Abstract
BACKGROUND
A new negative feedback loop has been proposed, which suggests connections between the circadian clock and SIRTUIN1 (SIRT1)-dependent functions associated with cell survival, development and metabolism.
OBJECTIVE
To develop a SIRT1 and circadian locomotor output cycles kaput (CLOCK) combined genotype and to assess its associations with the chronotype of subjects and their potential resistance to weight loss in a behavioral treatment for obesity based on a Mediterranean diet.
DESIGN
Overweight /obese subjects (n=1465), aged 20–65 years, who attended outpatient obesity clinics, were genotyped for SIRT1 (rs1467568) and CLOCK (3111T>C, rs1801260). Anthropometric, biochemical and dietary-intake variables were analyzed. Effectiveness of the program and weight loss progression during 30 weeks of treatment was assessed.
RESULTS
We found highly consistent associations between the morning/evening questionnaires across the different genotype categories. Subjects carrying minor alleles at SIRT1 and CLOCK loci (R group) displayed a higher resistance to weight loss and a lower weekly weight loss rate as compared with homozygotes for both major alleles (P group). Significant differences were found across genotypes in weight loss progression during the 30 weeks of treatment (P = 0.039). Dietary habits indicated that R carriers had a lower intake of total carbohydrates and monounsaturated fats, and a higher intake of saturated fats than those carrying the intermediate (M) and the P genotype (P = 0.02). Plasma ghrelin concentrations were also significantly higher in subjects carrying the R genotype.
CONCLUSION
Variants of both SIRT1 and CLOCK have an additive effect on resistance to weight loss that could be related to the chronotype of the subject, higher plasma levels of ghrelin and less adherence to Mediterranean diet patterns.
Keywords: Sirtuin1, clock, evening preference, weight loss
INTRODUCTION
Daily time keeping in many organisms depends on internal circadian clocks that temporally organize biological functions relative to each other as well as to the environment. These clocks generate physiological and behavioral rhythms by using circuits of gene expression that are organized in negative feedback loops.1
Recently, a new negative feedback loop has been proposed as an addition to this circuitry that involves the metabolite nicotinamide adenine dinucleotide and the protein SIRTUIN1 (SIRT1).2 The new loop suggests connections between the circadian clock and SIRT1-dependent functions associated with cell survival, development, inflammation and metabolism.
SIRT1 functions in heterochromatin formation and transcriptional repression through the deacetylation of histones H3 and H4.3 Histones are, however, not the only relevant target for deacetylation by SIRT1. Clock genes and different metabolic regulators are also deacetylated by SIRT1.4 In addition, circadian locomotor output cycles kaput (CLOCK), an essential element of the positive regulatory arm in the human biological clock, shows acetyltransferase activity toward histone H3 and brain and muscle Arnt-like protein-1.5 Both the acetyltransferase CLOCK and the deacetylase SIRT1 are associated with brain and muscle Arnt-like protein-1 as well as with each other.
Additionally, SIRT1 contributes to circadian control in vivo acting as an enzymatic rheostat of circadian function, transducing signals originated by cellular metabolites to the circadian clock. In this light, SIRT1 may be a point through which changes in cellular energy metabolism influence the functioning of the clock.6 Indeed, SIRT1 expression is reported to be required for high amplitude transcriptional rhythms of several core clock components and constitutes a reciprocal transcription loop linking metabolism with CLOCK oscillation.3,7 By associating with CLOCK, SIRT1 could affect the daily timing of molecular, cellular, metabolic, endocrine, physiological and behavioral functions.8
In addition, SIRT1 promotes lipid mobilization in white adipose tissue (WAT).9 Mobilization of free fatty acids from white adipose tissue in response to fasting is impaired in Sirt1−/− mice.9 Pharmacological inhibition or small interfering RNA-mediated knockdown of hypothalamic Sirt1 was shown to decrease food intake and body weight gain.10 Moreover, in population studies two common variants in SIRT1 were associated with a lower body mass index. Carriers of these variants had 13–18% decreased risk of obesity, and gained less weight over time.11 These findings suggest that sirtuins might provide important new targets for the treatment of obesity and related diseases.
Similarly, the CLOCK gene is implicated in obesity and weight loss.12 Previous studies in different populations have demonstrated the association of CLOCK gene polymorphisms with obesity,13 the particular role of CLOCK 3111T>C SNP in weight loss resistance 14 and the fact that sleep reduction, alterations in eating behaviors and evening preference that characterized CLOCK 3111C carriers could be affecting weight loss.15 Considering the tight connections between SIRT1 and CLOCK, and the role of SIRT1 in transducing signals originated by metabolic components of the circadian clock, the aim of the present study was to develop a SIRT1 and CLOCK combined genotype, and to assess its association with the chronotype of the subjects and their potential resistance to weight loss in a behavioral treatment for obesity based on a Mediterranean diet.
MATERIAL AND METHODS
Subjects and methods
We recruited overweight or obese subjects (body mass index>25 kg m−2 and <40 kg m−2) within the age range of 20 – 65 years, (n = 1465) who attended five outpatient obesity clinics during 2009 – 2010 in the city of Murcia, located in southeastern Spain. Around 82% of the population was women. Patients receiving thermogenic or lipogenic drugs, or those diagnosed with diabetes mellitus, chronic renal failure, hepatic diseases or cancer were excluded from the study. All procedures were in accordance with good clinical practice. Written consent was obtained from each patient before participation and the study principles were approved by the Research Ethics Committee of the Virgen de la Arrixaca Hospital. Patient data were codified to guarantee anonymity.
Characteristics of the treatment
The characteristics of the weight reduction program (Garaulet method) have been described elsewhere.16,17. Briefly, during the initial 4 months, subjects attended a weekly 60-min therapy session in support groups (n = 10), followed by a 5-month maintenance period. These sessions were conducted by a nutritionist and the treatment was based on the following: First, individual dietary energy requirements were assessed by calculating1 resting energy expenditure according to the Harris-Benedict formula and2 total energy expenditure according to the type and duration of physical activity estimated by the International Physical Activity Questionnaire. Second, about 600 kcal per day were subtracted from the total energy expenditure. The final dietary energy content ranged from 1200 to 1800 kcal per day for women and 1500 to 2000 kcal per day for men to induce an approximate loss of 0.5 –1 kg per week. The recommendations were consistent with the Mediterranean-type diet.18,19
Anthropometric and biochemical measurements
Subjects were weighed weekly and barefoot wearing light clothes, with a digital scale to the nearest 0.1 kg, at the same time each day to assess weight loss during the treatment. Height was measured using a Harpenden digital stadiometer (rank 0.7 – 2.05). The subject was positioned upright, relaxed and with the head in the Frankfurt plane. Body mass index was calculated according to these measurements as weight (kg)/(height(m))2. Total body fat was measured by bioelectrical impedance using TANITA TBF-300 (TANITA Corporation of America, Arlington Heights, IL, USA) equipment. Body fat distribution was assessed by the measurement of waist circumference, at the level of the umbilicus.19 All measurements were made with a flexible and inextensible tape measure.
Fasting blood samples, collected at 0800 hours were centrifuged at 4 °C, and the plasma was stored at −70 °C for subsequent analysis. Plasma ghrelin and leptin concentrations were measured in duplicates by radioimmunoassay (Linco Research, St Charles, MO, USA).
Morning–evening questionnaire
Subjects completed the morningness/eveningness questionnaire on a 19-item scale of Horne and Ostberg.20 Morningness/eveningness typology is a way to characterize subjects depending on individual differences of wake/sleep patterns and the time of day people feel or perform best. Some people like to stay up late at night and sleep late in the morning (evening type), whereas others prefer to go to bed at an early hour and arise with the break of dawn (morning type). Evening types were considered as scoring <53 and morning types >64. All subjects within the range of 53– 64 were classified as neutral type,21 which represents the largest category in most populations that has been evaluated.
Habitual dietary intake
To evaluate food habits, initial nutrient intake was determined by a 24-h dietary recall. Interviews were conducted from monday to friday, including 24 h recalls of food intake from weekends to weekdays. Total energy intake and macronutrient composition from the initial 24 h recalls were analyzed, with the nutritional evaluation software program Grunumur (University of Murcia, Murcia, Spain)22,23 on the basis of Spanish food composition tables.23 The intakes of fatty acids were calculated from Spanish food composition tables.24
DNA isolation and genotyping
DNA was isolated from blood samples using DNA isolation sets (Qiagen, IZASA S.A., Barcelona, Spain). We performed genotyping of SIRT1 (rs1467568) and CLOCK (3111T>C, rs1801260) because both single-nucleotide polymorphisms (SNPs) have been previously associated with body mass index.11,12 We used a TaqMan assay with allele-specific probes on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to the standardized laboratory protocols.13 The Applied Biosystems Assay ID were C___8746719_20 for CLOCK 3111T/ C and C___1340398_10 for SIRT1_rs1467568.
Statistical analyses
We used Pearson’s χ2 and Fisher’s exact tests to test differences in frequencies. We applied analysis of variance and the Student t-test to compare crude means across genotype groups. We tested different genetic inheritance models and a dominant model was applied in the final analyses for SIRT1 and CLOCK. Additive or interactive (synergistic or antagonistic) effects of the individual SNPs were first tested by examining the statistical significance of the main effects and interaction terms among the two SNPs. Homogeneity of genotypic effects according to sex was also tested by introducing the interaction term between the combined genotype variable and sex in the corresponding analysis of covariance model.
We performed multivariate adjustments of the associations by analysis of covariance and estimated adjusted means. We adjusted analyses for sex, age and study center. Statistical analyses were performed using SPSS 15.0 software (SPSS Inc., Chicago, IL, USA). A two-tailed P-value of <0.05 was considered statistically significant.
RESULTS
General characteristics of the studied population are presented in Table 1. Participants were overweight or obese, with an average weight loss of 9 kg. SNPs were in Hardy-Weinberg equilibrium in this population (P>0.05). SIRT1 (rs1467568) and CLOCK 3111T>C (rs1801260) SNPs were independent of each other in terms of linkage disequilibrium. Minor allele frequency was of 31.2 for SIRT1 (rs1467568) and of 27.6 for CLOCK 3111T>C. Following the dominant model carriers of one or two copies of the minor allele were grouped, and two categories were evaluated for each SNP. Interestingly, for both SNPs we found a significant association with weight loss. In both cases, subjects carrying the minor allele lost significantly less weight than non-carriers (P<0.05).
Table 1.
General characteristics of participants of the weight loss program
| n = 1465 | Mean | s.d. |
|---|---|---|
| Age, years | 39.37 | 12.29 |
| Females n, % | 1208 | 82.5 |
| Initial anthropometric and biochemical measurements | ||
| BMI, kg m−2 | 31.1 | 5.3 |
| Body fat, % | 37.3 | 6.6 |
| Waist, cm | 102 | 15 |
| Ghrelin, pmol l−1 | 313.3 | 294.9 |
| Leptin, nmol l−1 | 1.2 | 0.9 |
| Initial dietary intake | ||
| Total energy, kJ per day | 8650 | 2990 |
| Extra energy, kJ per day | 342 | 431 |
| Proteins, % total energy | 17.0 | 4.6 |
| Proteins, g per day | 85.8 | 32.8 |
| Carbohydrates, % total energy | 41.8 | 10.6 |
| Carbohydrates, g per day | 214.5 | 88.7 |
| Fiber, g per day | 18.8 | 11.1 |
| Fats, % total energy | 42.2 | 9.6 |
| Fats, g per day | 98.3 | 44.5 |
| MUFA, % total fat | 55.5 | 8.1 |
| PUFA, % total fat | 13.9 | 4.0 |
| SFA, % total fat | 29.8 | 8.5 |
| Initial chronobiological characteristics | ||
| Morning–evening score | 51.7 | 10.1 |
| Morning–evening classification | N | (%) |
| Neutral type | 864 | 59 |
| Morning type | 366 | 25 |
| Evening type | 235 | 16 |
| Weight loss | ||
| Weight loss, kg | 9.01 | 5.61 |
| Percentage of weight loss, % | 11.1 | 6.16 |
| Other characteristics | N | % |
| Obese | 791 | 54 |
| Central obesity | 1187 | 81 |
| Sedentary | 600 | 41 |
| SIRT1/CLOCK genotype (n = 1232) | N | % |
| R | 299 | 24.3 |
| M | 627 | 50.9 |
| P | 306 | 24.8 |
Abbreviations: BMI, body mass index; CLOCK, circadian locomotor output cycles kaput; M, subjects showing intermediate values in total weight loss (kg); MUFA: monounsaturated fatty acids; P, subjects showing a higher weekly weight loss rate; PUFA: polyunsaturated fatty acids; R, subjects displaying a greater resistance to weight loss; SFA: saturated fatty acid; SIRT1, SIRTUIN1 protein.
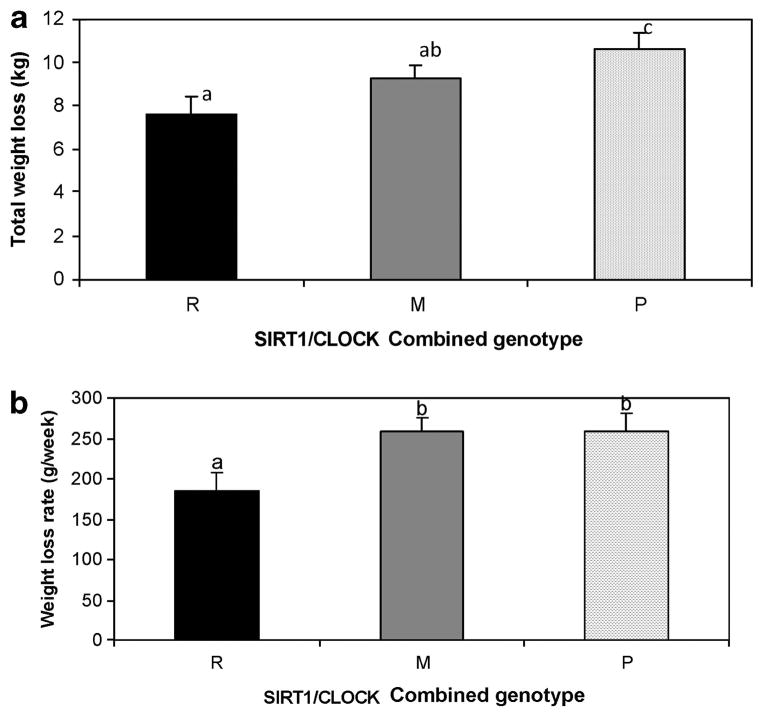
When we first tested additive or interactive effects of these two SNPs in analysis of covariance models (results not shown), we found no statistically significant interaction terms that suggested additive effects. Therefore, we defined four combined genotype groups based on these two SNPs. Next, we performed analyses to evaluate the effectiveness of the treatment, particularly for total weight loss (kg) and weight loss rate (g per week) across genotype categories, and we found an additive effect of the variants for total weight loss (Figures 1a and b). Indeed, one group of genotype subjects displayed a greater resistance to weight loss (we called them R for resistance) and a lower weekly weight loss rate as compared with another group, which was assigned P for protected subjects. Subjects who showed intermediate values in total weight loss (kg) were called M (medium) and the following classification was considered. (1) Carriers of the variant allele for the SIRT1 (AG or AA) and carriers of the variant allele at CLOCK SNP (TC or CC) after a first statistical approach with our data: this group (n = 299) was considered to be resistant to weight loss and was named R. (2) Homozygous for the major allele at the SIRT1 rs1467568 (GG) and carriers of the variant allele at CLOCK 1131T/C SNP (TC or CC): this group (n = 279) was considered to be intermediate in effects and was named M1. (3) Carriers of the variant allele at the SIRT1 rs1467568 (AG or AA) and homozygous for the major allele at the CLOCK 1131T/C SNP (TT): this group (n = 348) was also considered intermediate in effect and was named M2. (4) Homozygous for the major alleles for both SNPs (GG for rs1467568 at the SIRT1 gene and TT for CLOCK 1131T/C: (n = 306) was considered to be a protective combination and was termed the P group. Because the behavior of the intermediate groups M1 and M2 was similar and no significant differences were observed between these two groups, we pooled these two groups into one and renamed the genotype group M. Thus, three combined genotype groups were considered.
Figure 1.
Differences in total weight loss (a) and weight loss rate (g per week) (b) across SIRT1-CLOCK combined genotypes in the total treatment. Data are presented as mean ± s.e.m. based on analysis of variance (ANOVA) and after adjusting for sex, age and study center. R for subjects resistant in total weight loss and weight loss rate (kg), M for intermediate and P for protected subjects. We used different letters to indicate the differences between the groups (P<0.050).
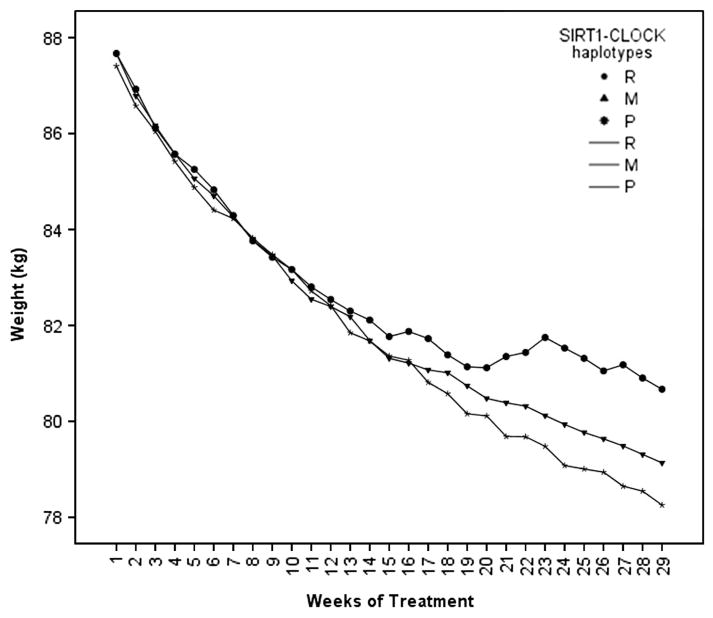
We also studied whether these genotypes were associated with weight loss progression during the 30 weeks of treatment (Figure 2). Significant differences were found across the three genotypes (P = 0.039). Interestingly, weight loss was significantly higher in carriers of the P genotype than in those carrying the R genotype (P = 0.02). Difficulty in losing weight was particularly evident after 12 weeks of treatment as evidenced by the analysis of repeated measurements (Figure 2).
Figure 2.
Weight loss progression during 30 weeks of treatment in subjects of different SIRT1-CLOCK combined genotypes (R, M and P) after adjusting for sex, age and study center. R for subjects resistant in total weight loss (kg), M for intermediate and P for protected subjects. Differences between the groups (P<0.050).
We found highly consistent associations with ME questionnaire scores across the genetic categories (P, M and R), with statistically significant P-values for linear trend. The percentage of evening type subjects was significantly higher in carriers of the R genotype whereas morning type subjects were more frequent in the P genotype group (Table 2). Particularly interesting were the differences in combined genotypes frequencies in response to specific questions from morningness/eveningness questionnaire. For example, in response to the question “At what time of the day do you think that you reach your ‘feeling best’ peak?”, R carriers were overrepresented in the ‘feeling best in the evening’ category compared with M and P subjects (34%, 28% 23%) for R, M and P, respectively (P = 0.034). Similar findings were obtained for the question: “At what time in the evening do you feel tired and as a result in need of sleep?”; 80% of R subjects felt less tired in the evening and needed to go to sleep later, whereas the percentages were significantly lower for M (77%) and P (64%) (P = 0.045). Moreover, R subjects tended to be less functional in the morning than M or P subjects (P = 0.07).
Table 2.
Differences in chronotype frequency across the SIRT1-CLOCK combined genotypes
| Genotype | Extreme morning | Moderate morning | Neutral type | Moderate evening | Extreme evening |
|---|---|---|---|---|---|
| R | |||||
| n | 3 | 41 | 137 | 32 | 10 |
| % | 1.3 | 18.4 | 61.4 | 14.3 | 4.5 |
| M | |||||
| n | 18 | 102 | 267 | 66 | 9 |
| % | 3.9 | 22.1 | 57.8 | 14.3 | 1.9 |
| P | |||||
| n | 9 | 56 | 142 | 23 | 4 |
| % | 3.8 | 23.9 | 60.7 | 9.8 | 1.7 |
| Total | |||||
| n | 30 | 199 | 546 | 121 | 23 |
| % | 3.3 | 21.7 | 59.4 | 13.2 |
2.5 P = 0.003* |
Abbreviations: R for subjects resistant in total weight loss (kg), M for intermediate and P for protected subjects.
Based on the Fisher test. Bold values represent significant differences among groups (P = 0.003).
Lastly, we examined the potential additive effect of the combined SIRT1 and CLOCK 1131T/C SNPs, on total food intake and two food intake hormones. Our results from dietary habits (Table 3) indicated that these genotype groups were significantly associated with the total intake of carbohydrate (g) and the number of portions derived from the bread group, which included foods rich in complex carbohydrates such as bread, potatoes and pasta. Genotypes were also associated with the quality of fat, specifically with monounsaturated fat (MUFA) intake and saturated fat intake, both as percentage of total fat. In general, carriers of the R genotype had a significantly lower intake of total carbohydrates (g), bread group portions, and total MUFA and a higher intake of saturated fat intake than those carrying the M and the P genotypes (P = 0.02). Similar relationships were detected for plasma ghrelin concentrations with significantly higher levels in subjects carrying the R genotype (1126±64 pg ml−1;(mean±s.e.)) than in P carriers (962±65 pg ml−1) (P = 0.048). No significant associations were found among genotype groups for leptin plasma values (data not shown).
Table 3.
Associations among genotypes and dietary habits
|
R
|
Genotype
|
P-value | |||||
|---|---|---|---|---|---|---|---|
| Mean | (s.e.m.) |
M
|
P
|
||||
| Mean | (s.e.m.) | Mean | (s.e.m.) | ||||
| Total energy, kJ per day | 8566 | 226 | 8755 | 151 | 8616 | 218 | 0.754 |
| Proteins, % total energy | 17.6 | 0.35 | 16.7 | 0.24 | 16.9 | 0.34 | 0.090 |
| Proteins, g per day | 87 | 2 | 85 | 1.6 | 85 | 2.42 | 0.825 |
| Carbohydrates, % total energy | 40.5 | 0.85 | 42.1 | 0.58 | 41.4 | 0.82 | 0.288 |
| Carbohydrates, g per day | 141a | 4 | 149a | 3 | 158b | 4 | 0.022 |
| Fats, % total energy | 42.8 | 0.7 | 42.5 | 0.5 | 42.2 | 0.7 | 0.853 |
| Fats, g per day | 99 | 3 | 100 | 2 | 98 | 3 | 0.855 |
| MUFA, % total fat | 53.9a | 0.60 | 55.8b | 0.40 | 56.3c | 0.6 | 0.009 |
| PUFA, % total fat | 13.6 | 0.29 | 13.6 | 0.19 | 13.7 | 0.3 | 0.883 |
| SFA, % total fat | 31.5a | 0.63 | 29.7b | 0.42 | 29.1b | 0.6 | 0.021 |
| Meat portions | 6.9 | 0.2 | 6.7 | 0.1 | 6.8 | 0.2 | 0.657 |
| Bread portions | 5.1a | 0.1 | 5.7b | 0.12 | 5.9c | 0.1 | 0.006 |
| Fat portions | 5.0 | 0.1 | 5.1 | 0.1 | 5.3 | 0.1 | 0.416 |
| Vegetables portions | 1.9 | 0.1 | 1.8 | 0.1 | 2.0 | 0.1 | 0.712 |
| Fruit portions | 1.3 | 0.1 | 1.3 | 0.05 | 1.3 | 0.08 | 0.923 |
Abbreviations: MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acid. R for subjects resistant in total weight loss (kg), M for intermediate and P for protected subjects. Data are presented as mean ± s.e.m. based on ANOVA, we used different letters to indicate the differences between the groups (P<0.050). Bold values represent significant differences among groups.
DISCUSSION
Results from previous association studies support the hypothesis that significant predictive value from genetic markers can be gained by combining genotype information from several loci.25 – 27 This concept can be applied to the ability to predict individual responses to weight loss treatment. In this context, we have shown in a total of 1465 participants in a weight loss program based on Mediterranean diet principles that genetic variants of both SIRT1 and CLOCK genes have an additive effect on resistance to weight loss. Moreover, the combination of both SNPs was associated with a particular chronotype of the subjects (evening type), with fewer adherence to Mediterranean food intake patterns and with higher plasma levels of ghrelin.
In the present population, the mean morningness/eveningness preference score showed normal distribution and was similar to that found in other adult populations,21,28 with a significantly higher proportion of morning type than evening type subjects. However, the proportion of subjects with evening preference among carriers of the SIRT1-CLOCK resistance (R) allele was higher than in the intermediate (M) and protective (P) allele subjects. Previous studies have evaluated the particular relevance of different clock genes to chronotype.29,30,15 ME preference in humans is affected by the free-running period, which is determined by circadian clock-relevant genes.1 Particularly for CLOCK SNP 3111T>C, located in the 3′-flanking region of CLOCK, previous studies similarly have reported that the 3111C (minor) allele was associated with evening preference in Caucasian15 and Asian populations, with a delayed shift of sleep onset time of 40–50 min compared with that in subjects with the 3111T allele.28,31 However, to our knowledge, this is the first study to analyze associations between ME preference and SIRT1, and we report a significant association with evening preference in minor allele subjects for SIRT1 (data not shown), and in SIRT1-CLOCK R combined genotype carriers.
Evening preference has been associated previously with obesity.32,33 Indeed, studies indicate a positive correlation between eating later in the day, especially in the afternoon and evening hours, and overall energy intake as well as body weight status.32,33 Our results indicate that subjects carrying minor alleles at both the SIRT1 and CLOCK loci (R group) had a significantly higher resistance to weight loss than did subjects homozygous for both major alleles (P group). Moreover, persons carrying minor alleles at either one of these loci (M group) had intermediate weight losses that were significantly different from both P and R subjects, and underscores the additive effect of the two polymorphisms comprising the combined genotype. Interestingly, the difficulty in losing weight was particularly evident after 12 weeks of treatment as evidenced by analysis of repeated measurements, which suggests that the relevance of genotype in weight loss increases with the length of the treatment. Possibly, psychological or behavioral habits influence these results. On the other hand, evening preferences in R allele carriers could influence sleep and dietary preferences toward a more obesogenic pattern and behaviors that are less effective in achieving weight loss.
We can hypothesize about cellular as well as behavioral mechanisms, which underlie resistance to weight loss observed in SIRT1minor allele carriers. It has been reported that sirtuins are implicated in shifting adipose tissue metabolism toward a more lipolytic and less lipogenic pattern.9,10 The current results suggest that the anti-obesogenic characteristics of SIRT1 could be impaired in R haplotype carriers promoting a greater resistance to weight loss. In this regard, mobilization of free fatty acids from adipose tissue in response to fasting is impaired in Sirt1−/− mice.9
One novelty of the present paper gives evidence that combinations of SIRT1 and CLOCK polymorphisms were associated with weight loss resistance. The mechanisms underlying the observed associations for these combinations need to be elucidated. However, SIRT1 was recently shown to modulate CLOCK expression,3 and it may thus form an intriguing link between sensing of cellular metabolism and the circadian clock.6 The coordinated recruiting of the CLOCK:brain and muscle Arnt-like protein-1 dimer and SIRT1 to circadian gene promoters suggests that these regulators may physically interact.10 Indeed, in coimmunoprecipitation experiments it has been revealed that SIRT1 interacts with CLOCK but not with PER2, and that the SIRT1-CLOCK interaction is mostly stable during the circadian cycle.10 Considered in this light, SIRT1 may be a transducer through which changes in cellular energy metabolism influence the functioning of the clock.6 By interacting with CLOCK, SIRT1 could affect the daily timing of metabolic, endocrine, physiological and behavioral functions associated with weight loss characteristics.8
Moreover, because SIRT1 has been found to control a number of nuclear receptors,34,35 we speculate that the CLOCK-SIRT1 interaction described represents a key event in the processes of fat and energy metabolism.7 SIRT1 expression as well as its activity are nutrient sensitive, and can translate nutritional information to clock genes.35 Although hypotheses regarding cellular mechanisms are largely theoretical, we were able to examine dietary patterns and nutrients more directly. Our dietary intake data indicated differences in composition of fat intake composition among SIRT1-CLOCK combined genotypes with a less Mediterranean dietary pattern among R carriers. Those subjects had significantly lower intakes of MUFA, higher saturated fat intake and a lower intake of complex carbohydrate than M or P carriers, and this effect was additive in most cases. The specific resistance to weight loss in R carriers could be related to a lower adherence to the Mediterranean principles that constitute the basis of this behavioral dietary intervention.
It has been described in experimental models that high intake of saturated fat modifies circadian synchronization to light and leads to metabolic abnormalities, which resemble human metabolic syndrome, including obesity and insulin resistance.36 For example, in the rat liver, the Sirt1 protein level is decreased by a high fat diet.37 Furthermore, we have shown previously the particular relevance of MUFA to the CLOCK gene.13 The circadian system regulates lipid metabolism throughout the expression and/or activity of some metabolic enzymes involved in fatty-acid metabolism.12
It is also possible that the effects on weight loss reported in the current study were caused by an influence of SIRT1 on appetite and energy intake because SIRT1 is highly expressed in brain.38 Several lines of evidence demonstrate that SIRT1 functions in the hypothalamus to regulate food intake and body weight. SIRT1 was shown to regulate the expression of Agouti Related Protein and Proopiomelanocortin.10,39 In the current population, when food intake hormones leptin and ghrelin were analyzed, genotypes showed a significant association with ghrelin levels, such that plasma concentrations were lower in P subjects, than in M and R carriers. It has been reported that SIRT1 is a metabolic sensor that can be modulated by adenosine monophosphate-activated protein kinase. Activation of hypothalamic adenosine monophosphate-activated protein kinase is mediated by ghrelin, leading to an increase in orexigenic neuropeptides and stimulation of food intake.40,41 We hypothesize that the impairment in SIRT1 in minor allele carriers could be related to a higher production of ghrelin and as a consequence to a greater sensation of hunger among R patients, with minor alleles in both SIRT1 and CLOCK genes. The lack of association with leptin plasma levels in the present study is consistent with the idea that leptin and Sirt1 might act through independent pathways to ultimately regulate S6K signaling.10 It has been shown that an increased expression of S6K in the mediobasal hypothalamus results in decreased feeding.42,43
In summary, the present study showed that the combination of minor alleles at the SIRT1 (rs1467568) and CLOCK loci (3111T>C) is associated with an evening preference, and that each minor allele exerts an additive effect on weight loss. Furthermore, we report allele-specific dietary habits and plasma ghrelin levels for the combined genotype, which suggest that subjects within the risk genotype group had higher plasma ghrelin concentrations and a lower adherence to a Mediterranean diet.
Acknowledgments
This work was supported by the Government of Education, Science and Research of Murcia (Project BIO/FFA 07/01-0004) and by the Spanish Government of Science and Innovation (projects AGL2008-01655/ALI) National Heart, Lung, and Blood Institute Grants HL-54776, National Institute of Diabetes and Digestive and Kidney Diseases, Grant Number DK075030 and by contracts 53-K06-5-10 and 58-1950-9-001 from the US Department of Agriculture Research.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Garaulet M, Madrid JA. Chronobiological aspects of nutrition, metabolic syndrome and obesity. Adv Drug Deliv Rev. 2010;62:967–978. doi: 10.1016/j.addr.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wijnen H. Circadian rhythms. A circadian loop asSIRTs itself. Science. 2009;324:598–599. doi: 10.1126/science.1174132. [DOI] [PubMed] [Google Scholar]
- 4.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 6.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechtold DA. Energy-responsive timekeeping. J Genet. 2008;87:447–458. doi: 10.1007/s12041-008-0067-6. [DOI] [PubMed] [Google Scholar]
- 9.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cakir I, Perello M, Lansari O, Messier NJ, Vaslet CA, Nillni EA. Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS One. 2009;4:e8322. doi: 10.1371/journal.pone.0008322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zillikens MC, van Meurs JB, Rivadeneira F, Amin N, Hofman A, Oostra BA, et al. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009;58:2828–2834. doi: 10.2337/db09-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sookoian S, Gemma C, Gianotti TF, Burgueño A, Castaño G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. 2008;87:1606–1615. doi: 10.1093/ajcn/87.6.1606. [DOI] [PubMed] [Google Scholar]
- 13.Garaulet M, Lee YC, Shen J, Parnell LD, Arnett DK, Tsai MY, et al. CLOCK genetic variation and metabolic syndrome risk: modulation by monounsaturated fatty acids. Am J Clin Nutr. 2009;90:1466–1475. doi: 10.3945/ajcn.2009.27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garaulet M, Corbalán MD, Madrid JA, Morales E, Baraza JC, Lee YC, et al. CLOCK gene is implicated in weight reduction in obese patients participating in a dietary programme based on the Mediterranean diet. Int J Obes (Lond) 2010;34:516–523. doi: 10.1038/ijo.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garaulet M, Sánchez-Moreno C, Smith CE, Lee YC, Nicolás F, Ordovás JM. Ghrelin, sleep reduction and evening preference: relationships to CLOCK 3111 T/C SNP and weight loss. PLoS One. 2011;6:e17435. doi: 10.1371/journal.pone.0017435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garaulet M, Pérez-Llamas F, Zamora S, Tebar FJ. Weight loss and possible reasons for dropping out of a dietary/behavioural programme in the treatment of overweight patients. J Hum Nutr Diet. 1999;12:219–227. [Google Scholar]
- 17.Corbalán MD, Morales EM, Canteras M, Espallardo A, Hernández T, Garaulet M. Effectiveness of cognitive-behavioral therapy based on the Mediterranean diet for the treatment of obesity. Nutrition. 2009;25:861–869. doi: 10.1016/j.nut.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Serra-Majem L, Aranceta J Spanish Society of Community Nutrition. Nutritional objectives for the Spanish population. Consensus from the Spanish Society of Community Nutrition. Public Health Nutr. 2001;4:1409–1413. doi: 10.1079/phn2001229. [DOI] [PubMed] [Google Scholar]
- 19.Ferrario VF, Sforza C, Schmitz JH, Miani A, Jr, Taroni G. Fourier analysis of human soft tissue facial shape: sex differences in normal adults. J Anat. 1995;187 (Part 3):593–602. [PMC free article] [PubMed] [Google Scholar]
- 20.Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- 21.Taillard J, Philip P, Chastang JF, Bioulac B. Validation of Horne and Ostberg morningness-eveningness questionnaire in a middle-aged population of French workers. J Biol Rhythms. 2004;19:76–86. doi: 10.1177/0748730403259849. [DOI] [PubMed] [Google Scholar]
- 22.Pérez-Llamas F, Garaulet M, Herrero F, Palma JT, Pérez de Heredia F, Marín R, et al. Multivalent informatics application for studies of the nutritional status of the population. Assessment of food intake. Nutr Hosp. 2004;19:160–166. (Article in Spanish) [PubMed] [Google Scholar]
- 23.Mataix J, Mañas M, Llopis J, Martínez M. Table of Composition Of Spanish Foods. Instituto de Nutrición y Tecnología, Universidad de Granada; Granada, Spain: 1996. Book in Spanish. [Google Scholar]
- 24.Moreiras O, Carvajal A, Cabrera L. Tables of composition of foods. Pirámide SA; Madrid: 1995. Book in Spanish. [Google Scholar]
- 25.Trichopoulou A, Yiannakouris N, Bamia C, Benetou V, Trichopoulos D, Ordovas JM. Genetic predisposition, nongenetic risk factors, and coronary infarct. Arch Intern Med. 2008;168:891–896. doi: 10.1001/archinte.168.8.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy M, Kane JP, et al. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities Study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 27.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 28.Mishima K, Tozawa T, Satoh K, Saitoh H, Mishima Y. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:101–104. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- 29.Dijk DJ, Archer SN. PERIOD3, circadian phenotypes, and sleep homeostasis. Sleep Med Rev. 2010;14:151–160. doi: 10.1016/j.smrv.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Mendlewicz J. Disruption of the circadian timing systems: molecular mechanisms in mood disorders. CNS Drugs. 2009;23 (Suppl 2):15–26. doi: 10.2165/11318630-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 31.Lee KY, Song JY, Kim SH, Kim SC, Joo EJ, Ahn YM, et al. Association between CLOCK 3111T/C and preferred circadian phase in Korean patients with bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:1196–1201. doi: 10.1016/j.pnpbp.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Eng S, Wagstaff DA, Kranz S. Eating late in the evening is associated with childhood obesity in some age groups but not in all children: the relationship between time of consumption and body weight status in US children. Int J Behav Nutr Phys Act. 2009;6:27. doi: 10.1186/1479-5868-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Striegel-Moore RH, Rosselli F, Wilson GT, Perrin N, Harvey K, DeBar L. Nocturnal eating: association with binge eating, obesity, and psychological distress. Int J Eat Disord. 2010;43:520–526. doi: 10.1002/eat.20735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 35.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 36.Mendoza J, Pévet P, Challet E. High-fat feeding alters the clock synchronization to light. J Physiol. 2008;586 (Part 24):5901–5910. doi: 10.1113/jphysiol.2008.159566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng XQ, Chen LL, Li NX. The expression of SIRT1 in nonalcoholic fatty liver disease induced by high-fat diet in rats. Liver Int. 2007;27:708–715. doi: 10.1111/j.1478-3231.2007.01497.x. [DOI] [PubMed] [Google Scholar]
- 38.Ramadori G, Lee CE, Bookout AL, Lee S, Williams KW, Anderson J, et al. Brain SIRT1: anatomical distribution and regulation by energy availability. J Neurosci. 2008;28:9989–9996. doi: 10.1523/JNEUROSCI.3257-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue B, Kahn BB. AMPK integrates nutrient and hormonal signals to regulate food intake and energy balance through effects in the hypothalamus and peripheral tissues. J Physiol. 2006;574 (Part 1):73–83. doi: 10.1113/jphysiol.2006.113217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korbonits M, Goldstone AP, Gueorguiev M, Grossman AB. Ghrelin--a hormone with multiple functions. Front Neuroendocrinol. 2004;25:27–68. doi: 10.1016/j.yfrne.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 43.Blouet C, Ono H, Schwartz GJ. Mediobasal hypothalamic p70 S6 kinase 1 modulates the control of energy homeostasis. Cell Metab. 2008;8:459–467. doi: 10.1016/j.cmet.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]