41].
Physiological or ‘normal’ concentration ranges of miRNAs in body fluids remain to be established. Methods are also needed to standardize isolation and sample preparation procedures, as well as establishing controls for normalizing detected miRNA abundance across multiple samples. Zampetaki et al. [38] analysed their qPCR data as unadjusted Ct values standardized to miR-454 and RNU6b (RNA U6B small nuclear) levels in plasma. The inclusion of a spiked-in control has been reported as being useful for sample preparation and analysis. Fichtlscherer et al. [34] supplemented the samples with 5 nmol/l Caenorhabditis elegans miR-39 (cel-miR-39) and used it for normalization of the RNA sample processing.
TRANSLATIONAL RESEARCH OF miRNA IN KIDNEY DISEASE AND HYPERTENSION
Several studies have reported the involvement of miRNA in renal development, function and disease in animal models [42–49]. However, very little is known about the function of miRNAs in normal renal physiological processes in humans. In healthy humans, miRNA profiles reveal many distinct tissue-specific differences in expression. A tissue miRNA profiling study by Sun et al. [50] found mouse and human kidney tissues to have an elevated expression of miR-192, miR-193, miR-204, miR-215 and miR-216, when compared with heart, lung, muscle or prostate tissue. Several studies have focused on the role of miR-192 in the development of fibrosis in the kidney [51–54].
Human tissue samples from kidney have been profiled in many diseases, including hypertensive nephrosclerosis, IgA nephropathy, diabetic nephropathy and renal tumours. Analysis of miRNA profiles generated from renal cell carcinoma tumours in humans resulted in a successful classification of distinct cancer subtypes and identified miRNA expression patterns that correlated with prognosis [55].
An exciting application for miRNA profiling has been in the prediction of acute renal allograft rejection in patients. Anglicheau et al. [56] reported that the level of miRNAs miR-142-5p, miR-155, miR-223, miR-10b, miR-30a-3p and let-7c obtained from biopsies of transplanted kidneys was highly predictive of acute rejection and even graft function [56].
Despite the limited human tissue availability, the significance of miRNA in the pathophysiology of human kidney disease and hypertension is becoming apparent through the merging of clinical and basic science with the help of analytical tools. One approach taken by several investigators is to combine studies in human kidney tissues, human kidney cell lines and animal models (Figure 1C). In our laboratory we have used a human renal tubular epithelial cell line (HK-2 cells) to identify miRNA–target pairs involved with phenotypic changes resulting from TGFβ1 (transforming growth factor β1) treatment. In this study [57], we compared proteomic and miRNA array data from TGFβ1- and vehicle-treated cells and identified miRNAs and their predicted target proteins showing reciprocal expression that was indicative of a regulatory interaction. We validated several of the identified miRNA–target interactions and confirmed that miR-382 contributed significantly to the TGFβ1-induced loss of epithelial characteristics in HK2 cells via regulation of target mnSOD (manganese superoxide dismutase) [57].
We have extended our study of miR-382 to animal models and human kidney tissues and demonstrated a significant role of miR-382 and its target genes in the development of renal interstitial fibrosis in mouse models and possibly human kidney disease [24a,58]. In mice, kidneys subjected to 3 days of unilateral urethral obstruction experienced a substantial up-regulation of miR-382 and medullary fibrosis. Intravenous administration of LNA-modified anti-miR-382 prevented the increase in miR-382 and completely inhibited the development of fibrosis in the inner medulla. Through an extensive set of in vitro and in vivo experiments, we discovered a completely novel pathway in which targeting of the (chymo)trypsin-like proteinase kallikrein-5 by miR-382 contributed to the fibrotic effect of miR-382 in the mouse model [24a]. We obtained fresh kidney samples from humans and found that the level of fibrosis in the inner medulla correlated positively with miR-382 expression and negatively with kallikrein-5 expression, supporting the relevance of the miR-382 and kallikrein-5 pathway to human kidney fibrosis [58].
Krupa et al. [52] treated HK-2 cells with high glucose to simulate a diabetic state; however, this treatment did not produce any changes in miRNA expression. Profiling of miRNA from human biopsies, however, uncovered expression differences between patients with diabetic nephropathy and healthy controls. Distinct miRNA profiles were identified for early- and late-progressing diabetic nephropathy. Overall, patients who were in an early stage of diabetic nephropathy had a higher expression of miR-192 than late presenters. In addition, the reduced amount of miR-192 correlated with the degree of tubulointerstitial fibrosis [52].
An example of the relevance of miRNAs to human hypertension involves regulation of the AT1R (angiotensin II type 1 receptor). Although several polymorphisms have been identified in this gene, 1166 A/C in particular was correlated with increased incidence of hypertension and cardiovascular disease in several studies [59–63]. Despite this, the mechanism underlying the association of the polymorphism to disease phenotypes was not clear because it resided within the non-coding 3′-UTR region of the gene. Martin et al. [64] used computer alignment tools to determine that this region would be predicted to bind miR-155. Subsequent analysis verified that the single nucleotide variation reduced the binding capability of miR-155 in the target region, preventing normal suppression of this gene, which appeared to contribute to the development of certain forms of human hypertension [64,65]. The role of miRNAs in genetic determination or control of disease is an exciting research area. For example, it has been reported that mutations in the seed region of miR-96 are responsible for a Mendelian form of hearing loss in human [66].
TECHNICAL CONSIDERATIONS FOR ANALYSING miRNAs IN HUMAN KIDNEY TISSUE
A major challenge to understanding the role of miRNA in human renal physiology and disease and the role of renal miRNAs in human hypertension is determining where the miRNAs are expressed within the kidney and how they regulate their targets. The kidney is a highly heterogeneous organ that can contain several tubule segments and cell types within a given plane (Figure 2). Tian et al. [67] found vastly different miRNA expression between the cortex and medulla in rats. It is expected that we will find distinct differences in miRNA expression between tubule segments and cell types within the same kidney region, each contributing to gene regulation in a unique way.
Because fresh human tissues are scarce, and most biopsied samples frequently exist in formalin-fixed paraffin-embedded archives, mechanistic studies in human tissue remain challenging. We do know that formalin-fixed paraffin-embedded kidney tissues from patients can be used to study miRNA expression [52,56,68–71]; however, the breadth of the studies is often limited with needle core biopsies containing mostly cortical tissue. Acquiring tissue from the renal medulla is uncommon, except in cases of partial or complete nephrectomy.
In situ hybridization has been used to visualize cellular localization of miRNA [72,73] (Figure 1B), but there are several obstacles in using this method to both quantify miRNA expression and to determine the cell location of the miRNA in kidney tissue. Ideally, in situ hybridization and immunohistochemistry could be combined to map the expression of the miRNA using fluorescent double labelling. However, the inherent autofluorescence of kidney tissue, combined with autofluorescence added by the formalin fixation and paraffin-embedding process, make it very difficult to tease apart low-abundance miRNA expression from background [74]. In our laboratory, we have opted to use an amplifying enzyme-based colorimetric stain to detect miRNA expression by in situ hybridization. The approach alleviates concerns about interference from autofluorescence. However, any subsequent colorimetric immunohistochemistry would mask some of the miRNA signal within a tissue, making visualization and relative quantification of either sample potentially difficult.
Another promising approach is miRNA expression analysis of laser-captured kidney tissue (Figure 1B). This approach can selectively isolate distinct renal tubules and cell types from sectioned kidney tissues. Many researchers have taken steps to optimize miRNA detection and quantification in formalin-fixed clinical samples [75,76]. Despite successful miRNA analysis in these samples, it has also been demonstrated that formalin fixation of human tissue may not preserve RNA optimally as well as ethanol fixation [77–79]. The impact of fixation, immunohistochemical labelling and processing reagents on laser-captured renal miRNA quality and quantity requires investigation.
CONCLUSIONS
The importance of miRNA regulation in human kidney disease and hypertension is becoming increasingly clear. The translational study of miRNA in kidney disease and hypertension has unique challenges. However, we can learn from experience, as well as from lessons obtained from translational miRNA research in other disease areas. New and improved tools for detecting, analysing and manipulating miRNAs in models and samples relevant to translational research are constantly being developed and improved. It will be important to carefully consider the limitations of each method or technique in the context of achieving valuable goals in translational miRNA research in kidney disease and hypertension. By merging complementary basic and clinical research we can work toward understanding miRNA regulation in kidney disease and hypertension and how best to manipulate miRNAs in relevant disease states.
Acknowledgments
FUNDING: Our own work relevant to the present paper was supported by the National Institutes of Health [grant numbers HL085267, DK084405, HL077263, HL082798, HL029587 (to M.L.)], a National Research Service Award, a Kern Family Foundation Innovation Award (to A.J.K.), and an American Heart Association Pre-Doctoral Fellowship (to D.M.).
Abbreviations
- AVV
adeno-associated virus
- CAD
coronary artery disease
- DM
diabetes mellitus
- LNA
locked-nucleic acid
- miRNA (miR)
microRNA
- qPCR
quantitative PCR
- TGFβ1
transforming growth factor β1
- UTR
untranslated region
References
- 1.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, non-coding RNA, is a major unexpected hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b.Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci USA. 2008;105:5874–5878. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olsen EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9d.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics. 2012 doi: 10.1152/physiolgenomics.0014.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, Scherr M, Eder M. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signalling pathways. Blood. 2011;117:4338–4348. doi: 10.1182/blood-2010-06-289058. [DOI] [PubMed] [Google Scholar]
- 11.Subramanyam D, Lamuille S, Judson RI, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rooj E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 14.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 16.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 19.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Krutzfield J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acid Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted
- 22.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 23.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 24.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Kriegel AJ, Liu Y, Cohen B, Usa K, Liu Y, Liang M. MiR-382 targeting of kallikrein 5 contributes to renal inner medullary interstitial fibrosis. Physiol Genomics. 2011;00173:2011. doi: 10.1152/physiolgenomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 26.Takakura Y, Mahato RI, Yoshida M, Hashida M. Uptake characteristics of oligonucleotides in the isolated rat liver perfusion system. Antisense Nucleic Acid Drug Dev. 1996;6:177–183. doi: 10.1089/oli.1.1996.6.177. [DOI] [PubMed] [Google Scholar]
- 27.Sawai K, Miyao T, Takakura Y, Hashida M. Renal disposition characteristics of oligonucleotides modified at terminal linkages in the perfused rat kidney. Antisense Res Dev. 1995;5:279–287. doi: 10.1089/ard.1995.5.279. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Shafer J, Lee ML, Schmittgen TD, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signaling. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 33.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 34.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 35.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-MiR45p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 36.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 39.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 40.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato M, Arce L, Natarajan R. MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol. 2009;4:1255–1266. doi: 10.2215/CJN.00520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saal S, Harvey SJ. MicroRNAs and the kidney: coming of age. Curr Opin Nephrol Hypertens. 2009;18:317–323. doi: 10.1097/MNH.0b013e32832c9da2. [DOI] [PubMed] [Google Scholar]
- 44.Wessely O, Agrawal R, Tran U. MicroRNAs in kidney development: lessons from the frog. RNA Biol. 2010;7:296–299. doi: 10.4161/rna.7.3.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhatt K, Mi QS, Dong Z. microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol. 2011;300:F602–F610. doi: 10.1152/ajprenal.00727.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang M. MicroRNA: a new entrance to the broad paradigm of systems molecular medicine. Physiol. Genomics. 2009;38:113–115. doi: 10.1152/physiolgenomics.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang M, Liu Y, Mladinov D, Cowley AW, Jr, Trivedi H, Fang Y, Xu X, Ding X, Tian Z. MicroRNA: a new frontier in kidney and blood pressure research. Am J Physiol Renal Physiol. 2009;297:F553–F558. doi: 10.1152/ajprenal.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karolia DS, Wintour EM, Bertram J, Jeyaseelan K. Riboregulators in kidney development and function. Biochimie. 2010;92:217–225. doi: 10.1016/j.biochi.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 49.Li JY, Yong TY, Michael MZ, Gleadle JM. The role of microRNAs in kidney disease. Neprology. 2010;15:599–608. doi: 10.1111/j.1440-1797.2010.01363.x. [DOI] [PubMed] [Google Scholar]
- 50.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-β-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol. 2010;21:438–447. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B, Herman-Edelstein M, Koh P, Burns W, Jandeleit-Dahm K, Watson A, Saleem M, Goodall GJ, Twigg SM, Cooper ME, Kantharidis P. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-β. Diabetes. 2010;59:1794–1802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R. A microRNA circuit mediates transforming growth factor-β 1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011;80:358–368. doi: 10.1038/ki.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, The BT. MicroRNA profiling of human kidney cancer subtypes. Int J Oncol. 2009;35:109–114. doi: 10.3892/ijo_00000318. [DOI] [PubMed] [Google Scholar]
- 56.Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA. 2009;106:5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor β 1: a novel role of miR-382. Nucleic Acids Res. 2010;38:8338–8347. doi: 10.1093/nar/gkq718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kriegel AJ, Mladinov D, Kelly H, Langenstroer P, See W, Liang M. Interstitial fibrosis in the renal inner medulla in humans is associated with up-regulation of miR-382 and down-regulation of kallikrein 5. FASEB J. 2011;25:663.12. [Google Scholar]
- 59.Bonnardeaux A, Davies E, Jeunemaitre X, Fery I, Charru A, Clauser E, Tiret L, Cambien F, Corvol P, Soubrier F. Angiotensin II (type-1) receptor locus: CA repeat polymorphism and genetic mapping. Hypertension. 1994;24:63–69. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- 60.Wang WY, Zee RY, Morris BJ. Association of angiotensin II type 1 receptor gene polymorphism with essential hypertension. Clin Genet. 1997;51:31–34. doi: 10.1111/j.1399-0004.1997.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 61.Osterop AP, Kofflard MJ, Sandkuiji LA, ten Cate FJ, Krams R, Schalekamp MA, Danser AH. AT1 receptor A/C1166 polymorphism contributes to cardiac hypertrophy in subjects with hypertrophic cardiomyopathy. Hypertension. 1998;32:825–830. doi: 10.1161/01.hyp.32.5.825. [DOI] [PubMed] [Google Scholar]
- 62.Benetos A, Gautier S, Ricard S, Topouchian J, Asmar R, Poirier O, Larosa E, Guize L, Safar M, Soubrier F, Cambien F. Influence of angiotensin-converting enzyme and angiotensin II type 1 receptor gene polymorphisms on aortic stiffness in normotensive and hypertensive patients. Circulation. 1996;94:698–703. doi: 10.1161/01.cir.94.4.698. [DOI] [PubMed] [Google Scholar]
- 63.Tiret L, Bonnardeax A, Pourier O, Ricard S, Marques-Vidal P, Evans A, Arveiler D, Luc G, Kee F, Ducimetiere P, et al. Synergistic effects of angiotensin-converting enzyme and angiotensin-II type 1 receptor gene polymorphisms on risk of myocardial infarction. Lancet. 1994;344:910–913. doi: 10.1016/s0140-6736(94)92268-3. [DOI] [PubMed] [Google Scholar]
- 64.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor + 1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;17:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interferacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 67.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Microarray analysis of micro-ribonucleic acid expression in primary immunoglobulin A nephropathy. Saudi Med J. 2008;29:1388–1393. [PubMed] [Google Scholar]
- 69.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2009;29:749–754. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 70.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest. 2010;90:98–103. doi: 10.1038/labinvest.2009.118. [DOI] [PubMed] [Google Scholar]
- 71.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am J Hypertens. 2010;23:78–84. doi: 10.1038/ajh.2009.208. [DOI] [PubMed] [Google Scholar]
- 72.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Viegas MS, Martins TC, Seco F, do Carmo A. An improved and cost-effective methodology for the reduction of autofluorescence in direct immunofluorescence studies on formalin-fixed paraffin embedded tissues. Eur J Histochem. 2007;51:59–66. [PubMed] [Google Scholar]
- 75.Andreasen D, Fog JU, Biggs W, Salomon J, Dahslveen IK, Baker A, Mouritzen P. Improved microRNA quantification in total RNA from clinical samples. Methods. 2010;50:S6–S9. doi: 10.1016/j.ymeth.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 76.Nonn L, Vaishnav A, Gallagher L, Gann PH. mRNA and microRNA expression analysis in laser capture microdissected-prostate biopsies: valuable tool for risk assessment and prevention trials. Exp Mol Pathol. 2010;88:45. doi: 10.1016/j.yexmp.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25:86–91. [PubMed] [Google Scholar]
- 78.Su JM, Perlaky L, Li XN, Leung HC, Antaliffy B, Armstrong D, Lau CC. Comparison of ethanol versus formalin fixation on preservation of histology and RNA in laser capture microdissected brain tissues. Brain Pathol. 2004;14:175–182. doi: 10.1111/j.1750-3639.2004.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S, Wang L, Zhu T, Gao X, Li J, Wu Y, Zhu H. Improvement of tissue preparation for laser capture microdissection: application for cell type-specific miRNA expression profiling in colorectal tumors. BMC Genomics. 2010;11:163. doi: 10.1186/1471-2164-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]


