Abstract
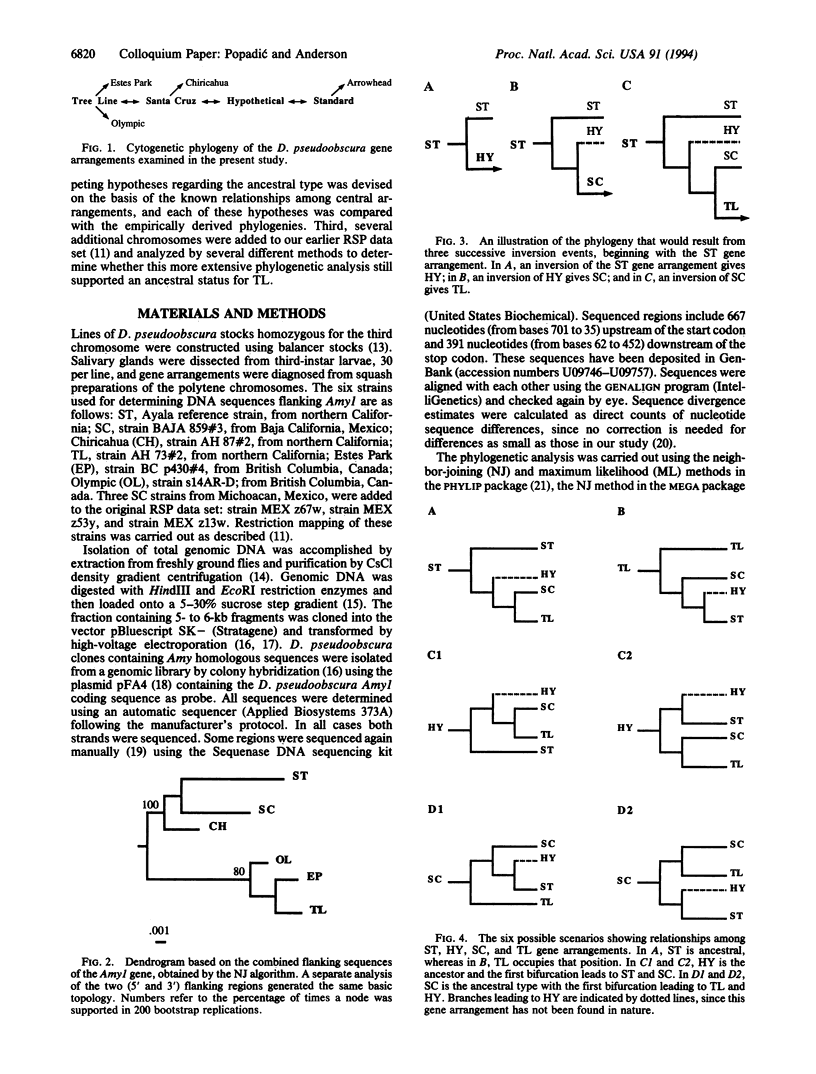
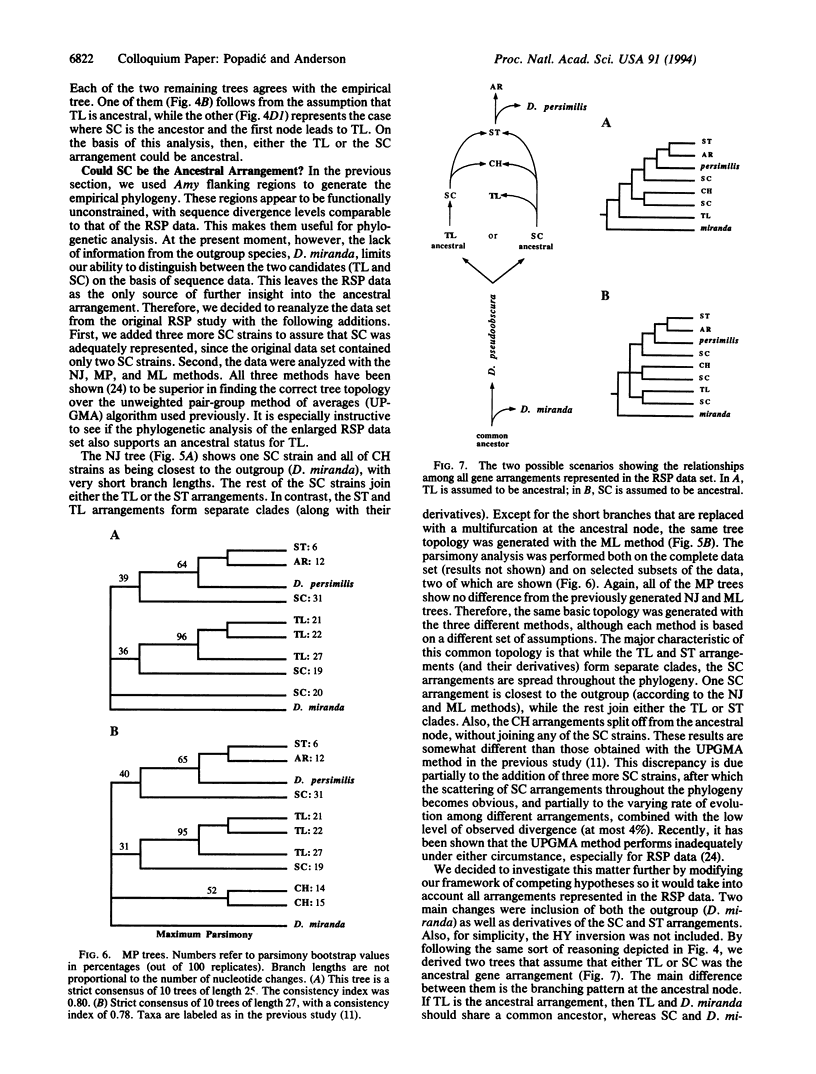
Although the chromosomal polymorphism for inversions in Drosophila pseudoobscura is one of the best studied systems in population genetics, the identity of the ancestral gene arrangement has remained unresolved for more than 50 years. There are more than 40 gene arrangements, and 4 of them (Standard, Hypothetical, Santa Cruz, and Tree Line) have been considered as candidates for the ancestral type. We propose a framework of competing hypotheses to distinguish among the alternatives. Two conclusions come from contrasting each hypothesis with the results from DNA sequencing and restriction mapping. First, not only Standard but also Hypothetical can be excluded as the ancestral gene arrangement. Second, although either Tree Line or Santa Cruz could be the ancestral type, the available data provide greater support for Santa Cruz.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aquadro C. F., Weaver A. L., Schaeffer S. W., Anderson W. W. Molecular evolution of inversions in Drosophila pseudoobscura: the amylase gene region. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):305–309. doi: 10.1073/pnas.88.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Aquadro C. F., Anderson W. W. DNA sequence evolution of the amylase multigene family in Drosophila pseudoobscura. Genetics. 1990 Sep;126(1):131–138. doi: 10.1093/genetics/126.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Sturtevant A H. Inversions in the Chromosomes of Drosophila Pseudoobscura. Genetics. 1938 Jan;23(1):28–64. doi: 10.1093/genetics/23.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Nei M. Relative efficiencies of the maximum-parsimony and distance-matrix methods of phylogeny construction for restriction data. Mol Biol Evol. 1991 May;8(3):356–365. doi: 10.1093/oxfordjournals.molbev.a040648. [DOI] [PubMed] [Google Scholar]
- Pavlovsky O., Dobzhansky T. Genetics of natural populations. XXXVII. The coadapted system of chromosomal variants in a population of Drosophila pseudoobscura. Genetics. 1966 May;53(5):843–854. doi: 10.1093/genetics/53.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Dobzhansky T. Inversions in the Third Chromosome of Wild Races of Drosophila Pseudoobscura, and Their Use in the Study of the History of the Species. Proc Natl Acad Sci U S A. 1936 Jul;22(7):448–450. doi: 10.1073/pnas.22.7.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B. In defense of verbal arguments. Perspect Biol Med. 1988 Winter;31(2):201–211. doi: 10.1353/pbm.1988.0049. [DOI] [PubMed] [Google Scholar]



