Abstract
In scattering specimens, multiphoton excitation and nondescanned detection improve imaging depth by a factor of 2 or more over confocal microscopy; however, imaging depth is still limited by scattering. We applied the concept of clearing to deep tissue imaging of highly scattering specimens. Clearing is a remarkably effective approach to improving image quality at depth using either confocal or multiphoton microscopy. Tissue clearing appears to eliminate the need for multiphoton excitation for deep tissue imaging.
Keywords: confocal, multiphoton, clearing, refractive index matching, scattering
Introduction
Depth of imaging is limited in light microscopy. A primary cause of image degradation with depth is refractive index discontinuity, resulting in spherical aberration. In studies of living cells in aqueous medium, when using an oil immersion objective, there is mismatch between the refractive index of the immersion media and the refractive index of the mounting media. This mismatch results in spherical aberration, seen as loss of brightness, loss of resolution, poorer optical sectioning, and poorer depth penetration even within single cells (Hell et al., 1993; Pawley, 2002). This issue has been addressed by the development of water immersion lenses, which has extended imaging depth for studies of living cells and tissues (Gerritsen & De Grauw, 1999).
Nevertheless, when the scale of imaging is extended from tens of microns for single cell layers to hundreds of microns for biological tissues, signal levels and resolution still deteriorate rapidly with the increased depth, even with use of both aqueous immersion media and mounting media. This is because refractive index discontinuities within the biological tissue still result in scattering of light (Tuchin, 2005b). Consequently, even when sample and immersion media are index matched, signal levels obtained with confocal microscopy rapidly decrease with depth.
Multiphoton microscopy improves imaging depth by a factor of 2 or more over confocal microscopy and is generally considered the best approach for deep tissue imaging. However, even multiphoton microscopy is ultimately limited by scattering. It is in this context that we examine the technique of clearing for deep tissue imaging of highly scattering tissues.
Methods
Sample Preparation
Rat kidneys were perfusion fixed using 4% paraformaldehyde in phosphate buffered saline (PBS) pH 7.4, kept in fixative at 4°C for at least 24 h, transferred to 0.25% paraformaldehyde in PBS, and stored at 4°C until sectioned. Kidney sections of 200–300 µm were cut using a vibratome (Technical Products International, Inc., St. Louis, MO, USA). Samples were blocked and permeabilized for at least 1 h at room temperature in 1% Triton-X 100, 1% bovine serum albumin, 2% serum, 1× PBS pH 7.4. Labeling steps were done overnight at room temperature with rotation in the same buffer as was used for blocking and permeabilization. Lens culinaris agglutinin-FITC (Vector Labs, Burlingame, CA, USA) was used at a dilution of 1:200. Hoechst was used at a dilution of 1:1,000 (10 mg/mL stock, Invitrogen, Carlsbad, CA, USA). Anti-vimentin (Sigma-Aldrich, St. Louis, MO, USA) was used at 1:100. Secondary antibody, Cy5 anti-mouse IgG, was used at a dilution of 1:50 (Jackson Immunoresearch, West Grove, PA, USA).
Labeled tissue was rinsed in PBS then incubated in a graded series of PBS-glycerol solutions with the final solution in the graded series composed of 20% PBS and 80% glycerol. Samples were incubated in each solution in the dark at room temperature with rotation for at least 2 h. The sample was judged to have equilibrated with the solution when it would eventually settle to the bottom rather than remain suspended or floating. Clearing solution (53% benzyl alcohol, 45% glycerol, 2% DABCO by weight) was prepared and refractive index of the clearing solution was measured as 1.511 using an Abbe refractometer (Sino Science & Technology Co., Ltd., ZhangZhou, FuJian, China). The refractive index of the clearing solution is tunable by altering the proportions of benzyl alcohol and glycerol. Samples were incubated in the clearing solution in the dark at room temperature with rotation for at least 2 h. When samples had equilibrated with the clearing solution, they were transferred to a fresh aliquot of clearing solution and incubated overnight. After overnight incubation samples were mounted on slides using fresh clearing solution. Spacers were used to prevent compression of the tissue. Prior to use for mounting, the thickness of each coverslip was measured using a micrometer. Mounts were sealed using rubber cement.
Antibody binding was not perturbed by tissue clearing with benzyl alcohol and glycerol, and the fluorescence of fluorophores used was not quenched. Likewise, Hoechst binding and DAPI binding were not perturbed, and their fluorescence was not quenched. Localization of phalloidin became diffuse after incubation in the clearing solution, although if imaged immediately after clearing adequate results could be obtained (not shown). Microtubule staining was unaffected by clearing (not shown). This clearing method does result in loss of eGFP fluorescence, but this problem could presumably be overcome by using an anti-GFP antibody for visualization. Variants of eGFP were not tested. Additionally, for most fluorophores, lower levels of excitation power were required to image cleared samples than uncleared samples, which minimized photobleaching even when using confocal microscopy.
Image Acquisition and Processing
Images were collected using an Olympus FV1000 confocal microscope system (Olympus America, Inc., Center Valley, PA, USA) adapted for two-photon microscopy. The system is equipped with a Mai-Tai titanium-sapphire laser pumped by a 10 W argon laser (Spectra-Physics, Santa Clara, CA, USA), a Pockels cell beam attenuator (Conoptics Inc., Danbury, CT, USA), and a Keplerian-style collimator/beam expander aligned to fill the back aperture of the objective. Image volumes of uncleared samples were collected using a 60× NA 1.2 water immersion objective. Image volumes of cleared samples were collected using a 60× NA 1.4 oil immersion objective or a 40× NA 1.3 oil immersion objective. Image volumes were rendered using Voxx software (Clendenon et al., 2002). Image volumes were segmented using Amira software (Visage Imaging, Inc., San Diego, CA, USA).
Results
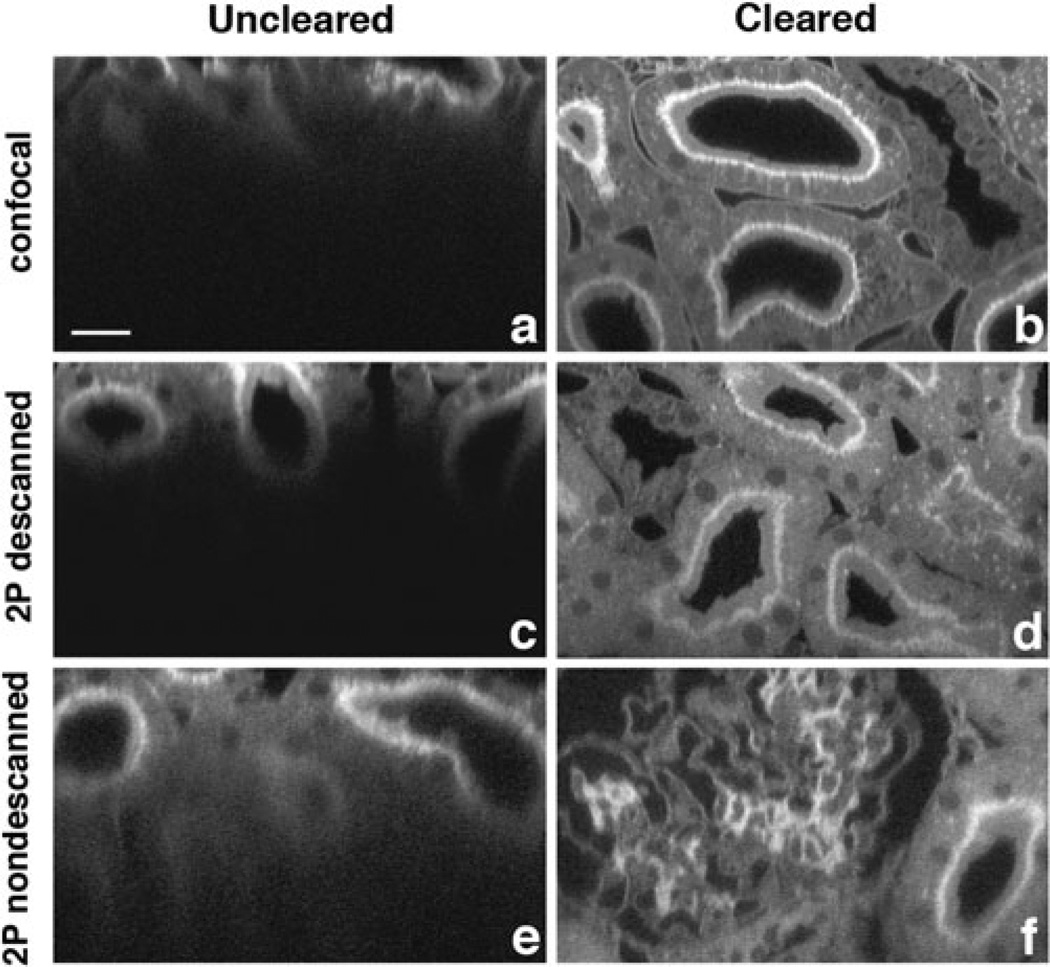
The drop of signal with depth is strikingly apparent in images of kidney tissue obtained by confocal fluorescence microscopy using a water immersion objective and aqueous mounting media (Fig. 1a). Signal levels in confocal microscopy are especially sensitive to scattering. In confocal microscopy the excitation light is attenuated by scattering and then fluorescence emission is scattered on its way back to the objective and is rejected by the confocal aperture, profoundly decreasing signal detection (Centonze & White, 1998). Since scattering decreases with the fourth power of wavelength, to some degree scattering losses can be addressed by using multiphoton microscopy, which uses longer excitation wavelengths, allowing greater imaging depth (Fig. 1c). Additionally, because multiphoton excitation only occurs at a single point in the sample, image collection can be conducted without a confocal aperture, capturing more of the scattered emission light and further increasing imaging depth (Fig. 1e). However, even with multiphoton fluorescence microscopy using nondescanned detectors, imaging depth is ultimately limited by scattering (Theer & Denk, 2006).
Figure 1.
Clearing enhances deep tissue imaging. (a–f) Shown are 100 µm xz-optical slices of fixed rat kidney tissue. Sections were cut to a thickness of 200 µm, stained with FITC-conjugated-lens culinaris, mounted in PBS, and imaged using a water immersion objective (a, c, e) or mounted in clearing solution and imaged using an oil immersion objective (b, d, f). Bar = 20 µm. Signal level at a depth of 100 µm using confocal microscopy was 22% of the maximum mean fluorescence intensity. Signal level at a depth of 100 µm using multiphoton microscopy and a nondescanned detector was 27% of the maximum mean fluorescence intensity.
To further improve the ability to image scattering tissues, scattering within the sample itself must be diminished. Scattering can be effectively controlled by optical clearing, which is accomplished by immersing the sample in a solution whose refractive index is near that of the cellular organelles and membranes within the tissue, displacing the aqueous cellular and interstitial fluid with the clearing solution (Tuchin, 2005b). In principal this has the effect of reducing refractive index discontinuities, thus reducing scattering within the tissue. We have applied the principal of tissue clearing using a solution of benzyl alcohol and glycerol with a refractive index of 1.51 to confocal imaging of thick sections of kidney, a highly scattering tissue with an average refractive index of 1.37 to 1.41 (Tuchin, 2005a). Tissue prepared in this way showed profoundly improved signal levels in images collected at depth by both confocal microscopy and multiphoton microscopy (Figs. 1b,d,f). Clearing had the most profound effect on images collected via confocal microscopy (compare Fig. 1a to Fig. 1b).
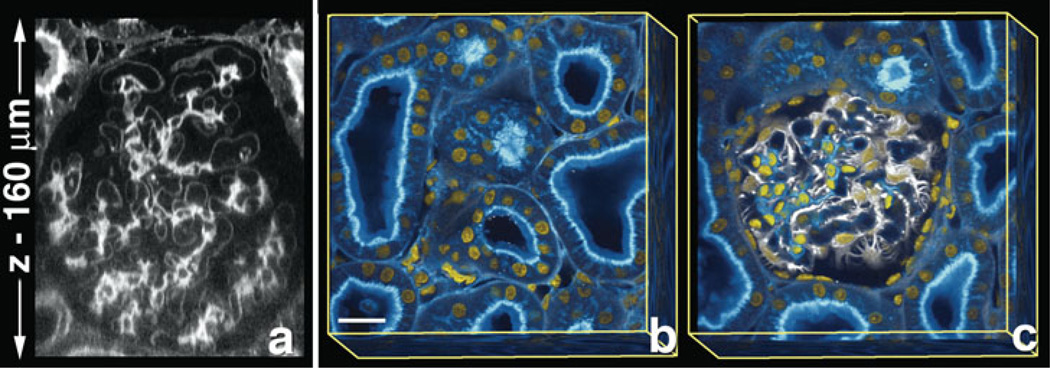
Further improvement of imaging at depth was obtained by increasing the level of laser illumination with depth of imaging into optically-cleared tissues. This allowed acquisition of image volumes whose depth was limited only by the working distance of the objective (Fig. 2a–c). This approach, combining optical clearing with the use of increased laser intensity with depth, has extended our ability to visualize three-dimensional (3D) structure at subcellular resolution in thick sections of adult rat kidney.
Figure 2.
Clearing allows visualization of intact 3D organization within tissues. Fixed adult rat kidney tissue was imaged using clearing plus increase of laser intensity with depth. (a) Single xz-section of rat kidney stained with FITC-conjugated-lens culinaris agglutinin. Rendered volume, 160 µm total depth (b, c), of rat kidney stained with FITC-conjugated-lens culinaris aggutinin (blue), vimentin labeled podocytes (white), and Hoechst labeled nuclei (yellow). (b) The surface of whole volume shows a network of kidney tubules. Superficial planes removed from panel b show the glomerulus within the network of tubules (c). (b, c) Bar = 20 µm.
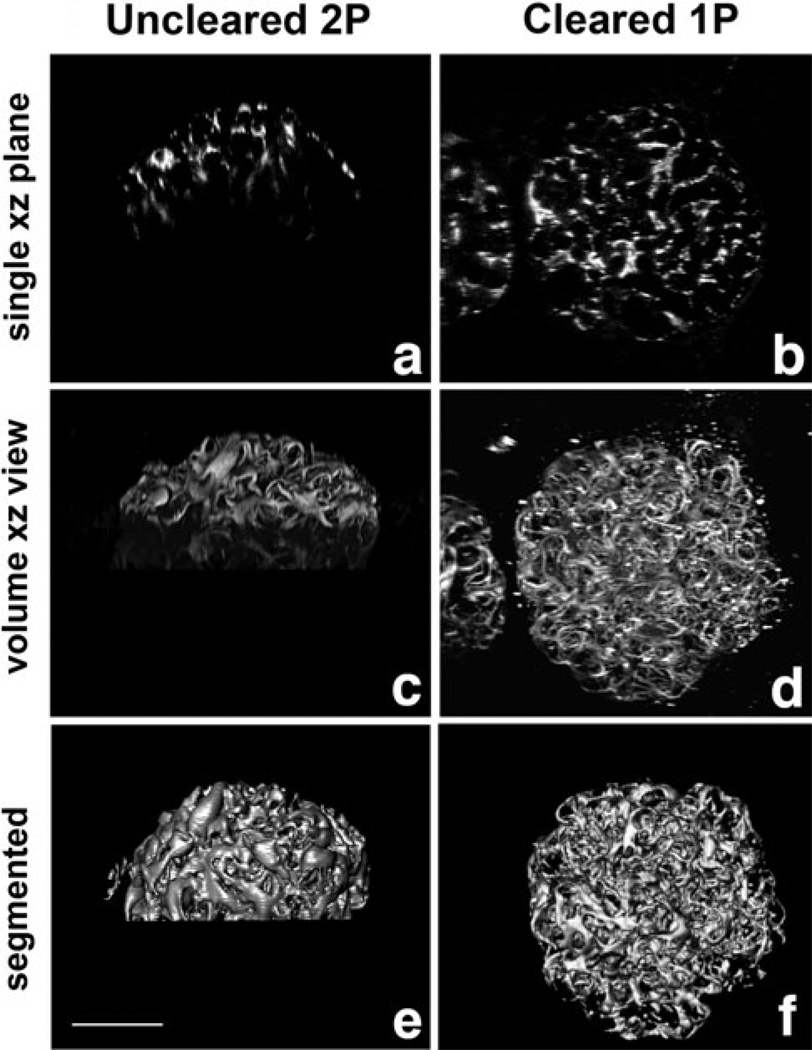
Without tissue clearing, it was not possible to capture images more than ~100 µm into kidney tissue, a depth corresponding to approximately half the thickness of a single rat glomerulus (Fig. 3a,c) even when using a water immersion objective, multiphoton excitation, and nondescanned detectors. These images were useful for morphological evaluation, but not for quantitative evaluation (Fig. 3e). Tissue clearing combined with increased laser intensity at depth enabled collection of high-quality 3D images of complete glomeruli (Figs. 2b,c, 3b,d). Image volumes of cleared kidney samples labeled for vimentin, a marker of the podocyte cell body, were easily segmented, using a simple thresholding approach (Fig. 3f). Total podocyte volume was measured to be 215,733 µm3 (±25,284, n = 4), a value similar to those previously obtained by more laborious stereometric methods (Amann et al., 1996).
Figure 3.
Clearing facilitates segmentation. Shown are xz views of fixed rat kidney tissue labeled with anti-vimentin antibody to visualize podocytes. Tissue mounted in aqueous solution and imaged using a water immersion objective and two-photon microscopy (a, c) is compared with tissue mounted in clearing solution and imaged using an oil immersion objective and confocal microscopy (b, d, e). (a, b) Single xz planes. (c, d) Alpha projections of 3D image volumes in xz orientation. (e, f) Surface rendering of segmentated podocytes from images c and d. Bar = 50 µm.
Discussion
Although the use of clearing methods in microscopy is not new (Gustafsson et al., 1999; Tuchin, 2005a, 2005b; Dickie et al., 2006; Tuchin et al., 2006; Appleton et al., 2009), it needs to be reconsidered in the context of multiphoton microscopy. Multiphoton microscopy has generally been considered the best approach for deep tissue optical sectioning microscopy. Here, we demonstrate that reduction of scattering by tissue clearing is such that deep tissue imaging can be conducted using confocal microscopy, making 3D tissue imaging accessible to a much broader range of researchers.
In addition to being less expensive, simpler, and more widely available, confocal microscopy has a distinct advantage over multiphoton microscopy. In multiphoton microscopy a single wavelength is used to excite all fluorophores simultaneously, often resulting in spectral bleedthrough. When deep tissue imaging is instead done using cleared tissue and confocal microscopy, fluorophores can be excited individually, minimizing bleedthrough from spectrally adjacent emission channels.
To our knowledge the benefits of an effective method of tissue clearing for high-resolution confocal and multiphoton fluorescence microscopy of thick tissues, without either initial methanol fixation or subsequent dehydration of the tissue, have not been explored. Clearing agents such as BABB are effective but are typically preceded by ethanol or methanol dehydration (Miller et al., 2005; Dickie et al., 2006), which may cause mislocalization or loss of some membrane proteins, particularly those found in small vesicles. Further, dehydration causes tissue shrinkage and has been reported to quench fluorescence of some fluorophores (Dickie et al., 2006). The technique of clearing using benzyl alcohol and glycerol does not require methanol fixation or dehydration and results in excellent preservation of morphology and antigenicity. We have applied this technique to rat kidney, mouse embryonic kidney (not shown), whole zebrafish embryos (not shown), and to whole mouse embryos (Ferkowicz & Yoder, 2011) with great success and expect that this technique can be applied to deep tissue imaging of other highly scattering tissues with similar benefits.
Summary
Tissue clearing provides a simple and inexpensive method for increasing the depth of imaging that can be achieved using fluorescence microscopy. In uncleared tissues and in living tissues, multiphoton microscopy is superior to confocal microscopy for deep tissue imaging. However, use of tissue clearing largely eliminates the benefits of multiphoton microscopy in fixed tissues, which, compared with confocal microscopy, requires more expensive equipment and is frequently more technically challenging. Use of clearing allows researchers who do not have access to multiphoton microscopes to perform high-resolution deep tissue imaging.
Acknowledgments
This project was supported by the National Institutes of Health (P50 DK 61594). Images were acquired at the Indiana Center for Biological Microscopy, Indiana University School of Medicine, Indianapolis, IN. We thank Bob Bacallao for providing the perfusion fixed rat kidney used in these studies.
Footnotes
Author contributions: S.G.C. designed this research and performed the experiments. P.A.Y., M.J.F., and K.W.D. aided development of the experiments. C.P. provided a raw image and aided development of the experiments. S.G.C. and K.W.D. wrote the manuscript.
References
- Amann K, Nichols C, Tornig J, Schwarz U, Zeier M, Mall G, Ritz E. Effect of ramipril, nifedipine, and moxonidine on glomerular morphology and podocyte structure in experimental renal failure. Nephrol Dial Transplant. 1996;11:1003–1011. [PubMed] [Google Scholar]
- Appleton PL, Quyn AJ, Swift S, Nathke I. Preparation of wholemount mouse intestine for high-resolution three-dimensional imaging using two-photon microscopy. J Microsc. 2009;234:196–204. doi: 10.1111/j.1365-2818.2009.03163.x. [DOI] [PubMed] [Google Scholar]
- Centonze VE, White JG. Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging. Biophys J. 1998;75:2015–2024. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendenon JL, Phillips CL, Sandoval RM, Fang S, Dunn KW. Voxx: A PC-based, near real-time volume rendering system for biological microscopy. Am J Physiol Cell Physiol. 2002;282:C213–C218. doi: 10.1152/ajpcell.2002.282.1.C213. [DOI] [PubMed] [Google Scholar]
- Dickie R, Bachoo RM, Rupnick MA, Dallabrida SM, DeLoid GM, Lai J, DePinho RA, Rogers RA. Three-dimensional visualization of microvessel architecture of whole-mount tissue by confocal microscopy. Microvasc Res. 2006;72:20–26. doi: 10.1016/j.mvr.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Ferkowicz MJ, Yoder MC. Whole embryo imaging of hematopoietic cell emergence and migration. In: Filippi M-D, Geiger H, editors. Stem Cell Migration. New York: Humana Press; 2011. pp. 143–155. [DOI] [PubMed] [Google Scholar]
- Gerritsen HC, De Grauw CJ. Imaging of optically thick specimen using two-photon excitation microscopy. Micros Res Tech. 1999;47:206–209. doi: 10.1002/(SICI)1097-0029(19991101)47:3<206::AID-JEMT6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG, Agard DA, Sedat JW. I5M: 3D widefield light microscopy with better than 100 nm axial resolution. J Microsc. 1999;195:10–16. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- Hell S, Reiner G, Cremer C, Stelzer HK. Aberrations in confocal fluorescence microscopy induced by mismatches in refractive index. J Microsc. 1993;169:391–405. [Google Scholar]
- Miller CE, Thompson RP, Bigelow MR, Gittinger G, Trusk TC, Sedmera D. Confocal imaging of the embryonic heart: How deep? Microsc Microanal. 2005;11:216–223. doi: 10.1017/S1431927605050464. [DOI] [PubMed] [Google Scholar]
- Pawley JB. Limitations on optical sectioning in live-cell confocal microscopy. Scanning. 2002;24:241–246. doi: 10.1002/sca.4950240504. [DOI] [PubMed] [Google Scholar]
- Theer P, Denk W. On the fundamental imaging-depth limit in two-photon microscopy. J Opt Soc Am A. 2006;23:3139–3149. doi: 10.1364/josaa.23.003139. [DOI] [PubMed] [Google Scholar]
- Tuchin VV. Optical Clearing of Tissues and Blood. Bellingham, WA: SPIE Publications; 2005a. [Google Scholar]
- Tuchin VV. Optical clearing of tissues and blood using the immersion method. J Phys D. 2005b;38:2497–2518. [Google Scholar]
- Tuchin VV, Altshuler GB, Gavrilova AA, Pravdin AB, Tabatadze D, Childs J, Yaroslavsky IV. Optical clearing of skin using flash lamp-induced enhancement of epidermal permeability. Lasers Surg Med. 2006;38:824–836. doi: 10.1002/lsm.20392. [DOI] [PubMed] [Google Scholar]