Abstract
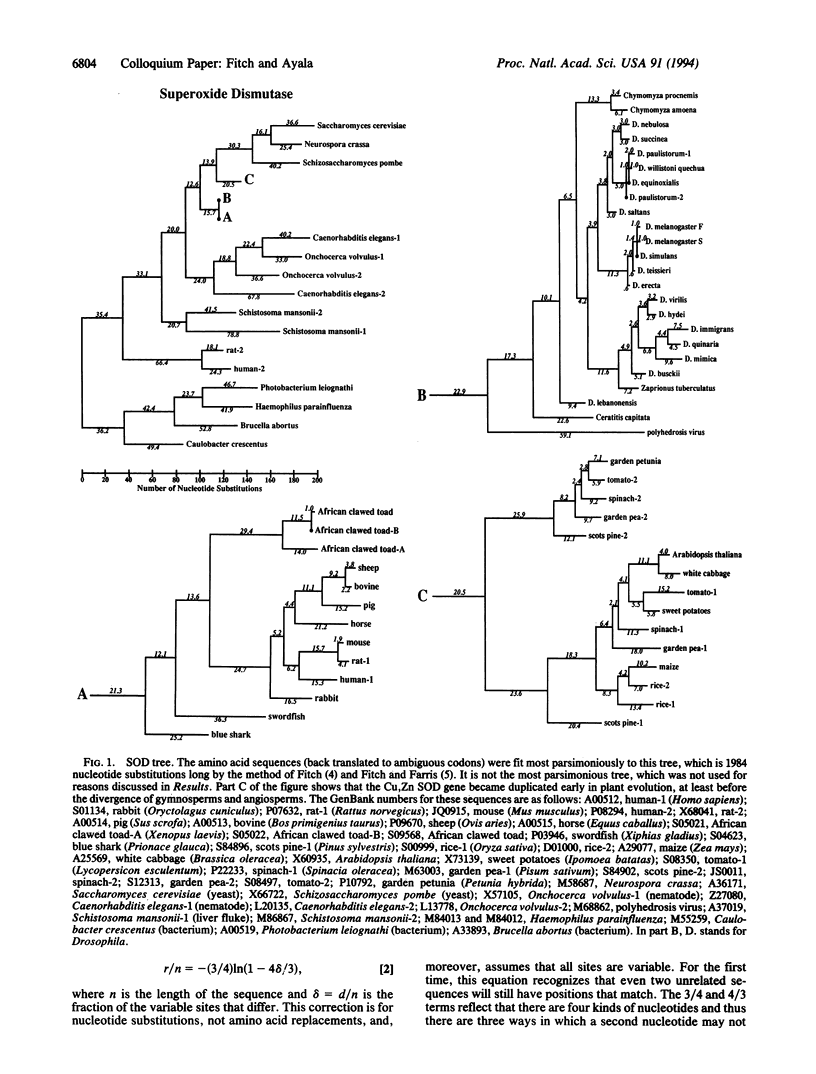
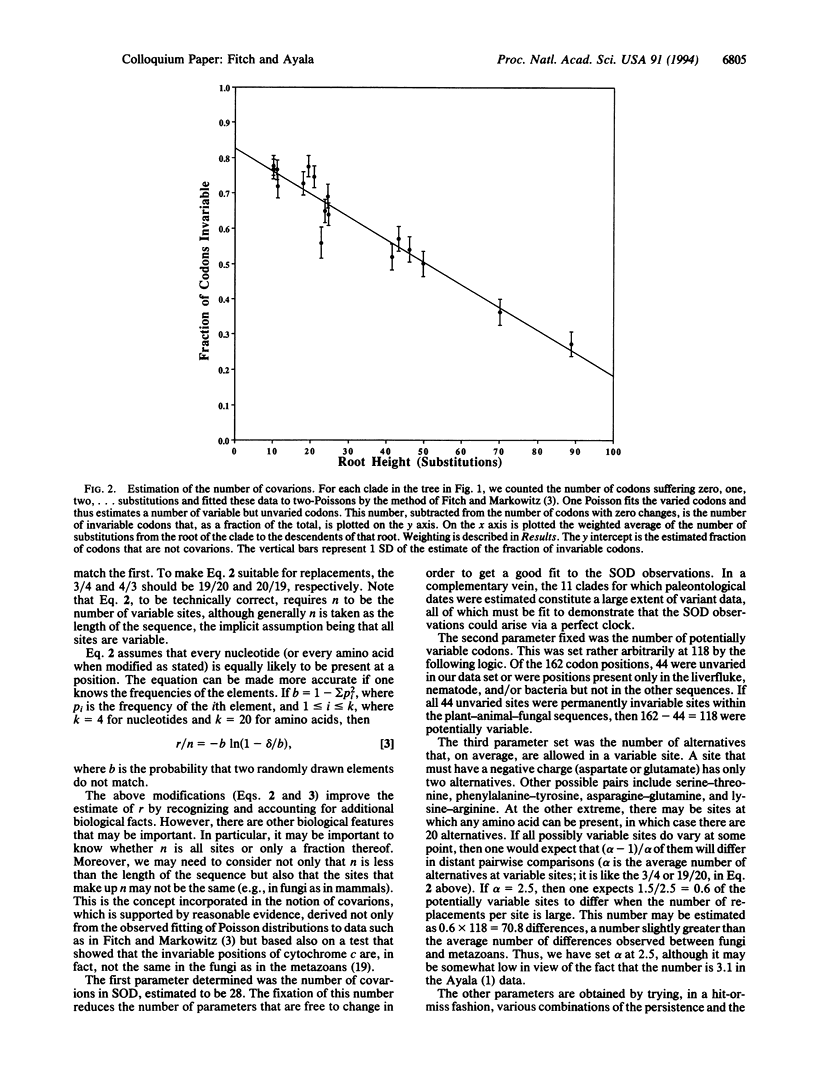
The Cu,Zn superoxide dismutase (SOD) was examined earlier and found to behave in a very unclock-like manner despite (accepted point mutation, or PAM) corrections for multiple replacements per site. Depending upon the time span involved, rates could differ 5-fold. We have sought to determine whether the data might be clock-like if a covarion model were used. We first determined that the number of concomitantly variable codons (covarions) in SOD is 28. With that value fixed we found that the observations for SOD could fit reasonably well a molecular clock if, given 28 covarions, (i) there are approximately six replacements every 10 million years, (ii) the total number of codons is 162, (iii) the number of codons that are permanently invariable across the range of taxa from fungi to mammals is 44, and (iv) the persistence of variability is quite low (0.01). Thus, the inconsistent number of amino acid differences between various pairs of descendent sequences could well be the result of a fairly accurate molecular clock. The general conclusion has two sides: (i) the inference that a given gene is a bad clock may sometimes arise through a failure to take all the relevant biology into account and (ii) one should examine the possibility that different subsets of amino acids are evolving at different rates, because otherwise the assumption of a clock may yield erroneous estimates of divergence times on the basis of the observed number of amino acid differences.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayala F. J. On the virtues and pitfalls of the molecular evolutionary clock. J Hered. 1986 Jul-Aug;77(4):226–235. doi: 10.1093/oxfordjournals.jhered.a110227. [DOI] [PubMed] [Google Scholar]
- Bannister J. V., Parker M. W. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci U S A. 1985 Jan;82(1):149–152. doi: 10.1073/pnas.82.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., Farris J. S. Evolutionary trees with minimum nucleotide replacements from amino acid sequences. J Mol Evol. 1974;3(4):263–278. doi: 10.1007/BF01796042. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 1970 Oct;4(5):579–593. doi: 10.1007/BF00486096. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. The molecular evolution of cytochrome c in eukaryotes. J Mol Evol. 1976 Jun 23;8(1):13–40. doi: 10.1007/BF01738880. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. The nonidentity of invariable positions in the cytochromes c of different species. Biochem Genet. 1971 Jun;5(3):231–241. doi: 10.1007/BF00485794. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Superoxide dismutases. Adv Enzymol Relat Areas Mol Biol. 1986;58:61–97. doi: 10.1002/9780470123041.ch2. [DOI] [PubMed] [Google Scholar]
- Gaut B. S., Muse S. V., Clegg M. T. Relative rates of nucleotide substitution in the chloroplast genome. Mol Phylogenet Evol. 1993 Jun;2(2):89–96. doi: 10.1006/mpev.1993.1009. [DOI] [PubMed] [Google Scholar]
- Getzoff E. D., Tainer J. A., Stempien M. M., Bell G. I., Hallewell R. A. Evolution of CuZn superoxide dismutase and the Greek key beta-barrel structural motif. Proteins. 1989;5(4):322–336. doi: 10.1002/prot.340050408. [DOI] [PubMed] [Google Scholar]
- Jollès J., Prager E. M., Alnemri E. S., Jollès P., Ibrahimi I. M., Wilson A. C. Amino acid sequences of stomach and nonstomach lysozymes of ruminants. J Mol Evol. 1990 Apr;30(4):370–382. doi: 10.1007/BF02101891. [DOI] [PubMed] [Google Scholar]
- Kwiatowski J., Hudson R. R., Ayala F. J. The rate of Cu,Zn superoxide dismutase evolution. Free Radic Res Commun. 1991;12-13 Pt 1:363–370. doi: 10.3109/10715769109145805. [DOI] [PubMed] [Google Scholar]
- Kwiatowski J., Skarecky D., Ayala F. J. Structure and sequence of the Cu,Zn Sod gene in the Mediterranean fruit fly, Ceratitis capitata: intron insertion/deletion and evolution of the gene. Mol Phylogenet Evol. 1992 Mar;1(1):72–82. doi: 10.1016/1055-7903(92)90037-h. [DOI] [PubMed] [Google Scholar]
- Shoemaker J. S., Fitch W. M. Evidence from nuclear sequences that invariable sites should be considered when sequence divergence is calculated. Mol Biol Evol. 1989 May;6(3):270–289. doi: 10.1093/oxfordjournals.molbev.a040550. [DOI] [PubMed] [Google Scholar]
- Steinman H. M. Bacterial superoxide dismutases. Basic Life Sci. 1988;49:641–646. doi: 10.1007/978-1-4684-5568-7_101. [DOI] [PubMed] [Google Scholar]
- Tainer J. A., Getzoff E. D., Beem K. M., Richardson J. S., Richardson D. C. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J Mol Biol. 1982 Sep 15;160(2):181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- Tainer J. A., Getzoff E. D., Richardson J. S., Richardson D. C. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983 Nov 17;306(5940):284–287. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]



