Abstract
As a tissue-specific stem cell for chondrogenesis, synovium-derived stem cells (SDSCs) are a promising cell source for cartilage repair. However, a small biopsy can only provide a limited number of cells. Cell senescence from both in vitro expansion and donor age presents a big challenge for stem cell based cartilage regeneration. Here we found that expansion on decellularized extracellular matrix (dECM) full of three-dimensional nanostructured fibers provided SDSCs with unique surface profiles, low elasticity but large volume as well as fibroblast-like shape. dECM expanded SDSCs yielded larger pellets with intensive staining of type II collagen and sulfated glycosaminoglycans compared to those grown on plastic flasks while SDSCs grown in ECM yielded 28-day pellets with minimal matrix as evidenced by pellet size and chondrogenic marker staining, which was confirmed by both biochemical data and real-time PCR data. Our results also found lower levels of inflammatory genes in dECM expanded SDSCs that might be responsible for enhanced chondrogenic differentiation. Despite an increase in type X collagen in chondrogenically induced cells, dECM expanded cells had significantly lower potential for endochondral bone formation. Wnt and MAPK signals were actively involved in both expansion and chondrogenic induction of dECM expanded cells. Since young and healthy people can be potential donors for this matrix expansion system and decellularization can minimize immune concerns, human SDSCs expanded on this future commercially available dECM could be a potential cell source for autologous cartilage repair.
Keywords: Decellularized extracellular matrix, Synovium-derived stem cell, Chondrogenesis, endochondral bone formation, Atomic force microscopy, Wnt signal
1. Introduction
Articular cartilage has a limited capacity for self-repair. Once damaged, the injured cartilage will develop defects. Despite promising results from autologous chondrocyte implantation [1], donor tissue availability is a challenge. Recent advances make adult stem cells an attractive cell source for cartilage regeneration, especially tissue-specific stem cells such as synovium-derived stem cells (SDSCs) [2,3]. Since in vitro expansion is a necessary step before in vivo application, accompanying cell senescence and dedifferentiation represents a formidable challenge for stem cell-based cartilage repair [4].
We found that decellularized extracellular matrix (dECM) deposited by mesenchymal stem cells could rejuvenate stem cells [5–11] and primary cells [12–14] in both proliferation and differentiation capacity. For instance, dECM deposited by SDSCs significantly promoted expanded porcine SDSCs (pSDSCs) in both proliferation and chondrogenic potential [5]. In vivo transplantation of dECM expanded pSDSCs demonstrated efficacy in promoting cartilage regeneration in a partial thickness cartilage defect porcine model [15].
Our recent reports suggested that this in vitro cell expansion system also benefits human SDSC (hSDSC) expansion and rejuvenation of chondrogenic potential [16,17], which brings hope for the potential use of this approach in clinical treatment [18,19]. However, a concomitant up-regulation of type X collagen (COL10A1) was also observed in chondrogenically differentiated hSDSCs that have undergone dECM expansion, indicating a tendency toward chondrogenic hypertrophy.
In this study, we fully characterized cell morphology, volume, elasticity, and surface phenotypes in hSDSCs following dECM expansion. We not only defined proliferation and chondrogenic potential in dECM expanded hSDSCs but also examined whether increased expression of COL10A1 could be a sign of endochondral bone formation. Since both the mitogen-activated protein kinase (MAPK) and Wnt signals are critical pathways for chondrogenesis and have crosstalk in stem cell mediated cartilage regeneration [20], these two signals were evaluated for their changes in both cell expansion and chondrogenic induction of hSDSCs after preconditioning using dECM and conventional plastic flasks, which might provide evidence for further investigation of potential mechanisms underlying the rejuvenation of hSDSCs by dECM expansion.
2. Materials and Methods
2.1 SDSC culture
Adult human synovial fibroblasts (4 donors, two male and two female, average 43 years old, all had no known joint disease), referred to as hSDSCs [16,17], were obtained from Asterand (North America Laboratories, Detroit, MI). Human SDSCs were plated and cultured in a growth medium [alpha minimum essential medium (αMEM) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL fungizone (Invitrogen, Carlsbad, CA)] at 37°C in a humidified 5% CO2 and 21% O2 incubator. The medium was changed every three days.
2.2 dECM preparation
The preparation of dECM was described in our previous study [16,17]. Briefly, plastic flasks (Plastic) were precoated with 0.2% gelatin (Sigma-Aldrich, St. Louis, MO) at 37°C for 1 h and seeded with passage 3 (P3) hSDSCs at 6,000 cells/cm2. After the cells reached 90% confluence, 50 μM L-ascorbic acid phosphate (Wako Chemicals USA, Inc., Richmond, VA) was added for 8 days. The medium was changed every other day. The deposited matrix was incubated with 0.5% Triton X-100 containing 20 mM ammonium hydroxide at 37°C for 5 min and stored at 4°C in phosphate-buffered saline (PBS) containing 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL fungizone.
2.3 SDSC expansion
P3 hSDSCs were cultured at 3000 cells/cm2 for one passage on two substrates: dECM or Plastic. The cell number was counted using a hemocytometer. Expanded cells were also evaluated for cell morphology using scanning electronic microscopy (SEM), and atomic force microscopy (AFM), cell number using a hemocytometer, and proliferation index and surface markers using flow cytometry.
2.4 Morphological observation using the SEM and AFM
Representative samples (n=2) were primarily fixed in 2.5% glutaraldehyde (Sigma-Aldrich) for 2 h, followed by secondary fixation in 2% osmium tetroxide (Sigma-Aldrich) for another 2 h. The samples were then dehydrated in a gradient ethanol series, in hexamethyldisilazane (HMDS, Sigma-Aldrich) at a ratio of 1:1 with ethanol twice for 1 h each time, in HMDS at a ratio of 1:2 with ethanol overnight, and in HMDS three times for 4 h each time. The samples were air-dried for 24 h and gold sputter was added. The images were recorded by an SEM (Hitachi, Model S 2400).
Morphology of culture substrates (dECM and Plastic) and both morphology and elasticity of expanded hSDSCs were performed using an MFP-3D-BIO AFM (Asylum Research, TE2000-U, Santa Barbara, CA) integrated with an inverted fluorescence microscope (Nikon Eclipse, Ti-U, Nikon Instruments Inc., Melville, NY) and Olympus TR400-PB cantilevers with manufacturer spring constant of 0.09 N/m. The samples were imaged in Petri dishes after they were fixed with 1% glutaraldehyde and washed once with PBS. The location of the cantilever on the sample was confirmed using a 10× microscopy objective. For the morphology imaging, each sample was mapped in PBS buffer using contact mode. An area of 90 μm by 90 μm was imaged with the pixel resolutions of 512 and scan rate at 0.2 Hz. For the quantitative nanomechanical analysis, a Sneddon’s modification of the Hertz model developed for a four-sided pyramid was employed. The fixed cell sample elasticity (Young’s modulus, E) was corrected with the indentation of the tip, δ, through the following equation: , where E is the elastic modulus, v is Poisson’s ratio with a value of 0.5 for ECM and cells, F is the force given by the cantilever deflection multiplied with the cantilever spring constant, α is the open angle used in this study which had a value of 36°, and lastly δ is the indentation depth [21]. The average height, projected cell area and the volume of the fixed cells were calculated using the MFP-3D Bio dedicated Igor Pro software (Asylum Research). The diameter of matrix fibers were determined by the measurement of full width of half maximum (FWHM) using ImageJ software.
2.5 Measurement of expanded cell proliferation index using flow cytometry
Before cell expansion, passage 3 hSDSCs were labeled with CellVue® Claret (Sigma-Aldrich) at 2×10−6 M for 5 min according to the manufacturer’s protocol. Expanded cells were collected and measured using a BD FACS (fluorescence activated cell sorting) Calibur ™flow cytometer (dual laser) (BD Biosciences, San Jose, CA). Twenty thousand events of each sample were collected using CellQuest Pro software (BD Biosciences) and cell proliferation index was analyzed by ModFit LT™ version 3.1 (Verity Software House, Topsham, ME).
2.6 Evaluation of cell surface phenotypes using flow cytometry
The following primary antibodies were used in flow cytometry to detect hSDSC surface phenotypes: integrin β1 (CD29) was purchased from Abcam (Abcam, Cambridge, MA); CD105 and the stage-specific embryonic antigen-4 (SSEA-4) were from Santa Cruz Biotechnology (Santa Cruz, CA); and CD90 was from BD Pharmingen (BD Biosciences). IgG1 and IgG2a (Beckman Coulter, Fullerton, CA) were used as the isotype controls. Samples (n=3) of each 0.2 × 106 expanded cells were incubated on ice in cold PBS containing 0.1% ChromPure Human IgG whole molecule (Jackson ImmunoResearch Laboratories, West Grove, PA) and 1% NaN3 (Sigma-Aldrich) for 30 min. Then, the cells were sequentially incubated in the dark in the primary antibodies for 30 min. The fluorescence was analyzed by a FACS Calibur (BD Biosciences) using FCS Express software package (De Novo Software, Glendale, CA).
2.7 Chondrogenic induction and evaluation of expanded hSDSCs
After in vitro expansion, 0.25×106 of hSDSCs from each group were centrifuged at 500 g for 5 min in a 15-mL polypropylene tube to form a pellet. After overnight incubation (day 0), the pellets were cultured for 35 days in a serum-free chondrogenic medium consisting of high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM), 40 μg/mL proline, 100 nM dexamethasone, 100 U/mL penicillin, 100 μg/mL streptomycin, 0.1 mM ascorbic acid-2-phosphate, and 1×ITS™ Premix [6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 μg/mL selenous acid, 5.35 μg/mL linoleic acid, and 1.25 μg/mL bovine serum albumin (BSA), from BD Biosciences] with supplementation of 10 ng/mL transforming growth factor beta 3 (TGF-β3, PeproTech Inc., Rocky Hill, NJ). Chondrogenic differentiation was evaluated using histology, immunostaining, biochemical analysis, microarray, and real-time polymerase chain reaction (PCR).
Representative pellets (n=3) were fixed in 4% paraformaldehyde at 4°C overnight, followed by dehydrating in a gradient ethanol series, clearing with xylene, and embedding in paraffin blocks. Five μm-thick sections from the center of representative pellets were histochemically stained with Alcian blue (Sigma-Aldrich; counterstained with fast red) for sulfated glycosaminoglycans (GAGs). For immunohistochemical analysis, the consecutive sections were immunolabeled with primary antibodies against type II collagen (II-II6B3; Developmental Studies Hybridoma Bank, Iowa City, IA) and type X collagen (Sigma-Aldrich) followed by the secondary antibody of biotinylated horse anti-mouse IgG and IgM, respectively (Vector, Burlingame, CA). Immunoactivity was detected using Vectastain ABC reagent (Vector) with 3,3′-diaminobenzidine as a substrate.
Representative pellets (n=4) were digested for 4 h at 60°C with 125 μg/mL papain in PBE buffer (100 mM phosphate and 10 mM ethylenediaminetetraacetic acid, pH 6.5) containing 10 mM cysteine, by using 200 μL enzyme per sample. To quantify cell density, the amount of DNA in the papain digestion was measured using the QuantiT™ PicoGreen® dsDNA assay kit (Invitrogen) with a CytoFluor® Series 4000 (Applied Biosystems, Foster City, CA). GAG was measured using dimethylmethylene blue dye and a Spectronic BioMate 3 Spectrophotometer (Thermo Scientific, Milford, MA) with bovine chondroitin sulfate as a standard.
Global gene expression was evaluated in hSDSCs after expanded cell chondrogenic induction. The pellets from dECM expanded cells were referred to as Ep while those from Plastic expanded cells were referred to as Pp. Total RNAs were isolated from the pellets using Trizol (Invitrogen) followed by additional purification using an RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The required amount of cDNA (5.5 μg) was processed for fragmentation and biotin labeling using the GeneChip® WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA). The entire reaction of fragmented and biotin-labeled cDNA (50 μL) with added hybridization controls was hybridized to the human GeneChip® 1.0 ST Exon Arrays (Affymetrix) at 45°C for 17 h in the GeneChip® Hybridization Oven 640 (Affymetrix). The raw data was uploaded into Partek (St. Louis, MO) software for initial analysis and Ingenuity Pathway Analysis (IPA, Redwood City, CA) for pathway and functional analysis. Briefly, raw intensity was background-subtracted, robust multi-array analysis (RMA) normalized, log-transformed, and fold changes determined. The batch effects (scan date) were removed before fold change calculation. Cluster analysis was used to compare the genes (the dECM group versus the Plastic group) annotated in IPA as affecting chondrodysplasia or bone ossification. For bone ossification, genes were filtered to 60 with the greatest differential expression. Clustering was done using heatmap.2 in R (Euclidean distance, complete linkage).
After extraction and purification of total RNA from samples (n=4), about 1 μg of mRNA was used for reverse transcriptase with a High-Capacity cDNA Archive Kit (Applied Biosystems) at 37°C for 120 min. Chondrogenic marker genes [type II collagen (COL2A1; assay ID: Hs00156568_m1), aggrecan (ACAN; assay ID: Hs00153935_m1), and SRY (sex determining region Y)-box 9 (SOX9; assay ID: Hs00165814_m1)], hypertrophic marker genes [COL10A1 (assay ID: H200166657_m1) and alkaline phosphatase (ALPL; assay ID: Hs01029144_m1)], Wnt signal genes [WNT3A (assay ID: Hs00263977_m1), WNT5A (assay ID: Hs00998537_m1), and WNT11 (assay ID: Hs00182986_m1)], and inflammation related genes [interleukin 1 beta (IL1B; assay ID: Hs01555410_m1), and a disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS5; assay ID: Hs00199841_m1)] were customized by Applied Biosystems as part of the Custom TaqMan® Gene Expression Assays. Eukaryotic 18S RNA (assay ID: Hs99999901_s1) was carried out as the endogenous control gene. Real-time PCR was performed with the iCycler iQ™ Multi-Color Real Time PCR Detection (Perkin-Elmer, Waltham, MA). Relative transcript levels were calculated as χ=2−ΔΔCt, in which ΔΔCt=ΔE−ΔC, ΔE=Ctexp−Ct18s, and ΔC=Ctct1−Ct18s.
2.8 Characterization of both MAPK and Wnt signals using Western blot
Adult hSDSCs from cell expansion (day 1, 3, and 5) and pellets before (day 0) and after (day 1 and 7) chondrogenic induction were dissolved in the lysis buffer (Cell Signaling, Boston, MA) with protease inhibitors. Total protein was quantified using the BCA™ Protein Assay Kit (Thermo Fisher Scientific). Thirty micrograms of protein from each sample were denatured and separated using NuPAGE® Novex® Bis-Tris Mini Gels in the XCell SureLock™ Mini-Cell (Life Technologies) at 120 V at 4°C for 3 h. Bands were transferred onto a nitrocellulose membrane using an XCell II™ Blot module (Life Technologies) at 15 V at 4°C overnight. The membrane was incubated with primary monoclonal antibodies in 5% bovine serum albumin (BSA) in TBST buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20) for 1 h (β-actin served as an internal control), followed by the secondary antibody of horseradish peroxidase–conjugated goat anti-mouse (Thermo Fisher Scientific) for 1 h. SuperSignal West Femto Maximum Sensitivity Substrate and CL-XPosure Film (Thermo Fisher Scientific) were used for exposure. The primary antibodies used in immunoblotting included the MAPK family antibody sampler kit [extracellular signal-regulated protein kinases 1 and 2 (Erk1/2), Jun N-terminal kinase (Jnk), and p38], phosphorylated (p-) MAPK family antibody sampler kit, and the Wnt signaling antibody sampler kit (Cell Signaling). Wnt11 polyclonal antibody was obtained from Thermo Fisher Scientific. ImageJ software was used to quantify immunoblotting bands.
2.9 Statistics
Numerical data are presented as the mean and the standard error of the mean. The Mann-Whitney U test was used for pair-wise comparison in biochemistry and real-time PCR data analysis. All statistical analyses were performed with SPSS 13.0 statistical software (SPSS Inc., Chicago, IL). A p value less than 0.05 was considered statistically significant. For microarray data analysis, a regulation z-score statistic was calculated to predict if the observed numbers of genes changed in the direction to produce the functional effect would occur by chance. A z-score > 2 or < −2 was considered significant.
3 Results
3.1 Expansion on dECM changed cell morphology and increased cell proliferation
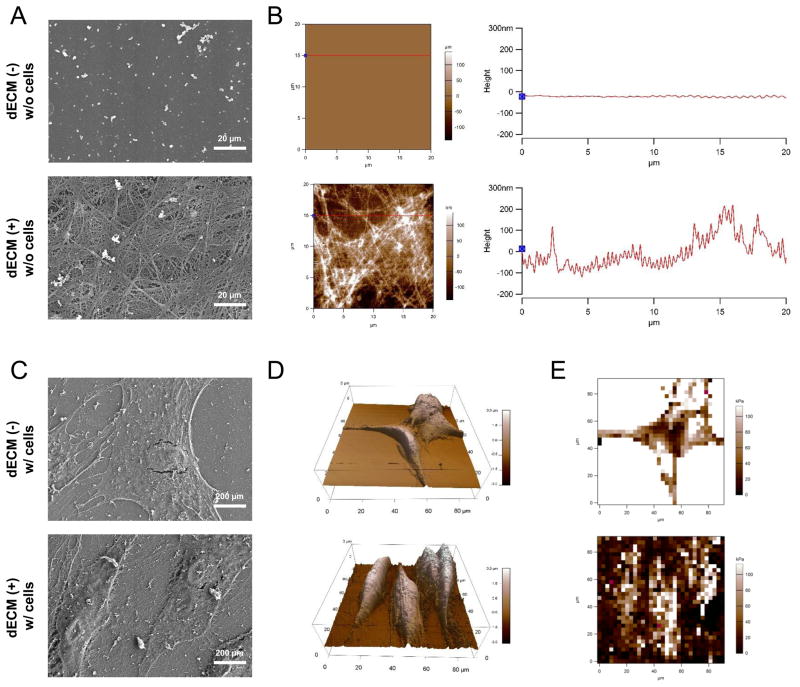
Before plating cells, SEM data showed that, compared with a flat surface on Plastic substrate, dECM coated flasks exhibited a three-dimensional structure with interconnected nanofibers (Fig. 1A) that was corroborated by morphological data from AFM (Fig. 1B), in which the diameter of matrix fibers was 109.5 ± 39.6 nm (n=82). SEM data also showed that hSDSCs displayed a well-organized fibroblast-like cell shape when expanded on dECM rather than a broad and flat cell shape when grown on Plastic substrate (Fig. 1C). This finding was also in line with cell morphology shown by AFM (Fig. 1D), in which several parameters were measured for fixed cells with glutaraldehyde after expansion on Plastic and dECM substrates, including average height (1.33 ± 0.57 μm versus 2.34 ± 0.44 μm), projected cell area (713.7 ± 154.4 μm2 versus 916.0 ± 190.9 μm2), and volume (962.3 ± 502.7 μm3 versus 2101.3 ± 354.1 μm3). Young’s moduli of the fixed cells were 91.22 ± 64.15 kPa after expansion on Plastic and 53.33 ± 53.47 kPa when expanded on dECM (Fig. 1E).
Fig. 1.
Morphological characterization of culture substrates and expanded cells. Surface topography of both substrates, Plastic and dECM, was characterized using SEM (A) and AFM (B). Scale bar for SEM (A): 20 μm. Expanded cells on either Plastic or dECM after fixation in glutaraldehyde were characterized using SEM (C) and AFM (D) for morphology and using AFM (E) for elasticity. Scale bar for SEM (C): 200 μm.
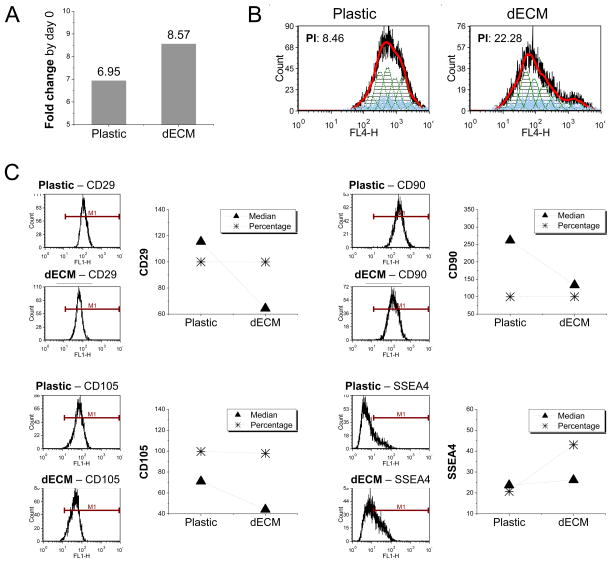
To determine whether dECM expansion also affected cell proliferation rate, 3000 cells per cm2 were incubated on both substrates for seven days. The data from cell counting showed that the cells grown on Plastic flasks increased 6.95-fold compared with 8.57-fold on dECM (Fig. 2A), which was confirmed by proliferation index data (8.46 versus 22.28) measured in expanded cells (Fig. 2B). To further characterize molecular phenotype changes following dECM expansion, flow cytometry was used to quantify both percentage and median fluorescence intensity of typical mesenchymal stem cell markers in expanded cells (Fig. 2C). Notably, despite almost 100% expression in the cells after expansion on both substrates, the median fluorescence intensity of CD29, CD90, and CD105 dramatically decreased in dECM expanded cells. In contrast, the percentage of SSEA4 doubled in dECM expanded cells concomitantly with a slight increase in median fluorescence intensity.
Fig. 2.
dECM promoted expanded hSDSCs’ proliferation and changed expression of stem cell surface markers. (A) Fold change of cell number by day 0 was used to indicate cell proliferation after 7-day-expansion on 175 cm2 flasks (n=6). (B) Flow cytometry was used to measure the proliferation index (PI) of expanded hSDSCs. (C) Flow cytometry was used to measure both percentage and median fluorescence intensity of mesenchymal stem cell surface markers (CD29, CD90, CD105, and SSEA4) of expanded hSDSCs.
3.2 Expansion on dECM enhanced cell chondrogenic potential
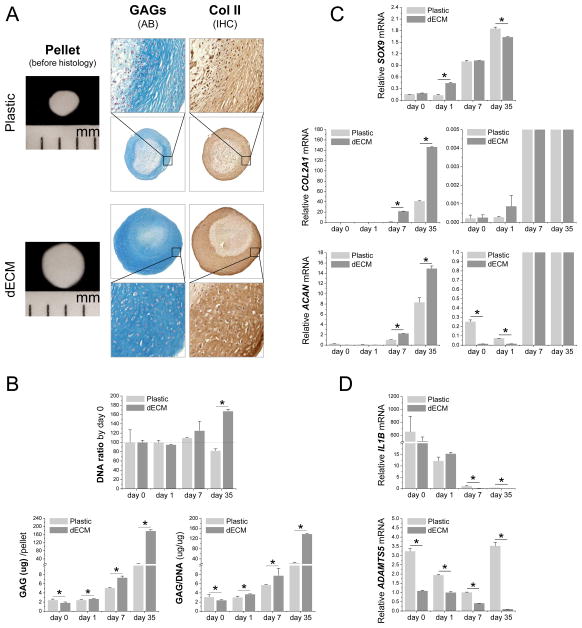
To determine whether dECM expansion could enhance hSDSC chondrogenic potential, a pellet culture system was used for chondrogenic induction. Histology data showed that dECM expanded cells yielded 35-day-pellets with a larger size despite starting with the same initial cell density as those from Plastic expanded cells. The increased pellet size was accompanied by similarly increased staining intensity of both sulfated GAG and type II collagen, two cartilage markers, in the pellets from dECM expanded cells (Fig. 3A). The staining data were further confirmed by the quantification data at both the protein and mRNA levels. Our biochemical analysis data showed that, after a 35-day-chondrogenic induction, expansion on dECM enhanced DNA ratio (167.35 ± 4.01 %, adjusted by day 0) while expansion on Plastic decreased this ratio (82.03 ± 4.06 %); despite the lower amounts at day 0, after chondrogenic induction, both GAG amount per pellet and ratio of GAG to DNA from the dECM group dramatically increased and were higher than those from the Plastic group (Fig. 3B).
Fig. 3.
dECM promoted expanded hSDSCs’ chondrogenic differentiation. Human SDSCs were grown on either Plastic or dECM for one passage followed by chondrogenic induction in a pellet culture system for 35 days. (A) Before histological staining, pellet size was measured with a scale bar as mm; Alcian blue (AB) was used to stain sulfated GAGs and immunohistochemistry staining (IHC) was used to detect type II collagen (Col II). (B) Biochemical analysis was used for DNA and GAG contents of pellets; cell viability in chondrogenic medium was evaluated using DNA ratio adjusted by day 0; a ratio of GAG to DNA indicated chondrogenic index. Real-time PCR was used to evaluate (C) chondrogenic marker gene expression [SOX9, ACAN and COL2A1 (the graph on the left with a large scale and on the right with a small scale)] and (D) inflammation related genes (IL1B and ADAMTS5). Data are shown as average ± standard deviation (SD) for n=4. *p < 0.05 indicated a statistically significant difference.
Real-time PCR data showed that all chondrogenic marker genes (SOX9, COL2A1 and ACAN) were up-regulated during chondrogenic induction, especially in cells from the dECM group (Fig. 3C). Interestingly, despite a dramatic increase 24 h after chondrogenic induction, SOX9 expression in dECM expanded cells reached a comparable level at day 7 and decreased afterwards compared with that from the Plastic group (Fig. 3C). Our data also showed that both IL1B and ADAMTS5, two typical inflammatory genes, were significantly down-regulated in chondrogenically induced hSDSCs from the dECM group (Fig. 3D).
3.3 Expansion on dECM promoted cell hypertrophy but not toward an osteogenic lineage
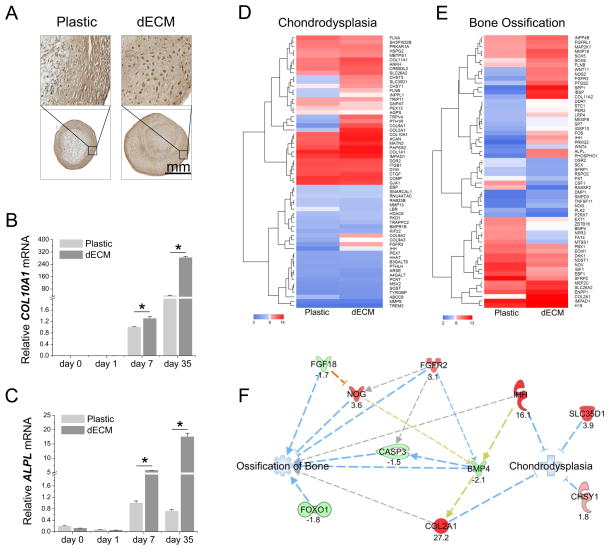
To determine whether expansion on dECM would enhance hSDSCs’ hypertrophy during chondrogenic induction, we examined markers of hypertrophy. Immunostaining data showed that the 35-day-pellets from dECM expanded hSDSCs were intensely stained with type X collagen compared with those from Plastic expanded cells (Fig. 4A). These data were further confirmed by our real-time PCR data in which both COL10A1 (Fig. 4B) and ALPL (Fig. 4C) were dramatically up-regulated in chondrogenically induced hSDSCs following dECM expansion.
Fig. 4.
dECM promoted expanded hSDSCs’ chondrogenic hypertrophy but not ossification of bone. Human SDSCs were grown on either Plastic or dECM for one passage followed by chondrogenic induction in a pellet culture system for 35 days. (A) Immunohistochemistry staining was used to detect type X collagen. (B and C) Real-time PCR was used to evaluate hypertrophic marker gene expression (COL10A1 and ALPL). Data are shown as average ± SD for n=4. *p < 0.05 indicated a statistically significant difference. (D) Expression (log2) of genes annotated in IPA to affect chondrodysplasia. (E) Expression (log2) of genes annotated in IPA to affect bone ossification. Genes were filtered to the 60 genes with greatest differential expression. (F) Microarray data were used to predict the effect of dECM expansion on subsequent chondrogenic induction, especially on ossification of bone and chondrodysplasia. Ossification of bone and chondrodysplasia were predicted to be decreased in the pellets from dECM expanded cells, relative to those from Plastic expanded cells. Expression results for the genes are shown as FC. The orange line denotes predicted activation of the target molecule (NOG) by the observed state of the upstream molecule (FGF18). Blue lines denote predicted inhibition of the molecule or function by the observed state (up-regulation or down-regulation) of the upstream molecule. Gray lines indicate that an effect on function was predicted or interaction between molecules was noted to occur, but existing data do not support prediction of direction of change. Gold indicates the effect on the downstream molecule is inconsistent with that predicted by direct interaction of the molecules.
Next, we wanted to determine whether expansion on dECM would promote hSDSCs toward bone ossification and/or chondrodysplasia. Pathway analysis of microarray data showed expression of 63 genes related to chondrodysplasia (Fig. 4D) and 377 genes related to bone ossification (the 60 genes most differentially expressed are shown in Fig. 4E). Compared with the pellets from Plastic expanded hSDSCs, chondrogenically induced hSDSCs from the dECM group exhibited an up-regulation in fibroblast growth factor receptor 2 (FGFR2) and noggin (NOG) and a down-regulation in FGF18, forkhead box O1 (FOXO1), bone morphogenetic protein 4 (BMP4), and caspase 3 (CASP3), suggesting that, after dECM expansion, ossification of bone is decreased (prediction z-score: −2.43) (Fig. 4F). An up-regulation of indian hedgehog (IHH), COL2A1, chondroitin sulfate synthase 1 (CHSY1), and solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter) member D1 (SLC35D1) indicated that, after dECM expansion, dysplasia of the skeleton (including chondrodysplasia) is decreased (prediction z-score: −2.00) (Fig. 4F).
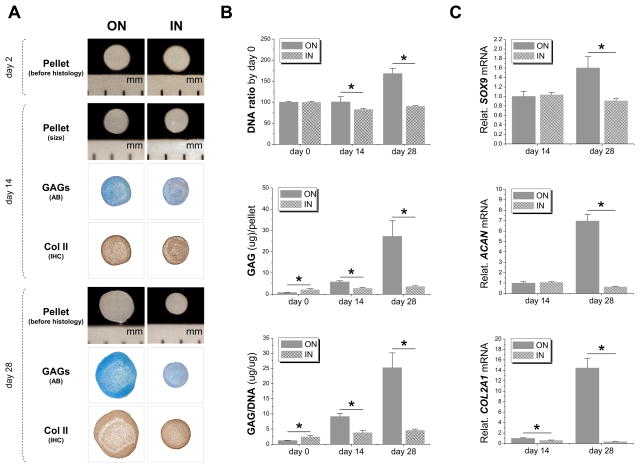
3.4 Growth in ECM dramatically decreased expanded cell chondrogenic capacity
To determine whether expansion on dECM (“ON”, deposited by other SDSCs) was superior to SDSCs that were grown in ECM (“IN”, deposited by expanded SDSCs) in chondrogenic capacity, expanded cells were incubated in a pellet culture system supplemented with chondrogenic induction medium for 28 days. Both groups exhibited a similar pellet size at day 2 but, at day 28, SDSCs expanded on dECM yielded pellets with dramatically increased size whereas SDSCs grown in ECM yielded pellets that had no change in size visible to the naked eye, which was accompanied by less staining intensity of both sulfated GAG and type II collagen compared to those expanded on dECM (Fig. 5A). Based on the DNA ratio adjusted by day 0, cell viability at day 28 in the IN ECM group was about 90% while in the ON dECM group, it was about 168%. Despite the higher GAG amount (2.73-fold) and ratio of GAG to DNA (1.99-fold) at the initial time point (day 0), 28-day chondrogenic induction yielded pellets from the IN ECM group with a lower GAG amount (0.13-fold) and ratio of GAG to DNA (0.18-fold) compared to those from the ON dECM group (Fig. 5B). Real-time PCR data showed that, after 28-day chondrogenic induction, all chondrogenic marker genes (SOX9, COL2A1 and ACAN) showed a similar trend in the pellets with lower mRNA levels from the IN ECM group compared to the ON dECM group (Fig. 5C).
Fig. 5.
hSDSCs grown in ECM (“IN”) exhibited a decline in chondrogenic potential compared with those expanded on dECM (“ON”). (A) Before histological staining, pellet size was measured with a scale bar as mm; Alcian blue (AB) was used to stain sulfated GAGs and immunohistochemistry staining (IHC) was used to detect type II collagen (Col II). (B) Biochemical analysis was used for DNA and GAG contents of pellets; cell viability in chondrogenic medium was evaluated using DNA ratio adjusted by day 0; a ratio of GAG to DNA indicated chondrogenic index. (C) Real-time PCR was used to evaluate chondrogenic marker gene expression (SOX9, ACAN, and COL2A1. Data are shown as average ± SD for n=4. *p < 0.05 indicated a statistically significant difference.
3.5 Both Wnt and MAPK pathways were actively involved in dECM mediated chondrogenesis
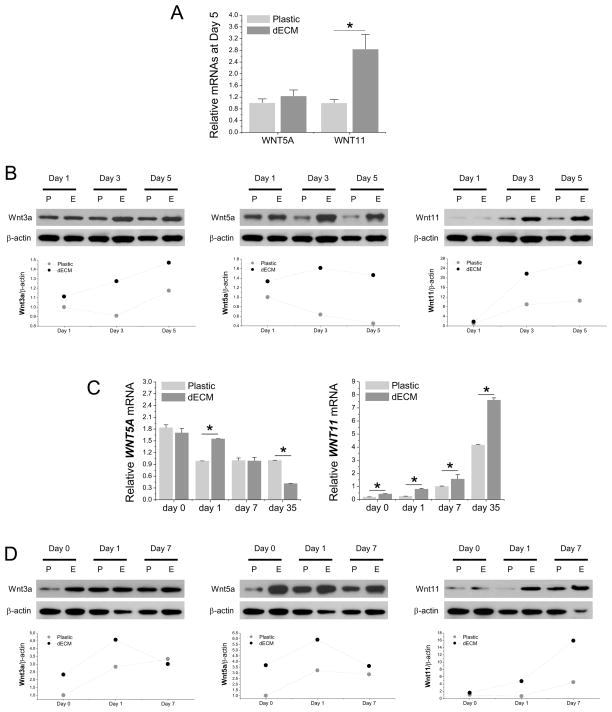
To determine whether Wnt signals were involved in dECM mediated cell expansion, real-time PCR was used to measure WNT3A (a canonical signal) and WNT5A and WNT11 (noncanonical signals) in hSDSCs following cell expansion. We found that WNT3A was undetectable but, compared to the Plastic group, both WNT5A and WNT11 were up-regulated in dECM expanded cells, particularly WNT11 (Fig. 6A). The above data were confirmed by Western blot data in which both Wnt5a and Wnt11 exhibited a higher level of expression in dECM expanded hSDSCs (Fig. 6B). Intriguingly, we also found that wnt3a protein was detectable and exhibited a higher level in dECM expanded hSDSCs compared to those grown on Plastic (Fig. 6B).
Fig. 6.
Wnt signals were actively involved in chondrogenesis of dECM expanded hSDSCs. Both real-time PCR (A, C) and Western blot (B, D) were used to evaluate the canonical Wnt signal (Wnt3a) and noncanonical Wnt signals (Wnt5a and Wnt11) in hSDSCs during cell expansion (A, B) and chondrogenic induction (C, D) at both mRNA and protein levels. The β-actin was used as an internal control. Data are shown as average ± SD for n=4. *p < 0.05 indicated a statistically significant difference. ImageJ software was used to quantify immunoblotting bands.
Similar to cell expansion, WNT3A mRNA was undetectable while Wnt3a protein exhibited a higher level in dECM expanded hSDSCs during the early stage of chondrogenic induction. Wnt5a and Wnt11 exhibited comparable expression profiles in both mRNA (Fig. 6C) and protein (Fig. 6D). Compared to an up-regulation of Wnt5a in the beginning followed by a decrease afterwards, a continuing increase of Wnt11 at both mRNA and protein levels was found in the pellets from both expanded cells, with dECM expanded hSDSCs exhibiting a significant advantage over those expanded on Plastic.
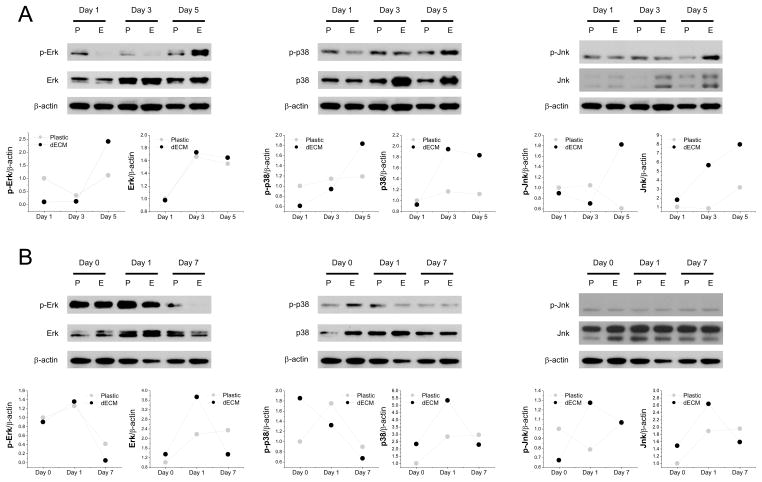
To determine whether MAPK signals were involved in dECM mediated cell expansion, Western blot was used to measure phosphor (p)-Erk, Erk, p-p38, p38, p-Jnk, and Jnk in hSDSCs after a five-day expansion. We found that p-Erk, p-p38, p-Jnk, p38, and Jnk exhibited higher levels in dECM expanded cells than those grown on Plastic (Fig. 7A). However, during chondrogenic induction, most MAPK signals increased at day 1 but decreased afterwards (Fig. 7B).
Fig. 7.
MAPK signals were actively involved in the chondrogenesis of dECM expanded hSDSCs. Western blot was used to evaluate Erk1/2, p38, and Jnk signals in both phosphorylated and nonphosphorylated formats in hSDSCs during cell expansion (A) and chondrogenic induction (B). β-actin was used as an internal control. ImageJ software was used to quantify immunoblotting bands.
4 Discussion
Our previous work found that dECM could rejuvenate pSDSCs for both proliferation and chondrogenic potential. We recently found this in vitro cell expansion system also applied to hSDSCs. In order to make this approach applicable to future clinical treatment, in this study, we characterized dECM and expanded cells using both SEM and AFM and compared them to the Plastic control; we also characterized proliferation and chondrogenic capacity of adult hSDSCs after expansion on dECM and determined whether dECM expanded cells exhibited a differentiation trend toward bone formation as well as potential involvement of Wnt and MAPK signals. We found that dECM expanded hSDSCs exhibited a fibroblast shape, a low level of elasticity, enhanced proliferation, unique changes in stem cell surface markers, and enhanced chondrogenic capacity and hypertrophy, which are consistent with our previous findings [16,17]. We also found, for the first time, that dECM expansion could dramatically decrease the potential differentiation toward bone and dysplasia in the skeleton. Additionally, we discovered that both Wnt and MAPK signals are actively involved in dECM expanded hSDSC chondrogenesis.
It is known that microenvironments play important roles in stem cell lineage specifications. Cell proliferation leads to an increase of cell volume while one of the hallmarks of apoptosis is cell shrinkage [22]. In this study, cells expanded on dECM showed more than a twofold increase in cell volume, suggesting strong mitotic activity; comparatively, the low volume value for those cells grown on Plastic substrate indicated low mitotic activity of cells that might experience senescence and apoptosis. This observation was supported by expanded cell proliferation index data (Plastic versus dECM: 8.46 versus 22.28). We also found that dECM was a nanostructured three-dimensional matrix, on which expanded cells possessed low elasticity and high chondrogenic potential compared to those grown on Plastic substrate, which might be explained by lower elasticity favoring expanded stem cells chondrogenic differentiation [16,23,24]. In this study, dECM expansion lowered cell elasticity, about 0.58-fold compared to those cells grown on Plastic substrate. This finding is consistent with our previous report, in which dECM expansion yielded cells with elasticity about 0.40-fold compared to those grown on Plastic substrate [16], despite the fact that the absolute values were much higher than previous ones due to the fixation of expanded cells using glutaraldehyde before measurement. Evidence has more recently emerged that fixation is considered the preferred method for reliable data analysis (reproducible) of both cell elasticity and cell morphology [25,26] since this method not only allows for longer scanning time but further limits changes associated with the inherent dynamics of living cells within the extended observation time of at least one hour per cell.
In this study, the increase in cell number from passage 4 to 5 in the dECM group was 1.23-fold that in the Plastic group while our previous study showed about an increase of 14.57-fold that in the Plastic group [5]. The chondrogenic index from chondrogenically induced pSDSCs in the dECM group was 1.36-fold that in the Plastic group while our previous study showed an increase of about 18.71-fold that in the Plastic group [5]. The discrepancy of nanofiber size is probably responsible for expanded SDSC proliferation rate and chondrogenic potential. Our data suggested that the diameter of dECM fibers measured by AFM was 109.5 ± 39.6 nm, which was about double the size of dECM fibers measured by SEM (45.1 ± 8.8 nm) from our previous study [5]. As described by Oh and colleagues, small (around 30-nm diameter) nanotubes promoted adhesion without noticeable differentiation, whereas larger (70- to100-nm diameter) nanotubes elicited a dramatic stem cell elongation (10-fold increase), which induced cytoskeletal stress and differentiation [27]. Another report by Park et al. indicates that a 15–20 nm spacing was better for integrin activation, while tube diameter larger than 50 nm severely impaired cell spreading, adhesion, and spacing of 100 nm resulting in dramatically reduced cell proliferation, migration, and differentiation [28]. Further investigation needs to determine whether donor age affects the diameter of dECM fibers.
For cell proliferation, we found that hSDSCs grown on dECM became fibroblast-like in shape and exhibited enhanced proliferation; CD29, CD90, and CD105 exhibited a decreased expression while SSEA4 was up-regulated, which is consistent with our recent report using hSDSCs [16]. Interestingly, despite a similar trend in cell morphology and proliferation capacity, our early report found that dECM expanded pSDSCs displayed an enhanced expression of CD90 [5] rather than a decreased level in this study, which is probably due to the source of SDSCs from different species (porcine versus human) and/or donor age (3-month-old versus 43-year-old). This discrepancy remains to be elucidated.
As we observed in a previous report [5], dECM expanded hSDSCs had a lower level of GAG and ACAN before chondrogenic induction (at day 0) which may be indicative of “stemness” because these pretreated SDSCs exhibited an enhanced chondrogenesis when incubated in a chondrogenic medium, as evidenced by an improved cell survival capacity (DNA amount adjusted by day 0) and enhanced chondrogenic differentiation. In contrast, SDSCs grown in ECM yielded pellets with the lowest levels in GAG/pellet, GAG/DNA, ACAN, and COL2A1, indicating the minimum chondrogenic matrix deposition.
The aim of using dECM for cell expansion is to provide a large quantity of high-quality stem cells for cartilage regeneration. In line with a recent report [17], an up-regulation of COL10A1 was observed in chondrogenically induced adult hSDSCs after expansion on dECM, but this outcome differed from the decreased level of hypertrophy in dECM expanded pSDSCs [5]. We expect the expanded cells to have high chondrogenic potential but less tendency to differentiate into bone. Our microarray data for pellets suggested that FGFR2 [29] and NOG [30] were up-regulated and FGF18 [31], FOXO1 [32], BMP4 [33], and CASP3 [34] were down-regulated during chondrogenic induction of dECM expanded hSDSCs. BMP signaling is critical for osteogenesis; as a potent BMP antagonist, reduction of noggin enhanced BMP signaling and in vitro osteoblast bone formation [30]; activating mutations of FGFR2 decreased the expression of NOG [35]. In addition, mice lacking FGF18 display delayed ossification and decreased expression of osteogenic markers [31]; as an early molecular regulator in the differentiation of mesenchymal cells into osteoblasts, silencing FOXO1 in a tibia organ culture model was demonstrated to decrease the expression of RUNX2 and impair bone formation [32]. Human BMP4 protein was reported to increase (in a dose dependent manner) ossification of endochondral bone [33]. Delayed ossification and decreased bone mineral density in caspase-3-deficient (Casp3−/− and Casp3+/−) mice suggested that caspase-3 is crucial for the differentiation of bone marrow stromal cells by influencing the TGF-beta/Smad2 pathway and cell cycle progression [34]. Our findings suggest that dECM expansion decreased the tendency of hSDSCs toward ossification of bone. We also found that IHH [36], COL2A1 [37], CHSY1 [38], and SLC35D1 [39] were up-regulated in dECM expanded hSDSCs, indicative of a down-regulation of dysplasia of the skeleton, including chondrodysplasia.
Wnt signaling molecules are involved in a multitude of developmental processes including chondrogenesis [40]. In this report, both canonical and noncanonical Wnt signals were activated in dECM expanded hSDSCs, indicating that Wnt signals are probably associated with enhanced cell proliferation. For example, Wnt3a promoted the proliferation of undifferentiated mesenchymal stem cells (MSCs) [41,42]. Wnt5a induced endothelial cell proliferation and enhanced cell survival under serum-deprived conditions [43]. Despite a continuing increase in both groups, especially in the dECM group, there is not sufficient evidence to support the role of Wnt11 in MSC proliferation. Intriguingly, overexpression of Wnt11 was recently demonstrated in suppressing MSC proliferation and arresting the cell cycle at the G0/G1 phase [44].
During chondrogenic induction, both WNT3A and WNT5A in dECM expanded hSDSCs exhibited a comparable trend, with an initial increase followed by a decrease; in contrast, Wnt11 expression exhibited a time-dependent increase in both groups, particularly for dECM expanded cells. This finding might indicate that the signals of both Wnt3a and Wnt5a are most related to cell expansion with a minor effect on the early stage of chondrogenic differentiation. Noncanonical Wnt signaling pathways such as Wnt5a and Wnt11 are known to promote chondrogenesis in vitro by increasing cartilage nodule formation [40,45]. Our data showed that Wnt5a and Wnt11 were up-regulated at both the mRNA and protein levels in dECM expanded hSDSCs except that Wnt5a exhibited a decrease at the later stage of chondrogenic induction. This outcome could possibly be explained by a previous report that Wnt5a specifically promotes entry into the prehypertrophic phase, whereas it conversely blocks chondrocyte hypertrophy, acting in a stage-specific context [46].
Wnt5a has been shown to have stage-dependent regulation of chondrocyte differentiation by promoting chondroprogenitor proliferation and inhibiting chondrocyte hypertrophy [47]; in addition, Wnt5a-infected limbs reveal a delay in the onset of type X collagen expression, a marker for hypertrophic chondrocytes [48]. It is possible that, in the early stage, the Wnt5a was highly activated in dECM expanded hSDSCs in order to boost chondrogenesis while the decreased expression of Wnt5a in the late stage contributes to enhanced hypertrophy. Interestingly, we also witnessed increased expression of Wnt11, which has been reported to greatly enhance chondrogenesis [44]. Although overexpression of Wnt11 did not appear to delay chondrocyte differentiation including type X collagen [40], more investigation is needed to confirm the correlation between Wnt11 expression and chondrogenic hypertrophy.
Similar to Wnt signals, all MAPK signals were up-regulated in dECM expanded hSDSCs, likely indicating a potential crosstalk between Wnt and MAPK signals during chondrogenesis. The MAPK pathway is one of the conserved signal transduction systems in cartilage, where it plays a crucial role in chondrogenic differentiation. Erk and p38 MAPK have central roles in mediating chondrocyte proliferation and related gene expression [49]. During the proliferation stage, dECM expanded cells showed enhanced expression of p-Erk and Erk; interestingly, Wnt3a can also promote Erk1/2 phosphorylation, and this signal was proposed to mediate proliferation of NIH3T3 cells [50]. Wnt5a also promotes Erk1/2 phosphorylation in endothelial cells and Wnt5a protects cells from apoptosis via Erk signaling [51].
During chondrogenic induction, the MAPK signals were observed to tentatively increase followed by a drop at day 7, indicating that the role of MAPK signals is mainly at the early stage of chondrogenic differentiation. This finding is in line with previous reports which maintain that the Erk pathway might be predominantly involved during early cartilage developmental stages in relaying signals down-steam of FGF actions [52,53] and Erk1/2 activities are observed to decrease as chondrogenesis proceeds and inhibition of Erk1/2 with PD98059 enhances chondrogenesis [54]. Jnk has been reported to have a minor role in chondrogenesis [55,56]. However, in this study, we did find Jnk signals to be increased during cell expansion and at the early stage of chondrogenic induction; the potential mechanisms are still under investigation.
5 Conclusion and Perspectives
Our study demonstrated that dECM deposited by hSDSCs formed a three-dimensional nanostructured matrix; expansion on dECM yielded a large-quantity of hSDSCs with enhanced chondrogenic potential; despite an increase of type X collagen in chondrogenically induced cells, dECM expanded cells had significantly lower potential for endochondral bone formation. Even though the composition of dECM has been characterized in a recent report [16], research on the crosstalk between expanded SDSCs and dECMs is still in its infancy. dECM expansion of hSDSCs and subsequent chondrogenic induction were accompanied by a significant change in both Wnt and MAPK signals, indicating potential mechanisms underlying dECM mediated hSDSC rejuvenation, which needs further in-depth investigation. Progress in understanding environment-oriented epigenetic regulatory mechanisms could benefit cartilage regeneration and engineering on a larger scale and provide more promising therapeutic applications [57]. For future clinical application, young and healthy donors can be selected for deposition of this ECM [16]; a decellularized process can minimize concerns about immune issues; and this dECM can be commercialized to make one-step in vitro expansion possible.
Acknowledgments
The authors thank Suzanne Danley for help in editing the manuscript. We also thank Ting Zheng and Dr. Cerasela Zoica Dinu for their help with the AFM instrument. This project was partially supported by research grants from the AO Foundation (S-12-19P) and the National Institutes of Health (NIH) (1 R03 AR062763-01A1 and P20 GM103434).
Footnotes
Competing Interest Statement
The authors do not have any conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karnes J, Zhang Y, Pei M. Cell Therapy for the Creation of Cartilage and Related Clinical Trials. In: Templeton NS, editor. Gene and Cell Therapy: Therapeutic Mechanisms and Strategies. 4. Taylor & Francis/CRC Press; 2014. pp. 1123–1135. [Google Scholar]
- 2.Jones B, Pei M. Synovium-derived stem cells: a tissue-specific stem cell for cartilage tissue engineering and regeneration. Tissue Eng Part B Rev. 2012;18:301–11. doi: 10.1089/ten.TEB.2012.0002. [DOI] [PubMed] [Google Scholar]
- 3.Pizzute T, Lynch K, Pei M. Impact of tissue-specific stem cells on lineage specific differentiation: a focus on musculoskeletal system. Stem Cell Rev Rep. 2014 doi: 10.1007/s12015-014-9546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JT, Pei M. Cell senescence: a challenge in cartilage engineering and regeneration. Tissue Eng Part B. 2012;18:270–87. doi: 10.1089/ten.TEB.2011.0583. [DOI] [PubMed] [Google Scholar]
- 5.He F, Chen XD, Pei M. Reconstruction of an in vitro tissue-specific microenvironment to rejuvenate synovium-derived stem cells for cartilage tissue engineering. Tissue Eng Part A. 2009;15:3809–21. doi: 10.1089/ten.TEA.2009.0188. [DOI] [PubMed] [Google Scholar]
- 6.He F, Pei M. Extracellular matrix enhances differentiation of adipose stem cells from infrapatellar fat pad toward chondrogenesis. J Tissue Eng Regen Med. 2013;7:73–84. doi: 10.1002/term.505. [DOI] [PubMed] [Google Scholar]
- 7.Li JT, He F, Pei M. Creation of an in vitro microenvironment to enhance human fetal synovium-derived stem cell chondrogenesis. Cell Tissue Res. 2011;345:357–65. doi: 10.1007/s00441-011-1212-8. [DOI] [PubMed] [Google Scholar]
- 8.Li JT, Pei M. Optimization of an in vitro three-dimensional microenvironment to reprogram synovium-derived stem cells for cartilage tissue engineering. Tissue Eng Part A. 2011;17:703–12. doi: 10.1089/ten.TEA.2010.0339. [DOI] [PubMed] [Google Scholar]
- 9.Pei M, He F, Kish VL. Expansion on extracellular matrix deposited by human bone marrow stromal cells facilitates stem cell proliferation and tissue-specific lineage potential. Tissue Eng Part A. 2011;17:3067–76. doi: 10.1089/ten.tea.2011.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei M, He F, Wei L. Three dimensional cell expansion substrate for cartilage tissue engineering and regeneration: a comparision in decellularized matrix deposited by synovium-derived stem cells and chondrocytes. J Tissue Sci Eng. 2011;2:104. [Google Scholar]
- 11.Pei M, Li JT, Zhang Y, Liu GH, Wei L, Zhang YY. Expansion on matrix deposited by nonchondrogenic urine stem cells strengthens repeated passage bone marrow stromal cells’ chondrogenic capacity. Cell Tissue Res. 2014;356:391–403. doi: 10.1007/s00441-014-1801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He F, Pei M. Rejuvenation of nucleus pulposus cells using extracellular matrix deposited by synovium-derived stem cells. Spine. 2012;37:459–69. doi: 10.1097/BRS.0b013e31821fcc64. [DOI] [PubMed] [Google Scholar]
- 13.Pei M, He F. Extracellular matrix deposited by synovium-derived stem cells delays chondrocyte dedifferentiation and enhances redifferentiation. J Cell Physiol. 2012;227:2163–74. doi: 10.1002/jcp.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pei M, Shoukry M, Li JT, Daffner S, France J, Emery SE. Modulation of in vitro microenvironment facilitates synovium-derived stem cell-based nucleus pulposus tissue regeneration. Spine. 2012;37:1538–47. doi: 10.1097/BRS.0b013e31825150bf. [DOI] [PubMed] [Google Scholar]
- 15.Pei M, He F, Li JT, Tidwell J, Jones A, McDonough EB. Repair of large animal partial-thickness cartilage defects using matrix rejuvenated synovium-derived stem cells. Tissue Eng Part A. 2013;19:1144–54. doi: 10.1089/ten.TEA.2012.0351. [DOI] [PubMed] [Google Scholar]
- 16.Li JT, Hansen K, Zhang Y, Dong CB, Dinu C, Dzieciatkowska M, et al. Rejuvenation of chondrogenic potential by young stem cell microenvironment. Biomaterials. 2014;35:642–653. doi: 10.1016/j.biomaterials.2013.09.099. [DOI] [PubMed] [Google Scholar]
- 17.Pei M, Zhang Y, Li JT, Chen DQ. Antioxidation of decellularized stem cell matrix promotes human synovium-derived stem cell-based chondrogenesis. Stem Cells Dev. 2013;22:889–900. doi: 10.1089/scd.2012.0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JT, Pei M. Decellularized stem cell matrix: a novel approach for autologous chondrocyte-based cartilage repair. In: Hayat MA, editor. Stem Cells and Cancer Stem Cells: Therapeutic applications in disease and injury. Springer Science + Business Media; Dordrecht: 2014. pp. 109–15. [Google Scholar]
- 19.Pei M, Li JT, Shoukry M, Zhang Y. A review of decellularized stem cell matrix: a novel cell expansion system for cartilage tissue engineering. Eur Cell Mater. 2011;22:333–43. doi: 10.22203/ecm.v022a25. discussion 343. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Pizzute T, Pei M. A review of crosstalk between MAPK and Wnt signals and its impact on cartilage regeneration. Cell Tissue Res. 2014;358:633–49. doi: 10.1007/s00441-014-2010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A-Hassan E, Heinz WF, Antonik MD, D’Costa NP, Nageswaran S, Schoenenberger CA, et al. Relative microelastic mapping of living cells by atomic force microscopy. Biophys J. 1998;74:1564–78. doi: 10.1016/S0006-3495(98)77868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang F, Ritter M, Gamper N, Huber S, Fillon S, Tanneur V, et al. Cell volume in the regulation of cell proliferation and apoptotic cell death. Cell Physiol Biochem. 2000;10:417–28. doi: 10.1159/000016367. [DOI] [PubMed] [Google Scholar]
- 23.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 24.Fernández-Muiños T, Suárez-Muñoz M, Sanmartí-Espinal M, Semino CE. Matrix dimensions, stiffness, and structural properties modulate spontaneous chondrogenic commitment of mouse embryonic fibroblasts. Tissue Eng Part A. 2014;20:1145–55. doi: 10.1089/ten.tea.2013.0369. [DOI] [PubMed] [Google Scholar]
- 25.Dong C, Kashon ML, Lowry D, Dordick JS, Reynolds SH, Rojanasakul Y, et al. Exposure to carbon nanotubes leads to changes in the cellular biomechanics. Adv Healthc Mater. 2013;2:945–51. doi: 10.1002/adhm.201200430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rheinlaender J, Geisse NA, Proksch R, Schäffer TE. Comparison of scanning ion conductance microscopy with atomic force microscopy for cell imaging. Langmuir. 2011;27:697–704. doi: 10.1021/la103275y. [DOI] [PubMed] [Google Scholar]
- 27.Oh S, Brammer KS, Li YS, Teng D, Engler AJ, Chien S, et al. Stem cell fate dictated solely by altered nanotube dimension. Proc Natl Acad Sci USA. 2009;106:2130–5. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, Bauer S, von der Mark K, Schmuki P. Nanosize and vitality: TiO2 nanotube diameter directs cell fate. Nano Lett. 2007;7:1686–91. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 29.Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129:3783–93. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- 30.Wan DC, Pomerantz JH, Brunet LJ, Kim JB, Chou YF, Wu BM, et al. Noggin suppression enhances in vitro osteogenesis and accelerates in vivo bone formation. J Biol Chem. 2007;282:26450–9. doi: 10.1074/jbc.M703282200. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–69. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teixeira CC, Liu Y, Thant LM, Pang J, Palmer G, Alikhani M. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J Biol Chem. 2010;285:31055–65. doi: 10.1074/jbc.M109.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnan V, Ma Y, Moseley J, Geiser A, Friant S, Frolik C. Bone anabolic effects of sonic/indian hedgehog are mediated by bmp-2/4-dependent pathways in the neonatal rat metatarsal model. Endocrinology. 2001;142:940–7. doi: 10.1210/endo.142.2.7922. [DOI] [PubMed] [Google Scholar]
- 34.Miura M, Chen XD, Allen MR, Bi Y, Gronthos S, Seo BM, et al. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. J Clin Invest. 2004;114:1704–13. doi: 10.1172/JCI20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol. 2005;168:1065–76. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18:641–59. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustafsson E, Aszodi A, Ortega N, Hunziker EB, Denker HW, Werb Z, et al. Role of collagen type II and perlecan in skeletal development. Ann N Y Acad Sci. 2003;995:140–50. doi: 10.1111/j.1749-6632.2003.tb03217.x. [DOI] [PubMed] [Google Scholar]
- 38.Wilson DG, Phamluong K, Lin WY, Barck K, Carano RA, Diehl L, et al. Chondroitin sulfate synthase 1 (Chsy1) is required for bone development and digit patterning. Dev Biol. 2012;363:413–25. doi: 10.1016/j.ydbio.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Hiraoka S, Furuichi T, Nishimura G, Shibata S, Yanagishita M, Rimoin DL, et al. Nucleotide-sugar transporter SLC35D1 is critical to chondroitin sulfate synthesis in cartilage and skeletal development in mouse and human. Nat Med. 2007;13:1363–7. doi: 10.1038/nm1655. [DOI] [PubMed] [Google Scholar]
- 40.Church V, Nohno T, Linker C, Marcelle C, Francis-West P. Wnt regulation of chondrocyte differentiation. J Cell Sci. 2002;115(Pt 24):4809–18. doi: 10.1242/jcs.00152. [DOI] [PubMed] [Google Scholar]
- 41.Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93:1210–30. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 42.Shang YC, Wang SH, Xiong F, Zhao CP, Peng FN, Feng SW, et al. Wnt3a signaling promotes proliferation, myogenic differentiation, and migration of rat bone marrow mesenchymal stem cells. Acta Pharmacol Sin. 2007;28:1761–74. doi: 10.1111/j.1745-7254.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- 43.Masckauchán TN, Agalliu D, Vorontchikhina M, Ahn A, Parmalee NL, Li CM, et al. Wnt5a signaling induces proliferation and survival of endothelial cells in vitro and expression of MMP-1 and Tie-2. Mol Biol Cell. 2006;17:5163–72. doi: 10.1091/mbc.E06-04-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu S, Zhang E, Yang M, Lu L. Overexpression of Wnt11 promotes chondrogenic differentiation of bone marrow-derived mesenchymal stem cells in synergism with TGF-β. Mol Cell Biochem. 2014;390:123–31. doi: 10.1007/s11010-014-1963-0. [DOI] [PubMed] [Google Scholar]
- 45.Ryu JH, Chun JS. Opposing roles of WNT-5A and WNT-11 in interleukin-1beta regulation of type II collagen expression in articular chondrocytes. J Biol Chem. 2006;281:22039–47. doi: 10.1074/jbc.M601804200. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Topol L, Lee H, Wu J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 2003;130:1003–15. doi: 10.1242/dev.00324. [DOI] [PubMed] [Google Scholar]
- 47.Hosseini-Farahabadi S, Geetha-Loganathan P, Fu K, Nimmagadda S, Yang HJ, Richman JM. Dual functions for WNT5A during cartilage development and in disease. Matrix Biol. 2013;32:252–64. doi: 10.1016/j.matbio.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;127:3141–59. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- 49.Krens SF, He S, Spaink HP, Snaar-Jagalska BE. Characterization and expression patterns of the MAPK family in zebrafish. Gene Expr Patterns. 2006;6:1019–26. doi: 10.1016/j.modgep.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Yun MS, Kim SE, Jeon SH, Lee JS, Choi KY. Both ERK and Wnt/beta-catenin pathways are involved in Wnt3a-induced proliferation. J Cell Sci. 2005;118(Pt 2):313–22. doi: 10.1242/jcs.01601. [DOI] [PubMed] [Google Scholar]
- 51.Almeida M, Han L, Bellido T, Manolagas SC, Kousteni S. Wnt proteins prevent apoptosis of both uncommitted osteoblast progenitors and differentiated osteoblasts by beta-catenin-dependent and -independent signaling cascades involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J Biol Chem. 2005;280:41342–51. doi: 10.1074/jbc.M502168200. [DOI] [PubMed] [Google Scholar]
- 52.Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–37. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- 53.Lunn JS, Fishwick KJ, Halley PA, Storey KG. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev Biol. 2007;302:536–52. doi: 10.1016/j.ydbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Oh CD, Chang SH, Yoon YM, Lee SJ, Lee YS, Kang SS, et al. Opposing role of mitogen-activated protein kinase subtypes, erk-1/2 and p38, in the regulation of chondrogenesis of mesenchymes. J Biol Chem. 2000;275:5613–9. doi: 10.1074/jbc.275.8.5613. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura K, Shirai T, Morishita S, Uchida S, Saeki-Miura K, Makishima F. p38 mitogen-activated protein kinase functionally contributes to chondrogenesis induced by growth/differentiation factor-5 in ATDC5 cells. Exp Cell Res. 1999;250:351–63. doi: 10.1006/excr.1999.4535. [DOI] [PubMed] [Google Scholar]
- 56.Stanton LA, Underhill TM, Beier F. MAP kinases in chondrocyte differentiation. Dev Biol. 2003;263:165–75. doi: 10.1016/s0012-1606(03)00321-x. [DOI] [PubMed] [Google Scholar]
- 57.Li J, Ohliger J, Pei M. Significance of epigenetic landscape in cartilage regeneration from the cartilage development and pathology perspective. Stem Cells Dev. 2014;23:1178–94. doi: 10.1089/scd.2014.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]