Abstract
We have ablated the cellular RNA degradation machinery in differentiated B cells and pluripotent embryonic stem (ES) cells by conditional mutagenesis of core (Exosc3) and nuclear RNase (Exosc10) components of RNA exosome and identified a vast number of long non-coding RNAs (lncRNAs) and enhancer RNAs (eRNAs) with emergent functionality. Unexpectedly, eRNA-expressing regions accumulate R-loop structures upon RNA exosome ablation, thus demonstrating the role of RNA exosome in resolving deleterious DNA/RNA hybrids arising from active enhancers. We have uncovered a distal divergent eRNA-expressing element (lncRNA-CSR) engaged in long-range DNA interactions and regulating IgH 3′ regulatory region super-enhancer function. CRISPR-Cas9 mediated ablation of lncRNA-CSR transcription decreases its chromosomal looping-mediated association with the IgH 3′regulatory region super-enhancer and leads to decreased class switch recombination efficiency. We propose that the RNA exosome protects divergently transcribed lncRNA expressing enhancers, by resolving deleterious transcription-coupled secondary DNA structures, while also regulating long-range super-enhancer chromosomal interactions important for cellular function.
Introduction
Recent advances in RNA biology have revealed a plethora of non-coding RNA transcripts whose identity and functions were previously unknown. It has been postulated that transcription control of coding genes is modulated by non-coding RNAs such as enhancer RNAs (eRNAs) (Kim et al., 2010) and long intergenic non-coding RNAs (lincRNAs) (Rinn and Chang, 2012). Of note, a significant number of non-coding RNAs are characterized as being expressed from regions proximal to the transcription start sites (TSSs) of coding genes. These transcripts include promoter-associated long RNAs (PALRs, >200 bp and bidirectional) (Kapranov et al., 2007), promoter-associated short RNAs (PASRs, 20-100 nt) (Kapranov et al., 2007), TSS-associated RNA (TSS-aRNA, small and divergently transcribed RNA) (Core et al., 2008; Seila et al., 2008), and transcription initiation RNAs (tiRNAs, 18 nt long and located 20 nt downstream of the coding TSS) (Taft et al., 2009). In addition, a large fraction of TSS-proximal transcriptional expenditure is dedicated to the production of unstable non-coding RNAs that are subject to RNA exosome-mediated degradation (PROMPTs, uaRNAs, xTSS-RNAs) (Flynn et al., 2011; Pefanis et al., 2014; Preker et al., 2008). While the characteristics of these new RNA species may overlap, it is abundantly clear that these non-coding RNAs function in the regulation of transcription initiation and transcription elongation by various mechanisms including control of RNA polII pausing and recruitment of chromatin modification factors (Flynn and Chang, 2012; Reyes-Turcu and Grewal, 2012; Shin et al., 2013).
Recently, some of these ncRNAs have been shown to be substrates of the RNA surveillance complex, RNA exosome (Andersson et al., 2014a; Andersson et al., 2014b; Pefanis et al., 2014; Wan et al., 2012). The eukaryotic RNA exosome complex functions in both the nucleus and the cytoplasm. Nuclear exosome is involved in 3′-5′ processing of rRNAs, sn/snoRNAs, degradation of hypomodified tRNAs and cryptic unstable transcripts (CUTs), whereas cytoplasmic exosome is responsible for the degradation of aberrant mRNA species subject to nonsense mediated decay, non-stop decay, or no-go decay (Schmid and Jensen, 2008); (Chlebowski et al., 2013). The eukaryotic exosome complex is comprised of a nine subunit core, consisting of six distinct proteins forming a ‘ring’ and three distinct RNA binding domain containing proteins forming a ‘cap’ structure required for the stabilization of the core structure. Enzymatic activity of the exosome complex is provided through two additional subunits: Rrp44 (Dis3) and Rrp6 (Exosc10) (Houseley et al., 2006; Januszyk and Lima, 2011; Liu et al., 2006; Lorentzen et al., 2008). Rrp6 is a nuclear specific 3′-5′ distributive exoribonuclease (Lykke-Andersen et al., 2009). Although in vitro Rrp6 and Dis3 bind the RNA exosome core (Exo9) independent of each other, Exo9 may interconnect the properties of the two RNase subunits in vivo (Schaeffer et al., 2009; Schaeffer and van Hoof, 2011; Wasmuth and Lima, 2012) so that different types of RNA substrates can be processed/degraded. Crystal structure analysis of an Rrp6-containing yeast RNA exosome complex suggests that Rrp6 may function in regulating the size of the central channel through which RNA traverses prior to degradation (Wasmuth et al., 2014). The true nature of Rrp6 function within the RNA exosome complex, via its distributive RNase activity and/or its contribution to central channel regulation, is incompletely understood. Moreover, mammalian RNA substrates of the RNA exosome complex with or without the Rrp6 component have not been systematically identified. The activity of the RNA exosome in co-transcriptionally degrading RNA plays a critical function in the nucleus, with recent observations in yeast and mammalian cells indicating a role for RNA degradation in early transcription termination (Colin et al., 2014; Hazelbaker et al., 2013; Lemay et al., 2014; Pefanis et al., 2014; Richard and Manley, 2009; Shah et al., 2014; Storb, 2014; Sun, 2013). As such, the role of RNA exosome in chromatin-associated events is a major focus of ongoing research.
In this study, we reveal and analyze the transcriptomes of Exosc3 and Exosc10-ablated ES cells and B cells, and identify a vast number of non-coding RNAs with emergent biological functionality. Strikingly, we find that the RNA exosome regulates the levels of divergently transcribed enhancer RNAs by promoting co-transcriptional silencing, thereby preventing the persistence of detrimental chromatin structures that can lead to genomic instability. Moreover, we provide evidence that RNA exosome substrate divergently transcribed loci may regulate interactions with super-enhancer loci. Thus, our study provides a mode of long-range chromatin regulation not previously described. As an example, we have identified the lncRNA-CSR expressing locus and report its regulation of immunoglobulin heavy chain DNA rearrangements by functionally interacting with the 3′ regulatory region super-enhancer sequence (3′RR).
Results
RNA exosome mutant ES cells and mouse models
To ascertain the role of the RNA exosome complex in the degradation of non-coding RNAs, we have generated mouse conditional alleles of Exosc10 (expressing the distributive nuclease subunit Rrp6) (Figs. S1A, S1B) and Exosc3 (expressing the RNA exosome core subunit Rrp40) (Pefanis et al., 2014). Using these two approaches, inducible RNA exosome deficiency was evaluated in either primary pluripotent embryonic stem cells or differentiated mature B cells. Exosc10 and Exosc3 allele schemes utilize Cre/lox conditional inversion (COIN) methodology to ablate normal gene expression upon exposure of the alleles to Cre recombinase activity (Economides et al., 2013; Pefanis et al., 2014). The salient feature of this approach, as utilized here, is the inversion of one or more endogenous coding exons resulting in the simultaneous “activation” of a fluorescent reporter terminal exon within the same locus (Figure 1A). Exosc10COIN/WT mice were crossed with mice heterozygous for a null allele of Exosc10 (Exosc10LacZ/WT) to derive ES cells and B cells of the genotype Exosc10COIN/LacZ. Similarly, we have generated Exosc3COIN/COIN ES cells and B cells (Pefanis et al., 2014). Both Exosc10COIN/LacZ and Exosc3COIN/COIN cells also contain the inducible ROSA26CreERt2 allele allowing for rapid ablation of RNA exosome activity upon tamoxifen treatment. When B cells from Exosc10COIN/LacZ mice were treated with 4-hydroxytamoxifen (4-OHT) ex vivo, inversion of the Exosc10COIN allele was observed in more than 90% of the cells (Figure 1B). Quantitative RT-PCR assays performed on total cellular RNA demonstrated nearly complete loss of Exosc10 mRNA in 4-OHT treated Exosc10COIN/LacZ B cells (Figure 1C). Western blotting of protein extracts from Exosc10COIN/LacZ B cells and ES cells demonstrated severe loss of Rrp6 protein following 4-OHT, indicating robust ablation of Exosc10 expression (Figure 1D). The RNA exosome previously has been implicated in catalyzing class switch recombination (CSR) in B cells by supporting the activity of activation-induced cytidine deaminase (AID) (Basu et al., 2011). Consistent with these observations, Exosc10 deficient B cells display reduced CSR efficiency as compared to wild type littermate control B cells (Figure S1C) despite comparable expression of AID (Figure S1D). Finally, RNA-seq analysis of Exosc10COIN/LACZ B cells and ES cells confirmed loss of Exosc10 transcripts in both cell types (Figure S1E). Similarly, and consistent with previously published characterization of Exosc3 ablation in Exosc3COIN/COIN B cells, RNA-seq analysis demonstrated a clear loss of Exosc3 transcripts in both Exosc3COIN/COIN B cells and ES cells (Figure S1F).
Figure 1. Generation of RNA exosome mutant ES cells and transcriptome analysis.
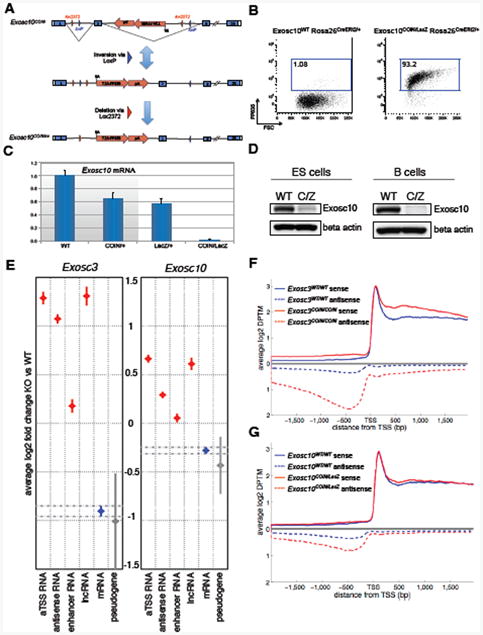
(A) Exosc10COIN allele and conversion to Exosc10COINinv. Cre-mediated inversion of loxP pair (blue triangles) and subsequent deletion via lox2372 pair (red triangles). FP635-expressing terminal exon represented by red arrow. SA, splice acceptor. (B) Induction of fluorescent reporter FP635 in Exosc10COIN/LacZ ROSA26CreERt2/+ B cells following 4-OHT treatment. (C) qRT-PCR analysis of Exosc10 mRNA expression in 4-OHT treated, LPS+IL-4-stimulated B cells. Indicated Exosc10 genotypes on ROSA26CreERt2/+ background. Expression levels normalized to cyclophilin A (Ppia) and plotted relative to Exosc10WT/WT. Splenic B cells were isolated and treated with 4-OHT for 24 hours and the cells were then washed. Total cellular RNA was isolated after 72 hours of B cell culture. Three technical replicates, error bars represent s.d. (D) Western blot detection of Rrp6 (Exosc10) from Exosc10WT/WT (WT) and Exosc10COIN/LacZ (C/Z) protein extracts obtained from 4-OHT treated ES cells and B cells. Indicated Exosc10 genotypes on ROSA26CreERt2/+ background. (E) Genome-wide differential expression level analysis of RNA subsets in Exosc3 (left) and Exosc10 (right) ablated mouse ES cells relative to wild-type littermate matched ES cells. The error bars represent confidence interval of mean value estimated by an improved version of the Tukey-Kramer method of different types of RNAs analyzed (see extended methods). (F, G) Genome-wide transcription start site proximal expression profile in Exosc3 (F) and Exosc10 (G) ablated mouse ES cells. Sense and antisense transcript levels 2 kb flanking the TSS of annotated coding transcripts are indicated. ES cells were treated with 4-OHT for 24 hours and then further cultured for an additional 48 hours before total RNA isolation. See Figure S1 and Supplementary tables 1,2, 3 and 4.
Transcriptome of RNA exosome mutant ES cells and B cells
We assembled the transcriptomes of littermate pairs of wild type control and Exosc10COIN/LacZ or Exosc3COIN/COIN B cells and ES cells using next-generation RNA sequencing technology. The bioinformatics pipeline used for transcriptome reconstitution is outlined in Figure S2A and described in further detail in Extended Methods. We find that in the exotomes (exosome deficient transcriptome) of Exosc3COIN/COIN (Figure 1E, left panel) and Exosc10COIN/LacZ ES cells (Figure 1E, right panel), relative levels of lncRNAs, antisense RNAs, and eRNAs are significantly increased genome-wide compared to wild type control ES cell transcriptomes. Comparing relative transcript accumulations of lncRNAs, antisense RNAs, and eRNAs indicates these non-coding RNA subsets experience greater stabilization within the Exosc3COIN/COIN exotome in comparison to the Exosc10COIN/LacZ exotome genome-wide. Transcription start site antisense divergent RNAs are well known substrates of the RNA exosome complex (Pefanis et al., 2014; Preker et al., 2008; Seila et al., 2008; Seila et al., 2009). Consistent with expectations, TSS-associated antisense RNAs are markedly stabilized within the Exosc3COIN/COIN ES cell transcriptome (Figure 1F). A list of antisense RNA in the body of the genes and around the genic TSS from B cell exotome and ES cell exotome are provided in Supplementary Tables 1 and 2, respectively. Relative to Exosc3-deficient cells, TSS-associated antisense transcripts are moderately stabilized within the Exosc10COIN/LacZ ES cell transcriptome (Figure 1G). Collectively, these results point toward a role for Exosc10 in the degradation of a subset of RNA exosome targeted lncRNAs (presumably fully represented via Exosc3 ablation).
RNA exosome substrate long non-coding RNA (x-lncRNA)
Previously, it has been shown that enhancers express bidirectional, divergently transcribed, RNA exosome sensitive, capped non-coding RNAs in human cell lines and primary mouse B cells (Andersson et al., 2014a; Andersson et al., 2014b; Pefanis et al., 2014; Wan et al., 2012). Taking clues from these studies, we evaluated whether our RNA exosome mutant mouse models could be utilized for identifying eRNAs in pluripotent ES or lineage-committed matured B cells. Following the analysis pipeline described in Extended Methods, we observed a subset of long non-coding RNAs were strong substrates of RNA exosome. We describe such transcripts here as x-lncRNA. As shown via heatmap representation, both in Exosc3WT/WT/Exosc3COIN/COIN and in Exosc10WT/WT/Exosc10COIN/LacZ RNA-seq analysis pairs, multiple x-lncRNA loci are revealed in RNA exosome deficient ES cells while weakly expressed in counterpart wild type control cells (Figure 2A) (details of expression and genome coordinates of these transcripts supplied in Supplementary Table 3). Next, we performed comparative expression analysis between Exosc3 and Exosc10 substrate x-lncRNAs and found that a significant number, although not all, Exosc3 x-lncRNAs also classify as Exosc10 x-lncRNAs (Figure 2B). Specifically, of a total of 2,729 Exosc3 x-lncRNAs in ES cells, 1,506 also fell within the cutoff for Exosc10 x-lncRNAs (Figure 2C, Figure S2B and Figure S2C; details in Supplementary Table 3). Surprisingly, only 59% of Exosc3 x-lncRNAs described here have been reported previously (Figures 2E and S2D). In fact, 236 of these identified x-lncRNAs are positioned close to enhancer sequences and thus may serve as RNA exosome target “x-eRNAs”. Moreover, the accumulation of x-lncRNAs mostly maps within 5-50 kb from the TSS of known coding genes, making it possible that these lncRNAs regulate gene expression of distal genes via long-range chromatin interactions (Figure 2D). As indicated earlier, there are substantial numbers of lncRNAs that are quite unstably expressed in wild type steady state ES cells, but their identity cannot be confidently evaluated due to weak detection. However, RNA-seq analysis of Exosc3COIN/COIN and/or Exosc10COIN/LacZ cells provides a methodology for the detection and characterization of highly unstable lncRNA species. One such example is provided as the sense/antisense x-lncRNAs in the Hoxa1 locus (Figure 2F). There are multiple species of antisense x-lncRNAs that are expressed in the Hoxa1 locus (Figure 2F), whose detection is amplified in the Exosc3COIN/COIN or Exosc10COIN/LacZ exotomes.
Figure 2. Identification and characterization of RNA exosome targeted lncRNAs (x-lncRNAs) in ES cells.
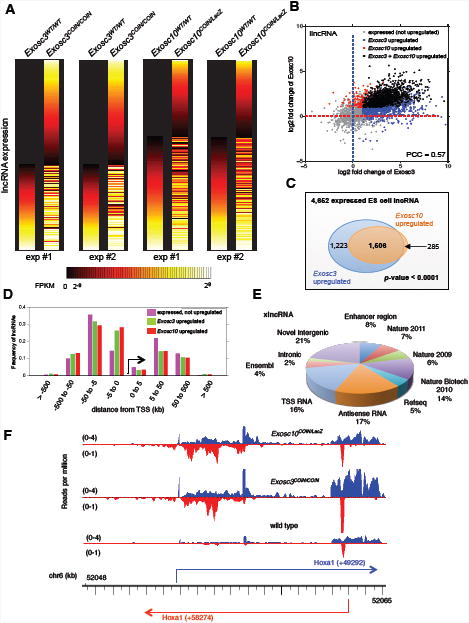
(A) Heat map of lncRNAs expressed in Exosc3WT/WT/Exosc3COIN/COIN and Exosc10WT/WT/Exosc10COIN/LacZ genotype pairs. Horizontal lines represent different lncRNAs, which were ranked by their expression level in matched wild type control. (B) Distribution of lncRNAs stabilized in the Exosc3-exotome (blue), Exosc10-exotome (red) and both Exosc3, Exosc10 exotomes (black). (C) Venn diagram demonstrating the distribution of overlapping lncRNAs in Exosc3 (blue) and Exosc10 (orange) exotomes. (D) Distribution of x-lncRNA TSS distances from closest neighboring coding gene TSS genome-wide. (E) The pie chart represents distribution of previously reported and newly identified lncRNAs from this study (Guttman et al., 2009; Guttman et al., 2011; Guttman et al., 2010). Each category represents RNAs unique to that category and non-overlapping with previous categories, with the initial category designated as “Enhancer region lncRNAs” and proceeding clockwise. (F) Expression of sense and antisense lncRNA expression profile at the Hoxa1 locus identified from Exosc3 and Exosc10 ablated ES cells. Based on RNA-seq read distribution, multiple lncRNAs are expressed in the sense and antisense directions. See Figure S2 and Supplementary table 3.
RNA exosome substrate enhancer RNA (x-eRNA)
Some enhancer RNAs (x-eRNAs) are predicted to form a subset of x-lncRNAs. Thus we analyzed eRNA stability and identity in both Exosc3 and Exosc10 exotomes and found overlapping as well as distinct requirements for these two RNA exosome subunits (Figure 3A). All eRNAs that could be identified from ES cells are listed in Supplementary Table 4. Of a total of 891 Exosc3 x-eRNAs in ES cells, a subset of 423 displayed a significant enrichment with Exosc10 loss (Figure 3B). In addition, 86% of the Exosc3 x-eRNAs reported here are previously unrecognized. Of the 37 Exosc3 x-eRNAs previously reported in VISTA, a subset of 18 were upregulated following Exosc10 depletion (not shown). In B cell exotomes, the degree of overlap between Exosc3 and Exosc10 x-eRNAs is reduced in comparison to ES cell exotomes (Figure 3C). Of the 870 identified B cell Exosc3 x-eRNAs, only 62 were Exosc10 targets (Figure 3D). Representative Exosc3 x-eRNAs within the Cd83 locus were significantly upregulated in Exosc3COIN/COIN B cells and modestly increased in Exosc10COIN/LacZ B cells (Figure 3E).
Figure 3. A subset of enhancer RNAs (x-eRNAs) are RNA exosome targets in ES cells and B cells.

(A) Distribution of upregulated ES cell x-eRNAs from Exosc3 and Exosc10 exotomes. Pearson correlation is indicated. (B) Overlap of identified x-eRNAs stabilized from degradation in ES cell Exosc3 (blue circle) and Exosc10 (orange circle) exotomes. (C) Distribution of upregulated B cell x-eRNAs from Exosc3 and Exosc10 exotomes. Pearson correlation is indicated. (D) Overlap of identified x-eRNAs stabilized from degradation in B cell Exosc3 (blue circle) and Exosc10 (orange circle) exotomes. (E) x-eRNA stabilization at annotated enhancers within the Cd83 locus in Exosc3WT/WT and Exosc3COIN/COIN B cells. Sense and antisense RNA indicated in blue and red, respectively. (F) Expression levels of x-lncRNAs in B cells identified from Exosc3WT/WT and Exosc3COIN/COIN transcriptomes. (G, H) Sequence conservation plot of coding genes (red), identified x-lncRNAs (blue) and random control (green) from B cells (G) and ES cells (H). To measure how conserved are the lncRNAs we have identified, we compared the lncRNAs with human genes (genome version hg19) by LiftOver tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). The percentage of genes that are conserved between human and mouse are shown according to different cutoffs. Same number of coding genes/random genomic regions with similar length are generated as controls. For each group of genes, the percentage of genes conserved between human and mouse (y-axis) is calculated based on UCSC liftover tool with given cutoff (x-axis). See Figure S3 and Supplementary table 4.
x-lncRNA (or x-eRNA) expression is detectable in wild type cells, although significantly stabilized in Exosc3COIN/COIN cells (Figure 3F). Moreover, in both B cells (Figure 3G) and ES cells (Figure 3H), the degree of conservation for x-lncRNAs genome-wide is greater than a random control set of sequences, albeit lower in amount than protein coding DNA sequences in the mouse genome. To determine the conservation of lncRNAs that we have identified in this study, we compared x-lncRNAs with human genes (genome version hg19) using the LiftOver tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). The percentage of genes that are conserved between human and mouse are shown distributed with different cutoffs. In Figures 3G and 3H, equivalent numbers of coding genes/random genomic regions with similar length were generated as controls. For each group of genes, the percentage that are conserved between human and mouse (y-axis) are calculated based on UCSC liftover tool with given cutoff (x-axis) (details in Extended Methods section). Taking these observations into account, it is likely that many x-lncRNAs (and their subset x-eRNAs) are biologically functional. The dependency of the RNA exosome complex on Rrp6 (Exosc10) to degrade various subsets of ncRNAs may vary based on the type of ncRNA and/or the cell type. For example, xTSS-RNAs (one type of antisense RNA) in B cells (Figure S3A) or in ES cells (Figure S3B) have markedly increased representation in Exosc3 exotomes in comparison to Exosc10 exotomes. In contrast, antisense RNA levels arising from gene bodies were similar between Exosc3 and Exosc10 B cell (Figure S3C) and ES cell exotomes (Figure S3D). Finally, to ascertain whether any major pathway was effected in the cells following RNA exosome activity depletion at the time points of RNA extraction, we performed gene set enrichment analysis (GSEA) in Exosc3WT/WT and Exosc3COIN/COIN ES cells. As would be expected, there were some perturbations in gene expression profiles in Exosc3COIN/COIN ES cells, with gene sets related to organic acid transport and carboxylic acid transport (for details for GSEA of upregulated and downregulated pathways in Exosc3COIN/COIN and Exosc3WT cells, see Supp. Tables 5 and 6, respectively.)
RNA exosome mediated degradation of eRNAs protects cells from genomic instability by preventing formation of DNA/RNA hybrids and by promoting heterochromatin marks at divergent enhancers
Regions of the B cell genome beyond the Ig loci are susceptible to hypermutation due to AID activity and may then undergo chromosomal translocations involving Ig genes. Genomic loci susceptible to AID-induced chromosomal translocation break-points may also accumulate x-eRNA reads in Exosc3COIN/COIN B cells in comparison to Exosc3WT/WT B cells. We observed that some IgH translocation partners identified through translocation capture techniques show x-eRNA expressing divergently transcribed enhancers as recurrent translocation hotspots. These include the Birc3 enhancer (Figure S4C), as well as the Ncoa3 enhancer (Figure S4D). These enhancer regions display overlapping sense and antisense RNA exosome substrate transcripts. Genomic overlaps between translocation breakpoints and x-eRNA expressing regions provide evidence that RNA exosome regulated enhancers in the B cell genome could be sensitive to DNA double strand breaks resulting from AID, a physiologically expressed DNA mutator. Indeed, recently it has been ascertained that Rrp6 (Exosc10) plays a role in DNA double strand break repair by affecting recruitment of ssDNA binding protein RPA (Manfrini et al., 2015; Marin-Vicente et al., 2015). In fact, multiple studies indicate that AID-induced chromosomal translocation sites in the B cell genome harbor RPA for DNA double strand break repair (Qian et al., 2014; Yamane et al., 2013).
Antisense RNAs that form co-transcriptional RNA/DNA hybrid structures called R-loops can initiate premature transcription termination and be a source of genomic instability (Bhatia et al., 2014; Pefanis et al., 2014; Skourti-Stathaki et al., 2014). In addition, such antisense RNAs can be substrates of the Dicer/Argonaute complex (Skourti-Stathaki et al., 2014) and RNA exosome (Pefanis et al., 2014). To investigate AID-independent DNA break formation in ES cells, we looked whether x-eRNA expressing regions are susceptible to genomic instability in RNA exosome deficient cells due to formation of persistent R-loop structures. ES cells were irradiated with ionizing radiation (20 Gy) and allowed to recover over a period of 30 minutes. We evaluated 3 x-eRNA expressing loci neighboring Klf6, Bcl6 and Cd38. x-eRNA arising from these enhancer loci display divergent transcription and are sensitive to Exosc3 function (Figures S4E-G). We evaluated the accumulation of DNA double strand break associated γ-H2AX foci at divergent x-eRNA expressing regions in Exosc3COIN/COIN and Exosc10COIN/LacZ cells. γ-H2AX accumulation at x-eRNA expressing sequences was significantly enhanced in both Exosc3 and Exosc10 ablated ES cell lines, implying a greater propensity for these sequences to undergo DNA double strand breaks in the absence of functional RNA exosome complex (Figure 4A). Using immunoprecipitation assays with anti-DNA/RNA hybrid S9.6 antibody, we found that in Exosc3COIN/COIN and Exosc10COIN/LacZ cells, x-eRNA expressing regions are significantly enriched for RNase H sensitive DNA/RNA hybrid structures (Figure 4B). In contrast, an enhancer region in the ES cell genome that does not demonstrate divergent transcription was not enriched for γ-H2AX foci or R-loops (Figure S4I and Figure S4J, respectively). These observations point towards the possibility that RNA exosome mutant ES cells are more prone to genomic instability insults at divergently transcribed enhancer sequences. Telomeric FISH assays performed on IR treated Exosc3COIN/COIN cells revealed a significantly greater frequency of chromosomal alteration in comparison to control Exosc3WT/WT cells (Figs. S4A and S4B). Taken together, RNA exosome mediated degradation of RNA in DNA/RNA hybrids at divergently transcribed enhancer sequences might serve as a mechanism for the maintenance of genomic integrity in mammalian cells.
Figure 4. Genomic instability in RNA exosome deficient ES cells along with accumulation of DNA/RNA hybrids and loss of chromatin silencing markers H3K9me2 and HP1γ at x-eRNA expressing sequences.

(A) γH2AX immunoprecipitation for DNA double strand breaks at enhancer sequences resident in the Bcl6 (left), Cd38 (middle), and Klf6 (right) loci in wild-type, Exosc10COIN/LacZ, and Exosc3COIN/COIN ES cells following ionizing radiation treatment. (B) DNA/RNA hybrid immunoprecipitation at Ncoa3 (left), Cd38 (middle), and Klf6 (right) enhancers in wild-type, Exosc10COIN/LacZ, and Exosc3COIN/COIN ES cells. (C) Immunoprecipitation for heterochromatin marker H3K9me2 at enhancer sequences resident in the Bcl6 (left), Cd38 (middle), and Klf6 (right) loci in wild-type, Exosc10COIN/LacZ, and Exosc3COIN/COIN ES cells. (D) Immunoprecipitation for heterochromatin marker HP1γ at enhancer sequences resident in the Bcl6 (left), Cd38 (middle), and Klf6 (right) loci in wild-type, Exosc10COIN/LacZ, and Exosc3COIN/COIN ES cells. Each plot is a representation of three independent experiments performed (*P<0.05; **P<0.01 by t-test). See Figure S4.
The established roles of H3K9me2 and HP1γ chromatin marks in the cellular processes of chromatin condensation and transcriptional repression have recently been identified to appear at sites of transcription termination of antisense non-coding RNAs (Skourti-Stathaki et al., 2014). Analysis of H3K9me2 (Figure 4C) and HP1γ (Figure 4D) occupancy revealed decreased levels of these repressive chromatin marks at x-eRNA expressing loci in Exosc3COIN/COIN and Exosc10COIN/LacZ cells. Thus, RNA exosome-mediated regulation of x-eRNAs levels in cells could occur via two distinct mechanisms, namely via post-transcriptional RNA degradation or possibly through repression of RNA synthesis by promoting early transcription termination. In summary, we provide evidence that x-eRNA expressing DNA sequences generate potentially deleterious DNA/RNA hybrids that might contribute to genomic instability.
x-eRNAs have biological function at super-enhancer sequences
Since enhancers are well known modulators of gene expression, we evaluated x-eRNAs that arose from our analyses for functionality in controlling gene expression. We observed two peaks of sense and antisense transcription at regions upstream of the Tgfbr2 gene (Figure S5A). Using CRISPR-Cas9 mediated deletion of these lncRNA expressing potential enhancer sequences in B cell line CH12F3, we observed a substantial decrease in the expression of Tgfbr2 mRNA by individually knocking out either of the two Tgfbr2 x-eRNA elements (Figure S5B).
We considered whether super-enhancer sequences, which are characterized by high density of individual enhancers and high regional enrichment for active chromatin marks, can generate RNA exosome substrate super-enhancer RNAs (x-seRNAs). As super-enhancer coordinates and functions can be identified in B cells using previously published bioinformatic pipelines (Loven et al., 2013; Meng et al., 2014), we evaluated the expression of x-seRNAs in these cells. Our analysis revealed a significant enrichment of x-seRNAs in both Exosc3 and Exosc10 exotomes (Figure 5A). Relative to Exosc3COIN/COIN cells, Exosc10-deficient cells retained significantly greater x-seRNA degradation activity, potentially due to RNA exosome complexes in these cells possessing the ability to utilize either the Exosc10-encoded Rrp6 or Dis3-encoded Rrp44 nuclease subunit in the degradation of x-seRNAs. We hypothesized that synthesis of antisense RNAs (either xTSS-RNA or those in the body of a gene) may functionally engage with super-enhancer elements to form higher-order chromosomal structures that may enable their local expression control. We sought such examples, i.e., super-enhancer sequences neighboring RNA exosome sensitive antisense RNA (x-asRNA) expressing genes, and illustrate two examples here. First, a super-enhancer (Chr 10SE)-enhancer (overlapping the Btg1 gene) pair separated by a distance of 232 kb from each other was found to express both x-seRNAs and xTSS-RNAs, respectively (Figure 5B). Accordingly, both the Chr 10SE x-seRNA and Btg1 xTSS-RNA are contained within the Exosc3 and Exosc10 exotomes. As a second example, we identified a Chr1 SE that closely paired with an x-asRNA arising within the Btg2 locus. In this case the separation of the SE and Btg2 was a mere 4 kb, with both the x-seRNA and the x-asRNA being part of the Exosc3 and Exosc10 exotomes (Figure 5C). A statistical analysis of the proximity between xTSS-RNA expressing genes and x-seRNA expressing super-enhancer sequences illustrates a remarkable correlation that genes less than 310 kb from a SE are statistically far more likely to express antisense xTSS-RNAs (p < 0.0001; Figure 5D). The 310 kb distance between xTSS-RNA and x-seRNA expressing sequences was set based on a genome-wide statistical analysis of distance between these elements in B cells. Beyond a distance of 310 kb from a super-enhancer, there is a consistent decrease in correlation of x-TSS-RNA expression (Figure S5C; details in extended methods section). These observations at individual loci such as Btg1 and Btg2, along with genome-wide analyses support a model whereby super-enhancer and counterpart gene interactions are controlled by expression and/or processing of RNA exosome substrate non-coding RNAs.
Figure 5. Super-enhancer sequences and neighboring conventional enhancers or coding genes express RNA exosome substrate antisense RNAs.
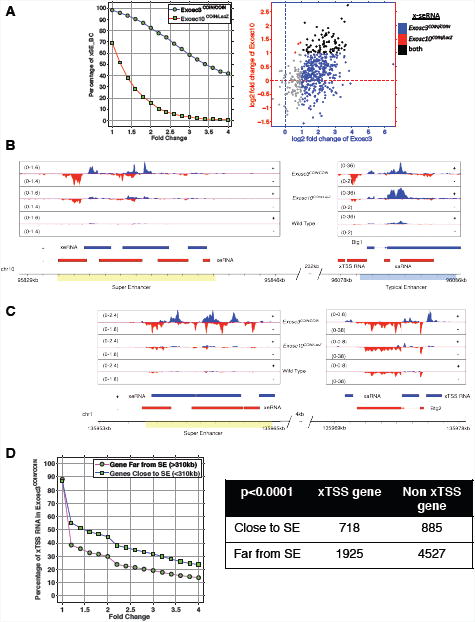
(A) Left panel: expression of super-enhancer RNAs at 200 annotated super-enhancers within Exosc3 (Exosc3COIN/COIN) and Exosc10 (Exosc10COIN/LacZ) exotomes. X-axis indicates the cutoff of fold change, and y-axis indicates the fraction of super-enhancers with higher expression (given x-axis) compared with wild type. Right panel: expression of Exosc3 (blue), Exosc10 (red), and overlapping (black) x-seRNAs in B cells. (B) A super-enhancer resident in chromosome 10 (SEChr10) and neighboring conventional enhancer element resident at the Btg1 locus express sense (blue) and antisense (red) x-eRNAs. (C) A super-enhancer resident in chromosome 1 (SEChr1) and neighboring a conventional enhancer element resident at the Btg2 locus express sense (blue) and antisense (red) x-eRNAs. (D) Genome-wide correlation between proximity of super-enhancer location and antisense x-TSS-RNA expression at neighboring genes in B cells. See Figure S5.
Molecular evidence that antisense RNA/super-enhancer RNA expression regulates long range IgH locus recombination
A pair of divergently transcribed x-lncRNAs were found to be expressed at a 2.6 Mb distal region downstream of the 3′ regulatory region (3′RR) of the IgH locus. Both members of this x-lncRNA pair--named here as B930059L03Rik and lncRNA-CSR--were significantly more stable in Exosc3COIN/COIN and Exosc10COIN/LacZ B cells, but also detectably expressed in wild type control B cells (Figure 6C). A detailed map of this lncRNA-locus is shown in Figure S6A; no transcription factor binding sites were computationally predicted to overlap this region (Figure S6A). We proceeded to delete the lncRNA-CSR locus in CH12F3 cells using CRISPR-Cas9 and demonstrated complete loss of expression of lncRNA-CSR (Figure 6A). We found that lncRNA-CSR homozygous deleted CH12F3 cells expressed similar levels of the IgH locus recombination catalyst enzyme AID (Figure S5D). When lncRNA-CSR deficient CH12F3 cells were assayed for CSR efficiency they showed substantial defect for isotype switching to IgA (Figure 6B and Figure S5E). Chromosome conformation capture (using lncRNA-CSR 3C primer Figure S6A and HS4 region primer Figure S6B) was performed to assess the interaction frequency of the lncRNA-CSR locus with regions of the IgH locus 3′RR super-enhancer (for details see supplementary methods.) Remarkably, we observed the HS4 region of the IgH locus 3′RR interacts with the lncRNA-CSR locus. Deletion of the lncRNA-CSR sequence substantially decreased the interaction frequency between the deleted locus and the 3′RR HS4 region, whereas the canonical 3′RR and Eμ interaction remained similar (Figure 6D). As can be seen from RNA-seq data, the antisense super-enhancer RNA peak corresponding to 3′RR HS4 (strongly visible in the Exosc3COIN/COIN track) also corresponds to the region of interaction with lncRNA-CSR based on DNA sequencing results from 3C assays (Figure 6C, bottom panel). The 3′RR HS4 region expresses multiple distinct x-seRNAs as can be seen from the non-overlapping RNA-seq reads from the Exosc3COIN/COIN transcriptome (Figure S6C). It is likely that the lncRNA-CSR element functions as a distal enhancer-like sequence and promotes the CSR stimulating activity of the 3′RR super-enhancer via the interaction of the antisense lncRNA-CSR and the HS4 x-seRNA expressing DNA regions. Thus, we provide functional evidence that RNA exosome substrate antisense RNA expressing elements can interact with super-enhancer RNA expressing regions to catalyze genomic rearrangement and organization.
Figure 6. Identification of divergently expressed lncRNA-CSR at an enhancer region controlling IgH recombination in B cells.

(A) lncRNA-CSR expression in parental and lncRNA-CSR-/- CH12F3 cells following CRISPR/Cas9 mediated deletion. (B) IgA class switch recombination efficiency of lncRNA-CSR deleted CH12F3 cells obtained from 18 independent lines of lncRNA-CSR-/- CH12F3 cells. (C) Upper panel: IGV expression profile of the lncRNA-CSR divergently transcribed enhancer locus that is stabilized in Exosc3COIN/COIN and Exosc10COIN/LacZ B cells. Middle panel: sense (blue) and antisense (red) tracks for 3′ regulatory region super-enhancer transcription in Exosc3COIN/COIN and Exosc10COIN/LacZ B cells. Bottom panel: DNA sequencing of the 3C-derived joint PCR product of the super-enhancer IgH 3′RR HS4 sequence with the lncRNA-CSR enhancer sequence. The SacI site is contributed from the lncRNA-CSR locus and the HS4 locus and demonstrates the joining of the two pieces of DNA in the 3C assay (D) Chromosomal conformation capture (3C) assay determination of relative interaction frequency of Eμ with 3′RR HS1.2 and lncRNA-CSR locus with 3′RR HS4. **P<0.01 and ***P<0.001 by t-test. See Figures S5 and S6.
We wanted to investigate the molecular mechanism of lncRNA-CSR transcription on the activity of 3′RR function in promoting class switch recombination (CSR). The 3′RR is known to regulate transcription of switch region germ line transcripts (GLTs) (Birshtein, 2014; Pinaud et al., 2011). IgSμ transcript levels were comparable between parental (WT) and ΔlncRNA-CSR CH12F3 clones (Figure 7A). On the other hand, we observed a significant suppression of IgA germline transcripts (IgSα) in the ΔlncRNA-CSR CH12F3 clones (Figure 7B). These observations point toward a role for lncRNA-CSR/HS4 interaction in regulating the transcription of downstream switch sequence transcripts at the Sα locus. Whether this transcription regulation is similarly enforced at other switch regions can only be determined by generating mouse models deleted of the lncRNA-CSR locus. There is accumulation of long-range DNA rearrangements between the IgH (Klein et al., 2011) and lncRNA-CSR loci in B cells that overexpress AID (Figure S7A). Deletion of the lncRNA-CSR locus (Figure S6A) is presumed to disrupt its divergent transcription. We find, at least in these cells where the transcription divergence is lost, H3K9me2 levels are decreased, raising the possibility that some level of heterochromatinization of these divergent sequences is important for their molecular activity to promote 3′RR interaction (Figure 7D). These observations are consisent with enhancer hetermochromatinization regulation in ES cells by RNA exosome, as shown in Figure 4C and 4D. Finally, we evaluated the effect on 3′RR HS4-lncRNA-CSR interaction in B cells deficient in RNA exosome activity (Exosc3COIN/COIN). We find that in the absence of Exosc3, B cells have increased HS4-lncRNA-CSR interaction frequency relative to wild type B cells (Figure 7C). However, increased interaction is not sufficient to promote CSR since RNA exosome also regulates AID's DNA deamination activity in B cells (Basu et al., 2011; Pefanis et al., 2014; Sun et al., 2013).
Figure 7. Mechanism of lncRNA-CSR mediated suppression of 3′RR superenhancer function.

(A) Germline transcripts at I μ in parental cells and two separate clones of lncRNA-cSR knockouts. Three independent sets of RNA were isolated for each cell line and assayed by quantitative RT-PCR. (B) Germline transcripts at I α in parental cells and two separate clones of lncRNA-cSR knockouts. Three independent sets of RNA were isolated for each cell line and assayed by quantitative RT-PCR. **P<0.01 and ***P<0.001 by t-test (C) Chromosomal conformation capture performed on Exosc3WT/WT and Exosc3COIN/COIN B cells that were stimulated for CSR with LPS+IL4 for 24 hrs. The frequency of 3′RRHS4 interaction with the lncRNA-CSR locus was measured by normalizing to an interaction downstream of the Calr gene locus. The experiment is a representation of three independently performed assays. **P<0.01 by t-test. (D) The accumulation of H3K9me2 marks (normalized to the presence of H3) in parental (CH12F3 cells), a random CRISPR/Cas9 mutated ΔPim1 (xTSS-RNA mutated) cell and ΔlncRNA-CSR. Experiment is a representation of three independently performed assays. The ChIP assay primer pairs for various regions surrounding the lncRNA-CSR locus are shown in the top panel *P<0.05 and **P<0.01 by t-test. (E) A model of RNA exosome substrate x-seRNA expressing superenhancer interaction with the divergently transcribing promoter of another enhancer or protein coding gene. We postulate that the activity of RNA exosome to process the x-seRNA and x-eRNAs has a role in titrating the proper level of interaction between regulatory elements that ultimately control gene expression. See Figure S7
Discussion
We envision the identification of vast numbers of RNA exosome targeted ncRNAs will enable the elucidation of their physiological roles in various developmental and gene expression regulatory pathways. Although many lncRNAs and their functions have been described (Bonasio and Shiekhattar, 2014; Rinn and Chang, 2012; Sauvageau et al., 2013), our study identifies a subclass targeted by RNA exosome (x-lncRNA), many of which have not been reported previously. To explore, visualize, and analyze the landscape of these x-lncRNAs, we have generated a public browser showing strand specific transcripts in the absence and presence of the RNA exosome complex subunits (see experimental methods). Such a tool may shed greater light on co-transcriptional processing dynamics at individual loci of interest and allow for generation of new hypothesis.
Recent findings have revealed the existence of vast numbers of intergenic and intragenic enhancer elements throughout the mammalian genome (Bonasio and Shiekhattar, 2014; Lam et al., 2014). How their activity is regulated is an exciting and open question. Enhancers generate eRNA transcripts whose biological role and regulation beyond chromatin remodeling are not well appreciated. In this study, we unravel the role of RNA exosome mediated degradation of eRNAs expressed from divergently transcribed loci. We demonstrate enhancer RNAs generate complexes with single-strand DNA that are protected from being converted to sites of genomic instability by the rapid action of the RNA exosome complex. The formation of R-looped DNA secondary structures can arise from failure to undergo proper transcriptional termination (Skourti-Stathaki et al., 2014). Early transcription termination serves as a mechanism for co-transcriptional RNA exosome recruitment (Lemay et al., 2014; Pefanis et al., 2014). Thus, in the absence of RNA exosome, x-eRNAs may accumulate not solely due to lack of RNA degradation but also due to failure of transiently forming R-loop structure induced termination at enhancer loci (Skourti-Stathaki et al., 2014). Divergent transcription can create enhanced negative DNA supercoiling that in turn promotes the generation of ssDNA structures surrounding enhancer TSSs (Rhee and Pugh, 2012), thereby promoting DNA double strand breaks and genomic instability (Pefanis et al., 2014). Such breaks could be caused by the activity of an endogenous DNA mutator such as cytidine deaminase AID or due to collisions of replication forks with stalled RNA polymerase complexes at these enhancer sequences (Kim and Jinks-Robertson, 2012). Sense/antisense x-eRNA pairs that form within the R-loop bubble may result in dsRNA that can be processed by RNA interference (RNAi) factors, eventually leading to local accumulation of chromatin condensation marks such as H3K9me2 and HP1γ (Skourti-Stathaki et al., 2014). Lack of RNA exosome activity may skew the ratio or abundance of sense and antisense eRNA transcripts, leading to impairment of RNAi pathway recruitment and heterochromatinization. Thus, RNA exosome may play an important role in promoting transcription termination-coupled silencing of divergent enhancer sequences genome-wide.
Super-enhancers are large, densely packed enhancer elements that are occupied by master regulators of transcription and mediator proteins (Hnisz et al., 2013; Whyte et al., 2013). These elements are responsible for controlling transcription of diverse sets of tissue specific gene expression programs. B cell super-enhancers have been found to overlap large regions of the human genome susceptible to mutations in diffuse large B cell lymphomas (Chapuy et al., 2013) (Meng et al; Qian et al Cell 2014). We evaluated super-enhancers for the presence of RNA exosome regulated transcripts and correspondingly identified x-seRNAs. Genes or canonical enhancers in proximity to super-enhancers express high levels of RNA exosome regulated antisense RNAs around their transcription start sites (xTSS-RNAs) or within gene bodies (x-asRNAs). We hypothesize that super-enhancers may interact with genes under their regulation via mechanisms that depend upon transcription of RNA exosome regulated transcripts. A test of this hypothesis was undertaken and we observed that the divergently transcribed lncRNA-CSR enhancer element interacts with the HS4 region of the 3′ regulatory region super-enhancer of the IgH locus to control class switch recombination. The dependence of a super-enhancer function on an interacting lncRNA expressing divergent enhancer provides a newly identified mechanism of gene expression regulation (see Figure 7E for a proposed model). Whether the interaction is dependent upon direct RNA-protein complexes that are co-transcriptionally generated at the cognate pairs of enhancer/promoter and super-enhancer loci is a question of immediate interest. Furthermore, the observation that 3′RR x-seRNAs and lncRNA-CSR are substrates of RNA exosome provides the possibility that RNA exosome regulates long distance genomic interactions either through its RNA degradation activities and/or through its ability to terminate transcription of ncRNAs at enhancers and super-enhancers.
Experimental Procedures
Details of ChIP experiments, DNA/RNA hybrid immunoprecipitation, and chromosome conformation capture (3C) can be found in extended experimental procedures.
Exosc10COIN allele design and construction
A mouse Exosc10 locus containing bacterial artificial chromosome (clone bMQ169f23) was modified using bacterial homologous recombination. Briefly, a lox2372-loxP array was inserted in the first intron of Exosc10. In a subsequent recombination event an inverted lox2372-loxP array, inverted FP635 expressing terminal exon (COIN module) in antisense orientation to Exosc10 transcription, and an FRT-flanked neor selection cassette were inserted within a non-conserved region of Exosc10 exon 2. The Exosc10 COIN module contains a 3′ splice acceptor sequence immediately followed by an in-frame T2A-FP635-pA cassette. Exosc10COINneo BAC recombinants were screened by PCR across all four modified junctions and confirmed using restriction digestion and pulse field electrophoresis. A 20-kb fragment containing the entire Exosc10COINneo modification was then subcloned into a plasmid containing a diphtheria toxin A (DTA) cassette. Exosc10COINneo homology arms in the DTA vector were 6.7 and 8.2 kb. Linearized Exosc10COINneo targeting vector was electroporated into ROSA26CreERt2/+, 129S6/SvEv × C57BL/6 hybrid ES cells. Correctly targeted ES cell clones were identified using external Southern blotting probes for both the upstream and downstream homology arms on HindIII or NsiI digested genomic DNA, respectively. Exosc10COIN/+ chimeric mice were created via blastocyst injection of targeted ES cells. Mice with the greatest ES cell derived coat color contribution were crossed with Tg(ACTB:FLPe) mice to delete the neor selection cassette and germline transmit the Exosc10COIN allele. The FLPe transgene was eliminated during back-crossing. All mouse experiments were conducted in accordance with approved Columbia University Institutional Animal Care and Use Committee protocols.
RNA-seq Analysis
rRNA depleted total RNA was prepared using the Ribo-Zero rRNA removal kit (Epicentre). Libraries were prepared with Illumina TruSeq and TruSeq Stranded total RNA sample prep kits, and then sequenced with 50-60 million of 2×100 bp paired raw passing filters reads on an Illumina HiSeq 2000 V3 instrument at the Columbia Genome Center. The details of generation of exotomes from Exosc3-deficient or Exosc10-deficient B cells and ES cells and their subsequent analysis are described in supplemental methods. All RNA-sequencing data are deposited in SRA (accession number #SRP042355).
Transcriptome reconstitution
Details of transcriptome reconsititution of the Exosc3 and Exosc10 exotomes from B cells and ES cells are described in detail in the extended methods section and the data are provided in Supplementary tables 1-4 and in the “Exotome browser” which can be accessed from (http://rabadan.c2b2.columbia.edu/cgi-bin/hgGateway).
Supplementary Material
Figure S1, related to Figure 1: Exosc10COIN allele gene targeting and analysis of class switch recombination in Exosc10 ablated B cells. (A) Schematic of the Exosc10COINneo targeted locus and screening strategy. The location of the external Southern blot probe is depicted in blue, Exosc10 exons 1-3 are depicted as white boxes, and lox2372 and loxP sites are depicted as red and violet triangles, respectively. HindIII restriction sites of the Exosc10WT and modified Exosc10COINneo loci are indicated. The inverted T2A-FP635-pA cassette is in red and the FRT neo-resistance cassette is in yellow. (B) Southern blot analysis of Exosc10COINneo allele targeted ES cell clones. Genomic DNA was digested using HindIII and probed as indicated in (A). The Exosc10WT and Exosc10COINneo alleles yield 12.2 and 16.6 kb HindIII restriction fragments, respectively. (C) Class switch recombination of Exosc10WT/WT ROSA26CreERt2/+ and Exosc10COIN/LacZ ROSA26CreERt2/+ B cells isolated from littermate pairs of mice. CD43 negative splenic B cells were isolated, treated with 4-OHT to invert the Exosc10COIN allele (to null form), and stimulated in culture using LPS and IL-4 for 72 hours to drive isotype switching to IgG1. Surface expression of B220 and IgG1 are indicated. (D) B cells from (C) were evaluated for expression of AID protein by Western blot using anti-AID antibody. The parallel loading control used anti-beta actin antibody. (E) RNA-seq track of Exosc10 expression in WT and Exosc10COIN/LACZ cells from transcriptomes generated in B cells and ES cells. (F) RNA-seq track of Exosc3 expression in WT and Exosc3COIN/COIN cells from transcriptomes generated in B cells and ES cells.
Figure S2, related to Figure 2: Transcriptome assembly of Exosc3 and Exosc10 ablated B cells and ES cells. (A) Stepwise depiction of bioinformatics pipeline and parameters utilized for analyzing the transcriptomes of Exosc3COIN/COIN ROSA26CreERt2/+ or Exosc10COIN/LacZ ROSA26CreERt2/+ B cells and ES cells. Detailed description in Extended Experimental Procedures. (B) The RNA length of the lncRNAs expressed in the Exosc3-exotome and that in both Exosc3 and Exosc10 exotomes from ES cells are shown. (C) Heatmap depicting the expression levels of 639 novel intergenic lncRNAs identified from the transcriptome analysis pipeline described in (A). (D) Summary of all 4652 expressed ES cell lncRNAs.
Figure S3, related to Figure 3: Expression of xTSS-RNA and x-asRNA in B cells and ES cells. (A, B) The fold change increase in expression of RNA exosome substrate xTSS-RNAs from B cells (A) and ES cells (B). Left: plot indicating percentage of xTSS-RNAs in a given fold change window. Right: plot indicating xTSS-RNAs exclusively upregulated in Exosc10-deficient (red) or Exosc3-deficient (blue) cells. xTSS-RNAs upregulated in both Exosc10- and Exosc3-deficient cells indicated in black. (C, D) The fold change increase in expression of RNA exosome substrate antisense RNAs (x-asRNAs) from B cells (C) and ES cells (D). Left and right are as above, but modified for x-asRNAs. In this analysis, all protein coding genes in the UCSC database were considered. asRNAs were defined as all transcripts in antisense of protein coding genes, while xTSS-RNA expression was measured by the number of reads 500 bp upstream of coding transcription start sites. Fold change was calculated using normalized read counts.
Figure S4, related to Figure 4: Expression of RNA exosome substrate enhancer RNAs (x-eRNAs) at AID-target sites in the B cell genome. (A, B) Representative examples of chromosomal instability in 4-OHT treated Exosc3COIN/COINROSA26CreERt2/+ cells analyzed via telomere fluorescence in situ hybridization (A). The frequency of chromosomal abnormalities in Exosc3COIN/COIN and wild-type control cells (WT), Exosc3COIN/+ (C/+), Exosc3COIN/COIN (C/C) are tabulated in (B). Close to 300 metaphases was analyzed for each genotype, obtained from 3 independent littermate mice sets for generating the plotted numbers. (C, D) B cell translocation capture sequencing (TCseq track) (Klein et al., 2011) identifies genome translocations utilizing IgH as the translocation partner. Blue and red peaks indicate expression of sense and antisense RNAs, respectively. Correlation between translocations and expression of RNA exosome substrate enhancer RNAs (x-eRNAs) are shown for the Birc3 enhancer sequence (C), and the Ncoa3 enhancer sequence (D). (E-H) Divergently expressed enhancer loci identified from the transcriptomes of Exosc3COIN/COIN and Exosc10COIN/LacZ ES cells residing close to the Cd38 (E), Bcl6 (F), Klf6 (G) and NcoA3 (H) loci. These loci were used for ChIP and DRIP experiments in Fig. 4. *P<0.05 and **P<0.01 by t-test. A non-divergent enhancer region (chrm4: 141,798,300-141,798,700) was identified that expresses eRNAs from a non-divergent promoter. This region was assayed for γH2AX foci formation (I) and for DNA/RNA hybrid formation (J). ns: non-significant difference.
Figure S5, related to Figure 5: Tgfbr2 expression is controlled by RNA exosome target enhancer sequences E1 and E2. (A) The expression pattern of sense (red) and antisense (blue) RNAs at the Tgfbr2 locus in Exosc3COIN/COIN, Exosc10COIN/LacZ, and wild-type control cells. Typical enhancers expressing x-eRNAs are illustrated in the map below in blue. (B) Tgfbr2 expression following CRISPR/Cas9 mediated deletion of the two divergently transcribed enhancer-like sequences E1 (Chr9: 116,152,511-116,155,370) E2 (Chr9: 116,128,150-116,130,790). The knockouts of E1 and E2 were accomplished in B cell line CH12F3 and the expression of the Tgfbr2 gene was evaluated using qRT-PCR. (C) Plot of the enrichment of xTSS-RNA genes close to superenhancer sequences that expresses x-seRNAs. The genomic distances of all expressed genes to their closest super enhancer (SE) regions are calculated. Given a cutoff of the genomic distance, genes are partitioned into the far and the close groups. A ranksum test is then performed to assess the difference between those two groups in terms of fold change of TSS RNA expression between Exosc3COIN/COIN and wild type. (D) Expression of AID mRNA levels in parental (WT) and lncRNA-CSR knockout CH12F3 cells using qRT-PCR. (E) The class switch recombination efficiency to IgA for CH12F3 cells (WT-parental and lincRNA-CSR-/-) stimulated in culture for 24 hrs or 52 hours with LPS, IL4, and TGFβ.
Figure S6, related to Figure 6: Maps of lncRNA-CSR and Igh 3′RR HS4 region on chromosome 12. (A) A schematic diagram displaying lncRNA-CSR (in red) divergently expressed from the known ncRNA B930059L03Rik. The region of lncRNA deletion is indicated. The primer sequence used for 3C experimentation in Fig. 6 is shown. (B) The Igh 3′RR HS4 region that interacts with the lncRNA-CSR region is shown. The 3C primer corresponding to the HS4 regions that is used in Fig. 6 is shown. The expression tracks of the 3′RR HS4 RNA in Exosc3COIN/COIN, Exosc10COIN/LacZ and WT transcriptomes are shown. (C) The expression of x-seRNAs in the 3′RR HS4 region is demonstrated. The blue boxes represent sense RNA reads; the red boxes, antisense. These RNA-seq tracks demonstrate that x-seRNAs are short RNAs transcribed on both strands of the superenhancer sequence.
Figure S7, related to Figure 7: Model for interaction of RNA exosome substrate expressing superenhancer sequences with divergently transcribed enhancer or genic promoter sequences. (A) The overlap of chromosomal rearrangement from the IgH locus (2.6 Mb away) to the lncRNA-CSR locus in B cells that overexpresses AID, as captured by TC-seq analysis (Klein et al, 2012). The top panel demonstrates tracks of translocations, the middle and bottom panels indicate the divergent transcription that is generated at the lncRNA-CSR locus. (B) The interaction frequency of Eμ with the HS1,2 sites of 3′ RR in the IgH locus in B cells obtained from Exosc3WT/WT and Exosc3COIN/COIN B cells is indicated. This interaction frequency is a control experiment for Fig. 7C. n.s.: not-significant. The graph represents data obtained from three independently performed 3C experiments.
Supplementary table S1, related to Figure 1: B cell specific RNA exosome substrate antisense RNAs from the body of the genes and from genic Transcription start sites.
Supplementary table S2, related to Figure 1: ES cell specific RNA exosome substrate antisense RNAs from the body of the genes and from genic Transcription start sites.
Supplementary table S3, related to Figure 2: ES cell specific RNA exosome substrate large non-coding RNAs (x-lncRNAs).
Supplementary table S4, related to Figure 3: ES cell specific RNA exosome substrate enhancer RNAs (x-eRNAs).
Supplementary table S5, related to Figure 1: Gene set enrichment analysis of all gene ontology gene sets that are upregulated in Exosc3COIN/COIN ES cells (4 samples).
Supplementary table S6, related to Figure 1: Gene set enrichment analysis of all gene ontology gene sets that are downregulated in Exosc3COIN/COIN ES cells (4 samples).
Acknowledgments
We thank Christopher Lima (MSKCC, New York), Frederic Chedin (University of California, Davis), Saul Silverstein, Stephen Goff, Sankar Ghosh, Lorraine Symington and members of the Basu lab for critical input and reagents. We thank Olivier Courrone of Columbia University genome center for RNA sequencing and Victor Lin of Herbert Irving Cancer Center Transgenic facility for targeting and generation of Exosc3COIN and Exosc10COIN allele ES cells and mouse models. This work was supported by grants from NIH (1DP2OD008651-01) and NIAID (1R01AI099195-01A1) (to U.B.); NIH (1R01CA185486-01; 1R01CA179044-01A1; 1U54CA121852-05) (to R.R.); (F31AI098411-01A1) (to D.K.). U.B. is a scholar of the Irma Hirschl Charitable Trust and the Leukemia and Lymphoma Society.
Footnotes
Author Contribution: E.P., J.W. and U.B. planned studies; E.P., J.W., G.R., R.R., U.B. interpreted data. Experiments were performed as follows: E.P., J.C., mouse model generation, RNA-seq studies; G.R., J. L., ChIP, DRIP and CRISPR/Cas9; J.W., bioinformatic studies and designing all the pipelines for determining various RNA exosome substrate non-coding RNAs; A.N.E advised on mouse model construct; R.R. oversaw bioinformatics and guided J.W.; D.K. and J.S. prepared and analyzed metaphases of RNA exosome mutant cells; J.W. and O.E. prepared exotome browser, A.F. and J.E.B identified the superenhancer sequences in B cells, and E.P., J.W. and U.B. wrote the manuscript which was further refined by all the other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014a;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Refsing Andersen P, Valen E, Core LJ, Bornholdt J, Boyd M, Heick Jensen T, Sandelin A. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nature communications. 2014b;5:5336. doi: 10.1038/ncomms6336. [DOI] [PubMed] [Google Scholar]
- Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- Birshtein BK. Epigenetic Regulation of Individual Modules of the immunoglobulin heavy chain locus 3′ Regulatory Region. Frontiers in immunology. 2014;5:163. doi: 10.3389/fimmu.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Shiekhattar R. Regulation of Transcription by Long Noncoding RNAs. Annual review of genetics. 2014 doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A. RNA decay machines: the exosome. Biochim Biophys Acta. 2013;1829:552–560. doi: 10.1016/j.bbagrm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Colin J, Candelli T, Porrua O, Boulay J, Zhu C, Lacroute F, Steinmetz LM, Libri D. Roadblock termination by reb1p restricts cryptic and readthrough transcription. Molecular cell. 2014;56:667–680. doi: 10.1016/j.molcel.2014.10.026. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economides AN, Frendewey D, Yang P, Dominguez MG, Dore AT, Lobov IB, Persaud T, Rojas J, McClain J, Lengyel P, et al. Conditionals by inversion provide a universal method for the generation of conditional alleles Proceedings of the. National Academy of Sciences of the United States of America; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev. 2012;22:172–178. doi: 10.1016/j.gde.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature biotechnology. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbaker DZ, Marquardt S, Wlotzka W, Buratowski S. Kinetic competition between RNA Polymerase II and Sen1-dependent transcription termination. Molecular cell. 2013;49:55–66. doi: 10.1016/j.molcel.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD. Structural components and architectures of RNA exosomes. Adv Exp Med Biol. 2011;702:9–28. doi: 10.1007/978-1-4419-7841-7_2. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nature reviews Genetics. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Larochelle M, Marguerat S, Atkinson S, Bahler J, Bachand F. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nature structural & molecular biology. 2014;21:919–926. doi: 10.1038/nsmb.2893. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Molecular cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. Journal of cell science. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- Manfrini N, Trovesi C, Wery M, Martina M, Cesena D, Descrimes M, Morillon A, d'Adda di Fagagna F, Longhese MP. RNA-processing proteins regulate Mec1/ATR activation by promoting generation of RPA-coated ssDNA. EMBO reports. 2015;16:221–231. doi: 10.15252/embr.201439458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Vicente C, Domingo-Prim J, Eberle AB, Visa N. RRP6/EXOSC10 is required for the repair of DNA double-strand breaks by homologous recombination. Journal of cell science. 2015 doi: 10.1242/jcs.158733. [DOI] [PubMed] [Google Scholar]
- Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, Amor C, Wasserman CR, Neuberg D, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014 doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud E, Marquet M, Fiancette R, Peron S, Vincent-Fabert C, Denizot Y, Cogne M. The IgH locus 3′ regulatory region: pulling the strings from behind. Advances in immunology. 2011;110:27–70. doi: 10.1016/B978-0-12-387663-8.00002-8. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, Resch W, Liang G, Tang Z, Mathe E, Benner C, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes & development. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nature structural & molecular biology. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2366–2371. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Shah S, Wittmann S, Kilchert C, Vasiljeva L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes & development. 2014;28:231–244. doi: 10.1101/gad.230177.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Wang HL, Lee J, Dinwiddie BL, Belostotsky DA, Chekanova JA. The role of the Arabidopsis Exosome in siRNA-independent silencing of heterochromatic loci. PLoS Genet. 2013;9:e1003411. doi: 10.1371/journal.pgen.1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014 doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U. Why does somatic hypermutation by AID require transcription of its target genes? Advances in immunology. 2014;122:253–277. doi: 10.1016/B978-0-12-800267-4.00007-9. [DOI] [PubMed] [Google Scholar]
- Sun J, Keim CD, Wang J, Oliver P, Rabadan R, Basu U. E3-ubiquitin ligase Nedd4 determines the fate of AID-associated RNA polymerase II in B cells. Genes & development. 2013;27:1821–1833. doi: 10.1101/gad.210211.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rothschild G, Pefanis E, Basu U. Transcriptional stalling in B-lymphocytes: A mechanism for antibody diversification and maintenance of genomic integrity. Transcription. 2013;4 doi: 10.4161/trns.24556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. Genome-wide measurement of RNA folding energies. Molecular cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, Lima CD. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014;511:435–439. doi: 10.1038/nature13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Molecular cell. 2012;48:133–144. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, Rommel PC, Brown EJ, Nussenzweig A, Nussenzweig MC, et al. RPA accumulation during class switch recombination represents 5′-3′ DNA-end resection during the S-G2/M phase of the cell cycle. Cell reports. 2013;3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1: Exosc10COIN allele gene targeting and analysis of class switch recombination in Exosc10 ablated B cells. (A) Schematic of the Exosc10COINneo targeted locus and screening strategy. The location of the external Southern blot probe is depicted in blue, Exosc10 exons 1-3 are depicted as white boxes, and lox2372 and loxP sites are depicted as red and violet triangles, respectively. HindIII restriction sites of the Exosc10WT and modified Exosc10COINneo loci are indicated. The inverted T2A-FP635-pA cassette is in red and the FRT neo-resistance cassette is in yellow. (B) Southern blot analysis of Exosc10COINneo allele targeted ES cell clones. Genomic DNA was digested using HindIII and probed as indicated in (A). The Exosc10WT and Exosc10COINneo alleles yield 12.2 and 16.6 kb HindIII restriction fragments, respectively. (C) Class switch recombination of Exosc10WT/WT ROSA26CreERt2/+ and Exosc10COIN/LacZ ROSA26CreERt2/+ B cells isolated from littermate pairs of mice. CD43 negative splenic B cells were isolated, treated with 4-OHT to invert the Exosc10COIN allele (to null form), and stimulated in culture using LPS and IL-4 for 72 hours to drive isotype switching to IgG1. Surface expression of B220 and IgG1 are indicated. (D) B cells from (C) were evaluated for expression of AID protein by Western blot using anti-AID antibody. The parallel loading control used anti-beta actin antibody. (E) RNA-seq track of Exosc10 expression in WT and Exosc10COIN/LACZ cells from transcriptomes generated in B cells and ES cells. (F) RNA-seq track of Exosc3 expression in WT and Exosc3COIN/COIN cells from transcriptomes generated in B cells and ES cells.
Figure S2, related to Figure 2: Transcriptome assembly of Exosc3 and Exosc10 ablated B cells and ES cells. (A) Stepwise depiction of bioinformatics pipeline and parameters utilized for analyzing the transcriptomes of Exosc3COIN/COIN ROSA26CreERt2/+ or Exosc10COIN/LacZ ROSA26CreERt2/+ B cells and ES cells. Detailed description in Extended Experimental Procedures. (B) The RNA length of the lncRNAs expressed in the Exosc3-exotome and that in both Exosc3 and Exosc10 exotomes from ES cells are shown. (C) Heatmap depicting the expression levels of 639 novel intergenic lncRNAs identified from the transcriptome analysis pipeline described in (A). (D) Summary of all 4652 expressed ES cell lncRNAs.
Figure S3, related to Figure 3: Expression of xTSS-RNA and x-asRNA in B cells and ES cells. (A, B) The fold change increase in expression of RNA exosome substrate xTSS-RNAs from B cells (A) and ES cells (B). Left: plot indicating percentage of xTSS-RNAs in a given fold change window. Right: plot indicating xTSS-RNAs exclusively upregulated in Exosc10-deficient (red) or Exosc3-deficient (blue) cells. xTSS-RNAs upregulated in both Exosc10- and Exosc3-deficient cells indicated in black. (C, D) The fold change increase in expression of RNA exosome substrate antisense RNAs (x-asRNAs) from B cells (C) and ES cells (D). Left and right are as above, but modified for x-asRNAs. In this analysis, all protein coding genes in the UCSC database were considered. asRNAs were defined as all transcripts in antisense of protein coding genes, while xTSS-RNA expression was measured by the number of reads 500 bp upstream of coding transcription start sites. Fold change was calculated using normalized read counts.
Figure S4, related to Figure 4: Expression of RNA exosome substrate enhancer RNAs (x-eRNAs) at AID-target sites in the B cell genome. (A, B) Representative examples of chromosomal instability in 4-OHT treated Exosc3COIN/COINROSA26CreERt2/+ cells analyzed via telomere fluorescence in situ hybridization (A). The frequency of chromosomal abnormalities in Exosc3COIN/COIN and wild-type control cells (WT), Exosc3COIN/+ (C/+), Exosc3COIN/COIN (C/C) are tabulated in (B). Close to 300 metaphases was analyzed for each genotype, obtained from 3 independent littermate mice sets for generating the plotted numbers. (C, D) B cell translocation capture sequencing (TCseq track) (Klein et al., 2011) identifies genome translocations utilizing IgH as the translocation partner. Blue and red peaks indicate expression of sense and antisense RNAs, respectively. Correlation between translocations and expression of RNA exosome substrate enhancer RNAs (x-eRNAs) are shown for the Birc3 enhancer sequence (C), and the Ncoa3 enhancer sequence (D). (E-H) Divergently expressed enhancer loci identified from the transcriptomes of Exosc3COIN/COIN and Exosc10COIN/LacZ ES cells residing close to the Cd38 (E), Bcl6 (F), Klf6 (G) and NcoA3 (H) loci. These loci were used for ChIP and DRIP experiments in Fig. 4. *P<0.05 and **P<0.01 by t-test. A non-divergent enhancer region (chrm4: 141,798,300-141,798,700) was identified that expresses eRNAs from a non-divergent promoter. This region was assayed for γH2AX foci formation (I) and for DNA/RNA hybrid formation (J). ns: non-significant difference.
Figure S5, related to Figure 5: Tgfbr2 expression is controlled by RNA exosome target enhancer sequences E1 and E2. (A) The expression pattern of sense (red) and antisense (blue) RNAs at the Tgfbr2 locus in Exosc3COIN/COIN, Exosc10COIN/LacZ, and wild-type control cells. Typical enhancers expressing x-eRNAs are illustrated in the map below in blue. (B) Tgfbr2 expression following CRISPR/Cas9 mediated deletion of the two divergently transcribed enhancer-like sequences E1 (Chr9: 116,152,511-116,155,370) E2 (Chr9: 116,128,150-116,130,790). The knockouts of E1 and E2 were accomplished in B cell line CH12F3 and the expression of the Tgfbr2 gene was evaluated using qRT-PCR. (C) Plot of the enrichment of xTSS-RNA genes close to superenhancer sequences that expresses x-seRNAs. The genomic distances of all expressed genes to their closest super enhancer (SE) regions are calculated. Given a cutoff of the genomic distance, genes are partitioned into the far and the close groups. A ranksum test is then performed to assess the difference between those two groups in terms of fold change of TSS RNA expression between Exosc3COIN/COIN and wild type. (D) Expression of AID mRNA levels in parental (WT) and lncRNA-CSR knockout CH12F3 cells using qRT-PCR. (E) The class switch recombination efficiency to IgA for CH12F3 cells (WT-parental and lincRNA-CSR-/-) stimulated in culture for 24 hrs or 52 hours with LPS, IL4, and TGFβ.
Figure S6, related to Figure 6: Maps of lncRNA-CSR and Igh 3′RR HS4 region on chromosome 12. (A) A schematic diagram displaying lncRNA-CSR (in red) divergently expressed from the known ncRNA B930059L03Rik. The region of lncRNA deletion is indicated. The primer sequence used for 3C experimentation in Fig. 6 is shown. (B) The Igh 3′RR HS4 region that interacts with the lncRNA-CSR region is shown. The 3C primer corresponding to the HS4 regions that is used in Fig. 6 is shown. The expression tracks of the 3′RR HS4 RNA in Exosc3COIN/COIN, Exosc10COIN/LacZ and WT transcriptomes are shown. (C) The expression of x-seRNAs in the 3′RR HS4 region is demonstrated. The blue boxes represent sense RNA reads; the red boxes, antisense. These RNA-seq tracks demonstrate that x-seRNAs are short RNAs transcribed on both strands of the superenhancer sequence.
Figure S7, related to Figure 7: Model for interaction of RNA exosome substrate expressing superenhancer sequences with divergently transcribed enhancer or genic promoter sequences. (A) The overlap of chromosomal rearrangement from the IgH locus (2.6 Mb away) to the lncRNA-CSR locus in B cells that overexpresses AID, as captured by TC-seq analysis (Klein et al, 2012). The top panel demonstrates tracks of translocations, the middle and bottom panels indicate the divergent transcription that is generated at the lncRNA-CSR locus. (B) The interaction frequency of Eμ with the HS1,2 sites of 3′ RR in the IgH locus in B cells obtained from Exosc3WT/WT and Exosc3COIN/COIN B cells is indicated. This interaction frequency is a control experiment for Fig. 7C. n.s.: not-significant. The graph represents data obtained from three independently performed 3C experiments.
Supplementary table S1, related to Figure 1: B cell specific RNA exosome substrate antisense RNAs from the body of the genes and from genic Transcription start sites.
Supplementary table S2, related to Figure 1: ES cell specific RNA exosome substrate antisense RNAs from the body of the genes and from genic Transcription start sites.
Supplementary table S3, related to Figure 2: ES cell specific RNA exosome substrate large non-coding RNAs (x-lncRNAs).
Supplementary table S4, related to Figure 3: ES cell specific RNA exosome substrate enhancer RNAs (x-eRNAs).
Supplementary table S5, related to Figure 1: Gene set enrichment analysis of all gene ontology gene sets that are upregulated in Exosc3COIN/COIN ES cells (4 samples).
Supplementary table S6, related to Figure 1: Gene set enrichment analysis of all gene ontology gene sets that are downregulated in Exosc3COIN/COIN ES cells (4 samples).


