To the Editor
Assignment of structure and function to all genes and gene products (such as proteins) of all organisms represents a major challenge in this postgenomic era. Here we present the I-TASSER Suite (http://zhanglab.ccmb.med.umich.edu/I-TASSER/download/), a stand-alone software package for protein structure and function modeling.
The gap between the number of proteins with known sequences and the number of proteins with experimentally characterized structure and function keeps increasing. One way to narrow this gap is by developing advanced computational approaches for modeling structure and function from sequences, where progress has been recently witnessed in community-wide blind experiments1,2.
I-TASSER3 was originally designed for protein structure modeling by iterative threading assembly simulations. It was recently extended for structure-based function annotation by matching structure predictions with known functional templates4,5. Here we introduce the I-TASSER Suite, a stand-alone package implementing the I-TASSER–based protein structure and function modeling pipelines. Although the on-line I-TASSER server is established and widely used in the community, limited computing resources from a single laboratory have prevented large-scale applications of these algorithms. We expect that the development of the stand-alone package will remove the computing resource barriers and therefore enable benchmarking of new structure and function modeling methods.
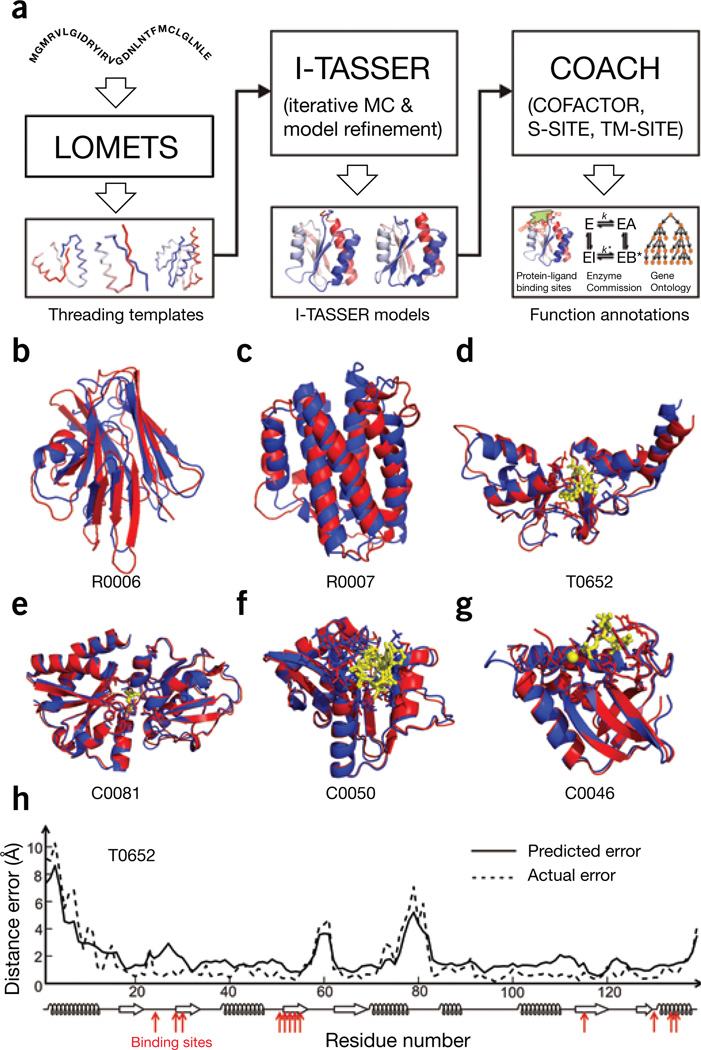
The I-TASSER Suite pipeline consists of four general steps: threading template identification, iterative structure assembly simulation, model selection and refinement, and structure-based function annotation (Fig. 1a). In the first step, the query is threaded by LOMETS through a nonredundant structure library to identify structural templates. LOMETS is a meta-threading method containing eight fold-recognition programs (PPAS, Env-PPAS, wPPAS, dPPAS, dPPAS2, wdPPAS, MUSTER and wMUSTER). These programs are generally based on sequence profile-to-profile alignments, but with various structural features combined (Supplementary Methods). Such variation is important for generating complementary alignments, which increase the coverage of template detections.
Figure 1.
Flow chart and illustrative examples of I-TASSER Suite for protein structure and function modeling. (a) Flowchart of the I-TASSER Suite pipelines that consist of LOMETS-based template identification, I-TASSER–based Monte Carlo (MC) structure assembly simulation and COACH-based function annotation. (b–g) Examples of protein structure and function prediction, where protein structures by I-TASSER and X-ray diffraction experiments are presented as blue and red cartoons, respectively, with ligand-binding residues highlighted by sticks over the cartoons. The ligand structures by COACH and experiments are shown as yellow stick-and-ball forms and sticks, respectively. Modeling parameters for all the targets are listed in Supplementary Table 3. Shown are a hypothetical protein BT_4147 from Bacteroides thetaiotaomicron (CASP ID: R0006; PDB ID: 4e0eA) (b), interleukin-34 protein from Homo sapiens (CASP: R0007; PDB: 4dkcA) (c), magnesium and cobalt efflux protein CorC from Escherichia coli bound to adenosine monophosphate (CASP: T0652; PDB: 4hg0A) (d), nutrient binding protein from Burkholderia cenocepacia bound to methionine (CAMEO: C0081; PDB: 4qhqA) (e), N-acyltransferase from Escherichia coli bound to acetyl–coenzyme A (CAMEO: C0050; PDB: 4qvtA) (f) and thermonuclease from Staphylococcus aureus bound to calcium ion and thymidine-3′,5′-diphosphate (CAMEO: C0046; PDB: 4qf4A) (g). (h) Illustrative example of the residue-level distance error estimations for T0652, where solid lines are predicted errors and dashed lines are actual errors of the I-TASSER model relative to the experimental structure. Bottom, secondary structure prediction; red arrows indicate the ligand-binding sites.
Following the query-to-template alignments, the sequence is divided into threading-aligned and threading-unaligned regions. The topology of full-length models is constructed by reassembling the continuously aligned fragments excised from templates, where the structure of unaligned regions is built from scratch by ab initio folding. The structure folding and reassembly are conducted by replica-exchange Monte Carlo simulations under the guidance of an optimized knowledge-based force field, consisting of three major components: (i) generic statistical potentials, (ii) hydrogen-bonding networks and (iii) threading-based restraints from LOMETS (Supplementary Methods).
The lowest free-energy conformations are identified by structure clustering. A second round of assembly simulation is conducted, starting from the centroid models, to remove steric clashes and refine global topology. Final atomic structure models are constructed from the low-energy conformations by a two-step atomic-level energy minimization approach. The correctness of the global model is assessed by the confidence score, which is based on the significance of threading alignments and the density of structure clustering; the residue-level local quality of the structural models and B factor of the target protein are evaluated by a newly developed method, ResQ, built on the variation of modeling simulations and the uncertainty of homologous alignments through support vector regression training.
For function annotation, the structure models with the highest confidence scores are matched against the BioLiP5 database of ligand-protein interactions to detect homologous function templates. Functional insights on ligand-binding site (LBS), Enzyme Commission (EC) and Gene Ontology (GO) are deduced from the functional templates. We developed three complementary algorithms (COFACTOR, TM-SITE and S-SITE) to enhance function inferences, the consensus of which is derived by COACH4 using support vector machines. Detailed instructions for installation, implementation and result interpretation of the Suite can be found in the Supplementary Methods and Supplementary Tables 1 and 2.
The I-TASSER Suite pipeline was tested in recent community-wide structure and function prediction experiments, including CASP10 (ref. 1) and CAMEO2. Overall, I-TASSER generated the correct fold with a template modeling score (TM-score) >0.5 for 10 out of 36 “New Fold” (NF) targets in the CASP10, which have no homologous templates in the Protein Data Bank (PDB). Of the 110 template-based modeling targets, 92 had a TM-score >0.5, and 89 had the templates drawn closer to the native with an average r.m.s. deviation improvement of 1.05 Å in the same threading-aligned regions6. In CAMEO, COACH generated LBS predictions for 4,271 targets with an average accuracy 0.86, which was 20% higher than that of the second-best method in the experiment.
Here we illustrate I-TASSER Suite–based structure and function modeling using six examples (Fig. 1b–g) from the community-wide blind tests1,2. R0006 and R0007 are two NF targets from CASP10, and I-TASSER constructed models of correct fold with a TM-score of 0.62 for both targets (Fig. 1b,c). An illustration of local quality estimation by ResQ is shown for T0652, which has an average error 0.75 Å compared to the actual deviation of the model from the native (Fig. 1h). The four LBS prediction examples (Fig. 1d–g) are from CASP10 (ref. 1) and CAMEO2; COACH generated ligand models all with a ligand r.m.s. deviation below 2 Å. COACH also correctly assigned the three- and four-digit EC numbers to the enzyme targets C0050 and C0046 (Supplementary Table 3).
In summary, we developed a stand-alone I-TASSER Suite that can be used for off-line protein structure and function prediction. The core programs have been extensively tested in benchmark and blind experiments that demonstrated its advantages over other state-of-the-art methods. In addition, the Suite contains a number of new developments, including six in-house threading algorithms, ResQ for local quality and B factor estimation, and EC and GO prediction algorithms; these developments are essential for making the I-TASSER Suite an efficient stand-alone tool for protein structure and function prediction. The package should enable large-scale applications by the biomedical community.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to many of the I-TASSER Suite users whose feedback helped improve the functionality and usability of the programs. The work was supported in part by the US National Science Foundation Career Award (DBI 0746198) and the US National Institute of General Medical Sciences (GM083107, GM084222).
Footnotes
AUTHOR CONTRIBUTIONS
Y.Z. conceived of the research; J.Y., R.Y., A.R., D.X. and J.P. performed research and created and tested the I-TASSER Suite package; J.Y., R.Y., A.R. and Y.Z. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper (doi:10.1038/nmeth.3213).
References
- 1.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Proteins. 2014;82(suppl. 2):1–6. doi: 10.1002/prot.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas J, et al. Database (Oxford) 2013;2013 doi: 10.1093/database/bat031. bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy A, Kucukural A, Zhang Y. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, Roy A, Zhang Y. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Roy A, Zhang Y. Nucleic Acids Res. 2013;41:D1096–D1103. doi: 10.1093/nar/gks966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y. Proteins. 2014;82(suppl. 2):175–187. doi: 10.1002/prot.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.