Abstract
The cytochrome P4501C (CYP1C) gene subfamily was recently discovered in fish, and zebrafish (Danio rerio) CYP1C1 transcript has been cloned. Here we cloned the paralogous CYP1C2, showing that the amino acid sequence is 78% identical to CYP1C1, and examined gene structure and expression of CYP1A, CYP1B1, CYP1C1, and CYP1C2. Xenobiotic response elements were observed upstream of the coding regions in all four genes. Zebrafish adults and embryos were exposed (24 hours) to 100 nM 3,3’,4,4’,5- polychlorinated biphenyl (PCB126) or 20 ppm acetone and subsequently held in clean water for 24 hours (adults) or 48 hours (embryos). All adult organs examined (eye, gill, heart, liver, kidney, brain, gut, and gonads) and embryos showed basal expression of the four genes. CYP1A was most strongly expressed in liver, whereas CYP1B1, CYP1C1, and CYP1C2 were most strongly expressed in heart and eye. CYP1B1 and the CYP1C genes showed an expression pattern similar to one another and to mammalian CYP1B1. In embryos CYP1C1 and CYP1C2 tended to have a higher basal expression than CYP1A and CYP1B1. PCB126 induced CYP1A in all organs, and CYP1B1 and CYP1C1 in all organs except gonads, or gonads and brain, respectively. CYP1C2 induction was significant only in the liver. However, in embryos all four genes were induced strongly by PCB126. The results are consistent with CYP1C1 and CYP1C2, as well as CYP1A and CYP1B1, being regulated by the aryl hydrocarbon receptor. While CYP1A may have a protective role against AHR agonists in liver and gut, CYP1B1, CYP1C1, and CYP1C2 may also play endogenous roles in eye and heart and possibly other organs, as well as during development.
Keywords: cytochrome P4501 (CYP1); 3,3’,4,4’,5- polychlorinated biphenyl (PCB126); real time PCR; zebrafish
Introduction
Cytochromes P450 (CYPs) are the principal enzymes catalyzing oxidative metabolism of toxicants, including important environmental chemicals. CYP1 family members (Nelson et al., 1996) are prominent in oxidation of polycyclic aromatic hydrocarbons, aromatic amines, and a number of drugs. In mammals, the paralogous CYP1A1 and CYP1A2 and related CYP1B1 catalyze detoxication as well as activation of protoxicants and promutagens (Conney, 1982; Nebert et al., 2004; Shimada and Fujii-Kuriyama, 2004). These CYP1s also catalyze oxidative steps in biosynthesis and degradation of endogenous regulatory molecules (e.g. steroids and eicosanoids; Nebert and Russell, 2002; Lewis, 2004). Exogenous chemicals can affect organisms by disrupting such endogenous processes directly, as well as after oxidation to more reactive metabolites. Differences in activity of the enzymes involved can influence susceptibility of organs, individuals, or species to toxicity, affecting the inference of mechanisms from experimental studies and possibly influencing clinical decisions.
Expression level is a major factor influencing the role of CYPs in substrate oxidation and effects. Knockout studies show that ligand-activated aryl hydrocarbon receptor (AHR) largely determines the induction of mammalian CYP1As and CYP1B1 (e.g., Shimada et al., 2002). AHR also regulates CYP1As in non-mammalian vertebrates; knock-down of AHR2 in zebrafish (Danio rerio) embryos abolishes induction of the single CYP1A by the potent AHR agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Prasch et al., 2003; Dong et al., 2004). TCDD also induces zebrafish CYP1B1 (Handley-Goldstone et al 2005), presumably via the AHR. Knockout and knockdown studies show that AHRs mediate TCDD toxicity in mammals and fish (Fernandez-Salguero et al., 1996; Prasch et al., 2003; Dong et al., 2004).
Zebrafish are becoming an important model in developmental and adult toxicological studies (e.g., teratogenesis and carcinogenesis). Developmental effects of TCDD have been studied extensively, and embryos exposed to TCDD show yolk sac and pericardial edema, slowed heart rate and blood flow, craniofacial malformations, swim bladder defects, neural degeneration, arrested gill development, and death (Henry et al., 1997; Handley-Goldstone et al., 2005; Carney et al., 2006). One hypothesis is that toxicity involves reactive oxygen species produced by uncoupling of CYP1A by TCDD or coplanar polychlorinated biphenyls (PCBs) (Schlezinger et al., 1999; Shertzer et al., 2004; Schlezinger et al., 2006). However, conflicting results regarding the role of CYP1A in TCDD toxicity (Teraoka et al., 2003; Carney et al., 2004), imply the involvement of other AHR regulated genes, possibly including other CYP1s. Understanding of the full complement of CYP1 genes is required to discern their role in embryonic, systemic, and organ-specific effects of AHR agonists. Knowledge of CYP1 genes is limited in zebrafish, especially in adults, where for many organs there is no information on any CYP1.
Recent discovery in fish of the new CYP1C subfamily (Godard et al., 2005; Itakura et al., 2005) highlights the lack of knowledge of CYP1s. There are two paralogs in fish, CYP1C1 and CYP1C2, but the regulation, substrates, and biological functions of the CYP1Cs in the zebrafish model are unknown. Our aim was to determine the patterns of expression of CYP1C1 and CYP1C2 as compared to CYP1A and CYP1B1 in adult organs and embryonic zebrafish. As the other known CYP1s are in part AHR regulated, we investigated both basal expression and the responses to two potent AHR agonists, 3,3’,4,4’,5-PCB (PCB126), a major component of the AHR agonist activity in the environment, and β-naphthoflavone (βNF), a commonly studied agonist. We addressed the hypothesis that the CYP1C genes respond to AHR agonists as do CYP1A and CYP1B1. In order to accomplish these studies, we cloned and compared the sequence of CYP1C2, not previously cloned from zebrafish. We also compared gene structure of the CYP1s, including possible response element motifs that might support involvement of AHR in regulation of the new CYP1s, as well as of CYP1B1 and CYP1A in zebrafish. The results indicate a broader AHR responsive set of CYP1s than previously known, and the expression patterns suggest that the new CYP1Cs may have physiological roles, apart from the involvement in xenobiotic responses.
Materials and methods
Animals
Sexually mature zebrafish of two wild-type varieties were used. Tubingen long fin (TL) zebrafish were progeny of TLs obtained from the laboratory of Mark Fishman, crossed with TLs raised from eggs obtained from the Zebrafish International Resource Center at the University of Oregon (Eugene, OR, USA). A second undefined variety was purchased at a local pet store (PS). Fish were maintained in the Woods Hole Oceanographic Institution zebrafish facility and the experimental procedures were approved by the Institutional Animal Care and Use Committee. The zebrafish were held in 2:1 female to male groups at a density of ≤5 fish/l in aerated, filtered and re-circulated system water (28.5 °C) in 3 or 10 l tanks in an Aquatic Habitat™ system (TL fish) or a 40-l glass tank (PS fish). The system water was composed of Instant Ocean™ (60 mg/l), sodium bicarbonate (50 mg/l), calcium sulfate (8.5 mg/l) and Kent's Freshwater Essentials™ (53 μl/l) in distilled water. Fish were fed 2× daily with brine shrimp (Artemia salina) and once daily with Omega One flakes (Omega Sea Ltd. Sitka, AK, USA). TL embryos were obtained from group breedings of 30 female and 15 male fish.
Cloning
CYP1C1 and CYP1C2 transcripts were cloned from gill tissue of both the TL and PS zebrafish varieties. Gill arches were dissected and placed in RNAlater™ (Ambion, Austin, TX, USA). RNA was prepared using RNA stat-60 (Tel-Test Inc. Friendswood, TX, USA), mRNA was isolated from this RNA using the MicroPoly(A)Pure kit (Ambion), and cDNA was subsequently synthesized using the PowerScript™ Reverse Transcriptase (Clontech Laboratories Inc., Mountain View, CA, USA) and oligo dT primers (Operon Biotechnologies Inc., Huntsville, AL, USA). Full-length CYP1C1 and CYP1C2 cDNAs were amplified by using PCR primers to the 3’ and 5’ untranslated regions (UTRs; Table 1; Operon Biotechnologies Inc.) and Advantage™ cDNA PCR Kit (Clontech Laboratories Inc.). CYP1C1 and CYP1C2 1600-base pair PCR products were resolved on a 1% agarose gel and then isolated, cloned, and sequenced using previously described procedures.
Table 1.
Primer sequences used for cloning of CYP1C1 and CYP1C2 transcripts and quantification of CYP1 gene expression by real time PCR.
| Primer | Sequences |
|---|---|
| Cloning | |
| CYP1C1 Utr F | 5’-ATCAGCACCGAACACCAGCG |
| CYP1C1 Utr R | 5’-CCCCATTCGACTGGATGTTTTAAC |
| CYP1C2 Utr F | 5’-ACCCTCCAACTGAAGAGGGCAGAAA |
| CYP1C2 Utr R | 5’-TCATAGGCAGTGGGTTAGACAGCACA |
| Real time PCR | |
| ZFCYP1A F1) | 5’-GCATTACGATACGTTCGATAAGGAC |
| ZFCYP1A R1) | 5’-GCTCCGAATAGGTCATTGACGAT |
| ZFCYP1B1 F | 5’-GCTCAGCTGGTCCATTGATACC |
| ZFCYP1B1 R | 5’-CATCAGCGACAGCAACACAC |
| ZFCYP1C1 F | 5’-AGTGGCACAGTCTACTTTGAGAG |
| ZFCYP1C1 R | 5’-CCAAACAAGAAAGACTTTTGAGC |
| ZFCYP1C2 F | 5’-GTGGTGGAGCACAGACTAAG |
| ZFCYP1C2 R | 5’-TTCAGTATGAGCCTCAGTCAAAC |
| β-actin F1) | 5’-CAACAGAGAGAAGATGACACAGATCA |
| β-actin R1) | 5’-GTCACACCATCACCAGAGTCCATCAC |
| ARNT2 F | 5’-CACCTTTGGATCACATCTCATTG |
| ARNT2 R | 5’-TCACCCTCCTTAGACGGACC |
Exposure of adult zebrafish
In three sets of experiments, fish were exposed via ambient water to 1 μM βNF, 100 nM PCB126, or a dose range of PCB126 (between 0.3 and 100 nM). The concentrations given are nominal.
βNF
Mixed groups of male and female fish of the TL and PS varieties (6 fish, i.e., 3.5–3.7 g biomass per liter) were placed in two cling-wrap-covered glass beakers filled with continuously aerated zebrafish system water (28 °C). Aliquots of βNF (Sigma-Aldrich Inc. St. Louis, MO, USA) dissolved in acetone or acetone only were added to the beakers, yielding 1 μM βNF and 20 ppm acetone. After 24 h of exposure, the dosing solutions were renewed, and 24 h later the fish were killed by decapitation. Sex was confirmed by gonad examination. Gill and liver were dissected and organs from individual fish were frozen in liquid nitrogen and stored at −80 °C.
PCB126
Male and female TL or PS zebrafish (6 fish, corresponding to 4.2–4.6 g and 1.6–2.5 g of biomass per liter, respectively) were exposed to PCB126 (Cambridge Laboratories Inc., Andover, MA, USA) as described above for βNF. Aliquots of PCB126 dissolved in acetone or acetone only were mixed into the water, yielding 100 nM PCB126 and 20 ppm acetone. Following 24 h of exposure, the dosing solutions were replaced with clean zebrafish system water, and 24 h later the fish were killed by decapitation and sex confirmed. Gill and liver from two similarly exposed TL fish of the same sex were pooled. Four pools were analyzed (2 male and 2 female). A wider range of organs was examined from PS fish, i.e., liver, gut, kidney, ovary, testes, heart, brain, eye, and gill. In this experiment organs from multiple (3–10) fish of the same sex were pooled to obtain sufficient amounts of cDNA for the analyses. Four replicate exposures were performed resulting in four male and four female pooled samples for each organ. The dissected organs were preserved in RNAlater.
PCB126 concentration-response
PS zebrafish were exposed to doses of PCB126 ranging from 0.3 nM to 100 nM (and 20 ppm acetone), or 20 ppm acetone alone, as above (6 fish, 2.0–2.7 g biomass per liter). Expression of the suite of CYP1 genes was analyzed in gills and livers from individual fish.
Exposure of zebrafish embryos
Fertilized TL zebrafish eggs (33±5) were placed in 10 cm glass Petri dishes containing 25 or 30 ml of 0.3× Danieau's solution. At 8 hours post-fertilization (hpf), aliquots (2.5 or 3.0 μl) of PCB126 in acetone, or acetone alone, were added to the dishes yielding 100 nM PCB126 and 100 ppm acetone (nominal concentrations). The embryos were incubated at 28 °C. At 32 hpf, the dosing solutions were replaced with fresh 0.3× Danieau's solution and any dead embryos were removed; mortality was normally 2 embryos or fewer per dish. No difference in mortality was observed between controls and exposed fish, and subsequent to 32 hpf no mortality was observed. The embryos were held for an additional 48 h with replacement of the Danieau's solution after 24 h. At 80 hpf the experiment was terminated. All embryos in a dish were pooled, frozen in liquid nitrogen, and stored at −80 °C. Ten biological replicates were analyzed.
Quantification of CYP1 mRNA
RNA was isolated as described above and the isolates were DNase treated (TURBO DNA-free™ kit, Ambion). The quantity and quality of the RNA were determined spectrophotometically (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE, USA). cDNA was synthesized using the Omniscript™ Reverse Transcriptase kit (Qiagen Inc., Valencia, CA, USA), random hexamer primers (Operon Biotechnologies Inc.) and the RNasin® RNase inhibitor (Promega, Madison, WI, USA).
Gene specific primers for CYP1A, CYP1B1, CYP1C1, CYP1C2, ARNT2 and β-actin cDNA were synthesized by Operon Biotechnologies Inc. (Table 1). The ARNT2 primers could amplify identical sequences in ARNT2a, ARNT2b, and ARNT2c. Real time PCR was performed using the iQ SYBR Green Supermix (according to the manufacturers instructions) and an iCycler iQ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The PCR reaction mixtures consisted of iQ SYBR Green Supermix, primer (5 pmol each of forward and reverse primer) and cDNA (derived from 0.1 μg total RNA). In each sample the genes were analyzed in duplicate with the following protocol: 95°C for 4.5 min; 95°C for 15 s and 62 °C for 1 min (40 cycles). To ensure that a single product was amplified, melt curve analysis was performed on the PCR products at the end of each PCR run.
Calculations and statistics
The cloned CYP1 zebrafish cDNA sequences were aligned and compared to sequences obtained from the Danio rerio genome assembly version 6 (Zv6) in Ensembl (http://www.ensembl.org/Danio_rerio/index.html) using Sequecher 4.5 (Gene Codes Corporation, Ann Arbor, MI, USA) and GCG 10 (Accelrys, San Diego, CA, USA). Long range genomic alignments were performed using Vista (Mayor et al., 2000). Promoter region mapping was performed using GCG and MatInspector (Cartharius et al., 2005) using the conservative consensus XRE (KNGCGTG) and the MatInspector database. Phylogenetic analyses were performed using MrBayes (Ronquist and Huelsenbeck, 2003).
No significant sex difference in CYP1 gene expression was detected. Therefore the data for male and female fish were combined in the calculations, with the exception that the testes and ovary data were treated separately. For each reaction, relative mRNA expression of the CYP1 genes was calculated according to the EΔΔCt-method (Livak and Schmittgen, 2001), using β-actin as a reference gene (unless otherwise indicated). PCR efficiency (E) was determined with the LinRegPCR program (Ramakers et al., 2003). Outliers were excluded based on the Grubbs test. Data were log-transformed and statistical differences in gene expression between groups were determined by one-way ANOVA followed by Tukey's, Bonferroni's, or Dunnet's multiple comparison tests (Prism 4 by GraphPad Software Inc., San Diego, CA, USA). The post-hoc test used for a specific data set is given in the figure legends. Lowest observed effect concentration (LOEC) values for PCB126 to induce the CYP1 genes were determined by one-way ANOVA followed by Dunnet's multiple comparison test. Values for EC50, i.e., the PCB126 concentration causing half maximal CYP1 gene induction, were calculated using the curve fitting routine of GraphPad Prism for nonlinear regression sigmoidal dose–response with variable slope.
Results
Cloned CYP1C1 and CYP1C2 transcripts
Using primers based on a predicted CYP1C2 sequence derived from the zebrafish genome (Zv6), we were able to clone sequences from gill of both the TL and PS zebrafish. In parallel, we repeated the cloning and sequencing of CYP1C1 from both TL and PS fish, using primers based on the CYP1C1 sequence published by Godard et al. (2005). The aligned zebrafish TL CYP1C1 and CYP1C2 protein sequences (Fig 1) had a 78 % amino acid sequence identity. Two amino acid sequences, one 43 and one 124 residues long (i.e., K45 to G87 and W327 to L450 in CYP1C2; Fig 1), and the heme binding signatures were identical in the two CYP1Cs.
Fig 1.
Alignment of CYP1C1 and CYP1C2 sequences derived from the Danio rerio genome assembly version 6 (Zv6; ENS 1C1 and ENS 1C2), and cloned CYP1C1 and CYP1C2 cDNA sequences from zebrafish Tubingen Longfin (TL) and zebrafish purchased in a local pet store (pet store-obtained zebrafish, PS). Small rectangles, amino acids that are identical in sequences cloned from both TL and PS zebrafish but differ from the corresponding Zv6 sequence; pentagons, amino acids in sequences cloned from either TL or PS zebrafish that differ from the other strain and from the Zv6 sequence; “heme bind” means heme binding site; shaded areas labeled SRS1-6 = Substrate binding sites 1–6.
Phylogenetic analysis showed that the zebrafish CYP1C2 is orthologous to CYP1C2 of other fish (pufferfish - Takifugu rubripes and scup - Stenotomus chrysops). The deduced TL CYP1C1 amino acid sequence differed from the Ensembl CYP1C1 sequence (ENSDARG00000058980) in four positions (E265, F266, P283, and D443), whereas the cloned TL CYP1C2 differed from the predicted zebrafish CYP1C2 by one amino acid (S444; Fig 1). Slight differences between the TL and PS sequences also were observed in both CYP1C1 and CYP1C2. The differing amino acids in the CYP1C1 sequences were I/V29 and D/N443 (TL/PS respectively) and those in the CYP1C2 sequences were L/M74, K/R223 and S/N259. K223 is located in a putative substrate recognition site in the F/G loop (SRS2: Fig 1).
Gene structure
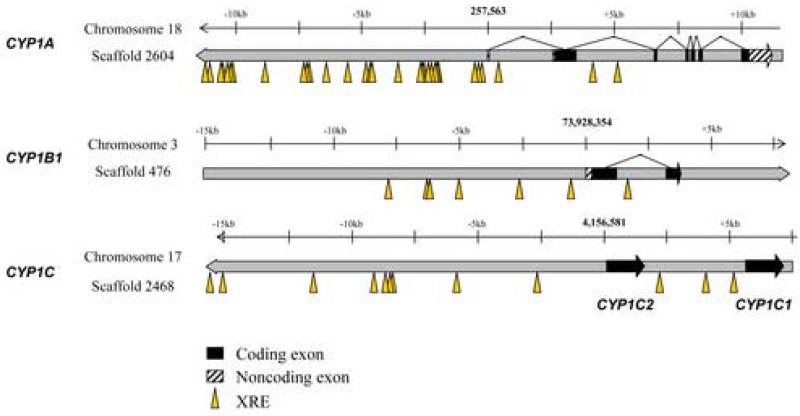
Analysis of the zebrafish genome shows that both CYP1C1 and CYP1C2 are single exon genes, located immediately adjacent to one another (4 kB apart) on chromosome 17 (Fig 2). Canonical xenobiotic response elements (XREs), which are binding sites for the AHR/ARNT heterodimer complex, were found in the promoter regions in all four of these zebrafish CYP1 genes(Fig 2; Table 2). In the CYP1C1 and CYP1C2 genes, XREs are located within two regions (approximately 1 kb each) upstream of the start sites, one upstream of CYP1C2, and one between the coding regions of the two genes. Searching genome databases showed that both regions are conserved across four fish species (zebrafish, pufferfish, medaka - Oryzias latipes, and three-spined stickleback - Gasterosteus aculeatus; data not shown). In zebrafish these conserved regions are located approximately 1.5 kB upstream of CYP1C1 and 8 kB upstream of CYP1C2. Other possible response elements in these conserved regions in CYP1C1 and CYP1C2, or in the 1-kB region immediately adjacent to the transcription initiation sites, include those binding E2F, nuclear respiratory factor 1 (NRF1) and cytokine-induced nuclear factor-kappa B (NFκB). In addition, putative binding sites for peroxisome proliferator-activated receptor/retinoid X receptor (PPAR/RXR), estrogen related receptor (ERR), retinoic acid receptor (RAR), RAR-related orphan receptor (ROR), and 1.25-(OH)2-vitamin D3/retinoid X receptor (VDR/RXR) all were found upstream of CYP1C2. Similar searches of the upstream proximal promoter regions of CYP1B1 and CYP1A revealed other putative response elements, including in particular estrogen receptor (ER).
Fig 2.
Xenobiotic response elements (XRE) and gene structure in the CYP1 gene promoter regions observed in the Danio rerio genome assembly version 6 (Zv6). The degenerate sequence KNCGGCGT was used to map the XREs.
Table 2.
Localization and number of putative xenobiotic response elements in the four CYP1 genes in zebrafish (Danio rerio).
| Gene | # exons | Number of consensus XRE's within | Notes | |||
|---|---|---|---|---|---|---|
| −1kb | −2kb | −5kb | −10kb | |||
| CYP1A | 7 | 3 | 3 | 15 | 21 | 5’ UTR length unknown intergenic region is 4kb |
| CYP1B1 | 2 | 0 | 1 | 2 | 6 | |
| CYP1C1 | 1 | 1 | 2 | 3 | 4 | |
| CYP1C2 | 1 | 0 | 0 | 2 | 6 | |
Basal and βNF-induced CYP1 expression in adult zebrafish
Initial studies examined expression of CYP1s in two organs (liver and gill) in TL and PS fish exposed to 1 μM βNF (or the carrier) via water. This βNF concentration has been shown to be high enough to induce ethoxyresorufin O-deethylase (EROD) activity in gills of various species of fish (Jönsson et al., 2002; Jönsson et al., 2003).
No difference was observed between the two zebrafish varieties in the basal expression of CYP1 genes in gill or liver (Fig 3 a–h). Both varieties showed induction of CYP1A and CYP1B1 in gills following exposure to βNF (Fig 3a and b). However, only the PS fish showed a significant induction of CYP1C1 by βNF in gills (Fig 3 c), and neither PS nor TL fish showed induction of CYP1C2 in gills (Fig 3 d). In the liver, none of the CYP1 genes were significantly induced by this treatment with βNF, in either the TL or the PS variety (Fig 3 e–h).
Fig 3.
CYP1A, CYP1B1, CYP1C1, and CYP1C2 expression in gills and liver of zebrafish TL (black bars) and PS (hatched bars) following exposure to β-naphthoflavone (1 μM) or the carrier (20 ppm acetone; mean ± SEM). For each CYP1 gene, basal and induced expression was calculated using the mean value of controls of both varieties as a calibrator. Differences between groups were determined by one-way ANOVA followed by Tukey's Multiple Comparison Test. A statistical difference between groups at p < 0.05 is indicated by differences in the letters above the bars.
Basal CYP1 gene expression in various tissues of zebrafish
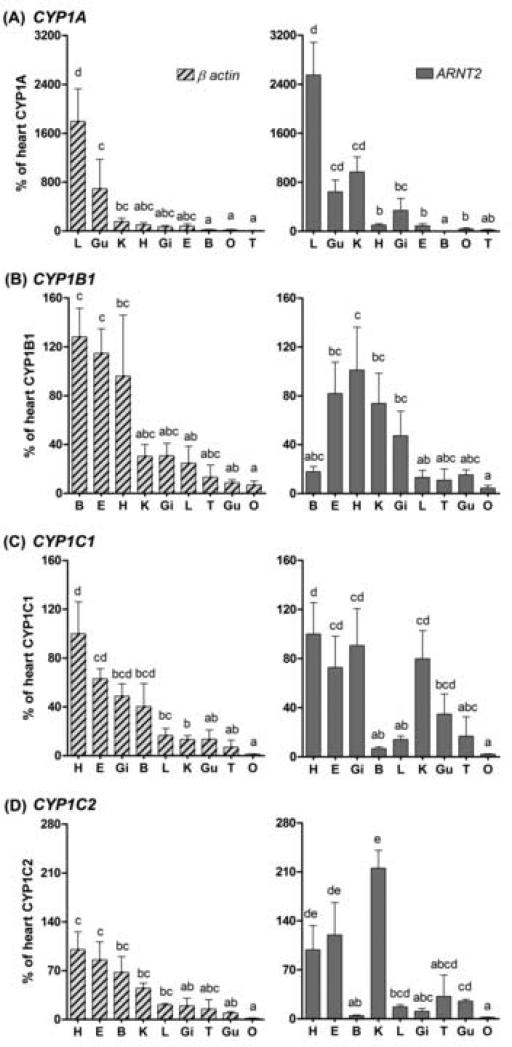
Expression of CYP1A, CYP1B1, CYP1C1, and CYP1C2 was detected in all organs examined in control zebrafish (PS), i.e., brain, eye, gill, heart, liver, kidney, gut, testes, and ovary. Comparing the relative expression of the four CYP1 genes in these nine organs requires a similar concentration of a reference transcript in all organs. The appropriateness of using β-actin as a reference gene in this way was assessed by comparing data normalized to β-actin with data normalized to ARNT2. This gave partially conflicting results, e.g., compared to other organs brain appeared to have higher CYP1C2 expression when β-actin was used as a reference gene and a lower expression when ARNT2 was used (Fig 4). This indicates that the mRNA concentration of either β-actin or ARNT2, or both, can vary in different tissues. However, irrespective of which reference gene was used, liver and gut were among the organs having the highest basal expression of CYP1A (Fig 4 a) and heart and eye tended to have the highest basal expression of CYP1B1 (Fig 4 b). Similarly, heart and eye were among the organs having the highest basal expression of both CYP1C1 and CYP1C2 (Fig 4 c and d). Gill exhibited a relatively high basal expression of CYP1C1 and kidney a relatively high basal expression of CYP1C2. Ovary was among the organs having the lowest expression of all CYP1 genes. The raw Ct values show higher transcript levels of β-actin than of ARNT2 in all adult organs as well as in whole embryos.
Fig 4.
Basal levels of CYP1 transcripts in various organs of the zebrafish PS as a percentage of the expression level in heart (i.e., heart was used as a calibrator organ). Data were normalized to both reference genes, β-actin and ANRT2. B, brain; E, eye; Gi, gill; Gu, gut; H, heart; K, kidney; L, liver; O, ovary; T, testes. Differences between groups were determined by one-way ANOVA followed by Tukey's Multiple Comparison Test after log-transformation. A statistical difference between groups at p < 0.05 is indicated by differences in the letters above the bars.
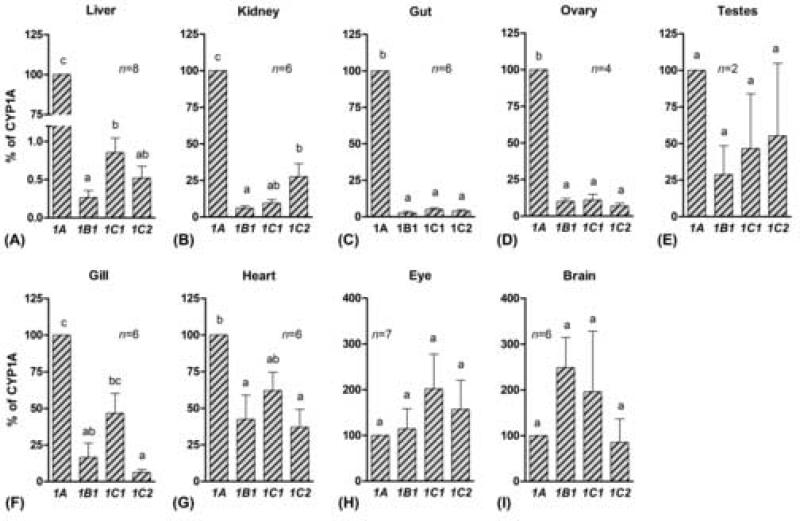
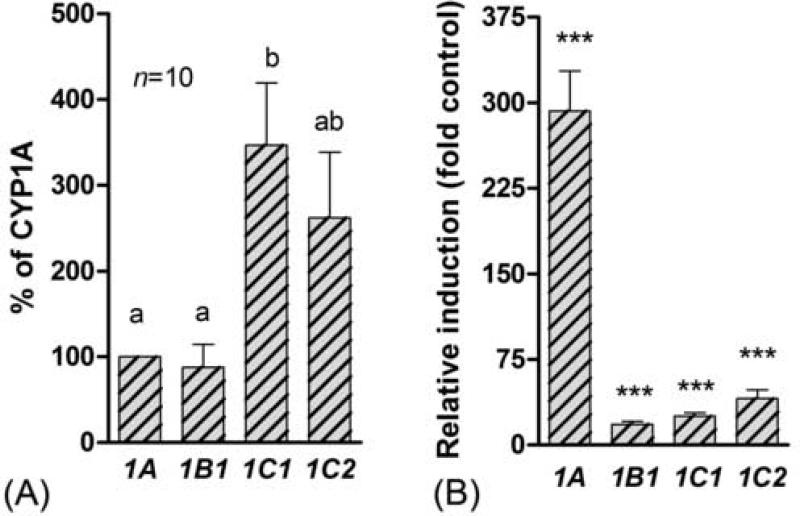
When the basal levels of the four CYP1 transcripts were compared within an organ in PS zebrafish, a higher level of CYP1A than of CYP1B1, CYP1C1, or CYP1C2 was observed in the organs of the abdominal cavity, i.e., liver, kidney, gut, and ovary (Fig 5 a–d). In liver the relative expression of CYP1A was more than 100-fold higher than the expression of the other CYP1 genes. In gills and heart the CYP1A expression was higher than that of CYP1B1 and CYP1C2 and similar to that of CYP1C1 (Fig 5 f and g), and in testes, eye, and brain no significant difference in basal level of the four CYP1 genes was observed (Fig 5 e, h, and i).
Fig 5.
Basal levels of CYP1 transcript in various organs in zebrafish PS (mean ± SEM). The relative expression of CYP1B1, CYP1C1, and CYP1C2 were calculated in each control sample using the mean values for CYP1A expression as a calibrator. Results are shown as a percentage of CYP1A expression. Differences in expression between CYP1 genes in an organ were determined by repeated measures one-way ANOVA followed by Tukey's Multiple Comparison Test after log-transformation. A statistical difference between groups at p < 0.05 is indicated by differences in the letters above the bars.
TL zebrafish exhibited patterns of basal expression of CYP1 genes in gill and liver that were similar to those in PS fish, i.e., the expression of CYP1B1, CYP1C1, and CYP1C2 was 19±8, 150±50, and 12±4 % of CYP1A in gills and 0.07±0.03, 1.5±1.0, and 0.3±0.1 % of CYP1A in liver (mean±SEM) in zebrafish TL.
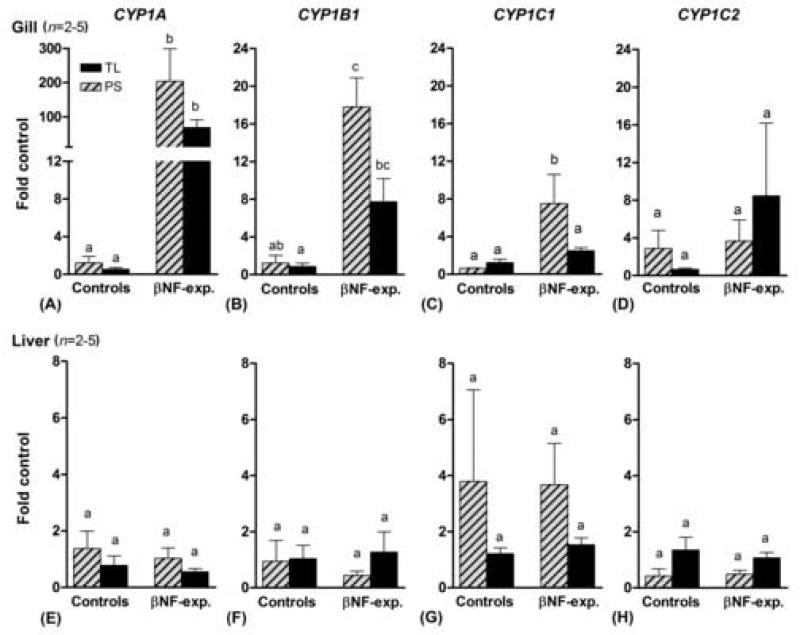
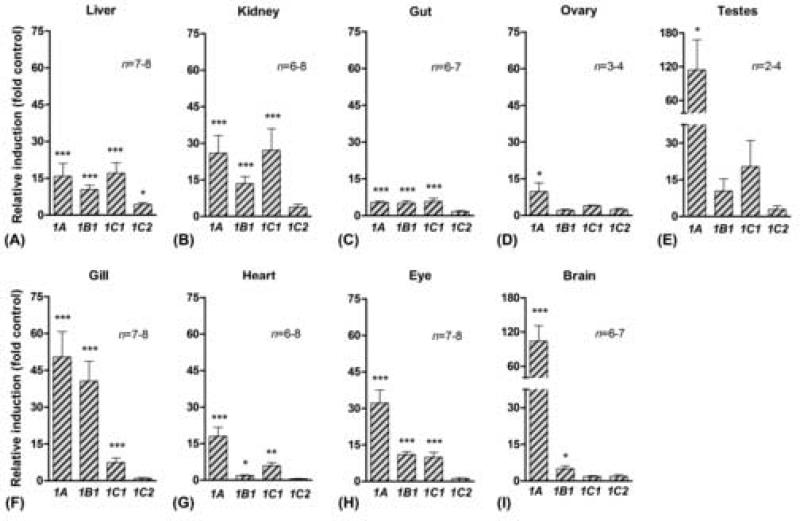
PCB126-induced CYP1 gene expression in various organs of zebrafish
To assess the organ distribution of CYP1 gene induction, fish were exposed to PCB126 at 100 nM in the water, a concentration we have seen to produce edema in embryos and thus likely high enough to ensure an effect on inducible genes. This concentration of PCB126 had no obvious effect on morphology or behavior of adult fish. CYP1A was induced by PCB126 (relative to the carrier control) in all organs examined (Fig 6), and CYP1B1 was induced in all organs except gonads in PS fish (Fig 6 a–c and f– i). Induction of CYP1C1 was observed in all organs except gonads and brain (Fig 6 a–c and f–h). In contrast, CYP1C2 was significantly induced by PCB126 only in the liver, although a tendency for induction (not significant) was also observed in kidney and possibly gonads (Fig 6 a, b, d, and e). The patterns of PCB126-induced CYP1 expression were basically similar in gills and liver of the two zebrafish varieties, except that the PS fish tended to show lower values for CYP1A and CYP1B1 and higher values for CYP1C1 and CYP1C2 than was observed in TL fish. In TL fish the fold induction of CYP1A, CYP1B1, and CYP1C1 was 160±50, 100±25, and 5±1 (all p<0.001), respectively, in gills, and 35±18, 35±4 (both p<0.001), and 6±1 (p<0.05) in liver (mean±SEM). In both gills and liver, CYP1C2 tended to show a slight increase (i.e., 2.0±0.5 fold of the control) after PCB126 exposure, although this was not significant in either organ (p>0.05).
Fig 6.
Fold induction of CYP1A, CYP1B1, CYP1C1, and CYP1C2 in various tissues in the zebrafish PS following exposure to PCB126 (100 nM; mean ± SEM). Mean values of control expression were used as calibrators (fold control). Significant differences as compared with the control (* = p<0.05, ** = p<0.01, and *** = p<0.001) were determined by one-way ANOVA followed by Bonferroni's multiple comparison of selected pairs.
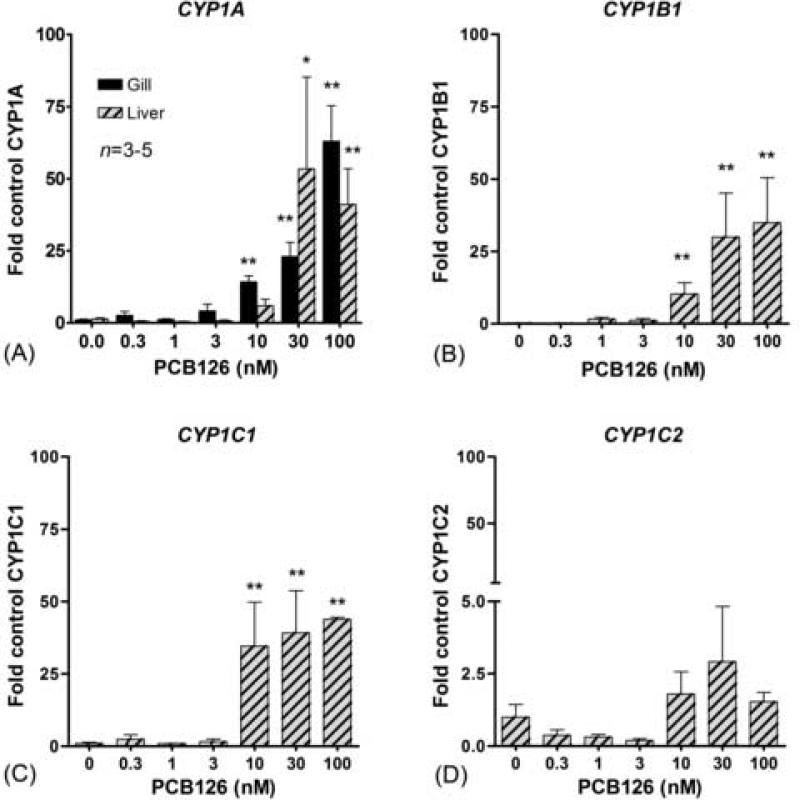
We also performed a dose response experiment, to determine whether responsiveness to PCB126 varies among the four CYP1 genes. The concentration-dependent effects of PCB126 on CYP1 expression were examined in gill (CYP1A expression) and liver (expression of all four genes). Induction of CYP1A and CYP1B1 showed similar concentration-dependence (Fig 7 a–b). CYP1C1 was strongly and similarly induced at the three highest doses of PCB26, implying a dramatically increased expression between 3 and 10 nM PCB126. The EC50 values for PCB126 induction of CYP1A, CYP1B1, and CYP1C1 in liver were 13, 14, and 7 nM, respectively. The EC50 for CYP1A induction in gill was 32 nM. The PCB126-EC50 values for CYP1 gene induction in the liver were not statistically different from one another, nor were the EC50 values for CYP1A induction in gill and liver significantly different. In liver the LOEC for CYP1A, CYP1B1 and CYP1C1 induction were 30, 10 and 10 nM, and in the gill the LOEC for CYP1A was 10 nM PCB126.
Fig 7.
PCB126 concentration-response of CYP1A, CYP1B1, CYP1C1, and CYP1C2 expression (fold control) in gills and liver in the zebrafish PS. Significant differences as compared with the control (* = p<0.05 and ** = p<0.01) were determined by one-way ANOVA followed by Dunnett's Test.
Basal and PCB126-induced CYP1 gene expression in zebrafish embryos
In addition to CYP1 gene expression in adult zebrafish, we also studied the expression of these genes in 80-hpf embryos exposed to the carrier (controls), or to 100 nM PCB126. In controls the CYP1C1 expression was significantly higher than the expression of CYP1A and CYP1B1 (Fig 8 a). CYP1C2 expression also tended to be higher than expression of CYP1A or CYP1B1, although this was not significant. Following exposure to PCB126 a strong induction of all four CYP1 genes was observed in the embryos (Fig 8b).
Fig 8.
Basal (A) and PCB126-induced (B) CYP1 gene expression in zebrafish embryos. (A) Basal expression is shown as percentage of the CYP1A expression and significant differences in basal expression between genes were determined by one-way repeated measures one-way ANOVA and Tukey's Multiple Comparison Test. A statistical difference between groups at p < 0.05 is indicated by differences in the letters above the bars. (B) Induced expression is shown as the multiple of the control (fold control) significant differences compared with the control (*** = p<0.001) were determined by one-way ANOVA followed by Bonferroni's multiple comparison of selected pairs.
Discussion
With the discovery of the CYP1Cs, the suite of CYP1 genes possibly involved in AHR agonist effects is expanding. Here we present a comprehensive analysis of the expression of the CYP1s in the zebrafish model, including the recently discovered CYP1Cs, showing that CYP1A, CYP1B1, CYP1C1 and CYP1C2 all are expressed in multiple organs of control adult zebrafish, and also appear to be constitutively expressed in zebrafish embryos. CYP1A has been widely studied in fish (e.g., Arinç et al., 2000) but there are very few studies of fish CYP1B (Leaver and George, 2000; El-kady et al., 2004b; El-kady et al., 2004a; Willett et al., 2006) or the CYP1Cs (Godard et al., 2005; Itakura et al., 2005; Wang et al., 2006). Our results showing appreciable basal levels of expression of the CYP1Cs and CYP1B1 in some organs and in embryos, suggest that there are endogenous roles for the CYP1B1 and CYP1Cs. Furthermore, all four CYP1s were induced by the potent AHR agonist PCB126. These results and the discovery of several putative XREs in the cis-regulatory regions of the CYP1C genes, suggest that they are AHR-regulated, as are the CYP1As and CYP1B1. However, whereas CYP1C2 showed a strong induction response in embryos, only a minor response was seen in adults, indicating that CYP1C2 has a difference in regulation from the other CYP1 genes.
Expression of CYP1 genes was studied in two different varieties of adult zebrafish, i.e., Tubingen Longfin (TL), which is widely used in developmental studies, and a more “common” zebrafish, purchased in a local pet store (PS). The two varieties exhibited largely similar patterns of basal and induced CYP1 gene expression, suggesting that the current results reflect responses that can be expected in zebrafish generally. However, there were some CYP1C sequence differences between the TL and PS fish (Fig. 1).
Basal expression of CYP1 genes in adult zebrafish
CYP1 genes likely play physiological functions in zebrafish during their whole lifespan, as all four genes showed a basal expression in embryos as well as in all organs we examined in adult controls. However, it seems likely they have different roles. In particular, there seems to be a distinct difference between CYP1A on one hand and CYP1B1 and the CYP1C genes on the other, as indicated by the difference in expression patterns and inducibility by PCB126. Thus, of the four CYP1 transcripts analyzed, CYP1A was the most abundant in organs in the abdominal cavity (i.e. liver, kidney, and gut), whereas other organs exhibited more comparable levels of expression of the four genes. The marked CYP1A expression in liver, kidney, and gut may reflect functions associated with the role of these organs in nutrient uptake and processing of body waste products, i.e., detoxification of endogenous metabolites and food-derived AHR agonists. Other studies suggest that CYP1A may play an important role in eliminating endogenous AHR ligands (Nebert et al., 2000).
CYP1B1, CYP1C1, and CYP1C2 all showed a marked similarity in organ distribution patterns. The similar expression patterns suggest that there may be functional similarities between the CYP1B and CYP1C genes. Phylogenetic analyses indicate that the CYP1B and CYP1C genes form a monophyletic clade, and thus are more closely related to each other than they are to the CYP1As. We observed a low coefficient of evolutionary functional divergence between the CYP1Bs and the CYP1Cs (Goldstone, unpublished) relative to the comparison with the CYP1As, calculated with the site-specific rates of amino acid substitution using the method of Gu and Vander Velden (2002). CYP1C1 and CYP1C2 are located immediately adjacent to one another on zebrafish chromosome 17, suggesting that the fish-specific CYP1C duplication is independent from, and likely more recent than, the separate whole genome duplication event in the fish lineage (Goldstone et al, unpublished results). Thus, CYP1C1 and CYP1C2 may have similar functions due to a short divergence time.
The basal expression patterns for zebrafish CYP1B1, CYP1C1, and CYP1C2 we observed are similar to that for CYP1B1 in mammals. Thus, the expression levels of the three fish genes were relatively high in heart and relatively low in ovary, like the expression of CYP1B1 in human (Choudhary et al., 2005). The stronger expression of CYP1A than CYP1B1, CYP1C1, and CYP1C2 in zebrafish liver also agrees with the relative expression of CYP1A1 and CYP1B1 observed in human liver (Choudhary et al., 2005). CYP1B1, CYP1C1, and CYP1C2 all showed high basal expression in zebrafish eye. CYP1B1 is essential for normal development of the mammalian eye (Libby et al., 2003). The cellular localization of CYP1B1, CYP1C1, or CYP1C1 in the fish eye has not been studied, but in mammals CYP1B1 is present in corneal and ciliary epithelia and in retina (Choudhary et al., 2006). The similar expression patterns of CYP1B1/1C genes in zebrafish and CYP1B1 in mammals suggest that endogenous functions of these genes may be served similarly in different vertebrate groups. Given the absence of CYP1C genes in mammals, it is possible that some of the CYP1C functions have been adopted by mammalian CYP1B1. The fact that the mammalian and fish CYP1B1s appear to diverge from one another may imply that the mammalian and fish CYP1B1s have evolved different functions.
Induced expression of CYP1 genes in adult zebrafish
CYP1A transcript was induced by PCB126 in all organs that we studied in zebrafish. In TCDD-exposed zebrafish increased immuno-staining for CYP1A protein was observed only in liver and kidney (Buchmann et al., 1993). However, although it is possible that some posttranscriptional regulation of CYP1A contributes to the differences between our findings and those of Buchmann et al, real time PCR is a more sensitive quantitative method than immunohistochemistry. Other studies using immunohistochemical detection have shown that CYP1A is induced at least in some cells (e.g., endothelial cells) in all organs of fish (e.g., Smolowitz et al., 1991; Van Veld et al., 1997). Our results thus support a ubiquitous expression of CYP1A but also indicate a large variation in the magnitude of induction among organs, i.e., from 5-fold in gut to over 100-fold in brain and testes.
CYP1B1 and CYP1C1 also were induced by PCB126 in most organs we studied in zebrafish. Previous studies have shown induction of CYP1B1 in a variety of organs in fish and mammals exposed to AHR agonists (e.g., Bhattacharyya et al., 1995; El-kady et al., 2004a; Willett et al., 2006). More recently, CYP1C1 was reported to be induced in many of the same organs we examined, in killifish exposed to benzo[a]pyrene (B[a]P) (Wang et al., 2006). The magnitude of induction of zebrafish CYP1B1 and CYP1C1 was relatively low in most organs, although gill showed a 41-fold induction of CYP1B1 and kidney a 27-fold induction of CYP1C1. To our knowledge, our results with CYP1C2 are the first to demonstrate induction of this new gene. However, in contrast to CYP1C1, which was strongly induced by PCB126 in many organs, only minor changes in the levels of expression of CYP1C2 were seen following exposure either to PCB126 or βNF, indicating a low responsiveness of CYP1C2 to AHR agonists in adult zebrafish. This difference in regulation suggests different metabolic roles for the two CYP1Cs.
CYP1 gene expression in zebrafish embryos
In control embryos CYP1C1 and CYP1C2 tended to have 2-fold higher basal expression than CYP1A and CYP1B1. Similar results showing higher expression for CYP1C1 relative to CYP1A were observed in killifish embryos (Wang et al., 2006). These results hint that the CYP1C genes play a role in fish embryo development. The finding that PCB126 caused a strong induction of CYP1C2 in embryos but only a small effect on CYP1C2 expression in adults suggests further that CYP1C2 is regulated differently in embryos than in adults.
The low basal or constitutive expression of CYP1A transcript in control zebrafish embryos is similar to results obtained previously (Andreasen et al., 2002). However, a remarkable induction (300-fold) was observed following exposure to PCB126 at 100 nM. In comparison, CYP1B1, CYP1C1, and CYP1C2 increased less (18-, 25- and 41-fold, respectively) in embryos exposed to PCB126. A higher responsiveness of CYP1A than of CYP1B1 was reported in a cDNA microarray study of zebrafish embryos exposed to TCDD, i.e., CYP1A and CYP1B1 were induced 63- and 5-fold, respectively (Handley-Goldstone et al., 2005). Killifish embryos exposed to B[a]P exhibited a 330-fold induction of CYP1A and a 18-fold induction of CYP1C1 (Wang et al., 2006). The large difference in relative induction between CYP1A and the CYP1B1 and CYP1C genes is in part a reflection of the lower basal level of CYP1A in embryos. Nevertheless, in PCB126-exposed embryos the CYP1A transcript seems to be more abundant than the CYP1C transcripts and much more abundant than the CYP1B1 transcript.
CYP1 induction by βNF and PCB126 in zebrafish liver
Somewhat surprisingly, CYP1A, CYP1B1, and CYP1C1 transcripts were induced in gills but not in liver of PS zebrafish exposed to βNF in the water, while similar exposure to PCB126 induced these genes in both organs. Toxicokinetic processes could be involved in the difference and in the lack of effect of βNF. PCB126 is considered a very slowly metabolized PCB congener, while βNF is well metabolized by CYP1A in fish (unpublished results). βNF conceivably could have been metabolized by CYP1A induced in gill and other organs such that the concentrations of parent compound reaching the liver were too low to induce transcription of CYP1A there. If there were induction early during the exposure that declined by sampling time, it would imply a rapid decay of CYP1A mRNA; human CYP1A1 transcript has a 7-hour half-life (Ciolino and Yeh, 1999). However, the 1 μM concentration of βNF we used (about 270 μg per liter ×2), with the 3.6 g of fish/liter would be equivalent to a dose of 150 mg/kg, assuming that all the βNF was accumulated. 100% accumulation is unlikely, yet even at 10 % accumulation, an extensive metabolism of βNF would be required to reduce the concentration to ineffective levels. By comparison, in studies with another rapidly metabolized inducer, B[a]P, Van Veld et al. (1997) found that Fundulus heteroclitus exposed to B[a]P in the water at a much lower dose (10 μg per liter = 40 nM) than our dose of βNF, was able to induce CYP1A protein in the liver at four days, and in rainbow trout (Oncorhynchus mykiss) an even lower dose, 1 nM of waterborne B[a]P, induces liver EROD 8-fold (Jönsson et al., 2006a). Troxel et al. (1997) observed rapid induction followed by a decline of EROD activity in the liver of zebrafish injected (ip) with 150 mg βNF/kg, i.e., the magnitude of EROD activity was halved from day 1 to day 2. While this might be interpreted as reflecting a rapid metabolism of βNF by the liver and decline in CYP1A, it also is possible that increasing βNF doses may have been somehow inhibitory to CYP1A expression, as well as to CYP1A activity. Jönsson et al. (2006b) saw that 10 μM βNF strongly suppressed EROD induction in primary gill cells in culture. Moreover, βNF at 3.6 μM was reported not to induce CYP1A activity in ZF-L cells, a TCDD-responsive zebrafish liver cell line (Miranda et al., 1993), while Evans et al. (2005), saw a strong induction of CYP1A transcript in ZF-L cells treated with 10 μM βNF. Determining actual rates of βNF metabolism, the content of βNF residues in liver of exposed fish, and whether βNF might suppress CYP1A transcript expression are required to resolve these issues.
CYP1 gene regulation
Consensus XRE sequences were identified upstream of the coding regions of both CYP1C1 and CYP1C2. The number of potential XREs upstream of the CYP1C genes is comparable to that in the CYP1B1 gene, but fewer than in the CYP1A gene, where there are a large number of upstream XREs (Fig 2; Table 2). Similar consensus XRE sequences have been shown to be functional in the eel (Anguilla japonica) CYP1A1 gene (Ogino et al., 1999). While the functionality of these XRE sequences in zebrafish needs to be confirmed, the number and position of the XREs in the CYP1C genes are consistent with regulation of these genes by the AHR. The fact that both CYP1C genes were induced in embryos by the potent AhR agonist PCB126 also indicates regulation via the AHR. Notably, the EC50 values we observed for PCB126 are of a similar order of magnitude as those reported for CYP1A mRNA induction in ZF-L cells (Henry et al., 2001). Furthermore, the EC50 value for PCB126-induced CYP1C1 expression in liver was similar to those of CYP1A and CYP1B1 suggesting a similar mechanism for induction of the three CYP1 genes, i.e., via the AHR. The concentration-dependent induction of CYP1C2 was not significant, although the calculated EC50 value for PCB126 was in the same range as those of the other three CYP1 genes, indicating that CYP1C2 also is AHR regulated. However it may also be controlled by additional factors.
In mammals, numerous CYP genes including CYP1s are expressed in embryos at various developmental stages (Choudhary et al., 2004). Nuclear receptors are known to modulate transcription of CYPs through complex mechanisms involving both activation and repression. In mammals, CYP1 enzymes have been suggested to function in the synthesis and elimination of nuclear receptor ligands, thus serving to regulate signal transduction (Choudhary et al., 2004). Changes in the level of various CYP1 enzymes could affect growth, morphogenesis, and homeostasis (Choudhary et al., 2004). The presence of putative response elements for various transcription factors (including E2F, NRF1, NFκB, PPAR/RXR, ERR, RAR, ROR, and VDR/RXR) upstream of the coding start sites in the CYP1C genes support the notion that the CYP1Cs could participate in a wide range of endogenous functions. These might include cell proliferation, respiration, inflammation, apoptosis, and fatty acid metabolism. Although the functionality of these possible response elements is not confirmed, they deserve further study.
Conclusion
Here we report the first comprehensive study of expression of the full suite of inducible CYP1 family genes, in zebrafish adults and embryos. CYP1A, CYP1B1, CYP1C1 and CYP1C2 transcripts were detected in all adult organs analyzed, as well as in embryos. Significant induction of CYP1A, CYP1B1 and CYP1C1 was detected in most adult organs studied, whereas CYP1C2 was less inducible than the other genes. Clearly the CYP1Cs are regulated at least in part via the AHR. CYP1C genes occur in amphibians and birds, as well as fish, but have been lost in mammals (Goldstone, unpublished). Thus, an understanding the regulation and functions of CYP1Cs becomes important to a general understanding of the mechanisms of toxic action of AHR agonists, as well as to understanding of possible endogenous functions of the CYP1 genes. Given their phylogenetic distribution, knowledge of the CYP1Cs in zebrafish or other non-mammalian models could point to physiological functions of the CYP1 family that may be more difficult to discern in mammals.
Acknowledgements
This study was financed by a grant from The Swedish Research Council Formas (to MEJ) and by NIH F32-ES012794 (to JVG), and the Superfund Basic Research Program at Boston University, NIH grant 5-P42-ES007381 (to JJS). Study sponsors had no involvement in the studies reported here or in the decision to submit this paper for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Andreasen EA, Spitsbergen JM, Tanguay RL, Stegeman JJ, Heideman W, Peterson RE. Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol Sci. 2002;68:403–419. doi: 10.1093/toxsci/68.2.403. [DOI] [PubMed] [Google Scholar]
- Arinç E, Sen A, Bozcaarmutlu A. Cytochrome P4501A and associated mixed function oxidase induction in fish as a biomarker for toxic carcinogenic pollutants in the aquatic environment. Pure Appl Chem. 2000;72:985–994. [Google Scholar]
- Bhattacharyya KK, Brake PB, Eltom SE, Otto SA, Jefcoate CR. Identification of a rat adrenal cytochrome P450 active in polycyclic hydrocarbon metabolism as rat CYP1B1. Demonstration of a unique tissue-specific pattern of hormonal and aryl hydrocarbon receptor-linked regulation. J Biol Chem. 1995;270:11595–11602. doi: 10.1074/jbc.270.19.11595. [DOI] [PubMed] [Google Scholar]
- Buchmann A, Wannemacher R, Kulzer E, Buhler DR, Bock KW. Immunohistochemical localization of the cytochrome P450 isozymes LMC2 and LM4B (P4501A1) in 2,3,7,8-tetrachlorodibenzo-p-dioxin-treated zebrafish (Brachydanio rerio). Toxicol Appl Pharmacol. 1993;123:160–169. doi: 10.1006/taap.1993.1233. [DOI] [PubMed] [Google Scholar]
- Carney SA, Chen J, Burns CG, Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol Pharmacol. 2006;70:549–561. doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- Carney SA, Peterson RE, Heideman W. 2,3,7,8-Tetrachlorodibenzo-p-dioxin activation of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator pathway causes developmental toxicity through a CYP1A-independent mechanism in zebrafish. Mol Pharmacol. 2004;66:512–521. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Sarfarazi M, Schenkman JB. Xenobiotic-metabolizing cytochromes P450 in ontogeny: evolving perspective. Drug Metab Rev. 2004;36:549–568. doi: 10.1081/dmr-200033447. [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Sarfarazi M, Schenkman JB. Physiological significance and expression of P450s in the developing eye. Drug Metab Rev. 2006;38:337–352. doi: 10.1080/03602530600570149. [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. Expression patterns of mouse and human CYP orthologs (families 1-4) during development and in different adult tissues. Arch Biochem Biophys. 2005;436:50–61. doi: 10.1016/j.abb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Ciolino HP, Yeh GC. The steroid hormone dehydroepiandrosterone inhibits CYP1A1 expression in vitro by a post-transcriptional mechanism. J Biol Chem. 1999;274:35186–35190. doi: 10.1074/jbc.274.49.35186. [DOI] [PubMed] [Google Scholar]
- Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- Dong W, Teraoka H, Tsujimoto Y, Stegeman JJ, Hiraga T. Role of aryl hydrocarbon receptor in mesencephalic circulation failure and apoptosis in zebrafish embryos exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2004;77:109–116. doi: 10.1093/toxsci/kfh023. [DOI] [PubMed] [Google Scholar]
- El-kady MA, Mitsuo R, Kaminishi Y, Itakura T. cDNA cloning, sequence analysis and expression of 3-methylcholanthrene-inducible cytochrome P450 1B1 in carp (Cyprinus carpio). Environ Sci. 2004a;11:231–240. [PubMed] [Google Scholar]
- El-kady MA, Mitsuo R, Kaminishi Y, Itakura T. Isolation of cDNA of novel cytochrome P450 1B gene, CYP1B2, from Carp (Cyprinus carpio) and its induced expression in gills. Environ Sci. 2004b;11:345–354. [PubMed] [Google Scholar]
- Evans BR, Karchner SI, Franks DG, Hahn ME. Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: structure, function, evolution, and AHR-dependent regulation in vivo. Arch Biochem Biophys. 2005;441:151–167. doi: 10.1016/j.abb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol Appl Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- Godard CA, Goldstone JV, Said MR, Dickerson RL, Woodin BR, Stegeman JJ. The new vertebrate CYP1C family: cloning of new subfamily members and phylogenetic analysis. Biochem Biophys Res Commun. 2005;331:1016–1024. doi: 10.1016/j.bbrc.2005.03.231. [DOI] [PubMed] [Google Scholar]
- Gu X, Vander Velden K. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18:500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- Handley-Goldstone HM, Grow MW, Stegeman JJ. Cardiovascular gene expression profiles of dioxin exposure in zebrafish embryos. Toxicol Sci. 2005;85:683–693. doi: 10.1093/toxsci/kfi116. [DOI] [PubMed] [Google Scholar]
- Henry TR, Nesbit DJ, Heideman W, Peterson RE. Relative potencies of polychlorinated dibenzo-p-dioxin, dibenzofuran, and biphenyl congeners to induce cytochrome P4501A mRNA in a zebrafish liver cell line. Environ Toxicol Chem. 2001;20:1053–1058. [PubMed] [Google Scholar]
- Henry TR, Spitsbergen JM, Hornung MW, Abnet CC, Peterson RE. Early life stage toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish (Danio rerio). Toxicol Appl Pharmacol. 1997;142:56–68. doi: 10.1006/taap.1996.8024. [DOI] [PubMed] [Google Scholar]
- Itakura T, El-Kady M, Mitsuo R, Kaminishi Y. Complementary DNA cloning and constitutive expression of cytochrome P450 1C1 in the gills of carp (Cyprinus carpio). Environ Sci. 2005;12:111–120. [PubMed] [Google Scholar]
- Jönsson EM, Abrahamson A, Brunström B, Brandt I. Cytochrome P4501A induction in rainbow trout gills and liver following exposure to waterborne indigo, benzo[a]pyrene and 3,3',4,4',5-pentachlorobiphenyl. Aquat Toxicol. 2006a;79:226–232. doi: 10.1016/j.aquatox.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Jönsson EM, Brandt I, Brunström B. Gill filament-based EROD assay for monitoring waterborne dioxin-like pollutants in fish. Environ Sci Technol. 2002;36:3340–3344. doi: 10.1021/es015859a. [DOI] [PubMed] [Google Scholar]
- Jönsson M, Abrahamson A, Brunström B, Brandt I, Ingebrigtsen K, Jørgensen EH. EROD activity in gill filaments of anadromous and marine fish as a biomarker of dioxin-like pollutants. Comp Biochem Physiol C. 2003;136:235–243. doi: 10.1016/j.cca.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Jönsson ME, Carlsson C, Smith RW, Pärt P. Effects of copper on CYP1A activity and epithelial barrier properties in the rainbow trout gill. Aquat Toxicol. 2006b;79:78–86. doi: 10.1016/j.aquatox.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Leaver MJ, George SG. A cytochrome P4501B gene from a fish, Pleuronectes platessa. Gene. 2000;256:83–91. doi: 10.1016/s0378-1119(00)00373-5. [DOI] [PubMed] [Google Scholar]
- Lewis DFV. 57 varieties: the human cytochromes P450. Pharmacogenomics. 2004;5:305–318. doi: 10.1517/phgs.5.3.305.29827. [DOI] [PubMed] [Google Scholar]
- Libby RT, Smith RS, Savinova OV, Zabaleta A, Martin JE, Gonzalez FJ, John SW. Modification of ocular defects in mouse developmental glaucoma models by tyrosinase. Science. 2003;299:1578–1581. doi: 10.1126/science.1080095. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Miranda CL, Collodi P, Zhao X, Barnes DW, Buhler DR. Regulation of cytochrome P450 expression in a novel liver cell line from zebrafish (Brachydanio rerio). Arch Biochem Biophys. 1993;305:320–327. doi: 10.1006/abbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and (Ah) gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Ogino Y, Itakura T, Kato H, Aoki J, Sato M. Functional analysis of promoter region from eel cytochrome P450 1A1 gene in transgenic medaka. Mar Biotechnol. 1999;1:364–370. doi: 10.1007/pl00011788. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol Sci. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schlezinger JJ, Struntz WD, Goldstone JV, Stegeman JJ. Uncoupling of cytochrome P450 1A and stimulation of reactive oxygen species production by co-planar polychlorinated biphenyl congeners. Aquat Toxicol. 2006;77:422–432. doi: 10.1016/j.aquatox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Schlezinger JJ, White RD, Stegeman JJ. Oxidative inactivation of cytochrome P-450 1A (CYP1A) stimulated by 3,3',4,4'-tetrachlorobiphenyl: production of reactive oxygen by vertebrate CYP1As. Mol Pharmacol. 1999;56:588–597. doi: 10.1124/mol.56.3.588. [DOI] [PubMed] [Google Scholar]
- Shertzer HG, Clay CD, Genter MB, Chames MC, Schneider SN, Oakley GG, Nebert DW, Dalton TP. Uncoupling-mediated generation of reactive oxygen by halogenated aromatic hydrocarbons in mouse liver microsomes. Free Radic Biol Med. 2004;36:618–631. doi: 10.1016/j.freeradbiomed.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Inoue K, Suzuki Y, Kawai T, Azuma E, Nakajima T, Shindo M, Kurose K, Sugie A, Yamagishi Y, Fujii KY, Hashimoto M. Arylhydrocarbon receptor-dependent induction of liver and lung cytochromes P450 1A1, 1A2, and 1B1 by polycyclic aromatic hydrocarbons and polychlorinated biphenyls in genetically engineered C57BL/6J mice. Carcinogenesis. 2002;23:1199–1207. doi: 10.1093/carcin/23.7.1199. [DOI] [PubMed] [Google Scholar]
- Smolowitz RM, Hahn ME, Stegeman JJ. Immunohistochemical localization of cytochrome P-450IA1 induced by 3,3',4,4'-tetrachlorobiphenyl and by 2,3,7,8-tetrachlorodibenzofuran in liver and extrahepatic tissues of the teleost Stenotomus chrysops (scup). Drug Metabol Dispos. 1991;19:113–123. [PubMed] [Google Scholar]
- Teraoka H, Dong W, Tsujimoto Y, Iwasa H, Endoh D, Ueno N, Stegeman JJ, Peterson RE, Hiraga T. Induction of cytochrome P450 1A is required for circulation failure and edema by 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish. Biochem Biophys Res Commun. 2003;304:223–228. doi: 10.1016/s0006-291x(03)00576-x. [DOI] [PubMed] [Google Scholar]
- Van Veld PA, Vogelbein WK, Cockran MK, Goksoyr A, Stegeman JJ. Route-specific cellular expression of cytochrome P4501A (CYP1A) in fish (Fundulus heteroclitus) following exposure to aqueous and dietary benzo(a)pyrene. Toxicol Appl Pharmacol. 1997;142:348–359. doi: 10.1006/taap.1996.8037. [DOI] [PubMed] [Google Scholar]
- Wang L, Scheffler BE, Willett KL. CYP1C1 messenger RNA expression is inducible by benzo[a]pyrene in Fundulus heteroclitus embryos and adults. Toxicol Sci. 2006;93:331–340. doi: 10.1093/toxsci/kfl072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett KL, Ganesan S, Patel M, Metzger C, Quiniou S, Waldbieser G, Scheffler B. In vivo and in vitro CYP1B mRNA expression in channel catfish. Mar Environ Res. 2006;62(Suppl):S332–336. doi: 10.1016/j.marenvres.2006.04.015. [DOI] [PubMed] [Google Scholar]