Abstract
The vascular network carries blood throughout the body, delivering oxygen to tissues and providing a pathway for communication between distant organs. The network is hierarchical and structured, but also dynamic, especially at the smaller scales. Remodeling of the microvasculature occurs in response to local changes in oxygen, gene expression, cell-cell communication, and chemical and mechanical stimuli from the microenvironment. These local changes occur as a result of physiological processes such as growth and exercise, as well as acute and chronic diseases including stroke, cancer, and diabetes, and pharmacological intervention. While the vasculature is an important therapeutic target in many diseases, drugs designed to inhibit vascular growth have achieved only limited success, and no drug has yet been approved to promote therapeutic vascular remodeling. This highlights the challenges involved in identifying appropriate therapeutic targets in a system as complex as the vasculature. Systems biology approaches provide a means to bridge current understanding of the vascular system, from detailed signaling dynamics measured in vitro and pre-clinical animal models of vascular disease, to a more complete picture of vascular regulation in vivo. This will translate to an improved ability to identify multi-component biomarkers for diagnosis, prognosis, and monitoring of therapy that are easy to measure in vivo, as well as better drug targets for specific disease states. In this review, we summarize systems biology approaches that have advanced our understanding of vascular function and dysfunction in vivo, with a focus on computational modeling.
1. INTRODUCTION
1.1 Motivation for a systems approach to the vasculature
Systems biology is an integrative approach that synthesizes our current understanding of molecular, physiological and pathological mechanisms to reconcile experimental data from multiple perturbations with the predictions of detailed computational models. By integrating detailed experimental data (e.g. from hi-throughput experiments) with mechanistic information (e.g. from multi-scale computational models and bioinformatics), we can formulate a more complete understanding of a system across multiple scales and at higher spatial and temporal resolution than would otherwise be possible. In addition, modeling the interconnectedness of the system from gene to protein to pathway, and from cell to tissue to organism, allows systems biology simulations to predict the system-wide response to perturbation, for example the change in blood supply to a tumor following delivery of drugs.
Systems biology is well-suited to studying vascular function and dysfunction because the vasculature and its regulation are highly complex. The insides of all blood vessels – from the smallest to the largest; arteries, veins, capillaries; newly sprouting or mature – are lined with endothelial cells (ECs). This cell type must therefore be sufficiently flexible to survive and thrive in diverse environments, and to perform different specialized functions in many tissues1. In particular, moving from in vitro systems in which perturbations to endothelial cues can be controlled to in vivo vascularized tissues necessitates a quantitative understanding of these complex systems. Whether following exercise2 or in a growing tumor3, there can be changes to the expression of many or all of the ligands and receptors regulating endothelial cell behavior, and not all in the same direction. The outcome of all of these changes would be impossible to calculate without a detailed quantitative model of the system.
Because of the number of potential levers and drivers of vascular changes, there are many possible quantitative metrics to measure, including potentially informative quantities that are difficult to measure in vivo. By incorporating detailed in vitro measurements, computational models can be validated and used to identify which in vivo measurements would be most informative – as diagnostics, prognostics, or as indicators of therapy effectiveness either before or after treatment.
1.2 Vascular development and remodeling
The vasculature supplies oxygen to tissues. Maintenance of homeostasis requires the vascular system to adapt in response to local stimuli (e.g. oxygen tension) sensed by endothelial and other cells. The smallest vessels, directly involved in delivery and transport of oxygen to tissues, develop new branches, expand in diameter, or are pruned as a result of these dynamic molecular, cellular, and tissue microenvironmental cues (Fig.1). Vascular network development, maintenance, and remodeling can occur through multiple distinct morphogenic processes. Each requires complex molecular and multicellular regulation, though the regulatory details are not completely understood for any of these forms of vascular remodeling.
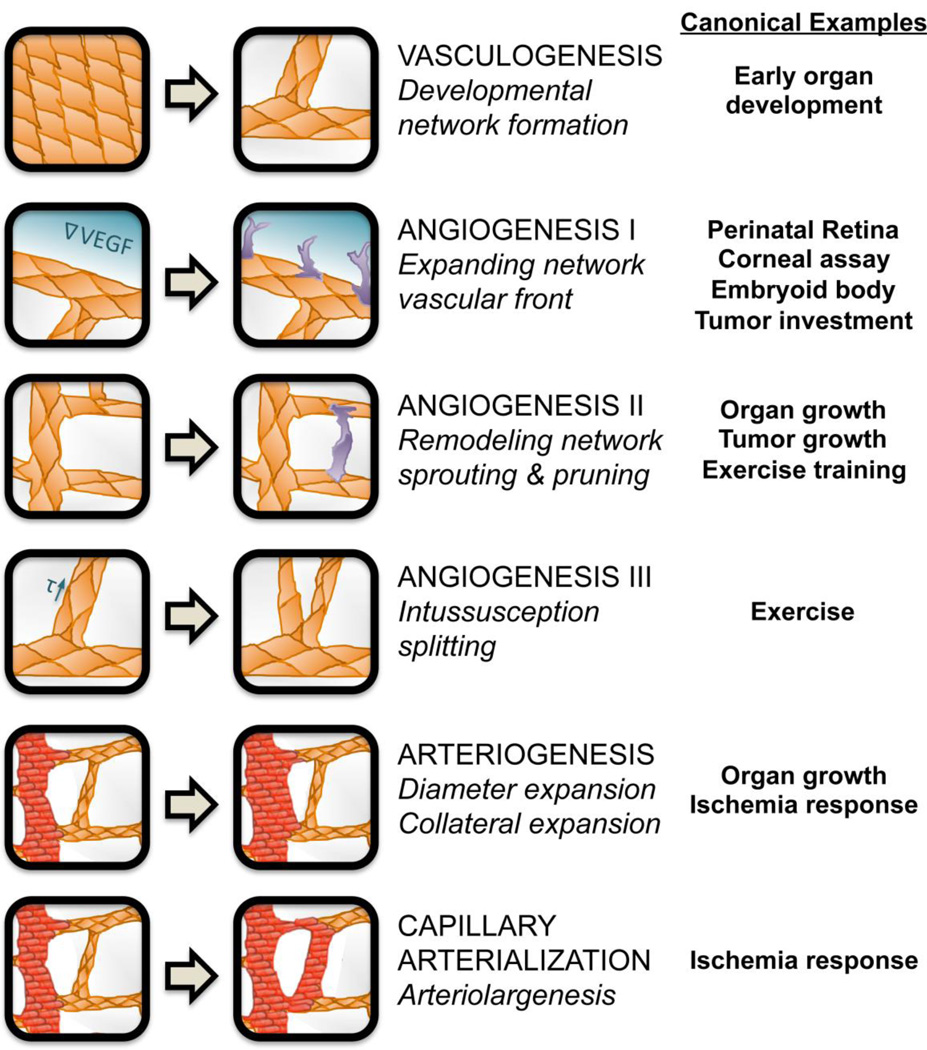
Figure 1. Vascular development and remodeling processes.
The six distinct types of in vivo blood vessel formation or remodeling, described in the text, are prevalent in different tissues and situations. Both the emergence and the dynamic adaptation of a functional hierarchical vascular system depend on the coordinated regulation of all these processes. Vasculogenesis results in de novo vessel formation, which is critical for development, while angiogenesis involves expansion of the existing network via sprouting or vessel splitting, and is required for network expansion. Arteriogenesis and capillary arterialization allow for remodeling of the vascular network in response to stressors such as ischemia, to alter blood flow within existing tissues. Examples of in vivo situations in which each process is particularly relevant are given.
Early in development, blood islands coalesce and lacunae form, resulting in a network of interconnected endothelial cords4. This process, by which whole networks can be formed simultaneously, is known as vasculogenesis. The tendency of ECs to coalesce and form cords in this way has been leveraged for in vitro assays5, and studied using computational models of early vascular network formation by the Glazier group4, 6.
Following vasculogenesis, the blood vessel networks in developing organs must be refined and expanded as tissues grow and differentiate. The process of angiogenesis increases vascular density by sprouting new vascular branches or splitting existing vessels in two. Sprouting angiogenesis takes two forms: first, expansion of vascular networks into currently avascular tissue – for example, the perinatal expansion of the retinal vasculature7, 8, or the investment of new vessels into small tumors; second, the dynamic sprouting and pruning/regression of vessels within an existing network9, for example due to exercise or within a growing organ. In both forms of sprouting angiogenesis, endothelial cells become activated by stimuli secreted from distant cells and undergo phenotypic differentiation to migratory, vessel-sprout-leading ‘tip’ cells. These cells degrade local extracellular matrix and lead proliferative stalk ECs to form sprouts that may ultimately anastamose and become part of the blood flow circuit. Intussusceptive angiogenesis is different to sprouting: existing endothelial tubes form internal pillars that lead to splitting of one vessel into two. This form of vascular expansion can result from changes to shear stress10, 11.
Vasculogenesis and angiogenesis are both typically processes of microvessel development. To obtain hierarchical vascular networks, growth (diameter expansion) is required. Arteriogenesis is the process of expansion of existing arterioles into larger vessels12, permitting the vessel to carry more blood flow. Capillary arterialization13, also known as arteriolargenesis14, is the process by which capillaries can, under specific circumstances, expand beyond typical capillary dimensions and acquire the characteristics of arterioles. Diameter expansion is typically accompanied by the acquisition of arterial/venous phenotype, including the investment of perivascular smooth muscle cells (SMCs)14.
Incorporating current understanding of the different vascular remodeling processes (Fig.1) into systems biology approaches is important for identifying proper strategies to promote or prevent vascularization in disease applications with distinct vascular network morphologies. The main drivers of these processes vary, including different local mechanical and chemical cues sensed by ECs. This suggests that multiple types of therapeutic targets may be combined to selectively activate or inhibit one or more of these remodeling processes. In this review, we focus primarily on non-developmental vascular remodeling, specifically discussing sprouting and intussusceptive angiogenesis. To date, arteriogenesis and capillary arterialization have not been the subjects of significant systems biology efforts; these provide opportunities for future work. Section 2 will provide more detail on the types of models used to study different vascular remodeling processes, and the components included in these models. Section 3 will discuss the use of systems biology to identify effective therapeutic approaches to stimulating or inhibiting the vasculature. Section 4 will highlight challenges and bottlenecks that must be addressed to translate advances in microvascular systems biology into improved clinical outcomes.
2. MICROVASCULAR SYSTEMS PHYSIOLOGY AND PATHOLOGY
Vascular development and remodeling in vivo comprises several multicellular, multi-scale morphogenic processes. A systems approach is required to understand these processes and the effect of physiological and pathological changes to the system. In this section, we will describe the multiple scales of integrated regulation involved in vascular remodeling (Fig. 2). While the goal is to improve clinical outcomes in disease, our ability to measure systems changes in vivo is often limited. As such, computational studies of molecular and cellular regulation rely heavily on in vitro experimental studies for validation. These results must then be interpreted or translated to an in vivo context to be used for biomarker development and prediction of therapeutic responses. Appropriate computational models can provide this bridge between in vitro and in vivo measurements. For a detailed review of the mathematics underlying many of the modeling techniques presented here, see15.
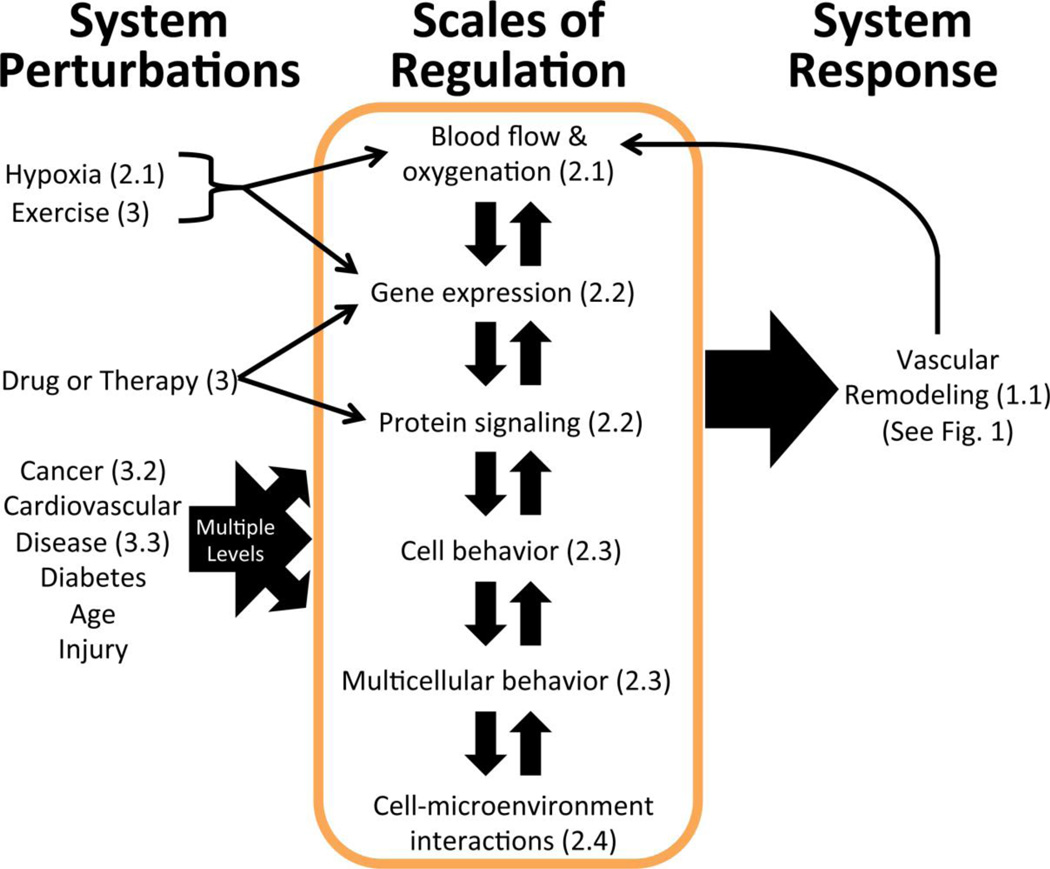
Figure 2. Vascular remodeling is a system-wide response to various perturbations at different scales.
Vascular homeostasis can be perturbed by disease, therapy, exercise, injury, or aging (left column). While some of the perturbations introduced by disease are relatively well-characterized (cancer, cardiovascular disease & hypoxia), others represent opportunities for future systems biology research (diabetes, age-related changes). These perturbations directly alter one or more of the scales regulating the vascular system (center box, discussed in Sections 2.1–2.4 as indicated), and propagate due to the connectedness of the system, inducing indirect changes at the other levels of regulation as well. As the vascular system adapts to the perturbation via remodeling (See Fig. 1), a new homeostasis is established (right column). This new homeostatic state may have different blood flow and gene expression than the pre-perturbation system, depending on the effectiveness of the physiological or therapy-induced remodeling. While perturbation/dysfunction can occur at any of the levels, most therapies target molecular regulation mechanisms (Section 2.2).
2.1 Blood flow and oxygen distribution: a system of delivery and consumption
The vasculature comprises a hierarchical network of interconnected endothelium-lined tubes. The flow of blood distributes oxygen to tissues, with local blood flow matching metabolic demand16. Mismatches in blood flow and tissue oxygen consumption can arise from normal processes such as growth and exercise, as well as pathological conditions including stroke, diabetes, respiratory disease, and myocardial infarction17. Mathematical models of blood flow fall into two categories: (a) three-dimensional models of blood flow, particularly potentially turbulent flow at sites of atherosclerosis in large vessels18, 19; (b) network models of blood flow in systems of smaller vessels, in which laminar flow permits the use of Poiseuille-based algebraic models. One example of the first category of models is work by the Diamond group, which integrates hemodynamics with signaling cascades in platelets20 and stochastic models of coagulation initiation21 to study the effect of hemodynamics on blood components, including red blood cells and platelets. These models allow for prediction of clot formation and drug sensitivity under varying platelet signaling and flow conditions22. The network models in the second category can incorporate experimental measurements of heterogeneous and dynamic microvessel diameters, pressure, flow rates, shear stress, and oxygen exchange23. Shear stress and local oxygen availability in particular are key stimuli for angiogenesis and remodeling of the vessel wall, for which predictive models have been developed by the Secomb and Pries groups24, 25. Combined experimental-computational systems studies such as these can produce interesting predictions with implications for in vivo physiology and pathology, such as that the vascular wall must be capable of sensing oxygen levels in order match experimental observations after changes in blood flow and oxygen distribution26.
The biomechanics of blood flow are important for intussusceptive angiogenesis. While this form of angiogenesis has not been studied as extensively as sprouting angiogenesis11, intussusception is thought to be the primary form of vascular remodeling in animal models with vascular endothelial growth factor (VEGF) overexpression27, chronic shear stress28, or colitis29. Computational models have demonstrated that hemodynamics and shear stress30–32, along with oxygen consumption33 contribute to vessel splitting and pillar formation, which are requirements for intussusceptive angiogenesis. Szczerba et al. generated the first model incorporating the combined effects of hemodynamics, chemical agents, and vessel wall stiffness on intussusceptive angiogenesis10. In this model framework, increasing vessel wall stiffness during development (a result of pericyte investment and/or basement membrane deposition) was required to produce realistic predictions of vessel splitting10. Interestingly, another computational model (of skeletal muscle) predicted that intussusceptive angiogenesis can more effectively maintain oxygen levels than sprouting angiogenesis when oxygen consumption is high33. Tumor vessels have also been shown to undergo intussusceptive angiogenesis after treatment with angiogenesis inhibitors, but a model of these processes has not yet been developed34.
Other computational models focus on oxygen distribution in tissues, which is regulated by blood flow, oxygen consumption, and by chemical signal molecules such as nitric oxide. By integrating a blood flow model with an oxygen diffusion/consumption model, the Popel group created a multi-scale model of oxygen transport in skeletal muscle, demonstrating the influence of muscle fiber type on oxygen distribution35. The simulations predicted that the distribution of muscle fiber sizes has a larger impact on O2 distribution than O2 consumption, myoglobin concentration or oxygen diffusivity35. Regulation of oxygen by nitric oxide, which stimulates vascodilation and is required for normal endothelial function, has been simulated36, but this has not been modeled in the context of angiogenesis. The effect of tissue oxygenation on wound healing has also been modeled37. More detail on the modeling of oxygen distribution in the microvascular circulation can be found in38 and39.
In a later section, we will discuss the importance of blood as a communication route for key proteins and drugs regulating vascular remodeling, as well as the centrality of blood measurements as clinically-relevant, reproducible biomarkers.
2.2 Molecular regulators of endothelial cell behavior and vascular remodeling
In the adult, mismatch of oxygen supply and demand can result in changes to the vascular network (Fig. 3), typically through the transcription factor hypoxia inducible factor (HIF) and the vascular endothelial growth factor (VEGF) family of extracellular ligands40, though other transcription factors and ligands are known to regulate vascular remodeling41, 42. Cancer, ischemia, diabetes, and other diseases alter gene regulation, protein expression, and signaling pathway function in angiogenesis, but these changes and their effects on vascular homeostasis are not yet completely understood3, 43. As examples, expression of cell surface receptors becomes heterogeneous in many solid tumors due to non-uniform oxygen pressure (resulting from structural abnormalities in tumor vessels)44; and changes in shear stress (e.g. due to elevated blood pressure) can alter endothelial gene expression11.
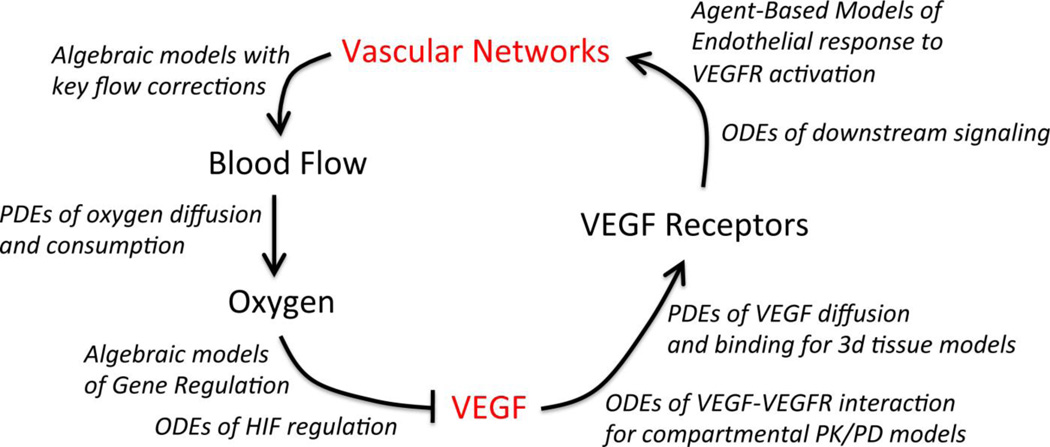
Figure 3. An example of vascular homeostasis and regulation by VEGF.
The many different computational model types employed to simulate the flow of information through the integrated multi-scale physiological models are indicated in italics. In general, models of in vivo pathology incorporate key elements of tissue physiology: vascular network geometry, blood flow, and/or oxygen distribution. Detailed models of molecular and cellular regulation, for example of the VEGF family, are often constructed and validated with in vitro experimental data, and then integrated into in vivo models and coupled to the other scales of regulation (Fig. 2) to predict the vascular remodeling and other physiological changes resulting from molecular perturbations (such as therapeutics). In diseases such as cancer, the homeostatic regulatory mechanisms can become non-functional or function in altered ways, leading to different vessel morphology than observed under physiological conditions.
Normal oxygen levels (normoxia) enable the hydroxylation of the transcription factor HIF1α by prolyl hydroxylases, resulting in HIF degradation40. Low oxygen (hypoxia) stabilizes HIF1α, which moves to the nucleus and activates transcription after binding HIF1β/ARNT45, 46. There are hundreds of downstream targets of HIF, notably members of the VEGF ligand and VEGF receptor families46. Multiple microenvironment-dependent HIF-1α signaling profiles (switch-like or gradual) have been demonstrated using computational models of HIF-1α regulation40, 47. Such divergent system behaviors are difficult to couch in a single framework without the use of computational methods. Models of HIF-1 have also been used to: determine the mechanisms through which HIF-1α senses oxygen40, 48; study the regulation of HIF-1α49, 50; and examine differences in HIF-1α regulation in cancer and ischemia51 with the goal of identifing promising therapeutic targets for different disease states.
While a wide variety of growth factors, adhesion molecules, and cell-cell communication proteins are involved in angiogenesis, including integrins, cadherins, Delta-Notch and semaphorins, we focus here on VEGF and fibroblast growth factor (FGF); as diffusible proteins that can be measured in the blood, they hold promise for validating predictive models of their transport and impact on vascular behavior.
The VEGF family of growth factors are critical regulators of both physiological and pathological angiogenesis, promoting endothelial cell survival, proliferation, and migration. There are five ligand genes, each with splice isoforms. These ligand genes and splice isoforms have varying affinity for the three VEGF receptors (which can hetero- or homo-dimerize upon ligand binding), two main VEGF coreceptors (the neuropilins), and the extracellular matrix (ECM)52. The VEGF receptors (VEGFRs) also exist as soluble and membrane-bound isoforms53. Recent work has demonstrated that post-translational modification (glycosylation, acetylation, methylation) can also modulate the activity of VEGFR254, 55. The multiplicative complexity of these ligands and receptors make understanding the spatial and temporal dynamics of the system and predicting reponse to VEGF-based therapies extremely difficult, as is highlighted by the lack of of success to date in VEGF-based pro-angiogenesis clinical trials56.
VEGF family members are secreted by parenchymal cells experiencing hypoxia, including: skeletal myocytes in exercise; neural and glial cells in retinal development; bone marrow-derived dendridic cells in wound repair; and hypoxic tumor cells57. VEGF isoforms diffuse through the extracellular matrix to bind VEGF receptors on endothelial cells. ECM-binding isoforms also become sequestered in tissues, where they can still activate VEGF receptors52, 58. The simulation of VEGF-VEGFR interactions and VEGFR-VEGFR coupling has been developed using biophysically-detailed ordinary differential equation models that are first vaildated against in vitro experimental data and then applied to in vivo scenarios. This allows for much more detailed understanding than would be possible using only in vivo data, which typically consists of plasma protein concentrations, plus some genetic and gene expression data. The scenarios examined to date include competition between ligands for binding to multiple receptors59; coupling and enhancement of VEGF binding by Neuropilin co-receptors60–62; dimerization of VEGF receptors63; downstream signaling of the Akt and ERK pathways64, 65; matrix-immobilized growth factors and VEGFR trafficking and phosphorylation66, 67. In addition to these detailed models of VEGF dynamics, models have been developed to directly predict VEGF production in skeletal muscle based on oxygen levels, both after exercise and in peripheral artery disease33, 68–70. These models allow comparison of disparate therapeutic strategies including exercise and VEGF delivery. Here, exercise was predicted to improve VEGFR ligation and VEGF gradients in ischemic tissue better than therapeutic delivery of VEGF; we will discuss the models of VEGF and exercise as therapies more in Section 3. More detail on the systems biology of VEGF can be found in52, 71.
The fibroblast growth factor (FGF) family has also been implicated in control of angiogenesis. FGFR signaling is complicated by the existence of multiple FGF ligands and the requirement for cell surface heparin sulfate proteoglycans (HSPGs) to stabilize FGF ligand-receptor complexes. A variety of computational models have been developed to study FGF ligand-receptor binding and regulation by HSPGs in vitro72–75, showing that HSPGs able to form active FGF2-HSPG-FGFR signaling complexes are required for effective downstream signaling76. FGF binding to EC receptors and to the vascular basement membrane under physiological flow conditions has also been simulated, both in vivo77, 78 and in the context of a bioreactor79, 80. These models have quantified variation in FGF-receptor binding as a function of flow conditions, FGF delivery method (bolus or continuous flow), HSPG and FGF receptor density, and binding affinities77, 79, 80. In particular, Filion et. al. showed that after intracoronary administration, myocardial deposition and retention of FGF2 is limited by the time required for FGF to bind cell surface receptors, and not by diffusion78. Additionally, they showed that the production and internalization rates of FGF receptors are important in regulating FGF distribution. These results have implications for the therapeutic delivery of FGF, and can be used to predict clinically relevant measurements that are difficult to obtain in vivo.
While the majority of systems biology techniques leverage computational methods, the use of systems biology principles in experimental data collection is increasing, and greatly enhances our understanding of the regulation of complex systems. In one example of such work, the lab of George Davis performs high-throughput experimental assays on endothelial cells cultured in the absence of serum81–83. This allows for the comparison of many experimental conditions in a well-controlled system, without the variability and background signaling generated by serum typical of most in vitro experiments. The angiogenesis and vasculogenesis assays performed by this group5 have clearly identified the minimal factors required for endothelial tube formation, identifying the key nodes in these complex regulatory networks. Such assays can be compared directly to computational models of in vitro sprouting angiogenesis and vasculogenesis, and then scaled to an in vivo context.
2.3 Vascular remodeling is a multicellular process
In translating extensive experimental results from ECs studied in vitro to understanding how endothelial cells behave in vivo, we must recognize the different environment that cells have in tissues – a multicellular environment where heterotypic neighbor interactions are key. Vascular remodeling requires the coordinated action of many endothelial cells and their neighbors. In sprouting angiogenesis, VEGF stimulation upregulates tip cell expression of Delta-like ligand 4 (Dll4)84. This results in activation of Notch in trailing stalk cells, reducing the sensitivity of these cells to VEGF by altering VEGF receptor expression85, and producing a non-uniform population of endothelial cells. This Delta-Notch system can be dysregulated in cancer86. Cell-cell adhesions (mediated by VE-Cadherin) can reduce VEGF-mediated EC migration87. Shear stress resulting from blood flow also regulates sprouting angiogenesis when blood flow is present23. Additionally, pericytes control angiogenesis and vessel stabilization by regulating EC proliferation and migration, along with contributing to formation of the vessel basement membrane88, 89. Pericytes express angiopoietin-1 (Ang1) and Ang2, which bind to Tie2 on endothelial cells88. Ang1 promotes vessel stabilization, while Ang2 destabilizes vessels. Endothelial cell-pericyte association is disrupted in many cancers, contributing to the formation of structurally and functionally abnormal vascular networks44.
Due to the critical coordination of cells during sprouting angiogenesis, agent-based models (ABMs) are commonly used to study the evolution of sprouting in space and time. ABMs represent each cell individually, with specific logic rules dictating cell behavior, which may be time- or location-dependent90, 91. Rule-based ABMs can also be coupled with ODE- or PDE- based models, for instance of VEGF distribution in tissues92, 93. Such models can recapitulate directional sprouting in response to VEGF gradients, and capture emergent differences in sprout morphology under varying conditions92. Cellular Potts Models (CPM), also known as Glazier-Graner-Hogeweg (GGH) models, are lattice-based ABMs in which each cell can evolve in shape, size, and interactions with other cells. As such, CPMs are used to study adhesion, cell elongation, and cell-cell signaling that alters EC behavior in angiogenesis and vasculogenesis4, 6, 94, 95.
ABM cell behavior rules can be relatively simple, such as growth and movement based directly on experimental observations of dynamic cell behavior data in zebrafish96. ABM rules can also be more complex, basing cell behavior on detailed ligand-receptor dynamics and signaling, e.g. filopodia extension, migration, and proliferation, leading to tip and stalk cell behaviors, based on the Dll4, Notch, and VEGFR2 network by Bentley and colleagues97, 98. This model predicted that ECs in a nascent sprout can continuously compete for tip position, resulting in dynamic changes in tip and stalk cell specification, which has been experimentally validated98. This and other models and experimental data indicate that the Notch system may be an interesting potential therapeutic target98–100. In another study, the Glazier group has shown using CPMs that contact inhibition of cell proliferation or migration in response to extracellular stimuli can regulate vascular patterning4. Other ABMs have studied sprouting in response to combinations of VEGF and brain-derived neurotropic factor101, and examined clean behavioral changes or knock-outs (e.g. tip and stalk cell proliferation and migration) that are not possible in vivo or even in vitro102, 103, which is a key advantage of computational modeling as a tool to enhance drug design. Taking an alternate approach, a Boolean model links activation of combinations of VEGF receptors, integrins, and cadherins to cell behaviors such as migration and proliferation104. Together, these models improve our understanding of how combinations of extracellular cues regulate vascular remodeling, allowing for identification of new ways to modulate these processes in vivo.
As angiogenesis progresses, sprouts form lumens and anastomose onto existing vessels, facilitating blood flow and introducing these ECs to shear stress. Anastomosis requires the tip cell to become quiescent, a transition that has been studied by Bentley and colleagues using a Spring-Agent model, a type of ABM where each agent is a collection of smaller entities connected by spring-like tensions105. This allows for cell shape and cell-cell contacts to change, altering Notch signaling between cells. A multi-scale model of exercise response in skeletal muscle from the Popel group includes sprout formation, branching, and anastomosis in a single framework integrating blood flow, oxygen distribution, and VEGF transport (continuous processes) with cell behavior (discrete ABM)106. In this model, anastomoses occur when tip cells come within close proximity to other vessels, but molecular detail of anastomoses is not included. Simulations of tumor angiogenesis and blood flow incorporating shear stress-induced vessel branching107, varying vessel morphology108, and vessel pruning in response to therapy109 suggest that vascular network morphology strongly influences delivery of both nutrients and chemotherapy drugs to tumors.
Some multi-scale models of vascular remodeling include other cell types, such as pericytes110–113. Pericytes must dissociate from vessels to permit sprouting, and their recruitment is required for vessel stabilization following remodeling. An ABM including pericyte recruitment in response to gradients of EC-secreted platelet-derived growth factor B (PDGF-B) and differentiation of interstitial cells into pericytes as a function of contact with endothelial sprouts can predict the portion of capillary coverage by smooth muscle α-actin-positive pericytes111. A separate computational model captured vessel stabilization and destabilization in response to VEGF, PDGF, Ang1, and Ang2 by integrating modules for tumor growth, endothelial angiogenesis, and vessel stabilization (by pericytes)112. Vessel stabilization was predicted to result in slower tumor growth. This growth model predicted that anti-VEGF therapy is more effective when the portion of immature vessels is high, and that co-application of anti-VEGF and anti-Ang1 resulted in prolonged inhibition of tumor growth112, in line with another model of metastatic ovarian cancer in vivo113.
While many of these agent-based models consider only a small number of cells, understanding the initiation, extension, and anastomosis of angiogenic sprouts is essential to predicting structural and functional characteristics of developing vascular networks in vivo. Even on this small scale, differences can be observed between the behaviors of sprouts forming due to physiological and pathological angiogenesis. The ABMs presented here describe angiogenesis in healthy tissue106, tumors92, 94, 107–110, 112, the cornea103, 113, and in vitro or developmental scenarios4, 6, 98, 101, as well as studying sprouting in a generalized context95, 97, 102, 104, 105, 111. Some incorporate expression levels of cell surface receptors or protein concentrations97, 98, 106, in order to understand how changes to these quantities alter sprout morphology in disease. Others integrate discrete models of angiogenesis with blood flow simulations106, 108, increasing our understanding of the crosstalk between these differing regulatory mechanisms.
2.4 Microenvironment of the microvasculature: high-resolution molecular biology
Not only do ECs receive guidance cues from soluble factors and neighboring cells, but also from mechanical and chemical interactions with their microenvironment114. Spatial and temporal patterning of these cues is required for formation of functional vascular networks that effectively oxygenate the surrounding tissue114. The extracellular matrix provides a scaffold for tissues; changes in its stiffness are sensed by endothelial and other cells. Additionally, EC signaling is altered by integrin adhesion to ECM proteins114. ECs alter their microenvironment by secreting ECM proteins and proteases that degrade ECM components, clearing a path for vessel growth and remodeling. One family of proteases implicated in angiogenesis are the matrix metalloproteinases (MMPs), inhibitors of which are also expressed by ECs115. In addition to degrading ECM, proteases can also cleave VEGF, releasing previously immobilized VEGF into the interstitial fluid114. The microenvironment in solid tumors is much different than in normal tissue, with perturbed ECM organization and high vascular permeability44, 116, while in peripheral artery disease the endothelial basement membranes are much thicker than in normoxic skeletal muscle117. Certain aspects of molecular regulation and cell-cell interactions can be studied in vitro, where detailed measurements are possible, but it is not feasible to exactly replicate the complete tissue microenvironment in which vascular remodeling occurs. Thus, multi-scale computational models are necessary to integrate the cues endothelial cells receive from their microenvironment and translate this information into predicted cellular behaviors.
A variety of modeling techniques have been used to study the influence of the microenvironment on vascular remodeling at higher spatial and temporal resolution than is feasible experimentally. For example, a CPM (Cellular Potts Model, discussed in the previous section) of tumor angiogenesis can predict vascular branching and anastomosis of adjacent sprouts using rules based on molecular, cellular, and local tissue environment dynamics (VEGF gradients, proliferation rates, ECM composition) instead of observed cellular behavior92. In this model by the Jiang group, inhomogeneities in the extracellular environment were required to obtain realistic predictions. Additional study with this model demonstrated regulation by ECM fiber density and orientation of sprout extension and branching, suggesting that the ECM itself is a therapeutic target95. Other computational models, ranging from ABMs to multi-phase models, have demonstrated regulation of vascularization by pore size in porous scaffolds118, collagen fiber orientation119, and a combination of expression of soluble and matrix-bound growth factors, EC proliferation rate, and MMP activity120.
In addition to the composition of the microenvironment, the local geometry surrounding an angiogenic sprout can significantly alter the availability of diffusible proteins to cell surface receptors. As such, the effect of distance between adjacent angiogeneic sprouts was studied in a 2D reaction-diffusion model by the Mac Gabhann group121. The model showed that decreased distance between two sprouts increased the probability that the sprouts would diverge. This study also demonstated that the VEGF-sequesting soluble VEGFR1 isoform, which is secreted by endothelial cells increases the gradient of VEGF-VEGFR2 along the length of sprouts121. These behaviors hold in extending the model to three-dimensional sprouts in tissues, and these models can provide molecular explanations for the observed behaviors of perturbed systems such as VEGFR1-knockouts122. These models are developed using high-resolution imaging of developing sprouts, enabling true image-based simulations that are specific to the different anatomical outcomes of the molecular perturbations.
Other computational models have focused on modification of the ECM due to endothelial secretion of proteases. Detailed models of the production, activation, and inhibition of several MMPs in the context of angiogenesis have been developed by the Popel group123–125. These models have been incorporated into larger 2D and 3D reaction-diffusion models of VEGF ligand-receptor binding and transport, and consider the release of HSPG-bound VEGF from the ECM via cleavage by proteases126, 127. It was shown that endothelial cells alone do not produce enough proteases to release a significant amount of VEGF, suggesting involvement of other neighboring cell types126. Additionally, simulation of the tissue distribution and gradient formation of HSPG-binding and non-HSPG-binding VEGF isoforms showed that isoform-specific degradation is necessary to match experimental measurements of VEGF localization, and is involved in vascular patterning127. These results are of particular relevance to tissue engineering, where the properties of the microenvironment can be tuned to promote proper vascular network formation. In addition to computatational modeling, high-throughput experiments and proteomic analysis have been used to understand the activity of MMPs and identify promising therapeutic targets128–130. The data generated by such studies can improve computational models of MMP activity in vascular remodeling and cancer127, 131, 132.
2.5 Homeostasis requires coordination of multiple scales of regulation
While we have presented distinct levels of vascular regulation in this section, it is vital for understanding in vivo physiology to recall that all of these levels are interconnected. Diseases can alter any of these regulatory mechanisms, while drugs typically target gene expression and/or protein signaling networks within cells. Systems biology can aid in identifying the regulatory levels perturbed in specific disease states, which are not fully established for many diseases. After any perturbation (Fig. 2), the system can adapt using the outlined regulatory mechanisms, resulting in vascular remodeling and reaching a new homeostatic state. A specific example of a homeostatic cycle relevant to altered blood flow/oxygenation is shown in Fig. 3, along with the types of computational models that are used to study each process in the system. An example of multiscale modeling applied to skeletal muscle to simulate this entire homeostatic cycle will be discussed in Section 3.3. Other tissue-specific multiscale models with multiple cell types are emerging, including a study of oxygen and growth factors in healing bone defects133–135. While it is not computationally feasible to unite all of these modeling techniques in a detailed 3D model of the complete human body, we use a subset of these tools (application-dependent), the insights resulting from other models, and quantities that are experimentally measureable (in vivo and in vitro) to understand regulation of vascular remodeling at multiple scales, and how perturbations at any of these levels alters both local and system-wide behavior. This is turn will lead to improved ability to identify biomarkers and potential therapeutic targets.
3. MICROVASCULAR SYSTEMS PHARMACOLOGY
Vascular remodeling plays key roles, beneficial or detrimental, in many diseases9. Angiogenesis is a hallmark of cancer136, 137, and ectopic vascularization drives retinopathies and other leading causes of blindness. In contrast, for diseases characterized by hypovascularization and/or ischemia, such as atherosclerosis, pre-eclampsia, Crohn's disease or hypertension, amelioration by the induction of angiogenesis or arteriogenesis continues to be an active area of therapeutic research. We focus here on cancer and peripheral artery disease as canonical diseases requiring anti-angiogenesis and pro-angiogenesis treatment, respectively.
Drugs, gene vectors, exercise, and other vascular-targeted therapeutic approaches can be studied using systems approaches. For example, the repeated lack of success in human clinical trials of proteins and genes encoding vascular-targeting growth factors suggests that scaling from mice and other pre-clinical models to humans is not trivial. The variability from person to person in responses to all drugs further complicates matters. Understanding the pharmacokinetics and pharmacodynamics of vascular-targeting agents is particularly difficult since the target cells for many of these – endothelial cells – have two active surfaces: one facing the blood stream where many of the drugs are delivered, and one facing the interstitial space of the tissue138. These two surfaces are not the same, and the effects of drugs at each surface are not the same.
Systems Pharmacology is crucial to improving the extremely low success rate in clinical trials generally. Clinical trials are very expensive, and using them we cannot try every target, drug combination, dose, or schedule. Systems Pharmacology enables us to virtually explore the therapeutic space. Thus, we call on computational models to test and compare multiple drugs, drug combinations, doses, schedules and routes of administration. We can also go further than drugs to include non-drug therapeutics, including mechanical and electrical stimulation, exercise, or the implantation of engineered or transplanted cells and tissues. In this way we can efficiently eliminate therapies unlikely to be successful and focus on optimizing approaches predicted to show success for at least a subset of the patient population.
Clinical data – gene and protein expression, but also height, weight and other measurements – can be incorporated into well-designed models to build individualized simulations and populations of predictive patient models. On the other side, predictive models need to make clinically testable and measureable predictions, for example the dynamics of change to concentrations of key molecules in the blood. Only by validating such pharmacological models can we hope to make them useful in the clinic. These models can also help in prospective design of clinical trials by identifying key biomarkers, including complex or nonlinear biomarkers that would not be obvious from a linear analysis of the data.
3.1 Whole-body compartment models: pharmacokinetics and pharmacodynamics
As a consistent framework for the analysis of therapies – not just small molecule drugs and biologics, but also gene therapies, physiological changes, and tissue transplants – we must integrate the molecular and cellular understanding outlined in Section 2 into a whole-body model that simulates the transport of key vascular regulatory proteins such as VEGF as well as their cellular targets. Clearly this cannot currently be done at the whole-body scale with the same level of three-dimensional anatomical detail and spatial resolution described in the models of Section 2.4; however, much of the anatomical specificity can be retained – for example, the multicellular nature of tissues; the heterogeneity of gene and protein expression between cell types; the volumes and surface areas associated with different cell types; the complex molecular interaction networks; and the dynamic nature of cells in responding to extracellular stimulus. By assuming each tissue compartment is well mixed, we can trade partial differential equations for ordinary differential equations71, 139, significantly speeding up computation without losing much of the key biology regulating vascular remodeling.
In Section 2, we discussed the importance of blood in delivering oxygen to tissues and the importance of computational models in building a quantitative understanding of tissue physiology and pathology. The blood compartment also plays a central role in any systems biology perspective of disease and treatment, because blood-based measurements are the most common type of in vivo data available for validation of computational models. Accessibility, reproducibility, low invasiveness and the ability to do sequential measurements make blood biomarkers highly sought after. Simple one-component blood-based biomarkers can have clear population-level changes in pathology, but not be informative for an individual140, suggesting that more complex biomarkers based on understanding of molecular mechanisms may be more informative. For example, a ratio of VEGF and sFlt1 protein levels in blood may be an important predictor of pre-eclampsia141, better than either metric alone. Going beyond detection and diagnosis, prediction of blood-based biomarkers for disease progression and therapeutic response is an area of high interest that opens the door to predictive, responsive and adaptive personalized medicine. Thus, understanding the relation between blood-based measurements (e.g. of soluble proteins) and disease state is an important goal that can be addressed using systems biology techniques.
3.2 Targeting angiogenesis in cancer: virtual clinical trials
Tumors can cause a perturbation to vascular homeostasis (Fig.2). At first, without vascular ingrowth, the tumor is oxygen limited. However, acquisition of pro-angiogenesis characteristics, such as the constitutive activation of HIF by oncogenic kRAS, can result in perfusion by new vessels. Because of the broken homeostatic cycle, hypervascularization and atypical vessels result – tortuous, inefficient and leaky. Tumor vascularization permits growth beyond the oxygen diffusion limit, and provides tumor cells a route for metastasis. Drugs developed to inhibit angiogenesis in cancer have targeted the key receptor tyrosine kinase pathways in vascular remodeling, including the VEGF receptors, EGF receptors (ErbB/HER) and FGF receptors on endothelial cells. These drugs include antibodies to ligands (e.g. bevacizumab) or to receptors (e.g. DC101) and tyrosine kinase inhibitors (e.g. sunitinib).
By building pharmacokinetic-pharmacodynamic (PK/PD) models of these growth factor-RTK systems, direct testing of multiple RTK-targeting drugs has been possible. These models can incorporate specific current drugs with known interactions and kinetics, but can also be used as a drug design tool by introducing molecules with different interactions. These models can give insight into whole classes of drugs and functions; for example, inhibiting receptor-receptor interactions has emerged from simulation of the VEGF/VEGFR system61, 142–145 as a strategy potentially superior to ligand targeting61. This is being borne out in recent experimental results for drugs targeting receptor dimerization146. More recently, the tendency of tumors to favor the expression of specific VEGF isoforms was identified using computational simulation to be a critical vulnerability and improve the predicted efficacy of anti-tumor VEGF-targeting144. The predicted impact of isoform-specific anti-VEGF agents are not as might be expected based on our understanding of physiological angiogenesis, in part because the regulation of isoforms is very different in tumors147.
An alternate model of VEGFR pharmacodynamics goes beyond the ligand-receptor interactions by incorporating VEGFR2’s downstream signaling pathways148. By doing this, the Birtwistle and Gallo groups were able to run sensitivity analyses of dosing for multiple drugs targeting VEGF, VEGF receptors and downstream signal molecules such as PLCγ. They then used optimization algorithms to define potential multidrug regimens with different dosing and scheduling148.
Validation of pharmacological models is crucial to developing helpful predictive simulations. For models of human pharmacology, the detail and complexity of the models results in many outputs that are not easily measurable, e.g. cell-type-specific activation of multiple receptor families, but also several that are. In particular, the models can predict the effect of multiple perturbations in different cells in different tissues on key proteins in the blood. For example, a multi-compartment PK/PD model of VEGF in humans was used to investigate dynamic changes in the tumor and in the blood following treatment with systemic infusion bevacizumab (anti-VEGF antibody). Counterintuitively, and without any fitting of data, the model predicted that the concentration of VEGF in blood would increase following anti-VEGF treatment145; this surprising effect has indeed been clinically observed149–151. Because of the highly detailed and mechanistic nature of the model, we could go further and determine that this emergent property resulted from a shuttling mechanism of the VEGF-antibody complex145. Such mechanistic hypothesis testing can result in strong and actionable therapeutic predictions.
Another key requirement of models – to be populated with high-quality, detailed experimental data – becomes a benefit of taking an integrated (experimental and computational) systems approach. Models can help us to identify which experimental measurements (target, type, location, spatial resolution and temporal resolution) are the most important or informative. For example, pharmacological models have identified that cell-specific receptor expression plays an important role in the response to therapy – many RTKs are expressed on multiple cell types and not just on the target cell type152 and the potential for synergistic or antagonistic side effects is clear. Model-based quantification of these multi-cellular (and multi-tissue) effects is clearly important to prediction of therapeutic outcome. Based on simulations, delivery of a VEGF-neutralizing agent can result in available VEGF in the tumor going either up or down depending on the variability in both ligand and receptor expression142, 144; even the difference between the apical and basolateral expression of VEGF receptors was predicted to play a major role in pharmacodynamics153 and this prediction of a systems biology model is now being borne out138.
3.3 Promoting vascularization in peripheral artery disease: from rodent to human
While therapies targeting hypervascularity in cancer and age-related macular degeneration have come to market, no pro-angiogenesis therapeutic agents have been approved. Indeed, multiple trials have failed56, 154, 155, including proteins or gene therapy targeting VEGF, HIF-1 or FGF. These failures occurred despite successes in pre-clinical animal models of ischemic disease. Thus, there is an urgent need for systems biology techniques to help predict which treatments would be successful, providing a better bridge from pre-clinical to human clinical trials.
To study the in vivo pharmacodynamics of angiogenesis-targeting treatments, we have developed two types of multi-scale models. First, a fully three-dimensional model, that uses image-based anatomical information to simulate a portion of tissue at micron resolution – for example, skeletal muscle (Fig.4A). While simulations using this model are confined to a particular volume of tissue, the pharmacodynamics of key treatments can still be tested – for example: local effects of gene delivery, which will alter the cell-specific expression rates in the model; or cell-based therapy, in which augmented stem cells can differentiate and integrate into the tissue; or exercise, which will impact gene expression but also blood flow and oxygen demand68, 69, 156, 157. These three-dimensional simulations identified key drivers of the VEGF concentration in the tissue as well as of VEGFR activation. Even at rest, without disease or external perturbation, there is heterogeneity in oxygen, VEGF expression, and VEGF and VEGFR concentration gradients. This was further studied using a more detailed anatomical model that included realistic muscle fiber type distributions35. The expression of VEGF receptors, and thus the location of the blood vessels, was identified as the key driver of VEGF gradients (which are thought to provide chemotactic guidance to nascent sprouts). We noted that exercise, which is encouraged therapeutically for PAD patients but is often difficult especially in more severe disease, results in up-regulation of both VEGF ligands and VEGF receptors. We were then able to identify using our models that therapeutics delivering only ligands are less effective at increasing the concentration gradients in tissues, and can induce these increases for a shorter time, than receptor expression changes. This, then, provides a possible path forward in developing the next generation of PAD therapeutics. Based on these models, we added an agent-based model of cell behavior to ‘complete the circle’ (Fig.3) and enable the simulation of chronic disease and treatment, or repeated bouts of exercise training93.
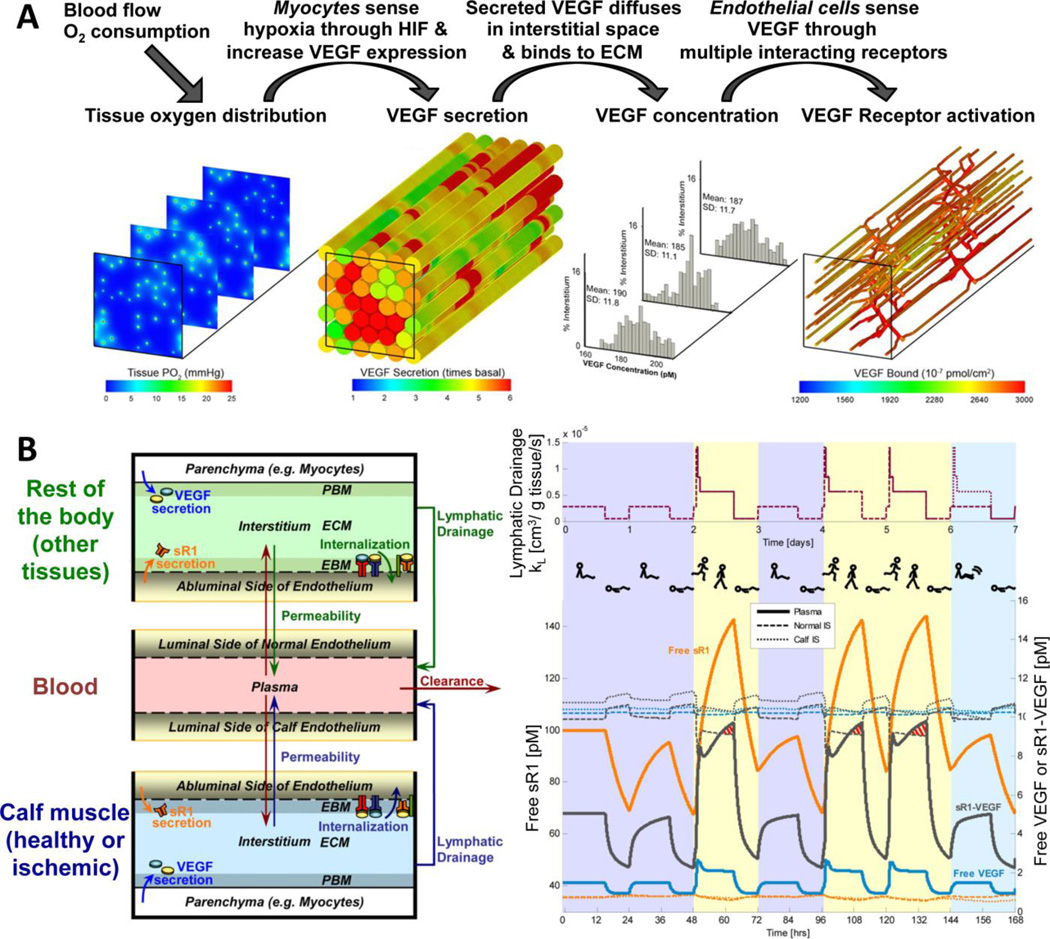
Figure 4. Multi-scale models of microvascular physiology and pathology in vivo. A, Three-dimensional multi-scale model of vascular regulation in skeletal muscle in vivo.
By integrating multiple model types (Fig.3), we can simulate the links from three-dimensional tissue anatomy and heterogeneity to blood flow, to oxygen distribution, to hypoxia-dependent VEGF secretion by parenchymal cells, to VEGF diffusion, to ligation of VEGF receptors on endothelial cells. The output is heterogeneous VEGF receptor activation across the vasculature, which can then be coupled to cell behavior models such as ABMs93, 106 to complete the homeostatic cycle and remodel the vascular network. This integrated model has been used to study peripheral ischemia disease and to test potential treatments. Simulation results figures adapted from157. B, Multi-compartment PK/PD model of the VEGF family. This model has multiple compartments, including calf muscle to enable studying the effects of PAD which results in significant pathological changes to that muscle. The model predicts the distribution of VEGF and soluble VEGFR1 and VEGF receptor activation throughout the body, including the blood concentrations of the diffusible proteins. The compartments of the PK/PD model can communicate via physiological processes such as vessel wall permeability and lymphatic drainage. An example application of the PK/PD model is also shown, a simulation of the dynamic effects of diurnal changes in lymphatic drainage (as a result of changes in posture and activity) on plasma soluble VEGFR1 and VEGF levels in a healthy patient. Purple background represents bed rest days, yellow represents active days, and aqua shows calf rest days. Models of this form allow for prediction of tissue VEGF concentrations, and net flows of VEGF between multiple tissues and the blood, and are also druggable – small molecule, protein and gene therapies can be added, as can therapeutic alterations to exercise scheduling. Schematic and simulation results figure adapted from159.
Building a whole-body three-dimensional model with the resolution needed to deal with the molecular gradients described above is not currently feasible. Instead, a second kind of model is needed – a compartmental PK/PD model158, similar to that described in the previous section for cancer, but now with a target ‘disease’ tissue of the ischemic calf muscle (Fig.4B). Although concentration gradients cannot now be simulated at this scale, we can test systemic organism-wide perturbations, such as sleep/wake and exercise cycles, which impact lymphatic flow as well as molecular expression70, 159 (Fig.4B), the impact of therapeutics on non-target not-diseased normal tissues, and the intravascular delivery of therapeutic molecules.
These two model types – 3D high-resolution models of tissue and the compartmental PK/PD models – can be directly compared because the interstitial concentrations in the compartments will be the same as the average concentrations adjacent to the interstitial surface of VEGFR-expressing ECs; the average VEGFR activation in the 3D model will be the same as the compartment-level VEGFR activation in endothelial cells.
Lastly, we note that a key issue in the treatment of peripheral artery disease is the failure in humans of treatments that work in rodents. This is a common problem and one for which systems biology is well suited. The parallel development of mouse-specific and human-specific computational models, with a common framework and species-specific parameters, will enable the translation of findings in one to predictions of successful approaches in the other.
4. CHALLENGES AND FUTURE DIRECTIONS
A wide variety of computational and experimental techniques have been harnessed to expand our knowledge of microvascular function in health and disease. Computational models are invaluable in their ability to integrate multiple experimental results into a single, often mechanistically-based framework. Progress has been made in integrating across multiple model types, biological regulation mechanisms, and geometric scales to provide a systems-level, dynamic view of the microvasculature and of its remodeling processes. And yet much remains to be done to meet the challenge of making these models, and our resultant understanding of this complex dynamic system, capable of bridging insights from the lab to the clinic.
Areas of potential growth include the development of species-specific and personalized models. Mouse-specific and human-specific models, parameterized with species-specific experimental data, can be used side-by-side to assist in successful translation from pre-clinical to clinical trials. Patient-specific models can incorporate not only individualized pharmacokinetic parameters but also the high variability in gene and protein expression that greatly affect pharmacodynamics. Such models can advance identification of biomarkers for specific subpopulations, and identify specific therapeutic strategies as being effective (or ineffective) for each group160.
It is crucial, as increasingly complex computational models are developed, to validate model outputs against quantities that are measurable in vivo, while leveraging non-measurable model outputs to predict changes in cellular signaling and behavior that may be important for disease prognosis and response to therapy. In parallel with continued model development, systematic collection of quantitative experimental measurements to characterize vascular growth and remodeling in healthy and diseased tissue, both before and after treatment, is critical to develop a sufficient mechanistic understanding of microvascular dynamics to provide meaningful clinical decision support. And as these models and experimental data are produced, it is essential to perform failure analysis – to probe the molecular mechanisms behind the failure of unsuccessful pro-angiogenic drugs. There is so much to learn from previous preclinical and clinical trials that can inform future therapeutic design.
There is also a need for further study of the less well-understood forms of vascular remodeling, such as arteriogenesis and capillary arterialization. In addition, more must be done to understand the layered and complex effects on vascular remodeling and therapeutics of key co-morbidities such as diabetes and hypertension. In the clinic, patient presentation is rarely single-factor, and a systems approach to multi-disease interactions could greatly improve outcomes.
While there remain many challenges to be met in microvascular systems biology, the progress of recent years highlights the value of systems computational and experimental approaches, and promises advances in clinical outcomes in the years to come.
Supplementary Material
The vascular network carries blood throughout the body, dynamically adjusting to maintain tissue oxygenation. The network and its constituent cells adapt in response to both acute and chronic stimuli. Controlling these key physiological and pathological processes is of great interest in many diseases. A systems biology approach is essential to unite our understanding of vascular physiology across the molecular, cellular, and tissue scales. Multiscale computational models can provide the bridge necessary to more effectively translate in vitro results to in vivo systems, and to translate pre-clinical animal models of disease and treatment to human therapy and clinical trials in the future. We present several examples of computational models that highlight the potential of systems biology approaches to generate novel insight into in vivo vascular biology.
ACKNOWLEDGMENTS
This work was supported by grants from the US National Institutes of Health (R00-HL093219 and R01-HL101200), American Heart Association (12BGIA12060154), and the US Department of Defense (PC111718), and by an NDSEG fellowship to LEC from the US Department of Defense.
Contributor Information
Lindsay E. Clegg, Institute for Computational Medicine and Department of Biomedical Engineering, Johns Hopkins University, Baltimore MD 21218
Feilim Mac Gabhann, Department of Materials Science & Engineering, Johns Hopkins University, Baltimore MD 21218.
REFERENCES
- 1.Chi JT, Chang HY, Haraldsen G, Jahnsen FL, Troyanskaya OG, Chang DS, Wang Z, Rockson SG, Van de Rijn M, Botstein D, Brown PO. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lloyd PG, Prior BM, Yang HT, Terjung RL. American Journal of Physiology-Heart and Circulatory Physiology. 2003;284:H1668–H1678. doi: 10.1152/ajpheart.00743.2002. [DOI] [PubMed] [Google Scholar]
- 3.Loureiro RMB, D'Amore PA. Cytokine & Growth Factor Reviews. 2005;16:77–89. doi: 10.1016/j.cytogfr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Merks RM, Perryn ED, Shirinifard A, Glazier JA. PLoS Comput Biol. 2008;4:e1000163. doi: 10.1371/journal.pcbi.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh W, Stratman AN, Sacharidou A, Davis GE. Angiogenesis: in Vitro Systems. 2008;443:83–101. doi: 10.1016/S0076-6879(08)02005-3. [DOI] [PubMed] [Google Scholar]
- 6.Merks RM, Brodsky SV, Goligorksy MS, Newman SA, Glazier JA. Dev Biol. 2006;289:44–54. doi: 10.1016/j.ydbio.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connolly SE, Hores TA, Smith LEH, Damore PA. Microvascular Research. 1988;36:275–290. doi: 10.1016/0026-2862(88)90028-3. [DOI] [PubMed] [Google Scholar]
- 8.De Smet F, Segura I, De Bock K, Hohensinner PJ, Carmeliet P. Arteriosclerosis Thrombosis and Vascular Biology. 2009;29:639–649. doi: 10.1161/ATVBAHA.109.185165. [DOI] [PubMed] [Google Scholar]
- 9.Logsdon EA, Finley SD, Popel AS, Mac Gabhann F. Journal of Cellular and Molecular Medicine. 2014;18:1491–1508. doi: 10.1111/jcmm.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szczerba D, Kurz H, Szekely G. J Theor Biol. 2009;261:570–583. doi: 10.1016/j.jtbi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Styp-Rekowska B, Hlushchuk R, Pries AR, Djonov V. Acta Physiol. 2011;202:213–223. doi: 10.1111/j.1748-1716.2011.02321.x. [DOI] [PubMed] [Google Scholar]
- 12.Schaper W, Scholz D. Arteriosclerosis Thrombosis and Vascular Biology. 2003;23:1143–1151. doi: 10.1161/01.ATV.0000069625.11230.96. [DOI] [PubMed] [Google Scholar]
- 13.Mac Gabhann F, Peirce SM. Microcirculation. 2010;17:333–347. doi: 10.1111/j.1549-8719.2010.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benest AV, Stone OA, Miller WH, Glover CP, Uney JB, Baker AH, Harper SJ, Bates DO. Arteriosclerosis Thrombosis and Vascular Biology. 2008;28:1462–1468. doi: 10.1161/ATVBAHA.108.169375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scianna M, Bell CG, Preziosi L. Journal of Theoretical Biology. 2013;333:174–209. doi: 10.1016/j.jtbi.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 16.Zakrzewicz A, Secomb TW, Pries AR. News in Physiological Sciences. 2002;17:197–201. doi: 10.1152/nips.01395.2001. [DOI] [PubMed] [Google Scholar]
- 17.Semenza GL. Trends in Molecular Medicine. 2001;7:345–350. doi: 10.1016/s1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- 18.Antonova N, Dong X, Tosheva P, Kaliviotis E, Velcheva I. Clinical Hemorheology and Microcirculation. 2014;57:159–173. doi: 10.3233/CH-141827. [DOI] [PubMed] [Google Scholar]
- 19.Xiong G, Figueroa CA, Xiao N, Taylor CA. International Journal for Numerical Methods in Biomedical Engineering. 2011;27:1000–1016. doi: 10.1002/cnm.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purvis JE, Chatterjee MS, Brass LF, Diamond SL. Blood. 2008;112:4069–4079. doi: 10.1182/blood-2008-05-157883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo K, Denney WS, Diamond SL. Pathophysiology of Haemostasis and Thrombosis. 2005;34:80–90. doi: 10.1159/000089929. [DOI] [PubMed] [Google Scholar]
- 22.Flamm MH, Colace TV, Chatterjee MS, Jing H, Zhou S, Jaeger D, Brass LF, Sinno T, Diamond SL. Blood. 2012;120:190–198. doi: 10.1182/blood-2011-10-388140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pries AR, Reglin B, Secomb TW. Int J Dev Biol. 2011;55:399–405. doi: 10.1387/ijdb.103218ap. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pries AR, Reglin B, Secomb TW. Hypertension. 2005;46:725–731. doi: 10.1161/01.HYP.0000184428.16429.be. [DOI] [PubMed] [Google Scholar]
- 25.Pries AR, Secomb TW, Gaehtgens P. Am J Physiol. 1998;275:H349–H360. doi: 10.1152/ajpheart.1998.275.2.H349. [DOI] [PubMed] [Google Scholar]
- 26.Reglin B, Secomb TW, Pries AR. Am J Physiol Heart Circ Physiol. 2009;297:H2206–H2219. doi: 10.1152/ajpheart.00348.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gianni-Barrera R, Trani M, Fontanellaz C, Heberer M, Djonov V, Hlushchuk R, Banfi A. Angiogenesis. 2013;16:123–136. doi: 10.1007/s10456-012-9304-y. [DOI] [PubMed] [Google Scholar]
- 28.Brown MD, Hudlicka O. Angiogenesis. 2003;6:1–14. doi: 10.1023/a:1025809808697. [DOI] [PubMed] [Google Scholar]
- 29.Konerding MA, Turhan A, Ravnic DJ, Lin M, Fuchs C, Secomb TW, Tsuda A, Mentzer SJ. Anatomical record. 2010;293:849–857. doi: 10.1002/ar.21110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filipovic N, Tsuda A, Lee GS, Miele LF, Lin M, Konerding MA, Mentzer SJ. Microvasc Res. 2009;78:286–293. doi: 10.1016/j.mvr.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee GS, Filipovic N, Miele LF, Lin M, Simpson DC, Giney B, Konerding MA, Tsuda A, Mentzer SJ. Journal of angiogenesis research. 2010;2:11–11. doi: 10.1186/2040-2384-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godde R, Kurz H. Developmental Dynamics. 2001;220:387–401. doi: 10.1002/dvdy.1118. [DOI] [PubMed] [Google Scholar]
- 33.Ji JW, Tsoukias NM, Goldman D, Popel AS. Journal of Theoretical Biology. 2006;241:94–108. doi: 10.1016/j.jtbi.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 34.Ribatti D, Djonov V. Cancer Lett. 2012;316:126–131. doi: 10.1016/j.canlet.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 35.Liu G, Mac Gabhann F, Popel AS. PLoS One. 2012;7:e44375. doi: 10.1371/journal.pone.0044375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kavdia M. Crit Rev Biomed Eng. 2011;39:461–472. doi: 10.1615/critrevbiomedeng.v39.i5.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schugart RC, Friedman A, Zhao R, Sen CK. Proc Natl Acad Sci U S A. 2008;105:2628–2633. doi: 10.1073/pnas.0711642105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman D. Microcirculation. 2008;15:795–811. doi: 10.1080/10739680801938289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toma-Dasu I, Dasu A. Comput Math Methods Med. 2013;2013:141087. doi: 10.1155/2013/141087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qutub AA, Popel AS. Journal of Cell Science. 2006;119:3467–3480. doi: 10.1242/jcs.03087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dow JK, White RWD. Urology. 2000;55:800–806. doi: 10.1016/s0090-4295(00)00457-x. [DOI] [PubMed] [Google Scholar]
- 42.Mizukami Y, Li JN, Zhang XB, Zimmer MA, Iliopoulos O, Chung DC. Cancer Research. 2004;64:1765–1772. doi: 10.1158/0008-5472.can-03-3017. [DOI] [PubMed] [Google Scholar]
- 43.Warren CM, Ziyad S, Briot A, Der A, Iruela-Arispe ML. Science Signaling. 2014;7 doi: 10.1126/scisignal.2004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmeliet P, Jain RK. Nature. 2000;407 doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 45.Powell FL. Annual Review of Physiology. 2003;65:203–230. doi: 10.1146/annurev.physiol.65.092101.142711. [DOI] [PubMed] [Google Scholar]
- 46.Semenza GL. Physiology. 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 47.Yu Y, Wang G, Simha R, Peng W, Turano F, Zeng C. Plos Computational Biology. 2007;3:1657–1668. doi: 10.1371/journal.pcbi.0030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dayan F, Monticelli M, Pouyssegur J, Pecou E. Journal of Theoretical Biology. 2009;259:304–316. doi: 10.1016/j.jtbi.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Yucel MA, Kurnaz IA. Biotechnology and Bioengineering. 2007;97:588–600. doi: 10.1002/bit.21247. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen LK, Cavadas MAS, Scholz CC, Fitzpatrick SF, Bruning U, Cummins EP, Tambuwala MM, Manresa MC, Kholodenko BN, Taylor CT, Cheong A. Journal of Cell Science. 2013;126:1454–1463. doi: 10.1242/jcs.119974. [DOI] [PubMed] [Google Scholar]
- 51.Qutub AA, Popel AS. Mol Cell Biol. 2008;28:5106–5119. doi: 10.1128/MCB.00060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mac Gabhann F, Popel AS. Microcirculation (New York, N.Y. : 1994) 2008;15:715–738. doi: 10.1080/10739680802095964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tugues S, Koch S, Gualandi L, Li X, Claesson-Welsh L. Mol Aspects Med. 2011;32:88–111. doi: 10.1016/j.mam.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Hartsough EJ, Meyer RD, Chitalia V, Jiang Y, Marquez VE, Zhdanova IV, Weinberg J, Costello CE, Rahimi N. Science Signaling. 2013;6 doi: 10.1126/scisignal.2004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahimi N, Costello C. Proteomics. 2014;00:1–10. [Google Scholar]
- 56.Mac Gabhann F, Qutub AA, Annex BH, Popel AS. Wiley Interdisciplinary Reviews-Systems Biology and Medicine. 2010;2:694–707. doi: 10.1002/wsbm.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Folkman J. Seminars in Oncology. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 58.Ferrara N. Mol Biol Cell. 2010;21:687–690. doi: 10.1091/mbc.E09-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mac Gabhann F, Popel AS. American Journal of Physiology-Heart and Circulatory Physiology. 2004;286 doi: 10.1152/ajpheart.00254.2003. [DOI] [PubMed] [Google Scholar]
- 60.Mac Gabhann F, Popel AS. American Journal of Physiology-Heart and Circulatory Physiology. 2005;288:H2851–H2860. doi: 10.1152/ajpheart.01218.2004. [DOI] [PubMed] [Google Scholar]
- 61.Mac Gabhann F, Popel AS. Plos Computational Biology. 2006;2 doi: 10.1371/journal.pcbi.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mac Gabhann F, Popel AS. American Journal of Physiology-Heart and Circulatory Physiology. 2007;292 doi: 10.1152/ajpheart.00637.2006. [DOI] [PubMed] [Google Scholar]
- 63.Mac Gabhann F, Popel AS. Biophysical Chemistry. 2007;128:125–139. doi: 10.1016/j.bpc.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan WH, Popel AS, Mac Gabhann F. Cellular Signalling. 2013;25:2496–2510. doi: 10.1016/j.cellsig.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan Hua T, Popel AS, Mac Gabhann F. Plos One. 2013;8 doi: 10.1371/journal.pone.0067438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clegg LEW, Mac Gabhann F. Angiogenesis. 2014;17:948–948. [Google Scholar]
- 67.Clegg L, Mac Gabhann F. PLOS Computational Biology. 2014 doi: 10.1371/journal.pcbi.1005445. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mac Gabhann F, Ji JW, Popel AS. Ann Biomed Eng. 2007;35:982–994. doi: 10.1007/s10439-007-9303-0. [DOI] [PubMed] [Google Scholar]
- 69.Mac Gabhann F, Ji JW, Popel AS. PLoS Comput Biol. 2006;2:e127. doi: 10.1371/journal.pcbi.0020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu FT, Stefanini MO, Mac Gabhann F, Kontos CD, Annex BH, Popel AS. Am J Physiol Heart Circ Physiol. 2010;298:H2174–H2191. doi: 10.1152/ajpheart.00365.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu FT, Stefanini MO, Mac Gabhann F, Popel AS. Methods Enzymol. 2009;467:461–497. doi: 10.1016/S0076-6879(09)67018-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Filion RJ, Popel AS. Annals of Biomedical Engineering. 2004;32:645–663. doi: 10.1023/b:abme.0000030231.88326.78. [DOI] [PubMed] [Google Scholar]
- 73.Forsten KE, Fannon M, Nugent MA. Journal of Theoretical Biology. 2000;205:215–230. doi: 10.1006/jtbi.2000.2064. [DOI] [PubMed] [Google Scholar]
- 74.Forsten-Williams K, Chua CC, Nugent MA. Journal of Theoretical Biology. 2005;233:483–499. doi: 10.1016/j.jtbi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 75.Kanodia J, Chai D, Vollmer J, Kim J, Raue A, Finn G, Schoeberl B. Cell Communication and Signaling. 2014;12 doi: 10.1186/1478-811X-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ibrahimi OA, Zhang FM, Hrstka SCL, Mohammadi M, Linhardt RJ. Biochemistry. 2004;43:4724–4730. doi: 10.1021/bi0352320. [DOI] [PubMed] [Google Scholar]
- 77.Shen W, Zhang C, Fannon MW, Forsten-Williams K, Zhang J. Ieee Transactions on Biomedical Engineering. 2009;56:2147–2155. doi: 10.1109/TBME.2008.2002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filion RJ, Popel AS. American Journal of Physiology-Heart and Circulatory Physiology. 2005;288:H263–H279. doi: 10.1152/ajpheart.00205.2004. [DOI] [PubMed] [Google Scholar]
- 79.Zhao B, Zhang C, Forsten-Williams K, Zhang J, Fannon M. Plos Computational Biology. 2010;6 doi: 10.1371/journal.pcbi.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel NS, Reisig KV, Clyne AM. Annals of Biomedical Engineering. 2013;41:154–171. doi: 10.1007/s10439-012-0622-4. [DOI] [PubMed] [Google Scholar]
- 81.Smith AO, Bowers SLK, Stratman AN, Davis GE. Plos One. 2013;8 doi: 10.1371/journal.pone.0085147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stratman AN, Davis MJ, Davis GE. Blood. 2011;117:3709–3719. doi: 10.1182/blood-2010-11-316752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bowers SLK, Meng C-X, Davis MT, Davis GE. Tissue Morphogenesis: Methods and Protocols. 2015;1189:171–189. doi: 10.1007/978-1-4939-1164-6_12. [DOI] [PubMed] [Google Scholar]
- 84.Hellstroem M, Phng L-K, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson A-K, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalen M, Gerhardt H, Betsholtz C. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 85.Jakobsson L, Bentley K, Gerhardt H. Biochemical Society Transactions. 2009;37:1233–1236. doi: 10.1042/BST0371233. [DOI] [PubMed] [Google Scholar]
- 86.Bray SJ. Nature Reviews Molecular Cell Biology. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 87.Dejana E. Nat Rev Mol Cell Biol. 2004;5:261–270. doi: 10.1038/nrm1357. [DOI] [PubMed] [Google Scholar]
- 88.Ribatti D, Nico B, Crivellato E. Int J Dev Biol. 2011;55:261–268. doi: 10.1387/ijdb.103167dr. [DOI] [PubMed] [Google Scholar]
- 89.Gaengel K, Genove G, Armulik A, Betsholtz C. Arterioscler Thromb Vasc Biol. 2009;29:630–638. doi: 10.1161/ATVBAHA.107.161521. [DOI] [PubMed] [Google Scholar]
- 90.Guidolin D, Rebuffat P, Albertin G. ScientificWorldJournal. 2011;11:1735–1748. doi: 10.1100/2011/586475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qutub AA, Mac Gabhann F, Karagiannis ED, Vempati P, Popel AS. IEEE Eng Med Biol Mag. 2009;28:14–31. doi: 10.1109/MEMB.2009.931791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bauer AL, Jackson TL, Jiang Y. Biophys J. 2007;92:3105–3121. doi: 10.1529/biophysj.106.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu G, Qutub AA, Vempati P, Mac Gabhann F, Popel AS. Theoretical Biology and Medical Modelling. 2011;8 doi: 10.1186/1742-4682-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shirinifard A, Gens JS, Zaitlen BL, Poplawski NJ, Swat M, Glazier JA. PLoS One. 2009;4:e7190. doi: 10.1371/journal.pone.0007190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bauer AL, Jackson TL, Jiang Y. PLoS Comput Biol. 2009;5:e1000445. doi: 10.1371/journal.pcbi.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shirinifard A, McCollum CW, Bolin MB, Gustafsson JA, Glazier JA, Clendenon SG. Dev Dyn. 2013 doi: 10.1002/dvdy.23946. [DOI] [PubMed] [Google Scholar]
- 97.Bentley K, Gerhardt H, Bates PA. J Theor Biol. 2008;250:25–36. doi: 10.1016/j.jtbi.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 98.Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, Schulte-Merker S, Gerhardt H. Nat Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- 99.Kuhnert F, Kirshner JR, Thurston G. Vascular Cell. 2011;3:20. doi: 10.1186/2045-824X-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gurney A, Hoey T. Vascular Cell. 2011;3:18. doi: 10.1186/2045-824X-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Long BL, Rekhi R, Abrego A, Jung J, Qutub AA. J Theor Biol. 2013;326:43–57. doi: 10.1016/j.jtbi.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 102.Qutub AA, Popel AS. BMC Syst Biol. 2009;3:13. doi: 10.1186/1752-0509-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jackson T, Zheng X. Bull Math Biol. 2010;72:830–868. doi: 10.1007/s11538-009-9471-1. [DOI] [PubMed] [Google Scholar]
- 104.Bauer AL, Jackson TL, Jiang Y, Rohlf T. J Theor Biol. 2010;264:838–846. doi: 10.1016/j.jtbi.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 105.Bentley K, Mariggi G, Gerhardt H, Bates PA. PLoS Comput Biol. 2009;5:e1000549. doi: 10.1371/journal.pcbi.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qutub AA, Liu G, Vempati P, Popel AS. Pacific Symposium on Biocomputing 2009. 2009:316–327. [PMC free article] [PubMed] [Google Scholar]
- 107.McDougall SR, Anderson AR, Chaplain MA. J Theor Biol. 2006;241:564–589. doi: 10.1016/j.jtbi.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 108.McDougall SR, Anderson ARA, Chaplain MAJ, Sherratt JA. Bull Math Biol. 2002;64:673–702. doi: 10.1006/bulm.2002.0293. [DOI] [PubMed] [Google Scholar]
- 109.Stephanou A, McDougall SR, Anderson ARA, Chaplain MAJ. Math Comput Model. 2005;41:1137–1156. [Google Scholar]
- 110.Plank MJ, Sleeman BD, Jones PF. Journal of Theoretical Biology. 2004;229:435–454. doi: 10.1016/j.jtbi.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 111.Peirce SM, Van Gieson EJ, Skalak TC. FASEB J. 2004;18:731–733. doi: 10.1096/fj.03-0933fje. [DOI] [PubMed] [Google Scholar]
- 112.Arakelyan L, Vainstein V, Agur Z. Angiogenesis. 2002;5:203–214. doi: 10.1023/a:1023841921971. [DOI] [PubMed] [Google Scholar]
- 113.Zheng X, Koh GY, Jackson T. Discrete and Continuous Dynamical Systems - Series B. 2013;18:1109–1154. [Google Scholar]
- 114.Eming SA, Hubbell JA. Experimental Dermatology. 2011;20:605–613. doi: 10.1111/j.1600-0625.2011.01309.x. [DOI] [PubMed] [Google Scholar]
- 115.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quail DF, Joyce JA. Nature Medicine. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baum O, Djonov V, Ganster M, Widmer M, Baumgartner I. Microcirculation. 2005;12:527–537. doi: 10.1080/10739680591003413. [DOI] [PubMed] [Google Scholar]
- 118.Artel A, Mehdizadeh H, Chiu YC, Brey EM, Cinar A. Tissue Eng Part A. 2011;17:2133–2141. doi: 10.1089/ten.tea.2010.0571. [DOI] [PubMed] [Google Scholar]
- 119.Edgar LT, Sibole SC, Underwood CJ, Guilkey JE, Weiss JA. Computer Methods in Biomechanics and Biomedical Engineering. 2013;16:790–801. doi: 10.1080/10255842.2012.662678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Travasso RD, Corvera Poire E, Castro M, Rodriguez-Manzaneque JC, Hernandez-Machado A. PLoS One. 2011;6:e19989. doi: 10.1371/journal.pone.0019989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hashambhoy YL, Chappell JC, Peirce SM, Bautch VL, Mac Gabhann F. Front Physiol. 2011;2:62. doi: 10.3389/fphys.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chappel JC, Cluceru JG, Nesmith J, Walpole J, Peirce SM, Mac Gabhann F. in review. [Google Scholar]
- 123.Karagiannis ED, Popel AS. J Theor Biol. 2006;238:124–145. doi: 10.1016/j.jtbi.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 124.Karagiannis ED, Popel AS. J Biol Chem. 2004;279:39105–39114. doi: 10.1074/jbc.M403627200. [DOI] [PubMed] [Google Scholar]
- 125.Vempati P, Karagiannis ED, Popel AS. J Biol Chem. 2007;282:37585–37596. doi: 10.1074/jbc.M611500200. [DOI] [PubMed] [Google Scholar]
- 126.Vempati P, Mac Gabhann F, Popel AS. PLoS One. 2010;5:e11860. doi: 10.1371/journal.pone.0011860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vempati P, Popel AS, Mac Gabhann F. BMC Syst Biol. 2011;5:59. doi: 10.1186/1752-0509-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, Lauffenburger DA. Integrative Biology. 2011;3:422–438. doi: 10.1039/c0ib00083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen CH, Miller MA, Sarkar A, Beste MT, Isaacson KB, Lauffenburger DA, Griffith LG, Han J. J Am Chem Soc. 2013;135:1645–1648. doi: 10.1021/ja307866z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Morrison CJ, Butler GS, Rodriguez D, Overall CM. Curr Opin Cell Biol. 2009;21:645–653. doi: 10.1016/j.ceb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 131.Ribba B, Saut O, Colin T, Bresch D, Grenier E, Boissel JP. J Theor Biol. 2006;243:532–541. doi: 10.1016/j.jtbi.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 132.Billy F, Ribba B, Saut O, Morre-Trouilhet H, Colin T, Bresch D, Boissel JP, Grenier E, Flandrois JP. J Theor Biol. 2009;260:545–562. doi: 10.1016/j.jtbi.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 133.Carlier A, van Gastel N, Geris L, Carmeliet G, Van Oosterwyck H. PLoS computational biology. 2014;10:e1003888–e1003888. doi: 10.1371/journal.pcbi.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carlier A, Geris L, van Gastel N, Carmeliet G, Van Oosterwyck H. Journal of Theoretical Biology. 2015;365:247–264. doi: 10.1016/j.jtbi.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 135.Carlier A, Geris L, Bentley K, Carmeliet G, Carmeliet P, Van Oosterwyck H. Plos Computational Biology. 2012;8 doi: 10.1371/journal.pcbi.1002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 137.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 138.Hudson N, Powner MB, Sarker MH, Burgoyne T, Campbell M, Ockrim ZK, Martinelli R, Futter CE, Grant MB, Fraser PA, Shima DT, Greenwood J, Turowski P. Developmental Cell. 2014;30:541–552. doi: 10.1016/j.devcel.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stefanini MO, Qutub AA, Mac Gabhann F, Popel AS. Mathematical Medicine and Biology-a Journal of the Ima. 2012;29:85–94. doi: 10.1093/imammb/dqq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kut C, Mac Gabhann F, Popel AS. British Journal of Cancer. 2007;97:978–985. doi: 10.1038/sj.bjc.6603923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rana S, Powe CE, Salahuddin S, Verlohren S, Perschel FH, Levine RJ, Lim K-H, Wenger JB, Thadhani R, Karumanchi SA. Circulation. 2012;125:911–U199. doi: 10.1161/CIRCULATIONAHA.111.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Finley SD, Engel-Stefanini MO, Imoukhuede PI, Popel AS. BMC Syst Biol. 2011;5:193. doi: 10.1186/1752-0509-5-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stoll BR, Migliorini C, Kadambi A, Munn LL, Jain RK. Blood. 2003;102:2555–2561. doi: 10.1182/blood-2003-02-0365. [DOI] [PubMed] [Google Scholar]
- 144.Finley SD, Popel AS. Jnci-Journal of the National Cancer Institute. 2013;105:802–811. doi: 10.1093/jnci/djt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stefanini MO, Wu FTH, Mac Gabhann F, Popel AS. Cancer Research. 2010;70:9886–9894. doi: 10.1158/0008-5472.CAN-10-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tvorogov D, Anisimov A, Zheng W, Leppanen V-M, Tammela T, Laurinavicius S, Holnthoner W, Helotera H, Holopainen T, Jeltsch M, Kalkkinen N, Lankinen H, Ojala PM, Alitalo K. Cancer Cell. 2010;18:630–640. doi: 10.1016/j.ccr.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 147.Vempati P, Popel AS, Mac Gabhann F. Cytokine & Growth Factor Reviews. 2014;25:1–19. doi: 10.1016/j.cytogfr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang XY, Birtwistle MR, Gallo JM. CPT: pharmacometrics & systems pharmacology. 2014;3:e92–e92. doi: 10.1038/psp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Segerstrom L, Fuchs D, Backman U, Holmquist K, Christofferson R, Azarbayjani F. Pediatr Res. 2006;60:576–581. doi: 10.1203/01.pdr.0000242494.94000.52. [DOI] [PubMed] [Google Scholar]
- 150.Willet CG, Boucher Y, Duda DG, di Tomaso E, Munn LL, Tong RT, Kozin S, Petit VL, Jain RK, Chung DD, Sahani D, Kalva SP, Cohen KS, Scadden DT, Fischman AJ, Clark JW, Ryan DP, Zhu AX, Blaszkowsky LS, Shellito PC, Mino-Kenudson M, Lauwers GY. J Clin Oncol. 2005;23:8136–8139. doi: 10.1200/JCO.2005.02.5635. [DOI] [PubMed] [Google Scholar]
- 151.Yang JC, Haworth L, Sherry RM, Hwu P, Schwartzentruber DJ, Topalian SL, Steinberg SM, Chen HX, Rosenberg SA. N Engl J Med. 2003;349:427–434. doi: 10.1056/NEJMoa021491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Imoukhuede PI, Popel AS. Cancer medicine. 2014;3:225–244. doi: 10.1002/cam4.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stefanini MO, Wu FTH, Mac Gabhann F, Popel AS. Plos Computational Biology. 2009;5 doi: 10.1371/journal.pcbi.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mac Gabhann F, Annex BH, Popel AS. Curr Opin Mol Ther. 2010;12:570–577. [PMC free article] [PubMed] [Google Scholar]
- 155.Annex BH. Nature Reviews Cardiology. 2013;10:387–396. doi: 10.1038/nrcardio.2013.70. [DOI] [PubMed] [Google Scholar]
- 156.Ji JW, Mac Gabhann F, Popel AS. Am J Physiol Heart Circ Physiol. 2007;293:H3740–H3749. doi: 10.1152/ajpheart.00009.2007. [DOI] [PubMed] [Google Scholar]
- 157.Mac Gabhann F, Ji JW, Popel AS. J Appl Physiol. 2007;102:722–734. doi: 10.1152/japplphysiol.00800.2006. [DOI] [PubMed] [Google Scholar]
- 158.Wu FTH, Stefanini MO, Mac Gabhann F, Kontos CD, Annex BH, Popel AS. Journal of Cellular and Molecular Medicine. 2010;14:528–552. doi: 10.1111/j.1582-4934.2009.00941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu FTH, Stefanini MO, Gabhann FM, Popel AS. Plos One. 2009;4 [Google Scholar]
- 160.Bender RJ, Mac Gabhann F. Plos One. 2013;8 doi: 10.1371/journal.pone.0061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.