Abstract
Loss of T cell homeostasis usually precedes the onset of AIDS. We hypothesized that rapid progressors may be transmitted with HIV-1 that is particularly able to perturb T cell homeostasis. To this end, we have tested two transmitted, syncytium-inducing (SI) viral isolates from a rapid progressor in two thymus models. One of the isolates (R3A) exhibited markedly rapid kinetics of replication and thymocyte depletion. These phenotypes mapped to the envelope, as a recombinant NL4-3 virus encoding the R3A envelope had similar phenotypes, even in the absence of nef. Notably, the viruses with high pathogenic activity in the thymus (R3A and NL4-R3A) did not show enhanced replication or cytopathicity in PHA-stimulated PBMCs. Furthermore, NL4-R3A did not enhance replication of the coinfected NL4-3 virus in the thymus, suggesting an intrinsic advantage of the R3 A envelope. The R3 A envelope showed higher entry activity in infecting human T cells and in depleting CD4+ thymocytes when expressed in trans. These data suggest that SI viruses with unique envelope functions which can overcome barriers to transmission may hasten disease progression by perturbing T cell homeostasis.
Keywords: HIV-1, Thymus, Replication, Pathogenesis, Envelope, Nef, Intravenous, Transmission, Homeostasis, Progression
Introduction
HIV-1 infection is characterized by progressive loss of CD4+ T cells, loss of T cell homeostasis, and eventual onset of systemic immunodeficiency (Margolick et al., 1995; Tersmette et al., 1989). The rate of disease progression is influenced by both host and viral factors. A strong CTL response, development of neutralizing antibodies, coreceptor polymorphism, HLA heterogeneity, and systemic T cell activation are all host factors shown to impact the kinetics of immune exhaustion and viral submission (Borrow et al., 1994; Letvin and Walker, 2003; Mackewicz et al., 1991; McCune, 2001; Sousa et al., 2002). In the absence of therapy, these host defense mechanisms ultimately fail to control replication and immunodeficiency ensues.
The nature of transmitted virus has also been shown to impact disease progression. A typical subtype B HIV-1 infection involves transmission of a macrophage tropic, CCR5-utilizing isolate across a mucosal or parenteral surface (Markham et al., 1995; Zhu et al., 1993). These transmitted viruses are typically not cytopathic or fusogenic to T cell lines in vitro, and are therefore termed non-syncytium inducing (NSI) (Fauci, 1996). In temporal correlation with the onset of AIDS, emergence of CXCR4-utilizing strains is observed in about 50% of patients and is thought to contribute to accelerated disease progression (Richman and Bozzette, 1994; Tersmette et al., 1989). CXCR4-utilizing variants are typically highly cytopathic and fusogenic in vitro and are thus termed syncytium-inducing (SI) (Fauci, 1996). While it is possible that such variants emerge opportunistically in the context of an exhausted immune system, results from SHIV-infected macaques clearly indicate that CXCR4 variants cause accelerated disease progression due, in part, to altered cellular tropism (Harouse et al., 1999). Furthermore, this disease progression has been correlated with distinct lesions in the thymus of newborn-infected macaques (Reyes et al., 2004). Factors governing the selective disadvantage of CXCR4-tropic viruses early in disease and their eventual emergence remain unclear.
The contributions of viral characteristics to rapid disease progression in humans are not fully understood. While host genetic and immune predisposition play a role, the nature of the transmitted virus could impact disease progression analogous to what is observed in the SHIV macaque model. During a normal HIV infection, CXCR4 variants take years to emerge and often arise before loss of T cell homeostasis and the onset of AIDS (Philpott, 2003). It is possible that variants which are cytopathic and have an enhanced ability to either perturb T cell homeostasis or cause elevated immune activation could cause accelerated disease progression if they are able to overcome intrinsic barriers to transmission (Yu et al., 1998).
The thymus contributes to T cell homeostasis and is responsible for production of naive T cells, particularly early in life. Contrary to expectations, the thymus continues to function throughout life and has shown the capacity to respond to T cell depletion (McCune et al., 1998; Smith et al., 2000). Numerous studies have demonstrated thymic parenchymal damage and involution during HIV-1 infection (Joshi and Oleske, 1985; Papiernik et al., 1992; Pekovic et al., 1987; Rosenzweig et al., 1993; Schuurman et al., 1989). Importantly, HIV infants with impaired thymus functions show accelerated disease progression (Kourtis et al., 1996). It is thus possible that disease progression could be affected by viruses that are particularly able to disturb the thymus, thereby hampering one arm of T cell homeostasis.
The ALIVE IV-drug user cohort identified six rapidly progressing patients who developed AIDS within 5 years of seroconversion (Yu et al., 1998). Three of these rapid progressors lost T cell homeostasis almost immediately after infection and were uniquely transmitted with SI viruses that were both macrophage and T cell tropic (Yu et al., 1998). To better understand the role of transmitted virus in rapid disease progression, we have characterized two transmitted SI HIV-1 isolates obtained from one rapid progressor in this cohort who demonstrated an early failure of T cell homeostasis. The viruses were obtained at the time of seroconversion and were previously shown to be macrophage-tropic and SI in vitro. We hypothesized that HIV-1 variants from this patient may be particularly effective at perturbing T cell homeostasis, thereby hampering the immune system’s ability to delay the onset of immunodeficiency. To test this hypothesis, we characterized the replication and pathogenesis of the isolates in PBMCs in vitro and in two human thymus models. We report that both isolates can utilize CCR5 and CXCR4 and are capable of efficient replication and CD4+ T cell depletion in PBMCs. One of the isolates, however, shows astounding levels of replication and thymocyte depletion in the thymus. The thymus-tropism of this variant maps to the envelope gene, which functions in the absence of nef, and intrinsically promotes viral replication in the thymus. Furthermore, this envelope shows an enhanced ability to mediate infection of the thymocyte-like SupT1 cell line and is efficient at depleting CD4+ primary human thymocytes when expressed in trans.
Results
Two early isolates from a rapid progressor are dual tropic, infect macrophages, and show normal kinetics of replication in stimulated PBMCs
Three of the rapid progressors (R1–3) from the ALIVE IV-drug user cohort with early loss of T cell homeostasis were uniquely transmitted with SI variants capable of replication in both macrophages and T cells (Yu et al., 1998). We obtained two different isolates (R3A and R3B) from one of these rapid progressors (R3) which were generated via coculture of patient PBMCs isolated at the time of seroconversion with PHA-stimulated allogenic PBMCs (Yu et al., 1998). Thus, the isolates are representative of the transmitted virus. We first confirmed macrophage tropism using peripheral blood monocyte-derived macrophages (Table 1). Using U373-Magi cells expressing CD4 and either CCR5 or CXCR4, we found that R3A and R3B could use both CXCR4 and CCR5 as coreceptors (Table 1). Both isolates also showed sensitivity to the CXCR4-specific inhibitor AMD3100 in PBMCs (Table 1 and data not shown). In addition, we analyzed their ability to use alternative chemokine receptors in vitro and found that neither R3A nor R3B was able to significantly use CCR1, CCR2b, CCR3, CCR4, V28, Bob, Bonzo, or CCR8 (data not shown).
Table 1.
HIV-1 isolates from a rapid progressor (R3) in the ALIVE cohort
MDM = monocyte derived macrophages.
Determined by infection of U373 cells expressing CD4 and CCR5 or CXCR4, and sensitivity to CXCR4- and CCR5-specific inhibitors in PBMCs and HF-TOC.
Standard laboratory reference strain used for comparison.
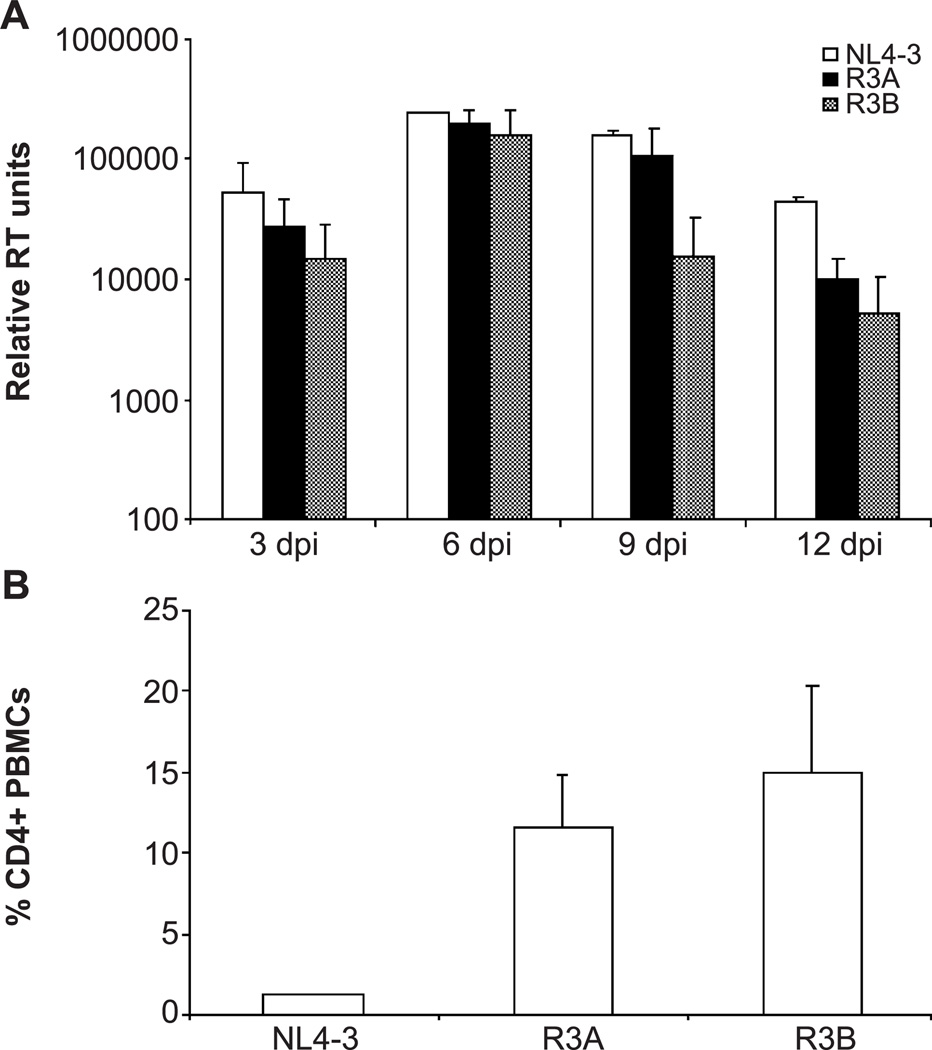
We next compared the replication kinetics and cytopathicity of R3A and R3B in PHA-activated PBMCs to the standard CXCR4-utilizing NL4-3 virus (Table 1 and Fig. 1). R3A and R3B replicated comparably to NL4-3 over a 2-week time course (Fig. 1A). Fourteen days post-infection, we measured the extent of CD4+ T cell depletion by FACS analysis and found that R3A and R3B were less capable of depleting CD4+ cells than NL4-3 (Fig. 1B), corresponding with slightly lower viral replication at late time points (Fig. 1A). Therefore, neither R3 isolate shows enhanced replication or cytopathicity when observed in the context of PHA-stimulated PBMCs relative to a typical CXCR4-utilizing virus or to 89.6 and 320.3.1 (Groenink et al., 1992), two other primary dual-tropic isolates (data not shown).
Fig. 1.
Replication and pathogenesis of the R3 isolates in activated PBMCs. (A) An equal MOI of virus was used to infect 10 million PHA-stimulated PBMCs. Supernatant was harvested every 3 days and quantitated by reverse transcriptase (RT) assay. Units for RT activity are derived from densitometric quantification and are arbitrary. Standard deviation was calculated from duplicate samples. Shown is a representative example from three independent experiments. (B) Depletion of CD4+ cells 14 days after infection. Cells were harvested and stained for CD4 and CD8. The percentage of cells gated live by scatter profiles which were CD4+ is shown. Shown is a representative experiment with duplicate samples.
R3A exhibits enhanced replication and cytopathicity in the human thymus
Unlike in vitro stimulated PBMCs, lymphoid micro-environments in vivo are composed of multiple cell types and more quiescent cell populations that require unique viral determinants for HIV replication and pathogenesis (Meissner et al., 2003; Su et al., 1997). To test if HIV-1 isolates from this rapid progressor may be particularly capable of replication and depletion of CD4+ cells in these environments, we characterized the R3A and R3B isolates using the human fetal-thymus organ culture (HF-TOC) and SCID-hu Thy/Liv mouse models.
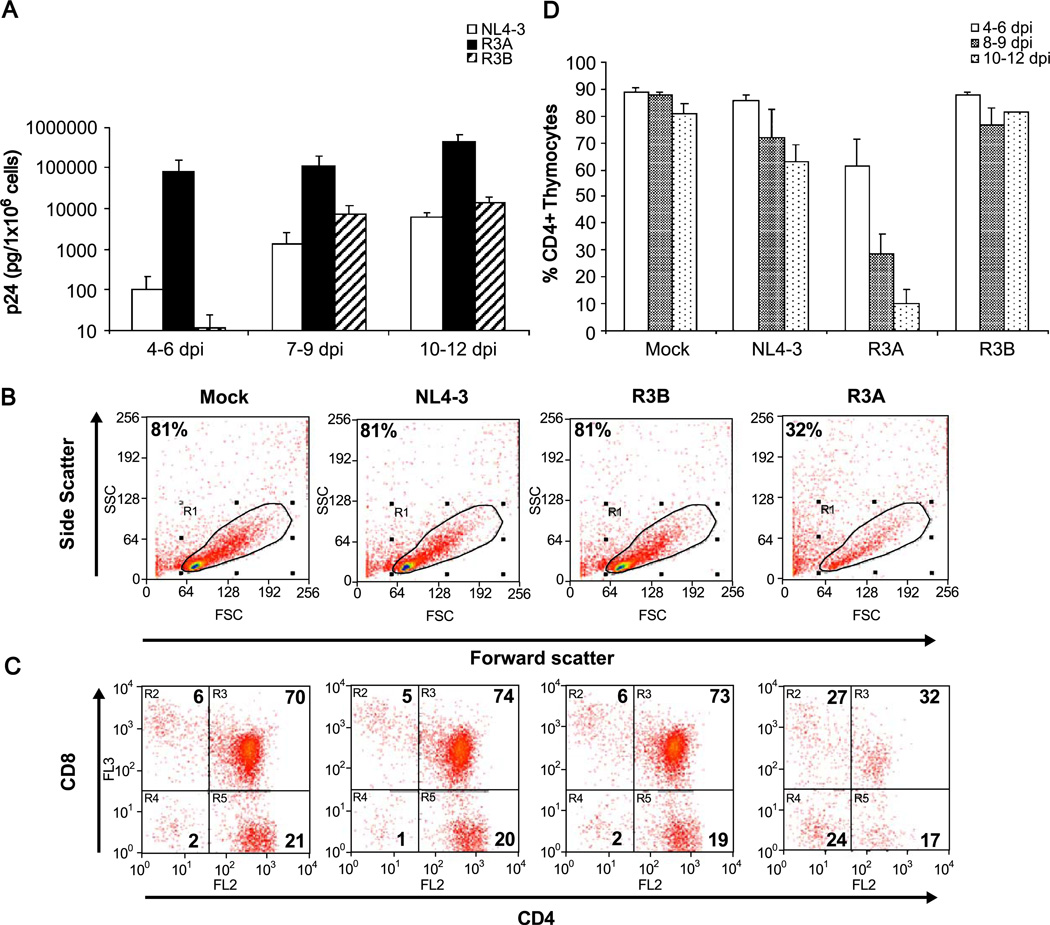
HF-TOC fragments were infected with 100 infectious units of R3A, R3B, NL4-3, or mock supernatant and were monitored for HIV replication and thymocyte depletion over 2 weeks. The R3B virus achieved levels of replication that were comparable to the standard NL4-3 strain. Surprisingly, R3A showed reproducibly higher levels of replication than both NL4-3 and R3B throughout the culture period (Fig. 2A). Pathogenesis was assessed by CD4 and CD8 FACS analysis to determine the extent of viral-mediated CD4 cell depletion (Figs. 2B–D). NL4-3 and R3B showed similar levels of depletion of CD4+ thymocytes relative to mock-infected fragments. In contrast, R3A induced almost complete depletion of CD4+ cells by 12 dpi, with significant depletion observable only 4–6 dpi. The percentage of cells in the live gate defined by scatter profiles paralleled depletion observed in the CD4+ population (Fig. 2B and data not shown).
Fig. 2.
Replication and cytopathicity of the R3 isolates in HF-TOC. In each experiment, fetal thymus fragments were infected with mock supernatant, NL4-3, R3A, or R3B. (A) Thymocytes were isolated and cell-associated p24 was measured by ELISA. The results from at least three independent experiments are summarized. (B–C) Isolated thymocytes were stained for CD4 and CD8 and gated by scatter profiles. A representative FACS plot showing pathogenesis mediated by R3A at 9 days post-infection relative to mock, NL4-3, and R3B is shown. Shown are the percentage of cells gated live or in each quadrant of CD4/ CD8 staining. (D) Pathogenesis of the R3 isolates was summarized by measuring total CD4+ thymocyte depletion. The total percent CD4+ cells was calculated by adding the % CD4+CD8+ immature thymocytes with the % CD4+CD8- thymocytes of cells gated live by forward and side scatter. The results from at least three independent experiments are summarized.
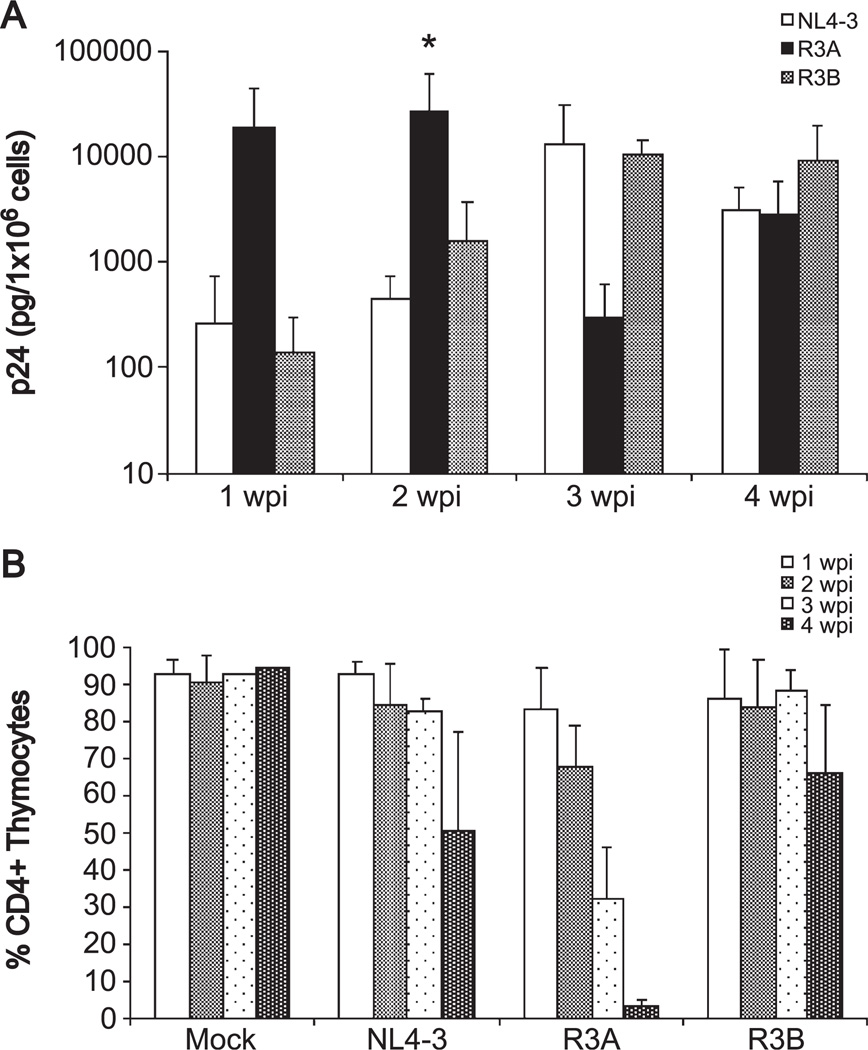
To verify these results, we assessed the replication and pathogenesis of the R3 isolates in the SCID-hu Thy/Liv model. SCID-hu Thy/Liv mice were infected with an equivalent amount of R3A, R3B, NL4-3, or mock supernatant. Thymocytes were harvested by biopsy each week for 4 weeks after infection, and the level of thymocyte-associated p24 was measured by ELISA. As in HF-TOC, R3B replicated comparably to the reference NL4-3 strain, while R3A replicated to extremely high levels at 1 and 2 weeks post-infection (Fig. 3A). When replication was normalized to levels of the reference strain NL4-3 within each experiment, R3A showed significantly higher replication than NL4-3 and R3B at 2 weeks post-infection (P < 0.05 by the Student’s t test, data not shown). The decline in viral load of R3A observed at 3 and 4 weeks post-infection was likely due to depletion of CD4+ thymocytes.
Fig. 3.
Replication and cytopathicity of the R3 isolates in SCID-hu Thy/Liv mice. In each experiment, three mice were infected with mock supernatant, NL4-3, R3A or R3B. (A) Thymocytes were harvested by biopsy each week for 4 weeks after infection and assessed for HIV replication by quantifying cell-associated p24 by ELISA. The combined results of three independent experiments are shown. When replication was normalized to NL4-3 within each experiment (data not shown), R3A showed a statistically significant increase in replication relative to R3B and NL4-3 at 2 wpi (*P < 0.05). (B) Pathogenesis of the R3 isolates was assessed by measuring CD4+ thymocyte depletion as in Fig. 2.
As shown in Fig. 3B, the depletion of thymocytes followed the peak of replication in the Thy/Liv organ. NL4-3 and R3B caused thymocyte depletion at 4 wpi, following their peak replication at 3 wpi. R3A, consistent with its earlier peak at 1–2 wpi, caused early and significant depletion at 2–3 wpi, with almost complete ablation of CD4+ thymocytes by four wpi. These results collectively indicate that the R3A isolate, which replicates normally in PBMCs with attenuated depletion of CD4+ cells, shows tropism for the in vivo thymic microenvironment with a striking ability to replicate in and deplete CD4+ thymocytes relative to other SI isolates.
The R3 envelopes recapitulate the replication and cytopathic phenotype of the parental R3 viruses in the absence of nef
While both R3A and R3B demonstrated the ability to replicate in the thymus, R3A was clearly unique in achieving the highest levels of replication and thymocyte depletion. We therefore sought to define the viral determinants responsible for this phenotype. We hypothesized that distinct features of the envelope may be responsible and therefore cloned and characterized the envelope from both R3A and R3B using PCR of genomic DNA extracted from the first passage of R3A or R3B-infected PBMCs in our laboratory (2nd passage overall). PCR products were subsequently cloned into a retroviral expression vector. One R3B and eight R3A clones were obtained.
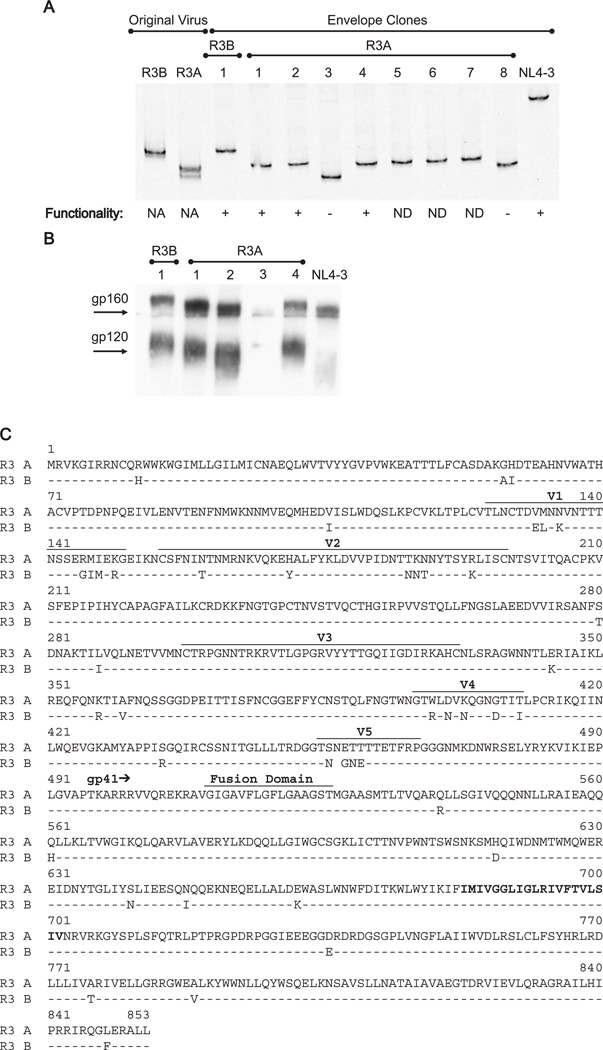
To assess whether the envelope clones were representative of the original viral isolates, we performed a heteroduplex tracking assay (HTA) using a radiolabeled probe homologous to a portion of the JR-FL V1/V2 region (Kitrinos et al., 2003). Relative migration of heteroduplexes is determined by the degree of homology between the query and probe sequence. The parental R3B isolate was found to contain one major variant and R3A to contain one major and one minor variant (Fig. 4A). HTA performed on the envelope clones verified that each major variant was represented by at least one clone.
Fig. 4.
Cloning and characterization of the R3 envelopes. (A) HTA analysis of the parental viruses and the clones obtained by nested PCR. PBMCs infected with the R3 viruses were used for nested PCR and products were cloned into a retroviral vector. PCR was performed on the V1/V2 region of the original viruses or of the clones. Resulting products were hybridized to the radiolabeled V1/V2 probe from the JR-FL virus. Heteroduplexes were visualized on polyacrylamide sequencing gels. Envelope functionality was determined by infecting SupT1 cells with pseudotyped virus. NA = not applicable, ND = not done. (B) Expression and processing of the envelope clones were assessed after transfection into A293T cells and probing of lysates with a polyclonal anti-gp120 antibody. (C) Sequence of the R3A and R3B envelopes. The V1-V5 regions and the fusion domains are indicated and the transmembrane domain of gp41 is shown in bold. The space in the V5 region of the R3B sequence indicates a gap.
The clones were next tested for envelope expression and processing. As shown in Fig. 4B, most clones produced the gp160 and gp120 envelope proteins, while several clones showed little (R3A #3) or abnormal (R3A #8) expression (Fig. 4B and data not shown). Clones 1 and 4 for R3A appeared identical by Western, while clone 2 appeared smaller, in spite of similar migration by HTA, suggesting the presence of different isoforms within the R3A population, which may vary by glycosylation sites.
The cloned envelope genes were next tested for function using a pseudotyping assay. NL4-luciferase pseudotyped with each env clone was used to infect the SupT1 cell line and productive infection was assessed by luciferase assay 48 h later. Each clone encoding a major isoform by HTA proved functional for infection in the pseudotyping assay, while the two misexpressing R3A clones (#3, #8) with faster HTA migration, possibly corresponding to the minor variant, proved non-functional for entry (Fig. 4A). We therefore obtained at least one functional clone for each R3 isolate that represented the major variant of the parental virus by HTA.
Clone 1 for R3B (GenBank accession no. AY608576) and clone 4 for R3A (GenBank accession no. AY608577) were chosen for sequence analysis based on HTA migration and functionality (Fig. 4C). We chose clone 4 as a representative species of R3A because it represented two of three functional clones obtained. Both chosen envelopes were capable of mediating infection via CCR5 and CXCR4 like the parental viruses in U373 cells (data not shown). As expected, the R3A and R3B envelope sequences were found to be extremely similar to each other. Their greatest variation in sequence was in the variable regions, and the slower migration of the R3B envelope by Western was consistent with its higher number of potential N-linked glycosylation sites relative to R3A (26 vs. 25 in gp120). Given their distinct phenotypes in the thymus, we were surprised that the V3 loop of R3A and R3B are identical in amino acid sequence. In contrast, the V1/V2 regions showed significant differences consistent with the difference observed in the HTA. Additionally, no differences were observed in the fusion domain, transmembrane domain, or in amino acids shown to contribute to the CD4 binding site (Kwong et al., 1998). The tat sequence in the overlapping region was identical for the R3A and R3B clones, while the rev sequences differed by three amino acids. The R3A and R3B vpu proteins differed at 4 amino acids which are known to be variant, while the major known functional motifs (McCormick-Davis et al., 2000) were conserved (data not shown).
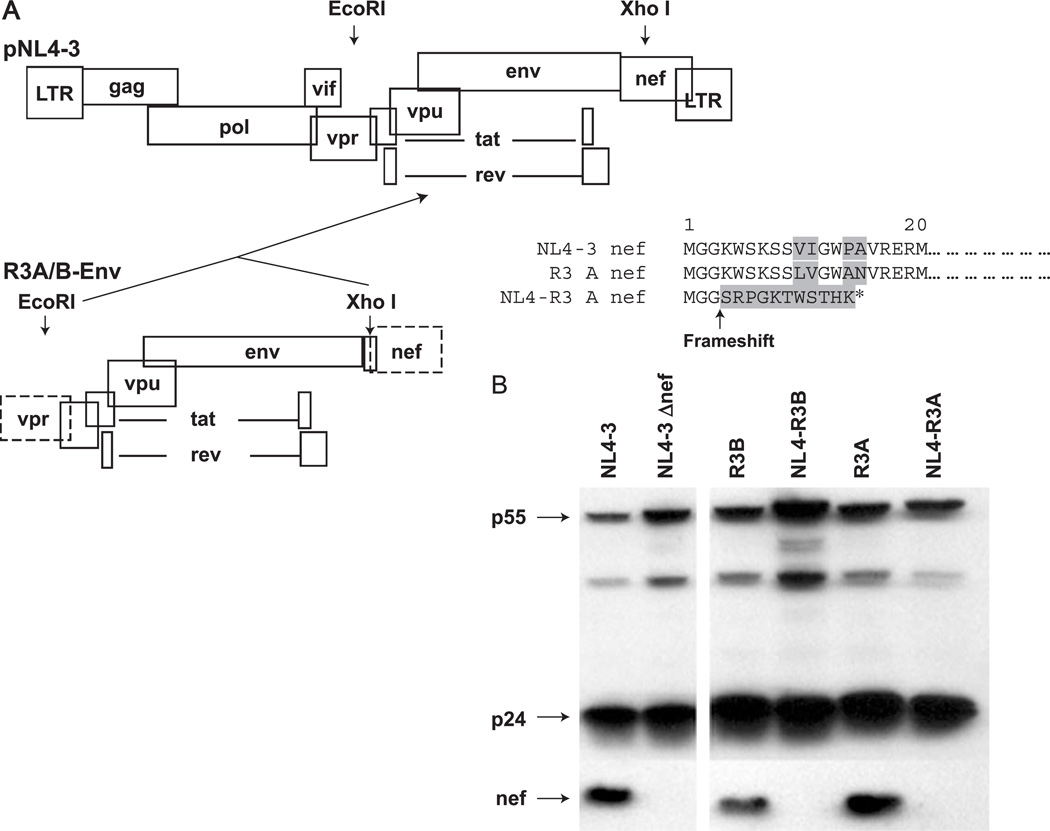
To determine whether the envelope of each R3 virus was responsible for the phenotype of the parental virus in the thymus, we tested the functional, sequenced R3A and R3B envelopes in the background of the NL4-3 proviral genome. As a result of the cloning strategy (Fig. 5A), nef was deleted from each recombinant virus, allowing for a strict characterization of envelope-dependent replication and pathogenesis. While both original R3A and R3B isolates express nef, neither of the NL4 recombinants (NL4-R3A and NL4-R3B) expresses nef due to the deletion and frameshift introduced during cloning (Figs. 5A–B).
Fig. 5.
Generation of NL4-R3 envelope recombinant viruses with deletion of the nef gene. (A) The env gene was digested with EcoR1 and Xho1, which was added directly at the end of the env ORF during cloning. Ligation of the Env fragment into the endogenous NL4-3 Xho1 site creates a deletion, resulting in a frameshift that eliminates expression of nef. (B) Infected PBMCs were lysed and probed by Western for gag and nef expression. While the parental R3 viruses encode nef, the recombinant NL4-R3 viruses do not.
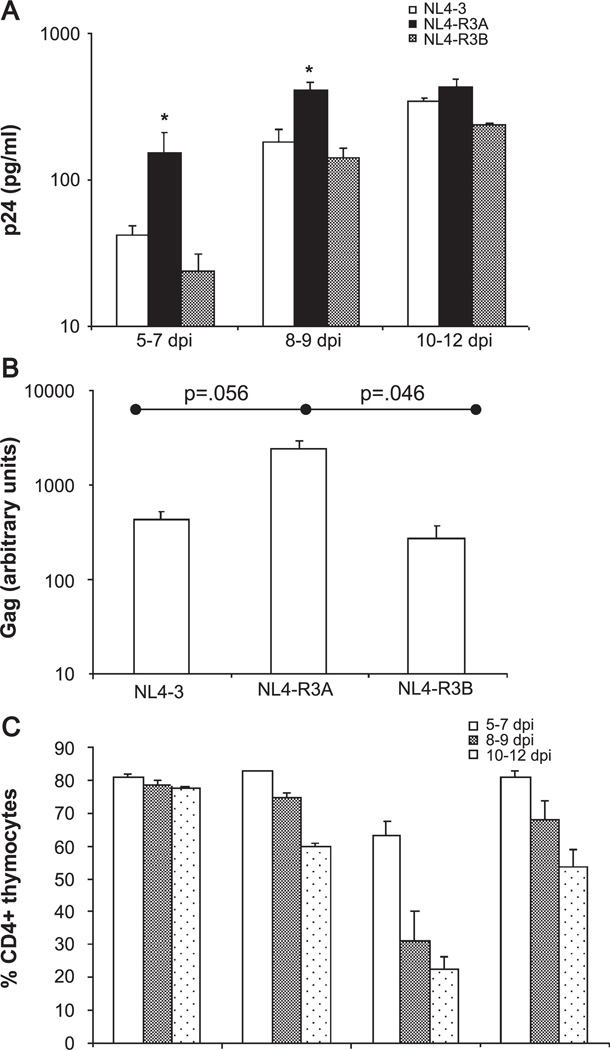
Replication and cytopathicity of the recombinants were first assessed in PHA-stimulated PBMCs. Like the parental viruses, NL4-R3A and NL4-R3B replicated comparably to NL4-3 and NL4-Δnef, and showed reduced depletion of CD4+ cells relative to NL4-3 (data not shown). Replication and pathogenesis of the recombinants in the thymus were then characterized in HF-TOC as above. As shown in Fig. 6A, replication of the recombinants NL4-R3B and NL4-R3A in HF-TOC was similar to the parental viruses R3B and R3A, respectively. NL4-R3B was found to replicate similarly to NL4-3, while NL4-R3A showed higher levels of replication. NL4-R3A showed significantly enhanced replication relative to NL4-3 and NL4-R3B at days 5–7 and days 8–9 when replication was normalized to NL4-3 replication (P < 0.05, data not shown). The high level of replication measured by p24 ELISA was paralleled by a greater extent of integrated HIV proviral DNA, as assessed by real time PCR of gag DNA 5 days post-infection (Fig. 6B). Finally, depletion of CD4+ thymocytes by the recombinants closely paralleled that observed with the parental viruses, as NL4-R3A caused rapid depletion of total CD4+ cells (Fig. 6C). This decrease in percent CD4+ cells was paralleled by a decrease in total cell number and the percent of cells gated live by scatter profiles (data not shown). In summary, the enhanced replication and cytopathicity of the R3A isolate can be accounted for by its unique envelope even in the absence of nef.
Fig. 6.
NL4-R3A shows enhanced replication and cytopathicity in HF-TOC. (A) Replication of NL4-3, NL4-R3A, and NL4-R3B in HF-TOC was measured by p24 ELISA of TOC supernatant. Shown are the combined results from four independent experiments. When replication was normalized to NL4-3 within each experiment (data not shown), NL4-R3A showed a statistically significant increase in replication relative to NL4-R3B and NL4-3 at 5–7 and 8–9 dpi (*P < 0.05 by the Student’s t test). (B) Increased replication of NL4-R3A as revealed by real time PCR analysis for proviral DNA. Thymocytes were harvested 5 days post-infection and DNA lysates was isolated. Real time PCR for the actin gene was used for normalization and a representative experiment is shown. Results are from one of four experiments with error bars derived from duplicate samples. (C) Pathogenesis was assessed as in Fig. 2. Shown are the cumulative results of four independent experiments.
Our in vitro studies suggested that the R3A and R3B envelopes were both capable of using CXCR4 and CCR5 to infect the U373 cell line. To address whether these coreceptors are partially or exclusively used in HF-TOC, we performed an infection in the presence of a CXCR4 (AMD3100) and/or a CCR5 antagonist (Tak-779). HF-TOC fragments were pretreated with AMD3100 and/or Tak-779 before infection and were replenished every other day for 7 days, at which time p24 replication was measured by ELISA. Either AMD3100 or Tak-779 inhibited replication of both R3A and R3B in HF-TOC. In addition, replication was cooperatively reduced (by at least 80%) for both NL4-R3A and NL4-R3B when both AMD3100 and Tak-779 were used (data not shown). These data suggest that both CXCR4 and CCR5 are used by the R3A and R3B envelopes in the thymus. Because complete blockage of viral infection was not observed, it is possible that the doses of coreceptor antagonist used were insufficient to block viral replication completely. Additionally, it remains possible that R3B and/ or R3A could use an alternate coreceptor in the thymus in addition to CXCR4 and CCR5, even though no additional known coreceptor usage was detected in our in vitro experiments.
The R3A envelope shows intrinsic, hyper-replicative activity in the thymus
As noted above, the R3 viruses and the NL4 recombinants did not show enhanced replication or cytopathicity when observed in activated PBMCs. However, R3A and NL4-R3A appeared hyperactive in replication and pathogenesis within the thymus. To test if the R3A envelope does intrinsically better in the thymus, independent of the secondary stimulatory events the R3A envelope may induce, we performed a series of competition experiments in both PBMCs and HF-TOC.
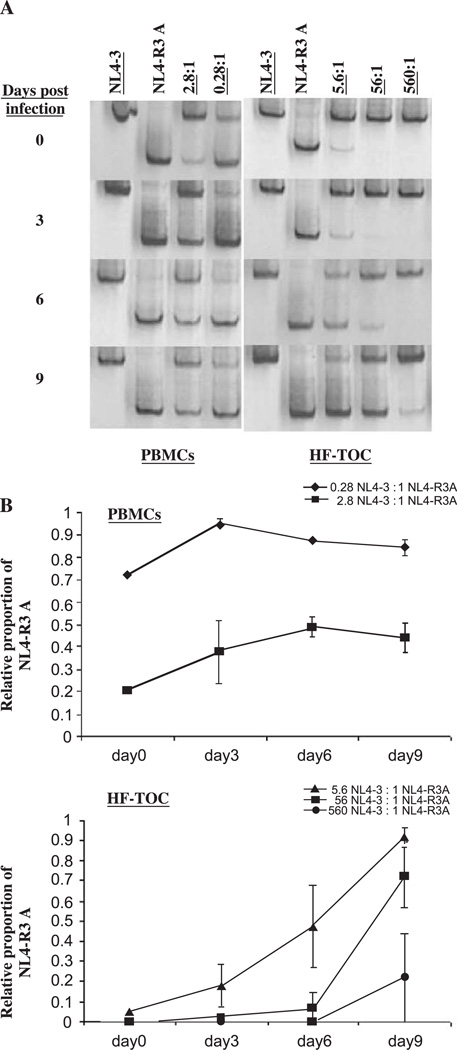
PBMCs and HF-TOC were infected with NL4-3 alone, NL4-R3A alone, or coinfected with a mixture of NL4-3 and NL4-R3A in various ratios as measured by virion-associated reverse transcriptase activity (Fig. 7). In the thymus, the most extreme competition gave NL4-3 a 560 to 1 starting advantage over NL4-R3A. Supernatant was harvested at 0, 3, 6, and 9 days post-infection and the relative amount of each virus was assessed by HTA followed by densitometric quantification (Figs. 7A–B). As seen in Fig. 7A, the relative NL4-R3A and NL4-3 levels were stable in coinfected PBMCs, indicating their similar replicative capacity. In stark contrast, NL4-R3A was able to efficiently outcompete NL4-3 in HF-TOC when applied at a 1:5.6 disadvantage. NL4-R3A even recovered when applied at a 1:560 disadvantage. Results from a similar competition experiment between NL4-R3B and NL4-R3A yielded similar results in HF-TOC with NL4-R3A outcompeting NL4-R3B (data not shown). These competition experiments clearly reveal that the R3A envelope is selectively capable of efficient replication within the context of the thymic microenvironment, an advantage that is not observed in activated PBMCs. In addition, these data indicate that the R3A envelope functions intrinsically to enhance NL4-R3A replication, but not the co-infected NL4-3 or NL4-R3B, in the thymus.
Fig. 7.
NL4-R3A is able to outcompete NL4-3 in the thymus, but not in activated PBMCs. (A) NL4-3 and NL4-R3A mixed in differing ratios were used to inoculate activated PBMCs and HF-TOC fragments. Viral supernatant was harvested every three days and the relative proportion of NL4-R3A to NL4-3 was determined by HTA (total proportion =1). Underlined numbers represent the relative amount of NL4-3 added. (B) The bands in (A) were quantitated by densitometry. The relative proportion of NL4-R3A in the total viral population (NL4-3 + NL4-R3A) in each sample was calculated. Error bars represent data from infections performed in duplicate.
The R3A envelope allows enhanced infection of SupT1 cells and demonstrates cytopathicity towards primary human CD4+ thymocytes when expressed in trans
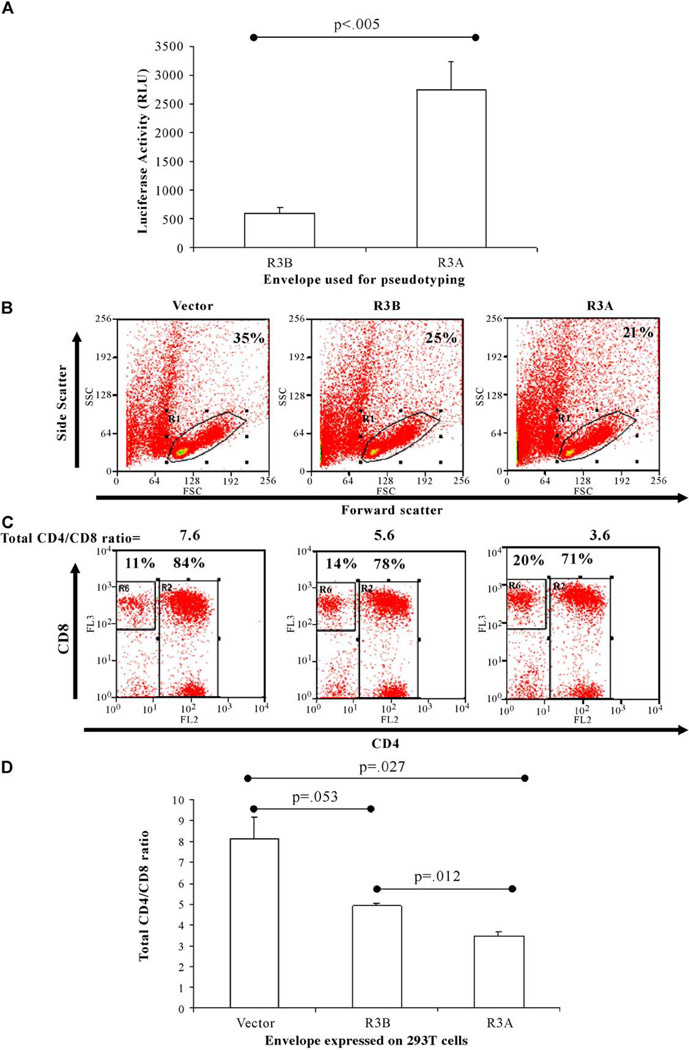
To address the potential mechanism of elevated replication and cytopathicity of the R3A envelope, we used NL4-luciferase pseudotyped with R3B or R3A envelope and infected SupT1 cells to measure their relative ability to mediate viral entry. When equal amounts of total virion were used, we found that R3A-pseudotyped virus was significantly more efficient at mediating entry in SupT1 cells than R3B-pseudotyped virus (Fig. 8A). This may explain, in part, how the R3A envelope is capable of achieving enhanced replication in the thymus.
Fig. 8.
The R3A envelope mediates enhanced entry into SupT1 cells and enhanced cytopathicity towards primary human thymocytes when expressed in trans. (A) NL4-luc was pseudotyped with either envelope and virus was used to infect SupT1 cells. Cells were lysed at 72 h post-infection and luciferase expression was measured. Shown is one of three experiments with infections performed in triplicate. Luciferase expression was normalized to input p24 (total virions). (B–D) 293T cells transfected with vector or an envelope construct were irradiated 24 h post-transfection and cocultured with freshly isolated human fetal thymocytes. Forty-eight hours post-coculture, cells were harvested and analyzed by FACS analysis. (B) Forward and side scatters were used to gate on live thymocytes, with the percentage of cells in the live gate shown. (C) Cytopathicity for CD4+ thymocytes is expressed as the ratio of total CD4+ thymocytes (CD4+CD8+ and CD4+CD8−) to CD4−CD8+ thymocytes. The percentages in each gate and the ratio are shown. (D) Shown is one of four experiments. Error bars represent standard deviation from samples run in duplicate.
We next used an in vitro coculture system to assess a possible mechanism for the enhanced cytopathicity of the R3A envelope. Expression of each envelope construct or a vector in 293T cells, followed by coculture with primary human thymocytes, allowed us to address the effect of each envelope when exposed in trans. We measured depletion of total CD4+ thymocytes (including CD4+CD8+ and CD4+CD8− thymocytes) 2 days after coculture relative to CD4−CD8+ thymocytes, which are relatively unaffected by envelope exposure (Figs. 8B–D). While R3B was capable of depleting CD4+ cells relative to vector-transfected cells, R3A was significantly more efficient in this depletion. Together, these data suggest that enhanced entry and/or fusion of the R3A envelope in thymocytes may contribute to its cytopathic phenotype. It will be interesting to determine whether the ability to enhance viral entry and the ability to deplete CD4+ thymocytes in trans are linked or are separable functions of the R3A envelope.
Discussion
Loss of T cell homeostasis evidenced by decreasing CD4 and CD8 T cells is strongly associated with the onset of AIDS (Margolick et al., 1995). To help maintain T cell homeostasis, the thymus functions throughout adult life to populate the periphery with naive CD4 and CD8 T cells (Douek et al., 1998). Furthermore, thymic production of naive cells is impaired in HIV-1 patients and suppression with HAART therapy allows a partial recovery of this production (Douek et al., 1998). In the ALIVE IV-drug user cohort, 3 of 105 seroconverters showed rapid loss of T cell homeostasis and progression to AIDS within 5 years, associated with transmission of SI HIV isolates (Yu et al., 1998). We hypothesized that these rapid progressors may be transmitted with isolates that are particularly capable of perturbing T cell homeostasis, including thymic potential. Herein, we characterize two independent isolates from a rapid progressor and identify a unique isolate, which shows a striking ability to selectively replicate in and deplete CD4+ thymocytes. Furthermore, this phenotype maps to the viral envelope and is observable in the absence of nef.
Both isolates from the rapid progressor could utilize both CXCR4 and CCR5. CXCR4-utilizing variants are usually not transmitted and typically emerge late in disease in correlation with the onset of AIDS. In cases where transmission of CXCR4 variants does occur, these variants are capable of productively infecting macrophages (Koning et al., 2001; Naif et al., 2002). Consistent with these findings, the isolates from patient R3 showed macrophage tropism in addition to their ability to use CXCR4 (Table 1 and (Scarlatti et al., 1997; Yi et al., 2003)). Because the R3 viruses can use CCR5 and CXCR4, it is unsurprising that these isolates should be capable of replication within the CXCR4-rich human thymus (Camerini et al., 2000; Glushakova et al., 1998; Taylor et al., 2001). However, our most intriguing finding was the identification of the highly thymus-tropic R3A isolate. This strain was particularly capable of replication and depletion within the thymus, an advantage that was not apparent when characterized relative to other strains in stimulated PBMCs or in the tonsil-lymph node organ culture (data not shown). Both stimulated PBMCs and the lymph node organ culture are permissive for replication by the tissue-culture adapted HXB2 virus, while the thymus is not, demonstrating the quiescent nature of the thymic microenvironment (Glushakova et al., 1998; Su et al., 1995). Additionally, R3A is unique because other X4/R5 dual tropic isolates which emerged during HIV infection do not show such enhanced pathogenesis in the thymus. For example, 89.6 replicated inefficiently in the thymus (unpublished observation) and 320.3–1, another primary X4/R5 isolate which emerged during the transition from R5 to X4 utilizing-variants (Groenink et al., 1992), demonstrates NL4-like levels of replication and pathogenesis in the thymus like R3B (unpublished observation). This suggests that R3A is particularly capable of efficient replication and pathogenesis within the thymus microenvironment, potentially including cell-to-cell spread. It will be interesting to assess whether R3A–like envelope species can be found in other rapid progressors with early loss of T cell homeostasis, or from patients with standard disease progression around the time of loss of T cell homeostasis.
The phenotype of the R3A virus was mapped to its envelope gene. While several different envelopes were obtained from the R3A isolate, the one chosen for analysis appears representative based on HTA analysis, functionality in vitro, and on its phenotype in the thymus. Although we are unable to completely rule out a unique effect of the R3A vpu or rev genes within the NL4-R3A recombinant, the R3A sequences were highly similar to the NL4-3 and R3B vpu and rev genes (data not shown).
Nef has previously been shown to be required for optimal replication and pathogenesis of the NL4-3 and JR-CSF isolates in the thymus (Jamieson et al., 1994). We have previously reported that a CXCR4-utilizing primary envelope which has evolved in vivo from the lab-adapted HXB2 strain is sufficient to allow replication in the thymus in the absence of nef, but with attenuated cytopathicity (Miller et al., 2001). Furthermore, we demonstrated that restoring nef in the virus enhances cytopathic activity without affecting its replication levels (Duus et al., 2001). We now demonstrate that an envelope without nef is capable of achieving astounding levels of replication and pathogenesis in the thymus. This suggests that inherent properties of the envelope, independent of coreceptor usage, can dramatically affect the success of viral replication and pathogenesis in the thymus organ. Furthermore, we demonstrate that coinfection of the thymus with NL4-R3A and NL4-3 led to efficient outgrowth of NL4-R3A and did not enhance NL4-3 replication. This suggests that inherent properties of the R3A envelope are responsible for efficient replication in the thymus, rather than a generally altered cytokine milieu.
To address the possible mechanism for the phenotype of the R3A envelope in the thymus, we performed several in vitro experiments to uncover any advantage the R3A envelope may provide for enhancing either replication or cytopathicity. We demonstrate that the R3A envelope allows for more efficient entry than R3B in the thymocyte-like SupT1 cell line using pseudotyped virus (Fig. 8A). To assess cytopathicity, we attempted to express envelope within thymocytes, but were unable to achieve sufficient levels of transduction with our retroviral envelope expression vector (data not shown). However, we did find that R3A is more cytopathic to CD4+ thymocytes when expressed in trans in a coculture experiment (Fig. 8B). When infectivity and cytopathicity were assessed in stimulated PBMCs, R3A was found to have a similar advantage over R3B, suggesting that this advantage is not specific to thymocytes in vitro (data not shown). These data point to an intrinsic advantage for the R3A envelope which may allow for more efficient or extensive interaction with T cells relative to the R3B envelope. Studies are ongoing to further elucidate how intrinsic properties of the R3A envelope may allow for its success in the thymus. In particular, we are interested in whether the R3A envelope shows an enhanced ability to mediate cell to cell transmission or activation of relatively quiescent thymocytes, two of the most obvious means by which the stimulated PBMC environment differs from the thymus.
The R3A and R3B env genes are highly similar in sequence yet show distinct phenotypes, despite both being primary, SI isolates from the same patient. The exact relationship of these two viruses from the perspective of evolution within the patient is unclear, although both were isolated from PBMCs obtained around seroconversion. Of particular interest is that the envelopes have identical V3 loops, can both utilize CXCR4 and CCR5, and do not appear utilize any of the alternative HIV coreceptors (Table 1 and data not shown). Study of these two envelopes should allow for fine dissecting of the region and mechanisms of enhanced HIV replication and pathogenesis in the thymus.
Materials and methods
Viral isolates and HIV production
The two R3 viruses were obtained as previously described (Yu et al., 1998). Both R3A and R3B were derived from cryopreserved blood obtained from patient R3 around the time of seroconversion.
The second PBMC passage of each R3 virus was amplified for further study using PHA-stimulated PBMCs cultured in 10 U/ml IL-2, with supernatant changed and collected daily as previously described (Su et al., 1995). For NL4-3 and the NL4-R3 recombinants, an equal amount of proviral DNA was transfected into A293T cells using Effectene (Qiagen) per the manufacturer’s protocol and the supernatant was used to infect PHA-stimulated PBMCs. Infectious titers were determined by MAGI assay using U-373-MAGI-CXCR4 glioblastoma cells as previously described (Miller et al., 2001; Vodicka et al., 1997). Reverse transcriptase activity for the competition experiments was quantitated using a previously described P33 based incorporation assay (Resch et al., 2002).
Coreceptor usage
Primary coreceptor usage was determined using U-373-MAGI cells expressing either CCR5 or CXCR4. To test for alternative coreceptor usage, we infected GHOST cells expressing either CCR1, CCR2b, CCR3, CCR4, CXCR4, Bob, Bonzo, V28, CCR8, or CCR5 with high titer virus (Morner et al., 1999). Forty-eight hours post-infection, cells were analyzed by flow cytometry (FACS) for GFP expression. While GFP+ cells were readily detectable using both CXCR4 and CCR5 expressing cells, we saw no significant signal above background when GHOST cells expressing the other chemokine receptors were used. For blocking experiments in the human thymus using AMD3100 (Bridger et al., 1995; De Clercq et al., 1994; Hendrix et al., 2000) and Tak-779 (Baba et al., 1999), thymus fragments were incubated with antagonists for 4 h before infection.
Human fetal-thymus organ culture (HF-TOC) assays
The procedure for HF-TOC has been previously described (Bonyhadi et al., 1995; Miller et al., 2001). Human fetal thymuses (19 to 24 gestational weeks) were dissected into approximately 2-mm fragments using a dissecting microscope. 5–6 fragments were placed on organotypic culture membranes (Millipore) underlaid by media (RPMI with 10% fetal bovine serum, 50 µg of streptomycin/ml, 50 U of penicillin G/ml, 1 × MEM vitamin solution [Gibco/BRL], 1× insulin-transferrin-sodium selenite medium supplement [Sigma], and betamercaptoethanol), in 6-well tissue culture plates. An equal amount of virus (100–800 IU) in 15 µl of supernatant from infected PHA-stimulated PBMCs or mock supernatant was applied to each fragment. Viral and mock supernatant produced from the same PBMC donor were used within each experiment. Fragments were cultured at 37 °C in 5% CO2 for 10 to 12 days with daily changes of culture media. Thymocytes were teased out of the fragments using pestles (Bellco Co.) and analyzed as described below by FACS. Supernatant was stored for assessment of p24 production.
Infection of SCID-hu Thy/Liv mice
SCID-hu Thy/Liv mice were constructed and infected as previously described (Krowka et al., 1991; McCune et al., 1988; Su et al., 1995). Briefly, HIV-1 produced from PHA-activated PBMCs or mock supernatant was used to infect each graft. Approximately 2000 IU in 50 µl was used for each infection. Each week after infection, a biopsy of the thymus was removed and assessed for viral replication by p24 ELISA and viral pathogenesis by FACS staining.
Quantification of viral replication
Cell lysates or HF-TOC supernatant were prepared in PBS-1% Triton X-100. p24 production was measured using an Alliance p24 ELISA kit (Perkin Elmer). Levels of proviral integration were assessed by TaqMan real-time PCR for Gag DNA using primers and probe which have been previously described (Douek et al., 2002). Approximately 10,000 thymocytes from infected HF-TOC fragments were pelleted and lysed in 50 µl of Florence Buffer (25 µl Florence A: 100 mM KCl, 10 mM Tris HCl pH 8.3, 2.5 mM MgCl2; 25 µl Florence B: 10 mM Tris HCl pH 8.3, 2.5 mM MgCl2, 1% Tween 20, 1% NP40, 200 µg Proteinase K added fresh). Lysis was performed at 60 °C for 1 h followed by 95 °C for 10 min. Input DNA was normalized using actin real-time PCR with the following primers and probes: actinF: (5′-TCACCCACACTGTGCCCATC-TACGA-3′), actinR: (5′-CAGCGGAACCGCTCATTGC-CAATGG-3′), and actin probe (5′-VIC-ATGCCCTCCC-CCATGCCATCCTGCGT-TAMRA-3′).
Flow cytometry analysis
Thymocytes isolated from either SCID-hu Thy/Liv biopsies or HF-TOC fragments were stained with CD4-PE and CD8-TC (Caltag) in PBS-2% fetal bovine serum, washed, and resuspended in PBS-1% formaldehyde. Viral pathogenesis was assessed by calculating the total percentage of cells that stained positive for CD4, including both CD4+CD8+ immature and CD4+CD8− mature thymocytes.
Cloning and sequencing of the envelope genes and construction of NL4-R3 recombinant viruses
Envelope genes were amplified from DNA prepared from an early passage of infected PBMCs as previously described (Kirchhoff et al., 1999). Nested PCR was performed to generate amplicons suitable for cloning. The outer primers have been previously described: VPRF (5′-ATGGAACAAGCMCCRGMAGACCA-3′; positions 5559 to 5581) and LTRR (5′-GACTACGGCCGTCTGAGG-GATCTCTAGYTACCA-3′; positions 9689 to 9657) (Kirchhoff et al., 1999). The first round of PCR was performed using high fidelity Pfu polymerase (Stratagene). Second round PCR was performed using the primers E5I (5′-TATGAAACTTATGGRGATACTTGGGCAGG-3′; positions 5697 to 5725) and EnvR (5′-CACTACTTTTT-GACCTCGAGCCACCCATCTTATAGC-3′; positions 8814 to 8779). Positions are listed relative to the NL4-3 genome. An artificially added Xho1 site added to EnvR is highlighted in bold. A 20:1 ratio of Taq polymerase (Sigma) to Pfu was used for the second round of PCR. Amplicons 3.1 kb in size were gel extracted (Qiagen), digested with EcoR1 and Xho1, and ligated into a retroviral expression vector (Coffield et al., 2003).
The HIV-1 env genes were sequenced entirely on both strands using a primer walking method on an automatic sequencer ABI 3100 (Applied Biosystems Inc., Foster City, CA)(Gao et al., 1996). Primary sequence contigs were assembled and edited with the Sequencher program (Version 4.0, Gene Codes Corp., Ann Arbor, MI).
To prepare NL4-3 recombinant viruses, the envelope expression vectors were digested with EcoR1 and Xho1 and ligated to the similarly digested NL4-3 backbone. Ligations were transformed into STBL2 competent cells (Invitrogen) and recombinant clone identity was verified by restriction digest and sequencing. Because of the Xho1 site engineered in the 3′ inner PCR primer (EnvR), nef was deleted from each recombinant virus as shown in Fig. 5A. Nef was detected by Western using a polyclonal rabbit antibody (kindly provided by Ronald Swanstrom, UNC) and gag was detected using a monoclonal anti-p24 hybridoma obtained from the NIH AIDS Research and Reference Reagent Program (Chesebro et al., 1992).
Heteroduplex tracking assay
The heteroduplex tracking assay was performed as previously described (Kitrinos et al., 2003). Briefly, viral RNA from the second PBMC amplification was purified using the Qiagen RNA viral miniprep kit and was used as substrate for RT-PCR using Titan reverse transcriptase. The V1/V2 region was amplified using the following primers: V1 (5′-TTATGGGATCAAAGCCTAAAGCCATGTGTA-3′), V2-(5′-CCTAATTCCATGTGTACATTGTACTG-TGCT-3′). The same fragment was also amplified from each cloned envelope. Amplified products were hybridized with the radiolabeled JR-FL probe and resolved on non-denaturing 6% polyacrylamide gels.
Envelope expression and pseudotyping assay
Each retroviral construct encoding an HIV envelope was transfected into A293T cells with NL4-luciferase in 6-well plates. NL4-luc contains a frameshift deletion in env and encodes the luciferase gene in the nef open reading frame (Connor et al., 1995). Twenty-four hours post-transfection, media were changed to DMEM-2% FBS and cells were cultured at 32 °C. Forty-eight hours post-transfection, virus was harvested and debris was removed by low-speed centrifugation (200 × g for 5 min). An equivalent amount of viral p24 was added to 500 µl of RPMI containing 200,000 SupT1 cells and 8 µg/ml of polybrene. Cells and virus were incubated at 25 °C for 20 min and were then centrifuged at 2000 × g for 3 h. Forty-eight hours post-transduction, cells were lysed and assayed using the Luciferase Assay System (Promega).
To detect envelope expression and processing, envelope constructs were transfected into A293T cells and harvested after 48 h. Lysates were prepared per standard protocol and 100 µg of protein was loaded onto a 6% SDS-polyacrylamide gel. Nitrocellulose membranes were probed with anti-gp120 SF-2 (Env 2–3) (Haigwood et al., 1990) followed by mouse anti-goat IgG-HRP (1:10,000 from Santa Cruz Biotechnology).
Expression of envelope in trans with primary human thymocytes
A293T cells in a 48 well plate (30,000 seeded the day before) were transfected with an equal amount of vector, R3B, or R3A retroviral expression vectors which also encode GFP under the PGK promoter. Twenty-four hours post-transfection, cells were irradiated with 3000 gy to stop cellular division. Equivalent levels of transfection were verified by assessing GFP expression by FACS. 200,000 freshly isolated human thymocytes were added to each well and coculture was allowed to proceed for 48 h. Wells were harvested and stained with CD4-PE and CD8-TC. The relative amount of CD4+ depletion was determined by gating on live thymocytes and normalizing to CD8+ cells as an internal control.
Acknowledgments
We thank Ron Swanstrom, Noah Hoffman, and Rhiannon D’Agostin for helpful discussion. We thank Gavin Henderson and Kim Ritola for assistance with the heteroduplex tracking assay. We thank the S. Fiscus lab for providing PBMCs and for assistance with the p24 ELISA. We thank Robin Hunt for assistance with the p24 ELISA. We thank the UNC Center for AIDS Research, NIAID, DHHS: P30-A150410 for institutional support. The following reagents were obtained through the AIDS Research and Reference Reagent Program, NIAID, NIH: U-373-MAGI-CXCR4CEM cells, and U373-MAGI-CCR5 cells from Dr. Michael Emerman, GHOST cells expressing CCR1, CCR2b, CCR3, CCR4, CXCR4, Bob, Bonzo, V28, CCR8, or CCR5 from Dr. Vineet N. KewalRamani and Dr. Dan R. Littman, HIV-1 p24 hybridoma (183-H12-5C) from Dr. Bruce Chesebro, Tak-779, bicyclam JM-2987 (hydrobromide salt of AMD-3100), and antiserum to HIV-1SF-2 gp120 from the Chiron Corporation. This work was supported by NIH grants AI041356 and AI53804. EM was supported by the “Molecular Biology of Viral Diseases” training grant, NIAID, DHHS, T32-AI07419.
References
- Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguxo K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. U.S.A. 1999;96(10):5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonyhadi ML, Su L, Auten J, McCune JM, Kaneshima H. Development of a human thymic organ culture model for the study of HIV pathogenesis. AIDS Res. Hum. Retroviruses. 1995;11(9):1073–1080. doi: 10.1089/aid.1995.11.1073. [DOI] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 1994;68(9):6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger GJ, Skerlj RT, Thornton D, Padmanabhan S, Martellucci SA, Henson GW, Abrams MJ, Yamamoto N, De Vreese K, Pauwels R, et al. Synthesis and structure-activity relationships of phenylenebis (methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication. Effects of macrocyclic ring size and substituents on the aromatic linker. J. Med. Chem. 1995;38(2):366–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]
- Camerini D, Su HP, Gamez-Torre G, Johnson ML, Zack JA, Chen IS. Human immunodeficiency virus type 1 pathogenesis in SCID-hu mice correlates with syncytium-inducing phenotype and viral replication. J. Virol. 2000;74(7):3196–3204. doi: 10.1128/jvi.74.7.3196-3204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 1992;66(11):6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffield VM, Jiang Q, Su L. A genetic approach to inactivating chemokine receptors using a modified viral proteinv. Nat. Biotechnol. 2003;21(11):1321–1327. doi: 10.1038/nbt889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- De Clercq E, Yamamoto N, Pauwels R, Balzarini J, Witvrouw M, De Vreese K, Debyser Z, Rosenwirth B, Peichl R, Datema R, et al. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob. Agents Chemother. 1994;38(4):668–674. doi: 10.1128/aac.38.4.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, McFarland RD, Keiser PH, Gage EA, Massey JM, Haynes BF, Polis MA, Haase AT, Feinberg MB, Sullivan JL, Jamieson BD, Zack JA, Picker LJ, Koup RA. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396(6712):690–695. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417(6884):95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- Duus KM, Miller ED, Smith JA, Kovalev GI, Su L. Separation of human immunodeficiency virus type 1 replication from nef-mediated pathogenesis in the human thymus. J. Virol. 2001;75(8):3916–3924. doi: 10.1128/JVI.75.8.3916-3924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci AS. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384(6609):529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G The WHO and NIAID Networks for HIV Isolation and Characterization. J. Virol. 1996;70(3):1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushakova S, Grivel JC, Fitzgerald W, Sylwester A, Zimmerberg J, Margolis LB. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat. Med. 1998;4(3):346–349. doi: 10.1038/nm0398-346. [DOI] [PubMed] [Google Scholar]
- Groenink M, Andeweg AC, Fouchier RA, Broersen S, van der Jagt RC, Schuitemaker H, de Goede RE, Bosch ML, Huisman HG, Tersmette M. Phenotype-associated env gene variation among eight related human immunodeficiency virus type 1 clones: evidence for in vivo recombination and determinants of cytotropism outside the V3 domain. J. Virol. 1992;66(10):6175–6180. doi: 10.1128/jvi.66.10.6175-6180.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigwood NL, Shuster JR, Moore GK, Lee H, Skiles PV, Higgins KW, Barr PJ, George-Nascimento C, Steimer KS. Importance of hypervariable regions of HIV-1 gp120 in the generation of virus neutralizing antibodies. AIDS Res. Hum. Retroviruses. 1990;6(7):855–869. doi: 10.1089/aid.1990.6.855. [DOI] [PubMed] [Google Scholar]
- Harouse JM, Gertie A, Tan RC, Blanchard J, Cheng-Mayer C. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science. 1999;284(5415):816–819. doi: 10.1126/science.284.5415.816. [DOI] [PubMed] [Google Scholar]
- Hendrix CW, Flexner C, MacFarland RT, Giandomenico C, Fuchs EJ, Redpath E, Bridger G, Henson GW. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob. Agents Chemother. 2000;44(6):1667–1673. doi: 10.1128/aac.44.6.1667-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson BD, Aldrovandi GM, Planelles V, Jowett JB, Gao L, Bloch LM, Chen IS, Zack JA. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J. Virol. 1994;68(6):3478–3485. doi: 10.1128/jvi.68.6.3478-3485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi VV, Oleske JM. Pathologic appraisal of the thymus gland in acquired immunodeficiency syndrome in children. A study of four cases and a review of the literature. Arch. Pathol. Lab. Med. 1985;109(2):142–146. [PubMed] [Google Scholar]
- Kirchhoff F, Easterbrook PJ, Douglas N, Troop M, Greenough TC, Weber J, Carl S, Sullivan JL, Daniels RS. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J. Virol. 1999;73(7):5497–5508. doi: 10.1128/jvi.73.7.5497-5508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitrinos KM, Hoffman NG, Nelson JA, Swanstrom R. Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J. Virol. 2003;77(12):6811–6822. doi: 10.1128/JVI.77.12.6811-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Schols D, Schuitemaker H. No selection for CCR5 coreceptor usage during parenteral transmission of macrophagetropic syncytium-inducing human immunodeficiency virus type 1. J. Virol. 2001;75(18):8848–8853. doi: 10.1128/JVI.75.18.8848-8853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis AP, Ibegbu C, Nahmias AJ, Lee FK, Clark WS, Sawyer MK, Nesheim S. Early progression of disease in HIV-infected infants with thymus dysfunction. N. Engl. J. Med. 1996;335(19):1431–1436. doi: 10.1056/NEJM199611073351904. [DOI] [PubMed] [Google Scholar]
- Krowka JF, Sarin S, Namikawa R, McCune JM, Kaneshima H. Human T cells in the SCID-hu mouse are phenotypically normal and functionally competent. J. Immunol. 1991;146(11):3751–3756. [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Walker BD. Immunopathogenesis and immunotherapy in AIDS virus infections. Nat. Med. 2003;9(7):861–866. doi: 10.1038/nm0703-861. [DOI] [PubMed] [Google Scholar]
- Mackewicz CE, Ortega HW, Levy JA. CD8+ cell anti-HIV activity correlates with the clinical state of the infected individual. J. Clin. Invest. 1991;87(4):1462–1466. doi: 10.1172/JCI115153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolick JB, Munoz A, Donnenberg AD, Park LP, Galai N, Giorgi JV, O’Gorman MR, Ferbas J. Failure of T-cell homeostasis preceding AIDS in HIV-1 infection. The Multicenter AIDS Cohort Study. Nat. Med. 1995;1(7):674–680. doi: 10.1038/nm0795-674. [DOI] [PubMed] [Google Scholar]
- Markham RB, Yu X, Farzadegan H, Ray SC, Vlahov D. Human immunodeficiency virus type 1 env and p17gag sequence variation in polymerase chain reaction-positive, seronegative injection drug users. J. Infect. Dis. 1995;171(4):797–804. doi: 10.1093/infdis/171.4.797. [DOI] [PubMed] [Google Scholar]
- McCormick-Davis C, Dalton SB, Singh DK, Stephens EB. Comparison of Vpu sequences from diverse geographical isolates of HIV type 1 identifies the presence of highly variable domains, additional invariant amino acids, and a signature sequence motif common to subtype C isolates. AIDS Res. Hum. Retroviruses. 2000;16(11):1089–1095. doi: 10.1089/08892220050075363. [DOI] [PubMed] [Google Scholar]
- McCune JM. The dynamics of CD4+ T-cell depletion in HIV disease. Nature. 2001;410(6831):974–979. doi: 10.1038/35073648. [DOI] [PubMed] [Google Scholar]
- McCune JM, Namikawa R, Kaneshima H, Shultz LD, Lieberman M, Weissman IL. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science. 1988;241(4873):1632–1639. doi: 10.1126/science.241.4873.1632. [DOI] [PubMed] [Google Scholar]
- McCune JM, Loftus R, Schmidt DK, Carroll P, Webster D, Swor-Yim LB, Francis IR, Gross BH, Grant RM. High prevalence of thymic tissue in adults with human immunodeficiency virus-1 infection. J. Clin. Invest. 1998;101(11):2301–2308. doi: 10.1172/JCI2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner EG, Duus KM, Loomis R, D’Agostin R, Su L. HIV-1 replication and pathogenesis in the human thymus. Curr. HIV Res. 2003;1(3):275–285. doi: 10.2174/1570162033485258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ED, Duus KM, Townsend M, Yi Y, Collman R, Reitz M, Su L. Human immunodeficiency virus type 1 IIIB selected for replication in vivo exhibits increased envelope glycoproteins in virions without alteration in coreceptor usage: separation of in vivo replication from macrophage tropism. J. Virol. 2001;75(18):8498–8506. doi: 10.1128/JVI.75.18.8498-8506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morner A, Bjorndal A, Albert J, Kewalramani VN, Liftman DR, Inoue R, Thorstensson R, Fenyo EM, Bjorling E. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 1999;73(3):2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naif HM, Cunningham AL, Alali M, Li S, Nasr N, Buhler MM, Schols D, de Clercq E, Stewart G. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Delta32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J. Virol. 2002;76(7):3114–3124. doi: 10.1128/JVI.76.7.3114-3124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papiernik M, Brossard Y, Mulliez N, Roume J, Brechot C, Barin F, Goudeau A, Bach JF, Griscelli C, Henrion R, et al. Thymic abnormalities in fetuses aborted from human immunodeficiency virus type 1 seropositive women. Pediatrics. 1992;89(2):297–301. [PubMed] [Google Scholar]
- Pekovic DD, Gornitsky M, Ajdukovic D, Dupuy JM, Chausseau JP, Michaud J, Lapointe N, Gilmore N, Tsoukas C, Zwadlo G, et al. Pathogenicity of HIV in lymphatic organs of patients with AIDS. J. Pathol. 1987;152(1):31–35. doi: 10.1002/path.1711520105. [DOI] [PubMed] [Google Scholar]
- Philpott SM. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 2003;1(2):217–227. doi: 10.2174/1570162033485357. [DOI] [PubMed] [Google Scholar]
- Resch W, Ziermann R, Parkin N, Gamarnik A, Swanstrom R. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 2002;76(17):8659–8666. doi: 10.1128/JVI.76.17.8659-8666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes RA, Canfield DR, Esser U, Adamson LA, Brown CR, Cheng-Mayer C, Gardner MB, Harouse JM, Luciw PA. Induction of simian AIDS in infant rhesus macaques infected with CCR5- or CXCR4-utilizing simian-human immunodeficiency viruses is associated with distinct lesions of the thymus. J. Virol. 2004;78(4):2121–2130. doi: 10.1128/JVI.78.4.2121-2130.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 1994;169(5):968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- Rosenzweig M, Clark DP, Gaulton GN. Selective thymocyte depletion in neonatal HIV-1 thymic infection. AIDS. 1993;7(12):1601–1605. doi: 10.1097/00002030-199312000-00009. [DOI] [PubMed] [Google Scholar]
- Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Liftman DR, Fenyo EM, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 1997;3(11):1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- Schuurman HJ, Krone WJ, Broekhuizen R, van Baarlen J, van Veen P, Golstein AL, Huber J, Goudsmit J. The thymus in acquired immune deficiency syndrome. Comparison with other types of immunodeficiency diseases, and presence of components of human immunodeficiency virus type 1. Am. J. Pathol. 1989;134(6):1329–1338. [PMC free article] [PubMed] [Google Scholar]
- Smith KY, Valdez H, Landay A, Spritzler J, Kessler HA, Connick E, Kuritzkes D, Gross B, Francis I, McCune JM, Lederman MM. Thymic size and lymphocyte restoration in patients with human immunodeficiency virus infection after 48 weeks of zidovudine, lamivudine, and ritonavir therapy. J. Infect. Dis. 2000;181(1):141–147. doi: 10.1086/315169. [DOI] [PubMed] [Google Scholar]
- Sousa AE, Carneiro J, Meier-Schellersheim M, Grossman Z, Victorino RM. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 2002;169(6):3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- Su L, Kaneshima H, Bonyhadi M, Salimi S, Kraft D, Rabin L, McCune JM. HIV-1-induced thymocyte depletion is associated with indirect cytopathogenicity and infection of progenitor cells in vivo. Immunity. 1995;2(1):25–36. doi: 10.1016/1074-7613(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Su L, Kaneshima H, Bonyhadi ML, Lee R, Auten J, Wolf A, Du B, Rabin L, Hahn BH, Terwilliger E, McCune JM. Identification of HIV-1 determinants for replication in vivo. Virology. 1997;227(1):45–52. doi: 10.1006/viro.1996.8338. [DOI] [PubMed] [Google Scholar]
- Taylor JR, Jr, Kimbrell KC, Scoggins R, Delaney M, Wu L, Camerini D. Expression and function of chemokine receptors on human thymocytes: implications for infection by human immunodeficiency virus type 1. J. Virol. 2001;75(18):8752–8760. doi: 10.1128/JVI.75.18.8752-8760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersmette M, Lange JM, de Goede RE, de Wolf F, Eeftink-Schattenkerk JK, Schellekens PT, Coutinho RA, Huisman JG, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;1(8645):983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233(1):193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- Yi Y, Chen W, Frank I, Cutilli J, Singh A, Starr-Spires L, Sulcove J, Kolson DL, Collman RG. An unusual syncytia-inducing human immunodeficiency virus type 1 primary isolate from the central nervous system that is restricted to CXCR4, replicates efficiently in macrophages, and induces neuronal apoptosis. J. Neurovirol. 2003;9(4):432–441. doi: 10.1080/13550280390218706. [DOI] [PubMed] [Google Scholar]
- Yu XF, Wang Z, Vlahov D, Markham RB, Farzadegan H, Margolick JB. Infection with dual-tropic human immunodeficiency virus type 1 variants associated with rapid total T cell decline and disease progression in injection drug users. J. Infect. Dis. 1998;178(2):388–396. doi: 10.1086/515646. [DOI] [PubMed] [Google Scholar]
- Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261(5125):1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]