Abstract
NK cells are promising effector cells for adjuvant immunotherapy of cancer. So far, several preclinical studies have shown the feasibility of gene-engineered NK cells, which upon expression of chimeric antigen receptors (CARs) are redirected to otherwise NK-cell resistant tumors. Yet, we reasoned that the efficiency of an immunotherapy using CAR-modified NK cells critically relies on efficient migration to the tumor site and might be improved by the engraftment of a receptor specific for a chemokine released by the tumor. Based on the DNAX-activation protein 12 (DAP12), a signaling adapter molecule involved in signal transduction of activating NK cell receptors, we constructed an EGFRvIII-CAR, designated MR1.1-DAP12 which confers specific cytotoxicity of NK cell towards EGFRvIII+ glioblastoma cells in vitro and to established subcutaneous U87-MGEGFRvIII tumor xenografts. So far, infusion of NK cells with expression of MR1.1-DAP12 caused a moderate but significantly delayed tumor growth and increased median survival time when compared to NK cells transduced with an ITAM-defective CAR. Notably, the further genetic engineering of these EGFRvIII-specific NK cells with the chemokine receptor CXCR4 conferred a specific chemotaxis to CXCL12/SDF-1α secreting U87-MG glioblastoma cells. Moreover, the administration of such NK cells resulted in complete tumor remission in a number of mice and a significantly increased survival when compared to the treatment of xenografts with NK cells expressing only the EGFRvIII-specific CAR or mock control. We conclude that chemokine receptor engineered NK cells with concomitant expression of a tumor-specific CAR are a promising tool to improve adoptive tumor immunotherapy.
Keywords: adoptive immunotherapy, engineered NK cell chemotaxis, chimeric antigen receptor, EGFRvIII, CXCR4, CXCL12/SDF-1α
Introduction
Despite ongoing developments in therapy, patients suffering from glioblastoma (GBM, WHO grade IV), the most common and malignant brain tumor in adults, still have a very poor prognosis.1 Due to its infiltrative behavior, GBM cannot be fully resected. In addition, glioma cells are resistant to chemo- or radiotherapy and have developed several strategies to evade immune surveillance.2 Despite multimodal treatment, the tumor most likely reappears after a short time resulting in median survival times not exceeding 14.6 months.3
In recent years, tumor-specific T cells and NK cells have emerged as promising tool for augmenting classical treatment of cancer.4 In particular, T cells genetically modified with chimeric antigen receptors (CARs) targeting glioma associated surface-antigens such as epidermal growth factor variant III (EGFRvIII), an oncogenic mutant form of the EGFR, have shown promising results in preclinical models and have entered clinical phase I/II studies (see http://clinicaltrials.gov/ct2/showNCT01454596).5-6
NK cells represent an important component of the innate immune system by killing virus-infected and transformed cells without the need for prior immune sensitization.7 However, recent insights into NK cell function have led to the understanding that these cells participate in innate as well as adaptive immune responses thereby increasing the scope for adoptive immunotherapy of cancer.8, 9 Unlike T cells, which depend on RAG-mediated somatic recombination of immune receptors, NK cells are characterized by simultaneous expression of various inhibitory and activating receptors of the lectin and immunoglobulin superfamily.10, 11 Interactions of inhibitory receptors with human leukocyte antigen (HLA) class I molecules on autologous normal cells induce dominant negative signals which override activating signals and therefore prevent cytotoxic activity.12 In the absence of negative signals and/or increased levels of activating ligands on target cells, NK cells become activated and kill target cells via the perforin-granzyme pathway, by action of death-receptor ligands or released cytokines such as IFN-γ.10, 13, 14 Previous studies have shown that retargeting lymphocytes to tumor antigens on solid tumors by the expression of CARs is promising but not in every case sufficient for clinical responses.15-17 NK cells have been shown to invade solid tumors in vivo, in particular gliomas, to a significant lesser extent than other leucocytes.18
Leukocyte trafficking to tumors essentially depends on chemokines and involves a complex interplay between activated endothelial cells and circulating lymphocytes characterized by subsequent steps of lymphocyte attachment to and rolling on endothelium, activation on the endothelial surface, secondary attachment and extravasation into the target tissue.19 Furthermore, it is most likely that ongoing neo-angiogenesis and immature tumor vessels of some tumor entities such as glioblastomas might lead to an enhanced extravasation of chemokine-attracted lymphocytes at the tumor site.20 We therefore reasoned that a combined expression of a CAR and of a chemokine receptor enabling chemotaxis to gliomas might expand the therapeutic range of CAR-modified NK cells.
As target on glioma cells for an experimental therapy using CAR-modified NK cells we chose EGFRvIII, the most common of several known oncogenic isoforms of the epidermal growth factor receptor (EGFR). EGFRvIII has been found in 30 – 40 % of malignant glioma. The constitutive activation of its kinase domain is the main reason for the significantly enhanced tumorigenicity of EGFRvIII positive tumors.21 Structurally, EGFRvIII shows an in frame deletion of amino acids 6 to 273 resulting in addition of a glycine and loss of exons 2 - 7. Therefore, this variant lacks most of its ectodomain but contains a neo-epitope at the fusion point, which specifically binds the single chain fragment variable (scFv) MR1.1 used for the CAR employed in this study.22
The chemokine CXCL12/SDF-1α belongs to the CXC chemokine family and binds the chemokine receptor CXCR4 and CXCR7.23, 24 The latter chemokine receptor is not expressed on peripheral blood leukocytes.25 CXCL12/SDF-1α has been originally described as pre-B-cell growth factor.26 Today it is known as an important homeostatic chemokine for T cells and bone marrow niches.23, 27 Upon maturation of CXCR4+-NK progenitors in the bone marrow, NK cells consecutively downregulate CXCR4 to undetectable levels and enter the blood stream and the lymphoid system.27 Interestingly, CXCL12/SDF-1α is also constitutively secreted in developing brain and mediates neuronal migration and angiogenesis.28, 29 It has been reported that CXCL12/SDF-1α is re-expressed in glioblastoma.30, 31 It increases proliferation and migration of tumor cells in an autocrine manner.29, 32, 33 Furthermore, its increased expression has been correlated with shorter progression free survival of patients with low grade glioma.34 In line with its role in developing brain, CXCL12/SDF-1α secreted by tumor cells has been reported to cooperatively act with VEGF and bFGF thereby guiding the developing tumor vasculature.35
In this study, we sought to express CXCR4 on NK cells to facilitate glioma homing on adoptive transfer of CAR-engineered NK cells specific for EGFRvIII. We proposed the use of CXCR4-engineered NK cells for adoptive immunotherapy of CXCL12/SDF-1α-secreting glioblastoma. To our best knowledge, this is the first report describing a genetically engineered redirected chemotaxis and combined gene-engineered specific cytotoxicity of NK cells towards glioblastoma xenografts.
Materials and Methods
Cell lines
The human embryonic kidney cell line HEK293T was maintained in DMEM (PAA, Cölbe, Germany) supplemented with 10 % v/v heat-inactivated FBS (PAA), 10 mM HEPES (PAA), 100 U/ml penicillin and 0.1 mg/ml streptomycin (PAA). The glioma cell line U87-MG and cell lines derived thereof were maintained in BME (PAA, Cölbe, Germany) supplemented with 10 % v/v heat-inactivated FBS (PAA), 10 mM HEPES (PAA), 2 mM L-glutamine (Biochrom, Berlin, Germany), 1 × MEM NEAA, 100 U/ml penicillin and 0.1 mg/ml streptomycin (PAA). The glioma cell line BS153 was maintained in DMEM (PAA, Cölbe, Germany) supplemented with 10 % v/v heat-inactivated FBS (PAA), 10 mM HEPES (PAA)36, 100 U/ml penicillin, 0.1 m/ml streptomycin (PAA), the medium for BS153resE additionally contained 10 μM Erlotinib (Santa Cruz). The NK cell line YTS was maintained in RPMI-1640 (PAA) with 10 % v/v heat-inactivated FBS (PAA), 2 mM L-glutamine (Biochrom, Berlin, Germany), 10 mM HEPES (PAA), 100 U/ml penicillin, 0.1 mg/ml streptomycin (PAA). Cell lines were cultivated at 37 °C and 5 % CO2 in a humidified incubator. All cell lines were authenticated (Multiplexion GmbH, Heidelberg, Germany) and confirmed.
Generation of lentiviral vectors
To generate DAP12-based CARs, a fragment consisting of the Igκ signal peptide (SP) and the anti-EGFRvIII single chain fragment variable scFv(MR1.1)37 were amplified by PCR using specific primers adding a C-terminal short peptide linker (Gly2Ser1)2. The resulting PCR-fragments were ligated into a p6NST1-derived38 self-inactivating lentiviral pHATtrick vector (manuscript in preparation) devoid of the WPRE,39 and containing an internal spleen focus forming virus (SSFV) U3 promoter followed by a multiple cloning site, a T2A and puromycine-resistance gene, resulting in pHATtrick-MR1.1-DAP12. The coding region of signaling adaptor protein DAP12 was established previously in our lab.40 Using PCR, appropriate restriction sites and a myc-tag were added flanked by short (Gly2Ser1) linkers. The signaling adaptor protein DAP12 was ligated in frame with scFv(MR1.1) to generate the lentiviral CAR vector pHATtrick-MR1.1-DAP12-puroR. To generate controls devoid of the antigen-binding moiety and only containing the signal adaptor the Igκ signal peptide was fused to myc-DAP12, respectively, and the resulting fragment was cloned into pHATtrick creating the lentiviral vector pHATtrick-myc-DAP12-puroR. A DAP12 with a phosphorylation-deficient ITAM was generated by PCR of pHATtrick-MR1.1-DAP12-puroR using the primers DAP12mut-For 5′-TTTTTGCATGCGTTAACGAACAAAAACTCATCT CAGAAGAGG-3′ and DAP12mut-Rev 5′-GAGTCGCCTTCTCAGGAGCTCCAGGGTC AGAGTCGGATGTCTCTAGCGACCTCAACACACAGAGGCCGTATACAAATGAGCGGCCGCTTTTT-3′ (Eurofins MWG Operon) resulting in DAP12mut fragment. This fragment was used to construct pHATtrick-MR1.1-DAP12mut-puroR. The cDNA encoding the CXCR4-chemokine receptor was a kind gift of Sebastian Brenner (Department of Pediatrics, University Clinic Carl Gustav Carus, TU Dresden, Germany). A PCR with sequence specific primers CXCR4-For 5′ - TTTTTaccggtgccaccATGGAGGGGATCAGTATATACAC – 3′ and CXCR4-Rev 5′ – AAAAgcggccgcggTccGCTGGAGTGAAAACTTGAAGACTCAGAC TCAGTGGAA – 3′ (both obtained from Eurofins MWG Operon) was used to add needed restriction sites for cloning the sequence into pHATtrick-puroR. For generating a pHATtrick-hygroR vector the puromycine-resistance gene was replaced via PCR cloning with a hygromycine-resistance gene. A full length cDNA of the EGFRvIII with the needed restriction sites was obtained from Eurofins MWG Operon and cloned into pHATtrick resulting in pHATRtrick-EGFRvIII-puroR. The vector p6NST61-DsRed was provided by Dirk Lindemann (Institute of Virology, TU Dresden, Germany) and was used to generate YTSDsRed and YTSDsRed/CXCR4 cells. The vector p6NST50-SDF-1α-IRES-EGFP-ZeoR and corresponding empty vector was a kind gift from Marc Cartellieri (Institute of Immunology, TU Dresden, Germany). All vectors were confirmed by DNA sequencing.
Virus production and transduction of cells
Lentiviral particles for transduction of cells were produced by a transient three vector packaging protocol.41 Briefly, 4 × 106 HEK293T cells were transfected using polyethylenimine (PEI) (Polysciences, Warrington, PA, USA), 5 μg pCD/NL-BH 41, pczVSV-G 42 and lentiviral vector, respectively. After 20 h HEK293T cells were incubated with 10 mM sodium butyrate (Sigma-Aldrich, Taufkirchen, Germany) for 6 h. 24 h after the replacement of sodium butyrate by fresh medium the lentiviral supernatant was removed from cells and passed through a 0.45 μm filter, mixed with 8 μg/ml polybrene (Sigma-Aldrich) and used for transduction. Transduction was repeated on two subsequent days using a spinoculation protocol. Plated cells were centrifuged at 805 × g for 1 h at 37 °C and further incubated at 37 °C, 5 % CO2 overnight. After the third transduction cells were maintained in supplemented cell culture media. The NK cell line YTS, which was kindly provided by Ricciarda Galandrini (University “La Sapienza”, Rome, Italy) was transduced with pHATtrick-CXCR4-hygroR and pHATtrick-puroR encoding for the different CARs. HEK293T were transduced with pHATtrick-EGFRvIII-puroR. U87-MG wt and U87-MGEGFRvIII were transduced with p6NST50-SDF-1α_IRES-EGFP-ZeoR or empty vector only encoding IRES-EGFP-ZeoR. In order to enhance the expression of transgenes, cells were selected in 150 mg/ml hygromycine, 15 mg/ml puromycine (Invitrogen, Karlsruhe, Germany) or 100 mg/ml Zeocin respectively for one week.
Western Blot analyses
Transduced YTS cells were lysed in lysis buffer (10 mM Tris-HCL pH 8.0; 140 mM NaCl; 1 % Triton-X-100). Cell lysates were cleared by centrifugation, separated by SDS-PAGE under reducing conditions and transferred on a Westran PVDF membrane (Whatman GmbH, Dassel, Germany). The protein loaded and blocked membrane was incubated with a monoclonal anti-c-myc antibody (1:2500; Invitrogen) or monoclonal anti-DAP12-antibody (specific for the DAP12 ITAM sequence QGQRSDVYSDLNTQRPYYK, clone 406288, 1:700, R&D) followed by a HRP-labeled rabbit anti-mouse secondary antibody (1:1000; Dako, Glostrup, Denmark). Equal loading of samples was determined by stripping and re-probing the membrane for total α-Tubulin (1:3000; Santa Cruz). Membranes were visualized and documented using the Luminata Forte Western HRP substrate (Millipore) and the LAS 3000 imager (FujiFilm Europe).
Flow cytometry analyses of tumor cells and YTS cells
The recombinant scFv(MR1.1) single chain antibody containing a C-terminal myc-tag was produced in our laboratory using HEK293T producer cells. The scFv(MR1.1), a secondary Biotin-labeled c-myc-tag specific antibody (1:15; Miltenyi Biotec and a tertiary anti-Biotin-PE antibody (1:15; Miltenyi Biotec) were used to detect EGFRvIII. Staining cells with scFv(AM1),43 which does not bind to the target cells, served as isotype control. For analysis of the cell surface expression of the CAR and controls, 3 × 105 modified YTS were centrifuged and stained with a Biotin-labeled c-myc-tag specific antibody (1:15; Miltenyi Biotec) and a secondary anti-Biotin-PE or Biotin-APC antibody, respectively (1:15; Miltenyi Biotec). As control, an IgG isotype was included. For analysis of the cell surface expression of CXCR4-chemokine receptor, 3 × 105 YTS cells were stained with anti-CXCR4-APC (0.5 μg; BD). IgG-isotype controls were included. To determine the cell surface expression of EGFRvIII, chemokine receptor, CAR and control constructs, stained cells were measured by FACS and analyzed by FlowJo software version 10 (TreeStar Inc., Ashland, OR, USA).
PCR for detection of EGFRvIII in tumor cell lines
The mRNA was prepared from whole cell tumor cell lysates using the Rneasy Plus Mini Kit (Qiagen) according to the instructions ofteh supplier. After incubation with DNase I for 10min a RT reaction was performed using the RevertAid cDNA Synthesis Kit (Thermo Fisher). To exclude gDNA contamination the cDNA was tested in a PCR reation with specific primers MECL1-GCA1-For 5′ – GCTTACTCCACCTCCATAGCC – 3′ and MECL1-YS1-Rev 5′ – CTGGCGGTGCTAGAAGACC – 3′. Primers for PCR of actin have been described previously.44 Primers for detection of EGFR wild type cDNA and EGFRvIII cDNA EGFR/EGFRvIII-For 5′-ATGCGACCCTCCGGGACGG-3′and EGFR/EGFRvIII-Rev 5′-ACCTTCTGGGATCCAGAGTCC-3′ were synthesized (Eurofins MWG Operon) and used to discriminate between EGFR wt and EGFRvIII amplicons by size of the resulting fragment (EGFR wt, 2.5 kb; EGFRvIII, 1.4 kb).
Chromium release assay
The antigen specific cytotoxicity of CAR-engineered YTS cells towards EGFRvIII positive tumor cells was tested by chromium release assays. Briefly, 2 × 106 target cells were labeled with 50 μCi sodium 51chromate (Perkin-Elmer, Ueberlingen, Germany) and incubated at 37 °C and 5 % CO2. After 1 h, cells were washed several times with PBS and seeded as triplicates in a round bottom 96-well plate (1 × 104 cells per well). CAR-modified YTS cells as well as controls were added to labeled target cells at various target to effector ratios. After 21 h, 25 μl of cell supernatant was mixed with 150 μl of scintillation solution OptiPhase SuperMix (Wallac Scintillation Products, Turku, Finland) in a 96-well plate by shaking for 3 - 5 min at room temperature. The chromium release was measured using a Wallace 1450 Microbeta Trilux Liquid Scintillation and Luminescence Counter (Perkin-Elmer). Maximal and minimal releases were measured by treating target cells with 5 % Triton X-100 (Serva, Heidelberg, Germany) and medium alone, respectively. Incubations of YTS cells with isogenic tumor wild type cells were included for comparison. Percentage of specific lysis was calculated using the standard formula: 100 × (cpm release target cells - cpm minimum release) / (cpm maximum release – cpm minimum release). The experiments were performed three to four times each with similar results.
CXCL12/SDF-1α-ELISA
Detection of secreted CXCL12/SDF-1α in the supernatant of transduced cells was accomplished using the DuoSet ELISA - huCXCL12/SDF-1α (R&D) as recommended. 25000 U87-MGEGFRvIII/SDF-1α cells were plated in 2 ml RPMI-1640 without FBS into 6-well-plates and the supernatant was collected after 24 h. As negative control U87-MGEGFRvIII/EGFP cells were used. To evaluate the capacity of the genetically altered U87-MG cells to secrete CXCL12/SDF-1α in an in vivo setting, 1 × 106 tumor cells were inoculated subcutaneously into female NMRI-Foxn1nu/Foxn1nu mice and passaged for 21 days. After re-cultivation secretion of CXCL12/SDF-1α was probed via ELISA again. Three independent experiments with similar results were performed.
Transwell assays
Migratory potential was tested in a Boyden-chamber (8 μm pores, Corning) using CXCL12/SDF-1α conditioned media (supernatant of 25000 U87-MGEGFRvIII/SDF-1α). As controls media without CXCL12/SDF-1α harvested from 25000 U87-MGEGFRvIII/EGFP cells and the small chemical compound AMD3100 (25 μg/ml; Sigma-Aldrich) known to inhibit CXCR4, were added. 50000 YTSCXCR4 and YTS wt cells, respectively, were starved for 48 h and added to the upper wells of the chamber in 100 μl FBS-free RPMI-1640 and incubated at 37 °C, 5 % CO2. After 3 h the number of YTS cells that migrated through the porous membrane was assessed by counting the viable cells in the lower wells. Three independent experiments with similar results were performed.
In vivo mouse tumor models
NMRI-Foxn1nu/Foxn1nu mice were obtained from the animal facility of the University of Dresden. Mice were held under standardized pathogen-free conditions with ad libitum access to food and water. Experiments were approved by the Landesdirektion Dresden under the auspices of the German Animal Protection Law. To establish tumors, 100 μl of PBS containing 1 × 106 U87-MGEGFRvIII or 1 × 106 U87-MGEGFRvIII/SDF-1α were subcutaneously injected into the left flank of female mice. After the tumor reached an average size of approximately 10 mm2, mice were intravenously injected with 100 μl PBS containing 4 × 106 YTSmyc-DAP12 cells, YTSMR1.1-DAP12mut cells, YTSMR1.1-DAP12 cells or YTSMR1.1-DAP12/CXCR4, respectively, via the tail vein every 48 h over a period of 40 days. As a control for in vivo tumor cell growth, one group was not treated. The experiments were repeated twice with in total N=10 mice per group for treating mice transplanted with U87-MGEGFRvIII and in total N=20 mice for treatment of U87-MGEGFRvIII/SDF-1α tumors. Tumors were measured in two dimensions two times per week by using a digital caliper. Once the tumor exceeded 18 mm in any of the 3 perpendiculars or animals appeared to be in distress mice were euthanized. The tumor area was calculated according to the formula of ellipse area (1/4 × π × (a × b)). For analysis of the migratory capacity of YTS cells, established tumors were treated with YTSDsRed and YTSDsRed/CXCR4, respectively, every 48 h over a period of for three weeks. Then tumors were extirpated, cryopreserved and cut into 10 μm slices using a microtome. Tumor slices were fixated with 4 % PFA, mounted with Vecta Shield Dapi Medium (Vector) and sealed before fluorescence microscopy analysis (LSM510, Zeiss). To calculate infiltrating cells five fields of view at 400× magnification were analyzed per treatment group. The experiment was performed twice with similar results.
Statistical analyses
The results of chromium release assays were expressed as mean ± and analyzed performing a one-way ANOVA (*p < 0.05) combined with a post-hoc Tukey's multiple comparison test (*p < 0.05). The results of the transwell-assay and results of the in vivo migration were analyzed using Student's t-test. A log-rank test was used for analyses of the survival data. All statistical analyses were performed with Prism software version 6.0 (GraphPad Software Inc., La Jolla, CA, USA).
Results
Generation of YTS cells expressing the chimeric antigen receptor MR1.1-DAP12
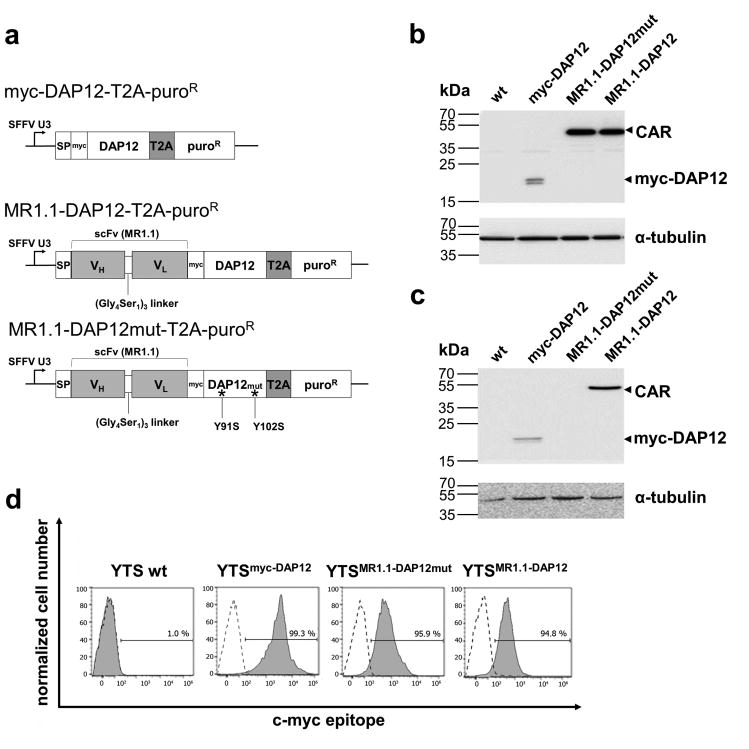
The design of the MR1.1-DAP12 chimeric NK cell antigen receptor and lentiviral vector regions is depicted in Figure 1a. The DAP12-based CAR was generated by fusion of the recently described EGFRvIII-specific MR1.1 single chain fragment variable scFv(MR1.1)37 to DAP12 cDNA devoid of its signal peptide. An internal SFFV U3 promoter in combination with a T2A-site allows concomitant expression of the CAR and of the pac gene. Furthermore, we included an extracellular c-myc-tag for detection of CAR surface expression (Fig. 1a). For our investigations we included a DAP12 control construct, designated myc-DAP12 in which in accordance with the CAR design, the DAP12 signal peptide was exchanged for the Igκ signal peptide. This control construct also contains a myc-epitope inserted between the Igκ signal peptide and the transmembrane domain of DAP12. As further control we constructed a CAR containing a phosphorylation-defective ITAM (MR1.1-DAP12mut).
Fig. 1. Engineering YTS-NK cells with the chimeric antigen receptor MR1.1-DAP12 with specificity for EGFRvIII.
a: Schematic representation of myc-DAP12, MR1.1-DAP12 and MR1.1-DAP12mut coding sequences in the lentiviral vector backbone. The MR1.1-DAP12mut harbors T91S and T102S mutations in its ITAM. The control construct myc-DAP12 lacks the EGFRvIII specific scFv(MR1.1). The internal SFFV U3 promoter of the provirus regulates the expression of the transgenes. Protein synthesis into the secretory pathway is provided by an Igκ signal peptide (SP) at the N-terminus. Expression of the pac-gene (PuroR) is controlled by a Thosea asigna T2A endoproteolytic cleavage site. b-c: Immunoblot analysis of total protein lysates of transduced YTS cells demonstrating expression of the MR1.1-DAP12 CAR and the control constructs. The detection of CAR and controls was accomplished using b: monoclonal c-myc-tag-specific antibody or c: monoclonal anti-DAP12 antibody followed by an HRP-labeled anti-mouse secondary antibody and ECL. Note that the DAP12 antibody cannot bind its epitope in MR1.1-DAP12mut due to mutagenesis in the ITAM. d: The proper surface expression of the functional anti-EGFRvIII-CAR and the controls as determined by flow cytometry analysis using anti-c-myc-biotin antibody and PE-labeled anti-biotin secondary antibody (filled histograms). Cells stained with IgG isotype antibody (dotted line) served as a control.
The NK cell line YTS was transduced with MR1.1-DAP12 CAR, MR1.1-DAP12mut control and myc-DAP12 control, respectively. After antibiotic selection the protein expression of the CAR as well as of the control constructs of the resulting cell lines was confirmed by Western blot analysis using anti-myc and anti-DAP12 antibodies (Fig. 1b, c). We detected bands with expected protein masses for the CAR (50 kDa) and myc-DAP12 control (20 kDa) confirming the proper T2A-mediated cleavage of the transgenes (Fig. 1b). As anticipated, the monoclonal DAP12 antibody, specific for the ITAM domain, failed to bind to the ITAM-mutated form of DAP12 but detected myc-DAP12 and MR1.1-DAP12 (Fig. 1c). Furthermore, a prominent surface expression of the CAR (≥ 95 %) and of the control constructs, respectively, was monitored in the transduced YTS cells (Fig. 1d) via flow cytometric analysis. We constantly observed that myc-DAP12-transduced in YTS cells gave a higher MFI when compared to MR1.1-DAP12 and MR1.1-DAP12mut, respectively. This decrease in MFI of the CAR constructs is likely due to a weakened binding of the anti-myc-antibody due to a possible steric hindrance by the fused scFv portion.
Specific killing of EGFRvIII+ tumor cell lines using YTS cells engineered with MR1.1-DAP12 CAR
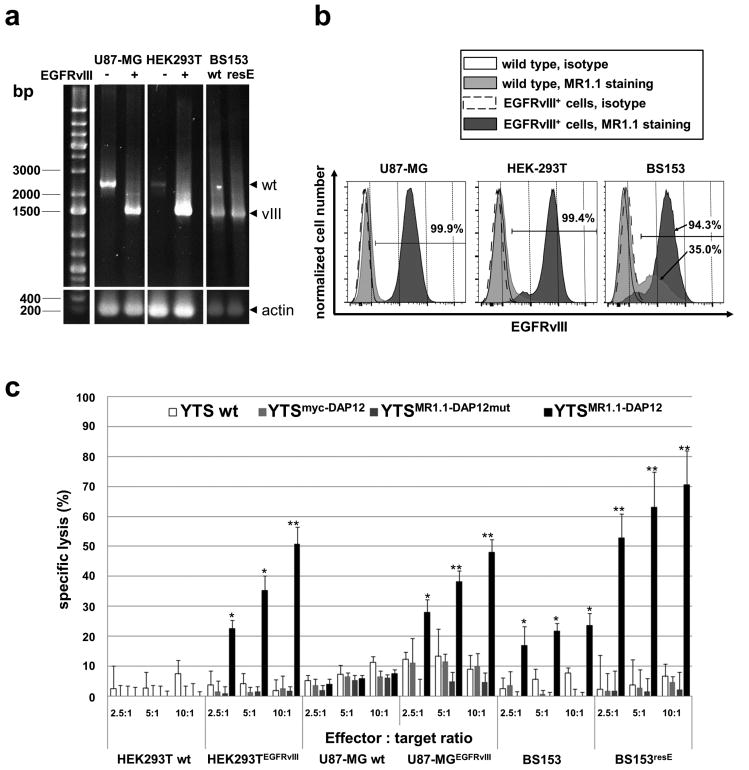
In order to investigate whether the engrafted anti-EGFRvIII CAR redirects NK cells to EGFRvIII+ target cells we utilized generated HEK293TEGFRvIII target cells and established U87-MGEGFRvIII with ectopic expression of EGFRvIII as well as BS153resE glioma cells with high endogenous EGFRvIII expression.45, 46 RT-PCR analyses of reverse transcribed mRNA and using specific primers for the simultaneous detection of both, EGFR wild type and EGFRvIII, revealed that all gene-engineered cells harbored the retroviral-derived EGFRvIII encoding provirus or in the case of BS153resE cells the endogenous EGFRvIII (Fig. 2a). Although the PCR reaction favored the amplification of an EGFRvIII-DNA fragment we were able to detect fragments representing the wild type EGFR gene in U87-MG, in HEK293T wild type cells and to a lesser extent in U87-MGEGFRvIII and HEK293TEGFRvIII cells (Fig. 2a). In our RT-PCR-analyses of BS153 glioblastoma cells with known EGFRvIII-genotype and derived BS153resE cells, which overexpress EGFRvIII due to acquired erlotinib resistance46, we detected, as expected, PCR-signals for genomic EGFRvIII in both cell lines. At the protein level, flow cytometry analyses demonstrated robust expression (94 - 99 % of cells) of EGFRvIII in U87-MGEGFRvIII, HEK293TEGFRvIII, and BS153resE cells when compared to control cells (Fig. 2b). The recombinant single chain antibody (scFv(MR1.1)) used for the analysis also stained a considerable EGFRvIII-positive fraction (mean 35 % EGFRvIII positive cells, SEM +/- 9 %) of BS153 cells which corroborates the results of our RT-PCR analysis. Furthermore, our results are also in line with a previous report describing EGFRvIII expression in a substantial fraction of BS153 wild type cells. 46
Fig. 2. Expression of EGFRvIII on target cell lines and specific cytotoxicity of MR1.1-DAP12 CAR-expressing YTS-NK cells against EGFRvIII+ tumor cells.
a: Agarose gel electrophoresis showing RT-PCR products using primers specific for EGFRvIII as well as EGFR wild type. The PCR reaction favored amplification of EGFRvIII (vIII) with approximately 1.4 kb. Note that also bands representing 2.5 kb EGFR (wt) are detected in U87-MG and HEK293T cells. As internal control an RT-PCR for actin is included. “-“ indicates wild type cells and “+” depicts cells transduced with lentiviral EGFRvIII expression vector. wt, wild type; resE, erlotinib resistant cells. b: FACS assisted analysis of EGFRvIII surface expression of U87-MGEGFRvIII, HEK293TEGFRvIII and BS153resE cells (dark grey histograms) and isogenic U87-MG and HEK293T as well as BS153 cells (light grey histograms). Staining was accomplished using c-myc-tagged scFv(MR1.1) recombinant single chain antibody. Isotype stainings using a prostate stem cell antigen (PSCA) specific scFv(AM1) are included for each isogenic cell line (open histograms). The scFvs were visualized by secondary biotin-labeled c-myc-tag specific antibody and a tertiary anti-biotin-PE antibody. Note the strong surface expression of EGFRvIII in the U87-MGEGFRvIII and HEK293TEGFRvIII target cell lines. Interestingly, BS153 parental cells contain a notable fraction of EGFRvIII-positive cells (see lower arrow) whereas BS153resE cells show a marked increase of the EGFRvIII-positive cell fraction after erlotinib treatment (94 %). c: Gene-engineered and parental YTS (wt) cells were co-cultured with 51chromuim-loaded EGFRvIII+ tumor cells and isogenic EGFRvIII- tumor control cells at different effector to target ratios for 21 h. The mean of specific tumor cell lysis and standard deviation of triplets of three chromium release assays is shown. Note the strong tumor cell lysis mediated by YTSMR1.1-DAP12 cells. (*p < 0.05, **p < 0.01).
In order to investigate the specific cytotoxic efficiency of MR.1-DAP12 CAR-modified YTS cells (YTSMR1.1-DAP12) towards EGFRvIII+ target cells we performed chromium release assays. In the experiments we included YTSMR1.1-DAP12 cells as well as the control cell lines YTSMR1.1-DAP12mut, YTSmyc-DAP12 and YTS wild type cells. As anticipated, YTSMR1.1-DAP12 cells significantly lysed U87-MGEGFRvIII, HEK293TEGFRvIII and BS153resE tumor cells at different effector to target ratios when compared with isogenic wild type or BS153 control cells (Fig. 2c). Of note, we observed a moderate cytotoxic reaction of YTSMR1.1-DAP12 cells against BS153 control cells. As depicted in Fig. 2 the BS153 cell line harbors genomic EGFRvIII and a notable fraction of BS153 cells expressed EGFRvIII, rendering them as a legitimate target for YTSMR1.1-DAP12 cells. Indeed, YTSMR1.1-DAP12 cells lysed between 17 % and 24 % of BS153 cells along with increasing effector target ratios of 2.5:1 to 10:1, which roughly correlates to the percentage of the EGFRvIII+ cell fraction.
YTSMR1.1-DAP12 cells showed an average tumor cell lysis depending on the EGFRvIII+ target cell line ranging from 20 % - 50 % at a effector to target ratio of 2.5:1 to about 50 % - 70 % at a effector to target ratio of 10:1. After normalization of the mean median fluorescence intensities of EGFRvIII staining, by subtracting the MFI of the corresponding isotype stainings, we noted that U87-MGEGFRvIII cells (normalized MFI of 2300) had similar amounts of EGFRvIII on the cell surface as BS153resE cells (normalized MFI of 1950). Yet, BS153resE cells were lysed by YTSMR1.1-DAP12 cells at higher efficiency. Furthermore, although having the highest EGFRvIII expression (normalized MFI of 5420), HEK293TEGFRvIII cells were lysed by YTSMR1.1-DAP12 cells at comparable efficiency as U87-MGEGFRvIII cells, which displayed a considerable lower normalized MFI. Of note, BS154resE cells, having a lower expression of EGFRvIII than HEK293TEGFRvIII cells were significantly better killed by YTSMR1.1-DAP12 cells (p < 0.05). Altogether, there was no direct correlation between the levels of EGFRvIII expression in the different tumor cell lines and the strength of the cytotoxic reaction of YTSMR1.1-DAP12 cells. This result is in line with previous reports and our own results using CAR-modified NK92 and YTS cells, respectively, engaging several ErbB2/HER-2- and prostate stem cell antigen (PSCA)-positive target cell lines. 40, 47 Hence, we suggest that the difference in the strength of cytotoxic reaction against the target cell lines was not only due to EGFRvIII expression levels but also depends on the composition and expression level of other surface molecules, in particular cell adhesion molecules.
Incubation of YTSMR1.1-DAP12 cells with EGFRvIII- isogenic HEK293T and U87-MG cells did not lead to specific tumor cell lysis. Moreover, unmodified YTS wt cells, YTSmyc-DAP12 cells and YTSMR1.1-DAP12mut cells lysed neither EGFRvIII+ nor EGFRvIII- HEK293T and BS153 tumor cells, yet we observed a weak basal cytotoxicity of YTS wild type cells and YTSmyc-DAP12 cells when confronted with U87-MG and U87-MGEGFRvIII cells. Taken together, these results indicate that the MR1.1-DAP12 chimeric antigen receptor confers specific cytotoxicity against EGFRvIII+ target cells.
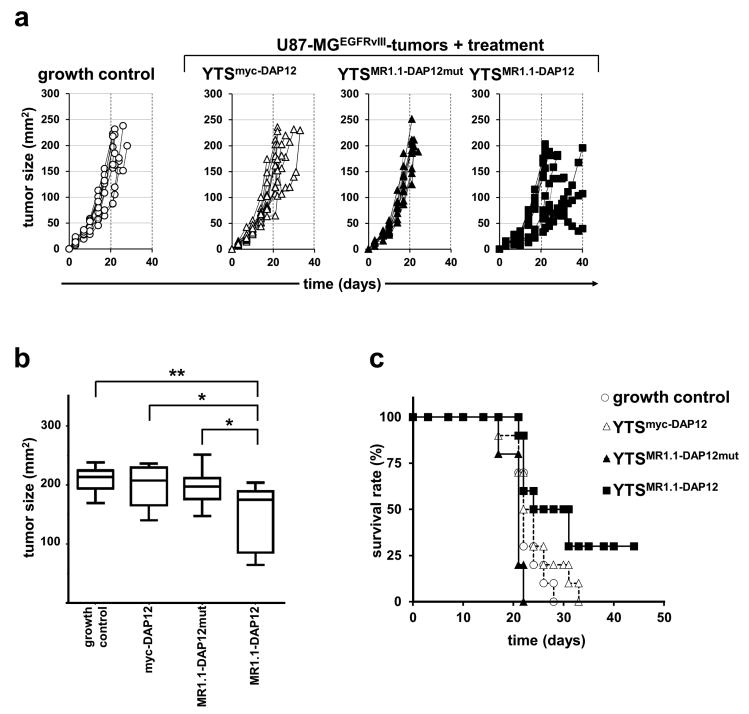
In a next step, we sought to evaluate the YTSMR1.1-DAP12 cells in an experimental immunotherapy using a xenograft model. For this, U87-MGEGFRvIII cells were subcutaneously injected into the left flank of NMRI-Foxn1nu/Foxn1nu mice. Five days after tumor cell injection and establishment of palpable tumors, YTSMR1.1-DAP12 and control cells (YTSMR1.1-DAP12mut, YTSmyc-DAP12), respectively, were injected via the tail vein every 48 h over a period of 40 days. Mice xenografted with U87-MGEGFRvIII cells and without YTS cell treatment served as internal tumor growth control group. We chose this treatment protocol, since a recent experimental immunotherapy using anti-PSCA-CAR armed YTS cells revealed that the cells were short lived and that an anti-tumoral effect was only achieved by sustained application.40 Remarkably, when looking at the individual tumor growth curves we observed that a notable fraction of mice treated with YTSMR1.1-DAP12 cells showed a delay of tumor growth when compared to all control groups (Fig. 3a). However, complete tumor remission was not observed. Besides this, also weak and non-responders to treatment were noted. Yet, the analysis of maximal tumor growth at day 30 showed a significant decrease in the mean tumor size of YTSMR1.1-DAP12 treated mice when compared to the controls in particular when compared to the group treated with NK cells modified with the phosphorylation-defective CAR MR1.1-DAP12mut (Fig. 3b). In line with this, treatment with YTSMR1.1-DAP12 cells caused a statistically significant increase of median survival time of 27.5 days in comparison with the tumor growth control group (tumor growth control 22 days; p < 0.05), and with the group of mice treated with YTSMR1.1-DAP12mut cells 21 days; p < 0.001). Furthermore, treatment with YTSMR1.1-DAP12 was accompanied by 30 % survivors at day 45 (Fig. 3c). Treatment of mice with YTSmyc-DAP12 cells resulted, as expected, in a median survival time shorter than observed with YTSMR1.1-DAP12-treated mice, but did not reach statistical significance (23 days; p = 0.109) which might account to the observed basal cytoxicity of YTSmyc-DAP12 cells towards U87-MG cells. Taken together, these data indicate a specific but so far only moderate anti-tumor effect of MR1.1-DAP12-redirected NK cells on established EGFRvIII+ tumors in a xenograft mouse model which we sought to improve by employment of an additional chemokine receptor for redirecting NK cells to the tumor site.
Fig. 3. MR1.1-DAP12 CAR-expressing YTS-NK cells attenuate tumor growth of EGFRvIII+ tumor cells and increase survival of treated mice.
U87-MGEGFRvIII cells were subcutaneously injected into NMRI-Foxn1nu/Foxn1nu mice. After tumor development, mice were treated with intravenous tail vein injections of YTSMR1.1-DAP12, YTSMR1.1-DAP12mut and YTSmyc-DAP12 NK cells. Untreated mice were included as an additional control (growth control). Depicted is the summarized data from two independent experiments (N=10 for each group) a: Tumor growth of mice in the different treatment groups was monitored up to 40 days and is depicted as individual tumor growth curves. b: Mean tumor size of mice injected with YTSMR1.1-DAP12 cells or control cells measured at day 30 post tumor cell injection. The box plot diagram shows means and 95 % confidence intervals. YTSMR1.1-DAP12-treated mice showed inhibited tumor growth when compared to controls. (*p < 0.05, **p < 0.01). c: Kaplan-Meier survival curve for overall survival of tumor bearing mice injected with YTSMR1.1-DAP12 NK cells or controls. Mice treated with the DAP12-based anti-EGFRvIII CAR showed statistically improved survival when compared to YTSMR1.1-yc-DAP12mut (p < 0.001) and growth control (p < 0.05). p = 0.109 for YTSMR1.1-DAP12 when compared to mice treated with YTSmyc-DAP12. (N = 10 for each group).
NK cells ectopically expressing CXCR4 exhibit chemotaxis towards CXCL12/SDF-1α
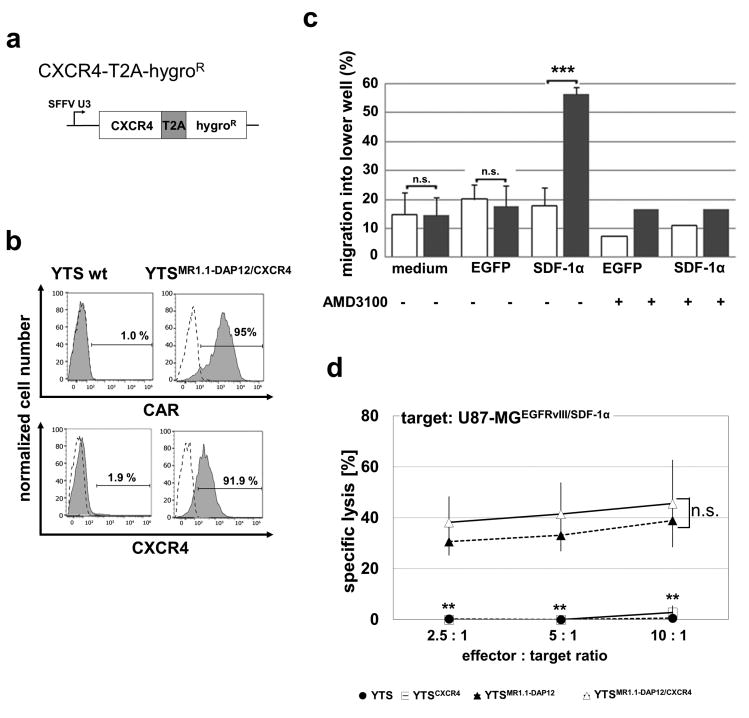
In order to enable a redirected migration of NK cells to the tumor site we focused on the CXCL12/SDF-1α / CXCR4 axis. For our experiments, we transduced U87-MGEGFRvIII cells with a lentiviral vector encoding SDF-1α-IRES-EGFP-ZeoR and with an IRES-EGFP-ZeoR empty vector, respectively. After antibiotic selection we performed proliferation analysis and revealed a significant decrease of 25 % in clonal survival and proliferation of U87-MGEGFRvIII/SDF-1α and U87-MGEGFRvIII/EGFP cells when compared to parental U87-MGEGFRvIII and U87-MG wild type cells. Proliferation rates of U87-MGEGFRvIII/SDF-1α and U87-MGEGFRvIII/EGFP cells were in the same range (data not shown). When we tested the cell culture supernatant of the U87-MGEGFRvIII/SDF-1α cells by ELISA we measured 2 +/- 0.228 ng/ml secreted CXCL12/SDF-1α whereas CXCL12/SDF-1α-levels of U87-MGEGFRvIII/EGFP cells were below the detection limit. Even after passaging for 21 days in mice and re-cultivation the U87-MGEGFRvIII/SDF-1α cells still secreted more than 1 +/- 0.292 ng/ml CXCL12/SDF-1α into the cell culture supernatant indicating the long term capability of these cells to attract chemosensitive cells in an in vivo setting (data not shown). In a next step we engineered YTSMR1.1-DAP12 cells to express CXCR4. The lentiviral construct used for transduction is depicted in Fig. 4a. After transduction and antibiotic selection we revealed YTSMR1.1-DAP12/CXCR4 cells displaying strong surface expression of CXCR4 (over 90 % positive cells) and concomitant robust MR1.1-DAP12 expression (95 % positive cells) (Fig. 4b). When the YTSMR1.1-DAP12/CXCR4 cells were tested in a transwell assay we furthermore observed a significantly increased migration capacity towards CXCL12/SDF-1α-containing conditioned medium from U87-MGEGFRvIII/SDF-1α cells when compared to the migration towards U87-MGEGFRvIII/EGFP conditioned medium and medium control, respectively (Fig. 4c). Furthermore, inhibition of the CXCR4 receptor by the small chemical compound AMD3100 reduced migration of YTSMR1.1-DAP12/CXCR4 cells to baseline level confirming that engraftment of CXCR4 confers a redirected chemotaxis to CXCL12/SDF-1α. When YTSMR1.1-DAP12/CXCR4 were compared to YTSMR1.1-DAP12 in a cytotoxicity assay no significant differences in specific lysis of EGFRvIII+ target cells were detected (supplementary Fig. S1) which indicates that besides CAR expression an additional ectopic CXCR4 expression does not impair the cytotoxic capacity of YTS cells. On the other hand, it was of special interest if simultaneous CAR and CXCR4 signaling might lead to an enhanced cytotoxic response of NK cells confronted with double positive EGFRvIII/SDF-1α target cells. As demonstrated in Fig. 4d cytotoxicity assays using U87-MGEGFRvIII/SDF-1α as target cell line displayed an only slightly increased cytotoxic reaction of YTSMR1.1-DAP12/CXCR4 cells when compared to YTSMR1.1-DAP12 cells. As expected, we did not observe a specific cytotoxicity of YTS wild type cells and YTSCXCR4 cells towards U87-MGEGFRvIII/SDF-1α target cells.
Fig. 4. Efficient transduction of CXCR4 and chemotaxis of YTSMR1.1-DAP12/CXCR4 cells to CXCL12/SDF-1α-conditioned medium.
a: Schematic representation of the CXCR4 coding sequences in the lentiviral vector backbone. b: FACS-assisted analysis of CAR and surface expression of double-transduced YTSMR1.1-DAP12/CXCR4 cells. CAR surface expression was verified using a monoclonal biotin-labeled c-myc-tag specific antibody and a secondary anti-Biotin-APC antibody. CXCR4 expression was detected using a monoclonal anti-CXCR4 antibody coupled to APC. Isotype staining as well as staining of parental YTS cells with anti-c-myc are included as controls. c: Depicted is the result from one representative transwell experiment using CXCL12/SDF-1α-conditioned medium derived from U87-MGEGFRvIII/SDF-1α cells (SDF-1α). As control we used normal medium and cell culture supernatant from U87-MGEGFRvIII/EGFP controls (EGFP). The CXCL12/SDF-1α-conditioned medium (SDF-1α) led to a significant increase in the percentage of YTSMR1.1-DAP12/CXCR4 cells (grey columns) reaching the lower wells of the Boyden chambers when compared to YTSMR1.1-DAP12 cells (white columns). Note that treatment with the CXCR4 inhibitor AMD3100 completely abolished the directed migration of YTSMR1.1-DAP12/CXCR4 cells towards the CXCL12/SDF-1α-conditioned medium. ***p < 0.001. d: Gene-engineered YTSMR1.1-DAP12/CXCR4, YTSMR1.1-DAP12, YTSCXCR4 and YTS wild type cells were co-cultured with 51chromium-loaded U87-MGEGFRvIII/SDF-1α cells at different effector to target ratios for 21 h. The mean of specific tumor cell lysis and standard deviation of three chromium release assays is shown. Note that the YTSMR1.1-DAP12/CXCR4 cells react with a slightly increased but statistically not significant (n.s.) extent to the U87-MGEGFRvIII/SDF-1α target cells when compared to the YTSMR1.1-DAP12 cells. Note the significant CAR-mediated specific cytotoxicity towards target cells when compared to YTSCXCR4 and YTS wild type cells (for both **p < 0.01).
YTSMR1.1-DAP12/CXCR4 with engrafted chemotaxis for CXCL12/SDF-1α improve survival of mice with U87-MGEGFRvIII/SDF-1α xenografts
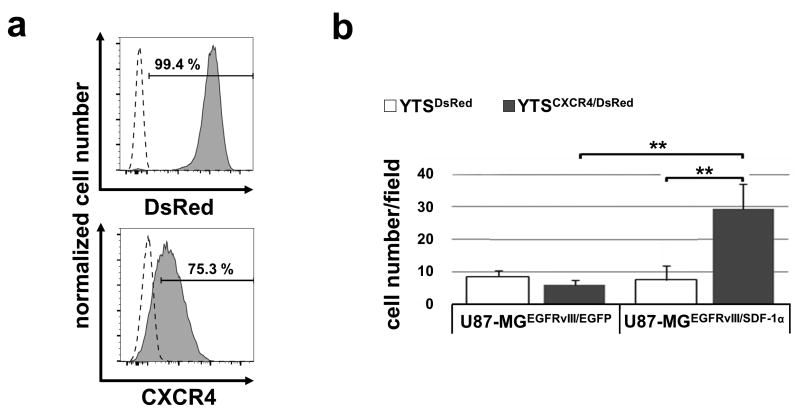
To pursue our chemotaxis-approach, YTSMR1.1-DAP12/CXCR4 cells were tested in an in vivo setting. First, the intra-tumoral accumulation of intravenously administered DsRed reporter gene-marked YTSCXCR4/DsRed cells and YTSDsRed control cells (see Fig. 5a) were investigated. After excision and analysis of tumor slices using fluorescence microscopy we observed that significantly more YTSCXCR4/DsRed cells migrated into the U87-MGEGFRvIII/SDF-1α tumors than YTSDsRed cells. Furthermore, there was no increased intra-tumoral accumulation of YTSCXCR4/DsRed as well as YTSDsRed cells in U87-MGEGFRvIII gliomas devoid of CXCL12/SDF-1α, confirming a specific chemotaxis of CXCR4-modified YTS towards CXCL12/SDF-1α-secreting U87-MGEGFRvIII/SDF-1α gliomas (Fig. 5b).
Fig. 5. Improved intratumoral infiltration of CXCR4-engineered YTS-NK cells into U87-MGEGFRvIII/SDF-1α-xenografts.
a: YTS cells were transduced with DsRed and established YTSDsRed cells were further genetically modified to express CXCR4. Depicted are the flow cytometry histograms showing DsRed expression (filled histogram). CXCR4 expression was detected using a monoclonal anti-CXCR4 antibody coupled to APC (filled histogram). Wild type cells and isotype stained cells, respectively, are included as control (dotted line). b: CXCL12/SDF-1α-secreting U87-MGEGFRvIII/SDF-1α and U87-MGEGFRvIII/EGFP control cells were subcutaneously injected into the right flank of NMRI-Foxn1nu/Foxn1nu mice. After tumor development, mice were treated with intravenous tail vein injections with either YTSDsRed/CXCR4 or YTSDsRed cells and the number of infiltrated DsRed+ cells in tumors was analyzed per field of view (40× objective). YTSDsRed/CXCR4 cells showed an improved intra-tumoral accumulation in U87-MGEGFRvIII/SDF-1α tumors when compared to all controls. **p < 0.01; n.s., not significant. The experiment was repeated twice with similar results.
For an experimental immunotherapy of CXCL12/SDF-1α-secreting U87-MGEGFRvIII/SDF-1α gliomas were treated with gene-modified YTS-NK cells which were injected via the tail vein every 48 h over a period of 40 days. Mice xenografted with U87-MGEGFRvIII/SDF-1α cells and without YTS cell treatment served as control group.
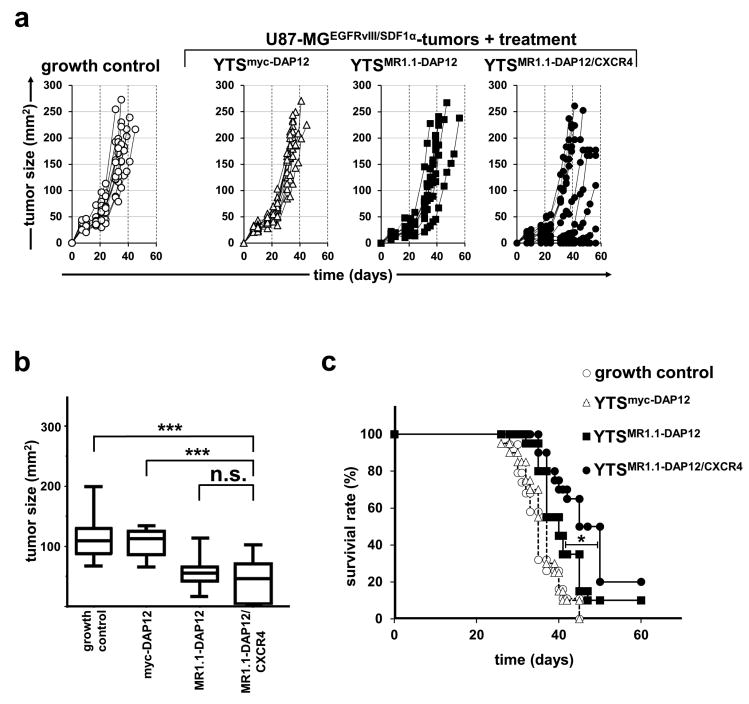
In line with our in vitro data we observed an attenuated growth of U87-MGEGFRvIII/SDF-1α tumors when compared to U87-MGEGFRvIII tumors (see Fig. 3) resulting in later death of tumor-xenografted mice (Fig. 6a, c). When we looked at individual responses to NK cell immunotherapy, we noted delay of tumor growth in a sizable number of mice treated with YTSMR1.1-DAP12/CXCR4 when compared to all control groups (Fig. 6a). Notably, 3 out of 20 mice treated with YTSMR1.1-DAP12/CXCR4 cells showed complete tumor eradication at days 15, 15 and 17 and remained tumor free during the whole experiment. We also observed some weak and non-responders as depicted in the individual tumor growth curves (Fig. 6a). However, analysis of maximal tumor size at day 30 showed a significantly reduced mean tumor growth of mice treated with YTSMR1.1-DAP12/CXCR4 when compared to the tumor growth control and mice treated with YTSmyc-DAP12 control cells. Yet, at day 30 no significant difference in mean tumor growth was reached when YTSMR1.1-DAP12/CXCR4-treated mice were compared to mice treated with YTSMR1.1-DAP12 (p = 0.165) (Fig. 6b). However, in the long run we observed a substantial increase of median survival time to 47.5 days of mice treated with YTSMR1.1-DAP12/CXCR4 when compared to treatment with YTSMR1.1-DAP12 (40 days,*p < 0.05) and when, compared to the control groups (tumor growth control 35 days; p < 0.0001, treatment with YTSmyc-DAP12 cells: 36 days; p < 0.0001) (Fig. 6c). Taken together, our data indicate an improved anti-tumoral performance of CAR-engineered NK cells when engrafted with an additional CXCR4 for targeting CXCL12/SDF-1α-secreting gliomas.
Fig. 6. CAR/CXCR4-engineered YTS-NK cells improve survival of mice with established U87-MGEGFRvIII/SDF-1α-xenografts.
U87-MGEGFRvIII/SDF-1α cells were subcutaneously injected into the right flank of NMRI-Foxn1nu/Foxn1nu mice. After tumor development, mice were treated with intravenous tail vein injections of YTSMR1.1-DAP12/CXCR4, YTSMR1.1-DAP12, and YTSmyc-DAP12 NK cells. Untreated mice were included as an additional control. a: Tumor growth of mice in the different treatment groups was monitored constantly and depicted as individual tumor growth curves (one representative experiment, N=10 for each group). b: Mean tumor size of mice injected with YTSMR1.1-DAP12 cells or control cells measured at day 30 post tumor cell injection. The box plot diagram shows means and 95 % confidence intervals. YTSMR1.1-DAP12/CXCR4-treated mice showed inhibited tumor growth when compared to YTSmyc-DAP12 treated tumors and when compared to the tumor growth control at day 30 (***p < 0.001). Differences in the tumor growth of YTSMR1.1-DAP12/CXCR4-treated mice at day 30 and YTSMR1.1-DAP12 treated tumors did not reach statistical significance (p = 0.165; n.s.). c: Kaplan-Meier survival curve for overall survival of tumor bearing mice injected with YTSMR1.1-DAP12/CXCR4 cells, YTSMR1.1-DAP12 cells or controls. Note that mice treated with the YTSMR1.1-DAP12/CXCR4 cells showed statistically improved survival when compared to YTSMR1.1-DAP12 as indicated (*p < 0.05), and to YTSmyc-DAP12 as well as growth control (both p < 0.0001). Mice treated with YTSMR1.1-DAP12 had a statistically increased survival when compared to YTSmyc-DAP12–treated mice (p < 0.05) and growth control (p < 0.001). (summarized data from two independent experiments, N = 20 for each group).
Discussion
Chemokines and chemokine receptors play an important role in cancer progression and lymphocyte migration to tumors.48, 49 So far previous studies using T cells genetically-engineered to express chemokine receptors CXCR2 and CCR2, respectively, provided first evidence that migration of T cells can be modulated towards a specific chemokine or chemokine secreting tumors,50 and can improve anti-tumor activity of CAR-modified T cells in preclinical xenograft models.51, 52 Yet, the genetic modification of NK cells with chemokine receptors had not been pursued so far.
In the present study, we used CXCL12/SDF-1α / CXCR4 axis as a generic chemotaxis model for redirecting NK cells to glioma cells. CXCL12/SDF-1α has been reported to be secreted by gliomas and is implicated in glioma progression.31, 32, 34 In particular we seek to investigate whether the anti-tumoral effects of an adoptive NK cell immunotherapy can be improved by a genetically engineered chemotaxis. U87-MG, which we selected as our target cell line, has been described to secrete CXCL12/SDF-1α,32 yet in our hands we were not able to detect significant amounts of the secreted chemokine by ELISA. Since it is tempting to speculate that this contradictory result was due to technical differences or just by a longer in vitro adaption of the U87-MG cell line we decided to generate U87-MG cells with ectopic expression of CXCL12/SDF-1α for our investigation. Interestingly, after transduction and selection of CXCL12/SDF-1α-positive U87-MGEGFRvIII/SDF-1α cells and U87-MGEGFRvIII/EGFP control cells we did not detect any significant differences in cell proliferation and clonogenic survival due to autocrine CXCL12/SDF-1α stimulation as previously reported.31-33 Yet, we observed a stable and continuous CXCL12/SDF-1α-secretion even after in vivo passaging of our modified U87-MG cell lines which rendered them suitable for our studies focusing on genetically engineered chemotaxis of NK cells.
As target structure for CAR-mediated activation of NK cells we chose the mutant EGFRvIII receptor. We therefore designed a specific CAR consisting of the scFv(MR1.1) and a fused DAP12 signal adaptor protein of NK cells containing one ITAM-motif. This study demonstrated, that YTS NK cells transduced with MR1.1-DAP12 readily lysed different EGFRvIII+-target cells with increasing efficiency at higher effector-to-target ratios whereas isogenic EGFRvIII- cells were not affected. That just the increased scFv(MR1.1)-mediated adhesion of the NK cells to U87-MGEGFRvIII target cells contributed to the significant increase in cytotoxicity towards EGFRvIII+-target cells was rendered unlikely since NK cells genetically modified with a phosphorylation defective MR1.1-DAP12mut did not attack EGFRvIII+-target cells. When we tested the feasibility of the CAR-modified NK cells in a subcutaneous glioma xenograft model we detected a moderate but significant inhibition of tumor growth and significantly increased survival after continuous systemic application of YTSMR1.1-DAP12 cells. In line with our recent data using anti-PSCA-DAP12 modified YTS cells to treat subcutaneous HEK293TPSCA xenografts, we monitored weak and non-responders which we hypothesized was linked to an insufficient NK cell migration to the tumors.40
In order to overcome insufficient NK cell migration into glioblastoma we established genetically modified YTS cells to express both CAR and CXCR4. At this point, it was of special interest whether a concomitant activation of the CAR and of the CXCR4 receptor on NK cells eventually results in an augmented cytotoxic response against EGFRvIII+ glioma cells, since it has been reported that binding of CXCL12/SDF-1α to CXCR4 can induce cell signaling through Akt and ERK1/2.32 In particular ERK-signaling is also involved in activation and degranulation of NK cells.53 Yet, in chromium release assays using YTSMR1.1-DAP12 and YTSMR1.1-DAP12/CXCR4 as effector cells no significant differences in the cytotoxic response against U87-MGEGFRvIII/SDF-1α cells were observed. In addition, YTS cell genetically modified with CXCR4 alone showed no cytotoxic reaction against U87-MGEGFRvIII/SDF-1α cells. It is therefore conceivable that CXCL12/SDF-1α exclusively enables chemotactic migration and does not lead to increased activation of NK cells. Considering NK cells as short-lived effector cells this result is of particular interest for future applications of CXCR4-modified NK cells. It indicates that CXCL12/SDF-1α signaling in NK cells does not lead to off-target effects in tissues described to express CXCL12/SDF-1α, in particular bone marrow, lung, and lymph nodes.54 Whether CXCR4-modified NK cells accumulate in these tissues and organs remains of special interest since a redirection of NK cells to these sites might be an attractive rationale for treatment of metastatic breast cancer and metastatic melanoma.54
That the NK cells engrafted with CXCR4 were indeed redirected to CXCL12/SDF-1α was proven in transwell-assays with CXCL12/SDF-1α-containing conditioned medium. Furthermore, analysis of CXCL12/SDF-1α-secreting U87-MGEGFR/SDF-1α xenografts revealed a statistically higher number of infiltrating NK cells modified with CXCR4 when compared to xenografted tumors treated with YTS cells devoid of the chemokine receptor. Finally, the genetic modification of NK cells with a CAR and additional chemokine receptor significantly improved survival of mice xenografted with U87-MGEGFRvIII/SDF-1α-tumors when compared to mice treated with only CAR-modified NK cells and lead to a complete tumor remission in a number of mice. Yet, we observed also weak and non-responders to therapy. We suggest that the anti-tumoral efficiency of YTSMR1.1-DAP12/CXCR4 NK cells in some animals was stalled by individual lower tumor perfusion rates and resulting insufficient CXCL12/SDF-1α gradients. In general, secreted chemokines, such as CXCL12/SDF-1α, are immobilized by heparin sulfate glycosaminoglycans (GAG) chains of proteoglycans, mostly bound to the surface of endothelial cells. This establishes a gradient with the highest concentration closest to the origin of chemokine secretion.55 So far most glioblastomas, as well as xenografted U87-MG display an unusual vessel formation which is accompanied by the random appearance of microthrombi which can impair perfusion of the tumor.56 In combination with an accumulation of matrix metalloproteinases (MMP-2 and-9),57 which cleave CXCL12/SDF-1α molecules,58 this might obstruct infiltration of CXCR4-modified NK cells at this site. In the future, it could be worthwhile to treat glioblastomas prior NK cell infusion with low doses of low molecular weight heparin, an anticoagulant drug which is regularly used in postoperative management of GBM, to improve chemotactic gradients as well as extravasation of CXCR4-modified NK cells at the tumor site.59
In the treatment of glioblastoma, a single agent may not work efficiently as a combination therapy. This could imply that other therapeutic interventions designed to synergize with redirected CAR-modified NK cells may be needed to achieve a better killing efficiency when targeting glioblastoma cells. Some examples of these approaches include: Aurora B and Survivin inhibitors that can increase the expression of TRAIL-R2/DR5 death receptors on glioblastoma cells and therefore increases killing efficiency of TRAIL+- NK cells, 60, 61 locked nucleic acids (LNAs) constraining tumor miRNAs and increasing expression of NKG2D ligands on glioma cells which can activate NK cells, 62 maintaining NK cell cytotoxicity by applying TGFβRI-inhibitors or soluble TGFβRII molecules to overcome the immunosuppressive milieu of glioblastomas. 63, 64
In summary, we have shown that NK cells genetically engineered with a tumor specific CAR and chemokine receptor can improve immunotherapy of solid tumors. The use of an EGFRvIII-specific CAR might be promising for the treatment of glioblastoma in particular when targeting EGFRvIII+ glioblastoma stem populations.65
In addition, to the use of chemokine receptors for CAR approaches it seems also conceivable to employ haploidentical NK cells with engineered chemotaxis for immunotherapy of tumors. In particular the use of CXCR4-modified haploidentical NK cells might be a rationale to redirect graft versus tumor effects to residual CXCL12/SDF-1α-secreting glioma nests. One might also consider the use of CXCR4-modified haploidentical NK cells when targeting other tumor entities such as AML, in particular therapy-refractory AML blasts residing in the bone marrow.66 In conclusion, we believe that engineered chemotaxis of NK cells expressing CARs or used in a haploidentical setting represent a promising tool for improvement of adjuvant cellular immunotherapy of cancer.
Supplementary Material
Supplementary Fig. S1: Simultaneous CAR and CXCR4 expression does not impair the cytotoxic capacity of YTS-NK cells. YTSMR1.1-DAP12, YTSMR1.1-DAP12/CXCR4 and YTSCXCR4 cells were co-cultured with 51chromium-loaded EGFRvIII+ tumor cells and isogenic EGFRvIII- tumor control cells at different effector to target ratios for 21 h. The mean of specific tumor cell lysis and standard deviation of triplets of three independent chromium release assays is shown. Note the strong tumor cell lysis mediated by YTSMR1.1-DAP12/CXCR4 cells which is similar to the cytotoxic reaction of YTSMR1.1-DAP12 cells. Both are significantly increased when compared to the nearly undetectable specific lysis of YTSCXCR4 cells. (* p < 0.05, **p < 0.01).
Acknowledgments
This work was supported by the Deutsche Krebshilfe e.V. (Az.: 109377) (A.T.) and Doktor Robert Pfleger-Stiftung (A.T.). This research was also supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (I.P.), and in part by a grant from the Forschungs- und Wissenschaftsstiftung Hamburg (K.L.). We thank Ralf Wiedemuth for supporting FACS analyses and Bianca Goldberg, Elke Leipnitz and Katja Robel for excellent technical assistance.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007 Aug;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topfer K, Kempe S, Muller N, Schmitz M, Bachmann M, Cartellieri M, et al. Tumor evasion from T cell surveillance. J Biomed Biotechnol. 2011;2011:918471. doi: 10.1155/2011/918471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009 May;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Darcy PK, Neeson P, Yong CS, Kershaw MH. Manipulating immune cells for adoptive immunotherapy of cancer. Curr Opin Immunol. 2014 Feb 14;27C:46–52. doi: 10.1016/j.coi.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Sampson JH, Choi BD, Sanchez-Perez L, Suryadevara CM, Snyder DJ, Flores CT, et al. EGFRvIII mCAR-modified T cell therapy cures mice with established intracerebral glioma and generates host immunity against tumor-antigen loss. Clin Cancer Res. 2013 Dec 18; doi: 10.1158/1078-0432.CCR-13-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jena B, Dotti G, Cooper LJ. Redirecting T-cell specificity by introducing a tumor-specific chimeric antigen receptor. Blood. 2010 Aug 19;116(7):1035–1044. doi: 10.1182/blood-2010-01-043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001 Oct;1(1):41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 8.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011 Jan 7;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paust S, von Andrian UH. Natural killer cell memory. Nat Immunol. 2011 Jun;12(6):500–508. doi: 10.1038/ni.2032. [DOI] [PubMed] [Google Scholar]
- 10.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008 May;9(5):495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks AG, Boyington JC, Sun PD. Natural killer cell recognition of HLA class I molecules. Rev Immunogenet. 2000;2(3):433–448. [PubMed] [Google Scholar]
- 12.Karre K. Express yourself or die: peptides, MHC molecules, and NK cells. Science. 1995 Feb 17;267(5200):978–979. doi: 10.1126/science.7863341. [DOI] [PubMed] [Google Scholar]
- 13.Smyth MJ, Cretney E, Kelly JM, Westwood JA, Street SE, Yagita H, et al. Activation of NK cell cytotoxicity. Mol Immunol. 2005 Feb;42(4):501–510. doi: 10.1016/j.molimm.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 14.Oshimi Y, Oda S, Honda Y, Nagata S, Miyazaki S. Involvement of Fas ligand and Fas-mediated pathway in the cytotoxicity of human natural killer cells. J Immunol. 1996 Oct 1;157(7):2909–2915. [PubMed] [Google Scholar]
- 15.Shrikant P, Mescher MF. Control of syngeneic tumor growth by activation of CD8+ T cells: efficacy is limited by migration away from the site and induction of nonresponsiveness. J Immunol. 1999 Mar 1;162(5):2858–2866. [PubMed] [Google Scholar]
- 16.Rossig C, Bollard CM, Nuchtern JG, Merchant DA, Brenner MK. Targeting of G(D2)-positive tumor cells by human T lymphocytes engineered to express chimeric T-cell receptor genes. Int J Cancer. 2001 Oct 15;94(2):228–236. doi: 10.1002/ijc.1457. [DOI] [PubMed] [Google Scholar]
- 17.Lizee G, Cantu MA, Hwu P. Less yin, more yang: confronting the barriers to cancer immunotherapy. Clin Cancer Res. 2007 Sep 15;13(18 Pt 1):5250–5255. doi: 10.1158/1078-0432.CCR-07-1722. [DOI] [PubMed] [Google Scholar]
- 18.Kmiecik J, Zimmer J, Chekenya M. Natural killer cells in intracranial neoplasms: presence and therapeutic efficacy against brain tumours. J Neurooncol. 2014 Jan;116(1):1–9. doi: 10.1007/s11060-013-1265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madri JA, Graesser D. Cell migration in the immune system: the evolving inter-related roles of adhesion molecules and proteinases. Dev Immunol. 2000;7(2-4):103–116. doi: 10.1155/2000/79045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charalambous C, Hofman FM, Chen TC. Functional and phenotypic differences between glioblastoma multiforme-derived and normal human brain endothelial cells. J Neurosurg. 2005 Apr;102(4):699–705. doi: 10.3171/jns.2005.102.4.0699. [DOI] [PubMed] [Google Scholar]
- 21.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997 Jan 31;272(5):2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 22.Beers R, Chowdhury P, Bigner D, Pastan I. Immunotoxins with increased activity against epidermal growth factor receptor vIII-expressing cells produced by antibody phage display. Clin Cancer Res. 2000 Jul;6(7):2835–2843. [PubMed] [Google Scholar]
- 23.Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005 Oct 21;280(42):35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- 24.Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006 Sep 4;203(9):2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berahovich RD, Zabel BA, Penfold ME, Lewen S, Wang Y, Miao Z, et al. CXCR7 protein is not expressed on human or mouse leukocytes. J Immunol. 2010 Nov 1;185(9):5130–5139. doi: 10.4049/jimmunol.1001660. [DOI] [PubMed] [Google Scholar]
- 26.Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayol K, Biajoux V, Marvel J, Balabanian K, Walzer T. Sequential desensitization of CXCR4 and S1P5 controls natural killer cell trafficking. Blood. 2011 Nov 3;118(18):4863–4871. doi: 10.1182/blood-2011-06-362574. [DOI] [PubMed] [Google Scholar]
- 28.Luo Y, Lathia J, Mughal M, Mattson MP. SDF1alpha/CXCR4 signaling, via ERKs and the transcription factor Egr1, induces expression of a 67-kDa form of glutamic acid decarboxylase in embryonic hippocampal neurons. J Biol Chem. 2008 Sep 5;283(36):24789–24800. doi: 10.1074/jbc.M800649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Z, Zhou W, Guan S, Wang J, Liang Y. Contribution of SDF-1alpha/CXCR4 signaling to brain development and glioma progression. Neurosignals. 2013;21(3-4):240–258. doi: 10.1159/000339091. [DOI] [PubMed] [Google Scholar]
- 30.Sehgal A, Keener C, Boynton AL, Warrick J, Murphy GP. CXCR-4, a chemokine receptor, is overexpressed in and required for proliferation of glioblastoma tumor cells. J Surg Oncol. 1998 Oct;69(2):99–104. doi: 10.1002/(sici)1096-9098(199810)69:2<99::aid-jso10>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 31.Bajetto A, Barbieri F, Dorcaratto A, Barbero S, Daga A, Porcile C, et al. Expression of CXC chemokine receptors 1-5 and their ligands in human glioma tissues: role of CXCR4 and SDF1 in glioma cell proliferation and migration. Neurochem Int. 2006 Oct;49(5):423–432. doi: 10.1016/j.neuint.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Barbero S, Bonavia R, Bajetto A, Porcile C, Pirani P, Ravetti JL, et al. Stromal cell-derived factor 1alpha stimulates human glioblastoma cell growth through the activation of both extracellular signal-regulated kinases 1/2 and Akt. Cancer Res. 2003 Apr 15;63(8):1969–1974. [PubMed] [Google Scholar]
- 33.Schulte A, Gunther HS, Phillips HS, Kemming D, Martens T, Kharbanda S, et al. A distinct subset of glioma cell lines with stem cell-like properties reflects the transcriptional phenotype of glioblastomas and overexpresses CXCR4 as therapeutic target. Glia. 2011 Apr;59(4):590–602. doi: 10.1002/glia.21127. [DOI] [PubMed] [Google Scholar]
- 34.Salmaggi A, Gelati M, Pollo B, Marras C, Silvani A, Balestrini MR, et al. CXCL12 expression is predictive of a shorter time to tumor progression in low-grade glioma: a single-institution study in 50 patients. J Neurooncol. 2005 Sep;74(3):287–293. doi: 10.1007/s11060-004-7327-y. [DOI] [PubMed] [Google Scholar]
- 35.Salcedo R, Wasserman K, Young HA, Grimm MC, Howard OM, Anver MR, et al. Vascular endothelial growth factor and basic fibroblast growth factor induce expression of CXCR4 on human endothelial cells: In vivo neovascularization induced by stromal-derived factor-1alpha. Am J Pathol. 1999 Apr;154(4):1125–1135. doi: 10.1016/s0002-9440(10)65365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones G, Machado J, Jr, Merlo A. Loss of focal adhesion kinase (FAK) inhibits epidermal growth factor receptor-dependent migration and induces aggregation of nh(2)-terminal FAK in the nuclei of apoptotic glioblastoma cells. Cancer Res. 2001 Jul 1;61(13):4978–4981. [PubMed] [Google Scholar]
- 37.Kuan CT, Wikstrand CJ, Archer G, Beers R, Pastan I, Zalutsky MR, et al. Increased binding affinity enhances targeting of glioma xenografts by EGFRvIII-specific scFv. Int J Cancer. 2000 Dec 15;88(6):962–969. doi: 10.1002/1097-0215(20001215)88:6<962::aid-ijc20>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 38.Amer DA, Kretzschmar G, Muller N, Stanke N, Lindemann D, Vollmer G. Activation of transgenic estrogen receptor-beta by selected phytoestrogens in a stably transduced rat serotonergic cell line. J Steroid Biochem Mol Biol. 2010 Jun;120(4-5):208–217. doi: 10.1016/j.jsbmb.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999 Apr;73(4):2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topfer K, Cartellieri M, Michen S, Wiedemuth R, Muller N, Lindemann D, et al. DAP12-Based Activating Chimeric Antigen Receptor for NK Cell Tumor Immunotherapy. J Immunol. 2015 Mar 4; doi: 10.4049/jimmunol.1400330. [DOI] [PubMed] [Google Scholar]
- 41.Mochizuki H, Schwartz JP, Tanaka K, Brady RO, Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J Virol. 1998 Nov;72(11):8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalajzic I, Stover ML, Liu P, Kalajzic Z, Rowe DW, Lichtler AC. Use of VSV-G pseudotyped retroviral vectors to target murine osteoprogenitor cells. Virology. 2001 May 25;284(1):37–45. doi: 10.1006/viro.2001.0903. [DOI] [PubMed] [Google Scholar]
- 43.Morgenroth A, Cartellieri M, Schmitz M, Gunes S, Weigle B, Bachmann M, et al. Targeting of tumor cells expressing the prostate stem cell antigen (PSCA) using genetically engineered T-cells. Prostate. 2007 Jul 1;67(10):1121–1131. doi: 10.1002/pros.20608. [DOI] [PubMed] [Google Scholar]
- 44.Temme A, Geiger KD, Wiedemuth R, Conseur K, Pietsch T, Felsberg J, et al. Giant cell glioblastoma is associated with altered aurora b expression and concomitant p53 mutation. J Neuropathol Exp Neurol. 2010 Jun;69(6):632–642. doi: 10.1097/NEN.0b013e3181e4c06e. [DOI] [PubMed] [Google Scholar]
- 45.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulte A, Liffers K, Kathagen A, Riethdorf S, Zapf S, Merlo A, et al. Erlotinib resistance in EGFR-amplified glioblastoma cells is associated with upregulation of EGFRvIII and PI3Kp110delta. Neuro Oncol. 2013 Oct;15(10):1289–1301. doi: 10.1093/neuonc/not093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schonfeld K, Sahm C, Zhang C, Naundorf S, Brendel C, Odendahl M, et al. Selective Inhibition of Tumor Growth by Clonal NK Cells Expressing an ErbB2/HER2-Specific Chimeric Antigen Receptor. Mol Ther. 2014 Nov 6; doi: 10.1038/mt.2014.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zlotnik A. Chemokines and cancer. Int J Cancer. 2006 Nov 1;119(9):2026–2029. doi: 10.1002/ijc.22024. [DOI] [PubMed] [Google Scholar]
- 49.Franciszkiewicz K, Boissonnas A, Boutet M, Combadiere C, Mami-Chouaib F. Role of chemokines and chemokine receptors in shaping the effector phase of the antitumor immune response. Cancer Res. 2012 Dec 15;72(24):6325–6332. doi: 10.1158/0008-5472.CAN-12-2027. [DOI] [PubMed] [Google Scholar]
- 50.Kershaw MH, Wang G, Westwood JA, Pachynski RK, Tiffany HL, Marincola FM, et al. Redirecting migration of T cells to chemokine secreted from tumors by genetic modification with CXCR2. Hum Gene Ther. 2002 Nov 1;13(16):1971–1980. doi: 10.1089/10430340260355374. [DOI] [PubMed] [Google Scholar]
- 51.Moon EK, Carpenito C, Sun J, Wang LC, Kapoor V, Predina J, et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin Cancer Res. 2011 Jul 15;17(14):4719–4730. doi: 10.1158/1078-0432.CCR-11-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Craddock JA, Lu A, Bear A, Pule M, Brenner MK, Rooney CM, et al. Enhanced tumor trafficking of GD2 chimeric antigen receptor T cells by expression of the chemokine receptor CCR2b. J Immunother. 2010 Oct;33(8):780–788. doi: 10.1097/CJI.0b013e3181ee6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gumbleton M, Kerr WG. Role of inositol phospholipid signaling in natural killer cell biology. Front Immunol. 2013;4:47. doi: 10.3389/fimmu.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001 Mar 1;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 55.Handel TM, Johnson Z, Crown SE, Lau EK, Proudfoot AE. Regulation of protein function by glycosaminoglycans--as exemplified by chemokines. Annu Rev Biochem. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 56.Radaelli E, Ceruti R, Patton V, Russo M, Degrassi A, Croci V, et al. Immunohistopathological and neuroimaging characterization of murine orthotopic xenograft models of glioblastoma multiforme recapitulating the most salient features of human disease. Histol Histopathol. 2009 Jul;24(7):879–891. doi: 10.14670/HH-24.879. [DOI] [PubMed] [Google Scholar]
- 57.Wang M, Wang T, Liu S, Yoshida D, Teramoto A. The expression of matrix metalloproteinase-2 and -9 in human gliomas of different pathological grades. Brain Tumor Pathol. 2003;20(2):65–72. doi: 10.1007/BF02483449. [DOI] [PubMed] [Google Scholar]
- 58.McQuibban GA, Butler GS, Gong JH, Bendall L, Power C, Clark-Lewis I, et al. Matrix metalloproteinase activity inactivates the CXC chemokine stromal cell-derived factor-1. J Biol Chem. 2001 Nov 23;276(47):43503–43508. doi: 10.1074/jbc.M107736200. [DOI] [PubMed] [Google Scholar]
- 59.Perry JR. Thromboembolic disease in patients with high-grade glioma. Neuro Oncol. 2012 Sep;14 Suppl 4:iv73–iv80. doi: 10.1093/neuonc/nos197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, Anderson MG, Tucker LA, Shen Y, Glaser KB, Shah OJ. Inhibition of Aurora B kinase sensitizes a subset of human glioma cells to TRAIL concomitant with induction of TRAIL-R2. Cell Death Differ. 2009 Mar;16(3):498–511. doi: 10.1038/cdd.2008.174. [DOI] [PubMed] [Google Scholar]
- 61.Hendruschk S, Wiedemuth R, Aigner A, Topfer K, Cartellieri M, Martin D, et al. RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo. Neuro Oncol. 2011 Oct;13(10):1074–1089. doi: 10.1093/neuonc/nor098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Codo P, Weller M, Meister G, Szabo E, Steinle A, Wolter M, et al. MicroRNA-mediated down-regulation of NKG2D ligands contributes to glioma immune escape. Oncotarget. 2014 Sep 15;5(17):7651–7662. doi: 10.18632/oncotarget.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhl M, Aulwurm S, Wischhusen J, Weiler M, Ma JY, Almirez R, et al. SD-208, a novel transforming growth factor beta receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Res. 2004 Nov 1;64(21):7954–7961. doi: 10.1158/0008-5472.CAN-04-1013. [DOI] [PubMed] [Google Scholar]
- 64.Naumann U, Maass P, Gleske AK, Aulwurm S, Weller M, Eisele G. Glioma gene therapy with soluble transforming growth factor-beta receptors II and III. Int J Oncol. 2008 Oct;33(4):759–765. [PubMed] [Google Scholar]
- 65.Emlet DR, Gupta P, Holgado-Madruga M, Del Vecchio CA, Mitra SS, Han SY, et al. Targeting a glioblastoma cancer stem-cell population defined by EGF receptor variant III. Cancer Res. 2014 Feb 15;74(4):1238–1249. doi: 10.1158/0008-5472.CAN-13-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mangan JK, Luger SM. Salvage therapy for relapsed or refractory acute myeloid leukemia. Ther Adv Hematol. 2011 Apr;2(2):73–82. doi: 10.1177/2040620711402533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1: Simultaneous CAR and CXCR4 expression does not impair the cytotoxic capacity of YTS-NK cells. YTSMR1.1-DAP12, YTSMR1.1-DAP12/CXCR4 and YTSCXCR4 cells were co-cultured with 51chromium-loaded EGFRvIII+ tumor cells and isogenic EGFRvIII- tumor control cells at different effector to target ratios for 21 h. The mean of specific tumor cell lysis and standard deviation of triplets of three independent chromium release assays is shown. Note the strong tumor cell lysis mediated by YTSMR1.1-DAP12/CXCR4 cells which is similar to the cytotoxic reaction of YTSMR1.1-DAP12 cells. Both are significantly increased when compared to the nearly undetectable specific lysis of YTSCXCR4 cells. (* p < 0.05, **p < 0.01).