Abstract
Brassica napus seed is a lipid storage organ containing approximately 40% oil, while its leaves contain many kinds of lipids for many biological roles, but the overall amounts are less than in seeds. Thus, lipid biosynthesis in the developing seeds and the leaves is strictly regulated which results the final difference of lipids. However, there are few reports about the molecular mechanism controlling the difference in lipid biosynthesis between developing seeds and leaves. In this study, we tried to uncover this mechanism by analyzing the transcriptome data for lipid biosynthesis. The transcriptome data were de novo assembled and a total of 47216 unigenes were obtained, which had an N50 length and median of 1271 and 755 bp, respectively. Among these unigenes, 36368 (about 77.02%) were annotated and there were 109 up-regulated unigenes and 72 down-regulated unigenes in the developing seeds lipid synthetic pathway after comparing with leaves. In the oleic acid pathway, 23 unigenes were up-regulated and four unigenes were down-regulated. During triacylglycerol (TAG) synthesis, the key unigenes were all up-regulated, such as phosphatidate phosphatase and diacylglycerol O-acyltransferase. During palmitic acid, palmitoleic acid, stearic acid, linoleic acid and linolenic acid synthesis in leaves, the unigenes were nearly all up-regulated, which indicated that the biosynthesis of these particular fatty acids were more important in leaves. In the developing seeds, almost all the unigenes in the ABI3VP1, RKD, CPP, E2F-DP, GRF, JUMONJI, MYB-related, PHD and REM transcript factorfamilies were up-regulated, which helped us to discern the regulation mechanism underlying lipid biosynthesis. The differential up/down-regulation of the genes and TFs involved in lipid biosynthesis in developing seeds and leaves provided direct evidence that allowed us to map the network that regulates lipid biosynthesis, and the identification of new TFs that are up-regulated in developing seeds will help us to further elucidate the lipids biosynthesis pathway in developing seeds and leaves.
Introduction
Lipids have many important biological functions, including storing energy, signaling and acting as structural components of cell membranes [1, 2]. Lipids, which are important biological macromolecules, occur in different forms, including fats, waxes, sterols, fat-soluble vitamins (such as vitamins A, D, E and K), monoglycerides, diglycerides, triglycerides, phospholipids and glycolipids. In plants, the lipid type and contents differ between the leaf and seed: plant seed is the oil-rich organ, whereas leaf mainly contains glycolipids and phospholipids.
The leaf is where photosynthesis occurs, which provides plants with the large amounts of carbohydrates and energy needed for growth and development. Glycolipids and phospholipids are the main components of the photosynthetic membranes in plant chloroplast envelopes and stroma [3]. They are also involved in the formation of the photosynthetic membranes and are part of the photosynthetic complexes [4–7]. The main lipid in leaves is monogalactosyldiacylglycerol (MGDG), followed by digalactosyldiacylglycerol (DGDG), phosphatidylglycerol (PG) and phosphatidylcholine (PC). There is also a small amount of triacylglycerol (TAG) [8]. However, in the seed, as the storage organ, most of the lipid is triacylglycerol, which is stored in the oil body (OB). Neutral lipids account for a large percentage of the oil in the seeds of rape, mustard, cotton, flax, maize, peanut and sesame [9]. Furthermore, Arabidopsis studies have revealed that there is a difference in lipid content and type between leaves and seeds, with 52.46% glycerolipids, 24.60% chlorophyll, 4.92% cutin monomers, 3.28% sphingolipids, 3.28% wax and 11.48% others present in leaves, and 94% storage lipids, 5% membrane lipids and 1% surface lipids present in seeds [3]. The total lipid content in seeds was more than in leaves and accounted for 37% of the total dry weight of seeds, but only 6.1% of the total dry weight in leaves. There was a difference in storage lipid content between the leaves and seeds and a significant difference in fatty acid contents. In Arabidopsis, the highest fatty acid content in seeds was 18:2, followed by 20:1, 18:3, 18:1, 16:0 [10], whereas the highest fatty acid content found in leaves was 18:3, followed by 18:2, 16:0, 16:3 [11]. In Brassica napus, the highest fatty acid content in seeds was 18:1, followed by 18:2, 18:3, 16:0 and others [12]. This may be due to the different functions of fatty acids in leaves and seeds. The fatty acids in leaves may be involved in the formation of membrane lipid structure, whereas the fatty acids in seeds may act as storage lipids [3].
Brassica napus is one of the most important edible oilseed crops in the world and produces considerable amounts of edible oil for human consumption. Research by United States Department of Agriculture (USDA) showed that the rapeseed provided more and more oil for human consumption, ranging from 24916 thousand metric tons to 26946 thousand metric tons in three years (http://www.usda.gov/wps/portal/usda/usdahome). Rapeseed breeding [13, 14] has produced varieties with zero seed erucic acid and low seed glucosinolate levels. It has also produced rapeseed with high oleic acid contents (78% to 88%), which has nutritional and health benefits, and high oil contents (40% to 45%) [15]. Many genes related to oleic acid and oil biosynthesis and regulation have been elucidated. In the prokaryotic fatty acid biosynthesis pathway, fatty acid desaturase 2 (FAD2) is involved in the regulation of oleic acid, linoleic acid and linolenic acid biosynthesis [16, 17], and acetyl-CoA carboxylase (ACCase) and the fatty acid synthase complex (FAS) are the key enzymes in the fatty acid synthesis [18, 19]. In TAG biosynthesis pathway, glycerol-3-phosphate acyltransferase 4 (GPAT4) and acyl CoA binding protein (ACBP) were involved in the regulation of oil content and fatty acid composition [12, 20]. 1-acyl-sn-glycerol-3-phosphate acyltransferase (LPAT) [21, 22], diacylglycerol O-acyltransferase (DGAT) and phosphatidate phosphatase (PP) [23, 24] were involved in TAG synthesis [25, 26]. Some transcription factors (TFs) also play key roles in lipid biosynthesis. Previous studies have revealed that ABI3, LEC1, LEC2, FUS3, WRI1, AP2, GL2, EMF2, HSI2, L1L, PKL, FIE and SWN acted as the regulator in fatty acid and TAG synthesis [27, 28].
Next generation sequencing (NGS) enables us to obtain genetic information [29–31]. Many plants have been sequenced and annotated using NGS, such as B. rapa [32] and B. oleracea [33], which are the species from which the Brassica napus originated, and other oil producing plants, such as palm [34], peanut [35], sesame [30], safflower [36], rape [37, 38], jatropha [39] and yellow horn [40]. Lipid contents differ significantly between the leaves and seeds in Brassica napus and lipid biosynthesis and regulation are also different. Although the de novo biosynthesis of fatty acids and lipids is now well understood, much less is known about how plants produce the different amounts and types of fatty acid and lipids between seeds and leaves in B. napus through the regulation of gene expressions. In this study, we compared the developing seeds and leaves transcriptome in B. napus, which revealed how seeds were able to store so much TAG and offered us clues on how to improve the content of specific lipids in seeds.
Materials and Methods
Plant material
B. napus cv Ninyou 12 was used as the material. The leaves at the stage when plant had 4–5 leaves, at the top two position were collected as the sample and developing seeds at 25 days after pollination (DAP) were harvested in the field and immediately frozen in liquid nitrogen and stored at—70°C for RNA extraction. Hereafter, seeds refer to 25DAP seeds unless special illustration. The experimental material was planted in Jiangsu University, and was specially used as the experimental research. And Brassica napus, has been used as the research object with no dangerous and harmful to the land and crop. The measurement of the fatty acid and TAG contents in leaves followed a previously reported method [41, 42], and mature seeds oil contents were measured using near infrared-reflectance spectroscopy (NIRS) [43, 44].
RNA extraction, library construction and RNA-seq
Total RNA of the collected leaves and 25 DAP seeds were extracted using TRIzol Reagent (Life technologies, Shang hai, USA) according to the manufacturer’s instructions. The extracted RNA was qualified and quantified using a OneDrop OD-1000+ spectrophotometer (RockGene, Shanghai, China) and the samples showed a 260/280 nm ratio between 1.8 and 2.2, and an OD260/230 > 1.0, which were within the requirements of Beijing Biomarker Technologies (http://www.biomarker.com.cn/index.php).
The mRNA-seq library was constructed using Illumina’s TruSeq RNA Sample Preparation Kit (Illumina lnc, San Diego, CA, USA), and the isolation of mRNA, fragment interruption and RNA-Seq were performed by the company according to their standard protocol. Finally, the mRNA-seq library was constructed for sequencing using the Illumina HiSeqTM 2000 sequencing platform.
Analysis of transcriptome sequencing results
The raw reads were first filtered by discarding the reads with adaptor contamination, low-quality sequences (reads with ambiguous ‘N’ bases), and reads with more than 10% Q < 20 bases. Then the clean reads were assembled into contigs using the Trinity program [45], which efficiently reconstructed full-length transcripts across a broad range of expression levels and sequencing depths. Subsequently, the contigs were linked into transcripts according to the paired-end information of the sequences, and the transcripts were clustered based on nucleotide sequence identity. The longest transcripts in the cluster units were regarded as unigenes in order to eliminate redundant sequences, and then the unigenes were combined to produce the final assembly used for annotation. The unigenes information was deposited in the Sequence Read Archive (SRA) database in NCBI (Accession number, SRR1916242).
To understand their functions, the unigenes were annotated using BLASTx alignment, with an E-value cut-off of 10–5, against the NCBI non-redundant (NR) database, and the UniProt/Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), Cluster of Orthologous Groups of proteins (COG) and Gene Ontology (GO) databases.
The RPKM (Reads Per Kilobase per Million mapped reads) method was used to calculate unigenes’ expression [46]. The RPKM method is able to reflect the molar concentration of a transcript by normalizing for RNA length and for the total read number. We compared the unigenes expressions using their RPKM values.
Detection of TFs in the transcriptome data
To detect TFs, we performed a BLAST search for all unigenes against the AGRIS (Arabidopsis Gene Regulatory Information Server) database with an e-value cut off of 10–5 [47].
Quantitative real-time PCR analysis
The selected differentially expressed transcript factors were confirmed through qRT-PCR using ABI 7300 Real-Time PCR Detection System (Applied Biosystems, Foster City, CA, USA) with SYBR Premix Ex TaqTM II (TaKaRa, Tokyo, Japan). First, we used RNase-free DNase I to remove residual trace amounts of DNA before cDNA synthesis, according to the manufacturer’s instruction (Thermo Scientific, Waltham, MA, USA). The synthesis of first strand of cDNA was according to the manufacturer’s instructions of the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) from 2μg of total RNA in a 20μL reaction using oligodT primers. Then the cDNA sets were diluted 1:10 with nuclease-free water and used for qPCR analysis. Each 20μL reaction mixture contained 10μL of 2×SYBR Premix Ex TaqTM, 2μL of diluted cDNA, 2μL of each primer (2μM), 0.4μL of ROX Reference Dye (50×) and 3.6μL of double distilled water. The qPCR cycling conditions were as follows: 95°C for 30s; followed by 40 cycles of 95°C for 10s, the respective annealing temperature for 30s and 72°C for 27s in PCR strip tubes (Axygen, Union City, CA, USA). We employed probes specific for the TIP41 [48] as references to analyze the expression level of TFs between leaves and seeds, and each reaction was performed three repeats.
Results and Discussion
Comparison of fatty acid contents between seeds and leaves
Seeds and leaves have considerable morphological differences. The fatty acid contents in seeds and leaves are also different (Table 1). The seeds contained oleic acid (59.14%), linoleic acid (23.43%), linolenic acid (9.18%), saturated fatty acids (6.61%) and others (1.64%). However, in the leaves, the largest fatty acid component was linolenic acid (47.55%), followed by saturated fatty acids (20.15%), linoleic acid (10.62%), oleic acid (4.85%) and others (16.82%). Hence, these data showed the big difference between the components in leaves and seeds. We tried to discern the mechanism controlling lipid biosynthesis between the seeds and leaves by their transcriptome.
Table 1. The content of different fatty acids between leaves and 25DAP seeds.
| Component | oleic acid | linoleic acid | linolenic acid | saturated fatty acids | Others |
|---|---|---|---|---|---|
| Leaves(mol%) | 4.85 | 10.62 | 47.55 | 20.15 | 16.82 |
| Seeds(mol%) | 59.14 | 23.43 | 9.18 | 6.61 | 1.64 |
The raw data of leaves and seeds transcriptome sequencing
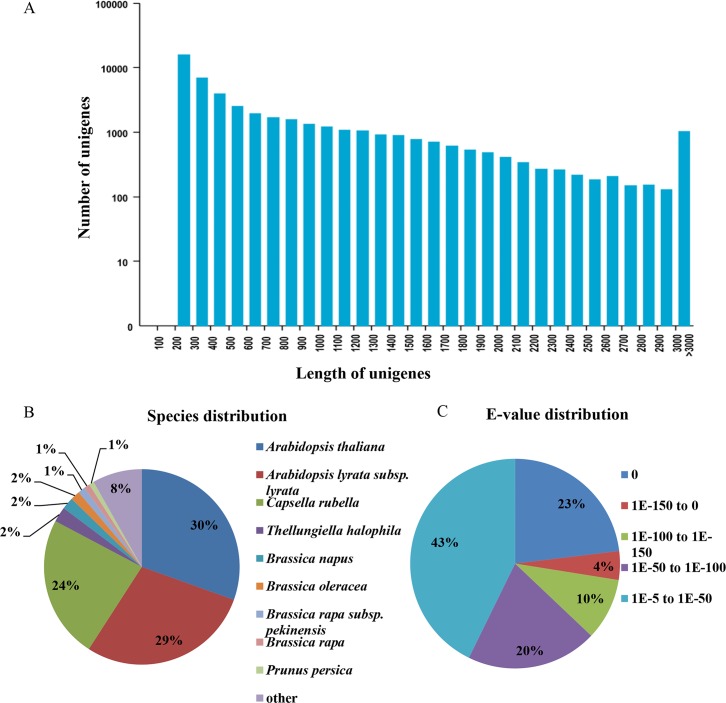
By sequencing, the leaves sample produced 27086293 reads and the seeds sample produced 40496936 reads (Table 2). The average quality value was ≥ 20 for 100% of the cycle with a near zero ambiguous “N”. The Q30 percentage exceeded 80% and the GC content 48.67% and 48.68% for the leaves and seeds, respectively, which suggested that the sequencing was highly accurate and reliable. After the removal of adaptor sequences and the exclusion of contaminated or short reads, the high-quality reads were assembled into 10949964 contigs with a mean length of 39.2 bp, 114337 transcripts with a mean length of 1008.59 bp and 47216 unigenes with a mean length of 755.65 bp (Table 3) using the Trinity de novo assembly program [45]. Out of these 47216 unigenes, 11528 unigenes were ≥ 1000 bp and accounted for 24.41% of the total. The size distribution of all the unigenes is shown in Fig 1A. These results showed that the throughput and sequencing quality were high enough for the following analyses.
Table 2. The raw data of transcriptome between leaves and 25DAP seeds in Brassica napus.
| SampleID | Total reads | Total nucleotides(bp) | GC(%) | N(%) | Q20% | CycleQ20% | Q30% |
|---|---|---|---|---|---|---|---|
| Leaves | 27086293 | 5457985744 | 48.67 | 0.08 | 89.83 | 100 | 80.74 |
| Seeds | 40496936 | 8098613808 | 48.68 | 0 | 89.3 | 100 | 80.58 |
Table 3. The de novo assembly of raw data.
| Length range | Contigs | Transcripts | Unigenes |
|---|---|---|---|
| 200–300 | 10910319(99.64%) | 21416(18.73%) | 15816(33.50%) |
| 300–500 | 16636(0.15%) | 18974(16.59%) | 10863(23.01%) |
| 500–1000 | 11316(0.10%) | 28213(24.68%) | 9009(19.08%) |
| 1000–2000 | 8465(0.08%) | 33325(29.15%) | 8192(17.35%) |
| 2000+ | 3228(0.03%) | 12409(10.85%) | 3336(7.07%) |
| Total number | 10949964 | 114337 | 47216 |
| Total length | 429283622 | 115318903 | 35678684 |
| N50 length | 37 | 1459 | 1272 |
| Mean length | 39.2 | 1008.59 | 755.65 |
Fig 1. Unigenes functional annotation results.
(A) Distribution of unigenes in length. (B) Top species distribution for BLASTx matches for B. napus unigenes using the following order of priority: NR and Swiss-Prot. (C) E-value distribution of top BLASTx hits for each unigene.
Functional annotation
According to the results of functional annotation (Table 4), there were 36176 (76.62%) unigenes homologous proteins in the Nr protein database and 26964 (57.11%) unigenes similar to proteins in the Swiss-Prot database. And, 29780 (63.07%), 9394 (19.90%) and 8123 (17.20%) unigenes had significant matches with sequences in the GO, COG and KEGG databases, respectively. In total, 36368 (77.02%) unigenes were successfully annotated using the Nr, Swiss-Prot, GO, COG and KEGG databases. However, the remaining unmapped unigenes (22.98%) were not annotated in these databases, which could be attributable to the short sequence reads generated by the sequencing technology or the relatively short sequences of the resulting unigenes lacked conserved functional domains [49].
Table 4. The functional annotation of unigenes in COG, GO, KEGG, Swissprot and Nr database.
| Anno Database | Annotated Number | 300< = length<1000 | length> = 1000 |
|---|---|---|---|
| COG_Annotation | 9394(19.90%) | 3130 | 4983 |
| GO_Annotation | 29780(63.07%) | 12642 | 9629 |
| KEGG_Annotation | 8123(17.20%) | 3215 | 2828 |
| Swissprot_Annotation | 26964(57.11%) | 10892 | 9902 |
| nr_Annotation | 36176(76.62%) | 15660 | 11154 |
| All_Annotated | 36368(77.02%) | 15722 | 11168 |
To identify the species specificity of the unigenes (36176) annotated in the Nr database, we matched these unigenes and found that all the unigenes were found in at least one species, with unigenes from Arabidopsis thaliana (11023, 30.47%), Arabidopsis lyrata subsp. Lyrata (10362, 28.64%), Capsella rubella (8553, 23.64%), Thellungiella halophile (905, 2.50%), Brassica napus (720, 1.99%), Brassica oleracea (567, 1.56%), Brassica rapa subsp. Pekinensis (431, 1.19%), Brassica rapa (345, 0.95%), Prunus persica (300, 0.83%) and other species (2970, 8.21%) (Fig 1B). The little proportion of unigenes belonging to B. napus might be the results of searching in public databases which contain few data of B. napus. We also found that 57.26% unigenes had an E-value of less than 1.0E-50, and there was a very strong homology among these aligned unigenes. The remaining 42.74% unigenes had an E-value of between 1.0E-5 to 1.0E-50 (Fig 1C).
GO, COG and KEGG classification analysis
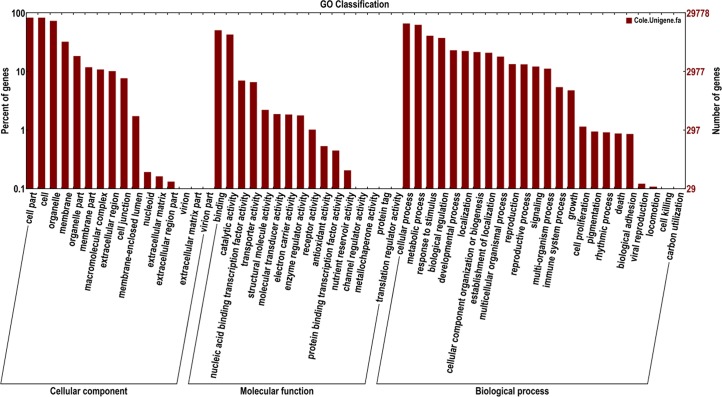
To further predict and classify the function of annotated unigenes, we used the sequences of these unigenes to search for genes with GO assignments, COG classifications and KEGG pathway assignments. First, we performed a Gene Ontology (GO) [50] analysis based on their Nr annotation, which revealed the cellular component, molecular function and biological process unigenes, based on sequence homology. Among the unigenes, 29780 were assigned into three main GO functional categories and then were divided into 56 sub-categories, among which many unigenes were assigned to one or more sub-categories (Fig 2). The largest category was biological process containing 112676 unigenes, followed by cellular component (99279) and molecular function (34902). The biological process category contained 24 sub-categories and two of the biggest sub-categories were “cellular process” and “metabolic process”, which contained 19705 and 18703 unigenes, respectively, which suggested that these unigenes were enriched in the B. napus transcriptome libraries. The second category, cellular component, was divided into 16 sub-categories and three of the largest sub-categories were “cell part”, “cell” and “organelle”, which contained 24824, 24784 and 21854 unigenes, respectively. The last category, molecular function, was categorized into 16 GO sub-categories and the two largest sub-categories were “binding” and “catalytic activity”, with 15115 and 12754 unigenes, respectively.
Fig 2. Gene Ontology (GO) categories assigined to the B. napus unigenes.
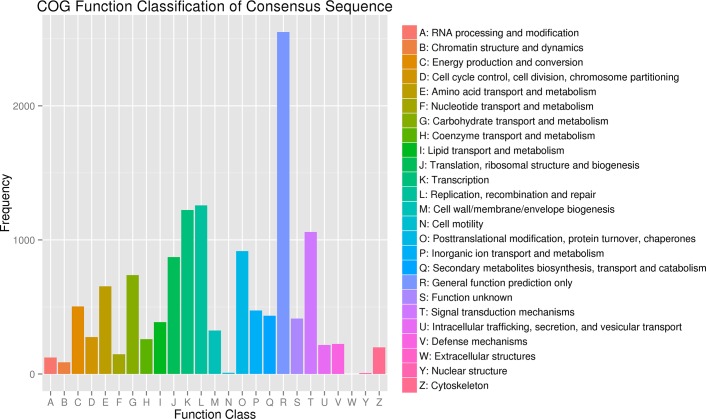
Then, we performed a COG analysis of all the unigenes for functional prediction and classification. A total of 9394 unigenes sequences showed a hit with the Nr database and could be assigned to COG classifications that were functionally clustered into 24 COG categories with no unigenes involved in the “Extracellular structures” category. Among these categories, the cluster for “General function prediction only” was the largest group containing 2550 unigenes (27.14%), followed by “Replication, recombination and repair” (1258, 13.39%), “Transcription” (1223, 13.02%), “Signal transduction mechanisms” (1059, 11.27%), “Post-translational modification, protein turnover chaperones” (917, 9.76%), “Translation, ribosomal structure and biogenesis” (872, 9.28%) and “Carbohydrate transport and metabolism” (737, 7.85%). Only a few unigenes were assigned to the two smallest categories, “Cell motility” and “Nuclear structure” (9 and 7 unigenes, respectively) (Fig 3).
Fig 3. Clusters of orthologous groups (COG) classifications of B. napus unigenes.
Finally, in order to better understand the biological pathways in B. napus, we used the KEGG [51] database to categorize gene functions with an emphasis on biological pathways. The results showed that a total of 8123 unigenes were assigned to 121 pathways (Table 4; S1 Table). The number of unigenes involved in these 121 pathways was 8515 instead of 8123, which suggested that some unigenes might be involved in more than one KEGG pathway, such as unigene “c18488.graph_c0” which is involved in the glycerolipid metabolism, galactose metabolism, sphingolipid metabolism and glycosphingolipid biosynthesis-globo series pathways. Among the 121 pathways, the largest pathway was Plant hormone signal transduction, which contained 384 unigenes, followed by Ribosome (334), Plant-pathogen interaction (252), Protein processing in endoplasmic reticulum (239) and RNA transport (234), etc. The smallest pathway was Anthocyanin biosynthesis, which only contained one unigene (S1 Table). When we concentrated on fatty acid and lipid biosynthesis and metabolism, we found that there were 95 unigenes for glycerophospholipid metabolism, 73 for fatty acid metabolism, 64 for glycerolipid metabolism, 56 for biosynthesis of unsaturated fatty acids, 36 for fatty acid biosynthesis, 35 for pantothenate and CoA biosynthesis, 27 for linoleic acid metabolism, 23 for arachidonic acid metabolism and 8 for fatty acid elongation in mitochondria. These results will provide precise and more targeted information for further analysis.
Differentially expressed genes (DEG) analysis
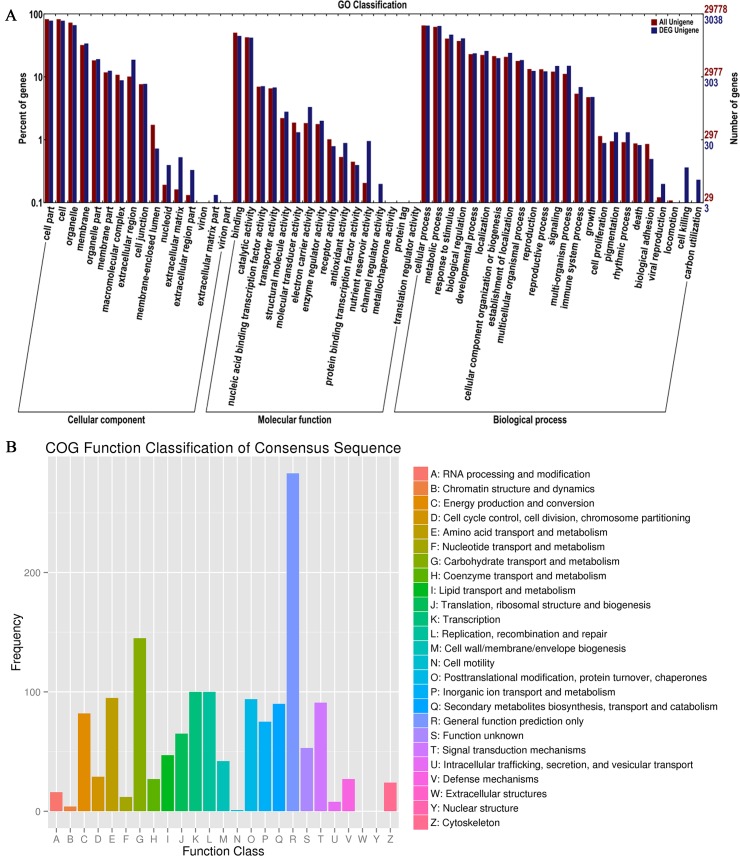
The difference in lipid content between seeds and leaves might be caused by different genes expression. So we performed a differentially expressed genes (DEG) analysis and found that there were 4544 unigenes that were differentially expressed. We then performed GO and COG classification analyses to identify the function of these differentially expressed genes (Fig 4). In the GO classification analysis, 4544 unigenes were assigned to three main GO functional categories and then were divided into 56 sub-categories, among which many unigenes were assigned to more than one sub-category. Then we calculated the percentage of DEG involved in each sub-category (S2 Table). The largest percentage of sub-category in the “cellular component” category was extracellular matrix part (DEG accounting for 80.00% of all unigenes involved in this category), followed by extracellular matrix (33.33%), extracellular region part (25.64%) and nucleoid (21.05%). The largest percentage of sub-category in “molecular function” was channel regulator activity (50%), followed by nutrient reservoir activity (47.54%) and protein tag (22.22%). The largest percentage of sub-category in “biological process” was cell killing (57.89%), followed by utilization (46.67%). These results indicated that the difference in lipid content between the seeds and leaves might be due to the differential expression of genes in these nine sub-categories, which would provide a direction for further analysis.
Fig 4. Function classification of Differential Expression Genes (DEG) in B. napus.
(A) Gene ontology (GO) classification of DEG. (B) Clusters of orthologous groups (COG) classifications of DEG.
The COG function classification analysis showed that the DEG were distributed across 24 COG categories. We also calculated the percentage of DEG in each category (S3 Table) and found that the largest percentage of category was “Secondary metabolites biosynthesis, transport and catabolism” (20.69%), followed by “Carbohydrate transport and metabolism” (19.67%), “Energy production and conversion” (16.27%) and “Inorganic ion transport and metabolism” (15.82%). Many unigenes were differentially expressed in these categories, which might result in the different lipid contents seen in the seeds and leaves.
Genes related to fatty acid biosynthesis in B. napus
Fatty acids are stored as a form of TAG and their biosynthesis pathway can be divided into three steps in nearly all oil plants [52]. The first step is de novo fatty acid synthesis. In plants, de novo fatty acid synthesis occurs in the plastid instead of the cytosol and is catalyzed mainly by the fatty acid synthase complex (FAS). Furthermore, biosynthesis is not restricted to specific tissues or organs, but occurs in every plant cell [3]. The second step is the synthesis of triacylglycerol (TAG) using the fatty acid and glycerol as substrates. This occurs in the endoplasmic reticulum (ER). Finally, TAG is combined with oil proteins, such as oleosin, caleosin and steroleosin, to form OBs (oil bodies), which are released from the ER into the cytoplasm [53, 54].
A manually repeated search based on the KEGG pathway assignment and functional annotation of the unigenes found that 36 unigenes were annotated as encoding ten key enzymes involved in fatty acid biosynthesis and ten unigenes encoding acyl carrier protein (ACP) (Table 5; S4 Table). Based on these identified enzymes, we reconstructed the fatty acid biosynthesis pathway by referencing previous reports (Fig 5) [3, 40]. The first committed step in fatty acid synthesis is the formation of malonyl-CoA from acetyl-CoA, which is catalyzed by acetyl-CoA carboxylase (ACCase, EC: 6.4.1.2) [18]. We identified ten unigenes that were involved in encoding four subunits of this enzyme (four for biotin carboxyl carrier protein, three for biotin carboxylase, one for α-carboxyltransferase and two for β-carboxyltransferase). Among these ten unigenes, six were up-regulated in seeds compared to leaves, two were down-regulated and two were unchanged, which suggested that this critical process would provide more substrates for the fatty acid synthesis in seeds than in leaves. Next, the fatty acids are grown by a series of condensation reactions with malony-CoA. The ACP-bound acyl chain grows by consecutively adding two carbon units per cycle over six or seven cycles to form 16:0-ACP or 18:0-ACP. This process is catalyzed by the fatty acid synthase complex (FAS), which is composed of five components encoded by 20 unigenes. Two unigenes encoded malonyl-CoA-ACP transacylase (MAT, EC: 2.3.1.39), ten encoded 3-oxoacyl-ACP-synthase (KAS; one encoded KAS I (EC: 2.3.1.41), six encoded KAS II (EC: 2.3.1.179), three encoded KAS III (EC: 2.3.1.180)), two encoded 3-oxoacyl-ACP reductase (KAR, EC: 1.1.1.100), five encoded hydroxyacyl-acyl-ACP dehydratase (HAD, EC: 4.2.1.-) and one encoded enoyl-ACP reductase (EAR, EC: 1.3.1.9). Besides the five components of FAS, the acyl carrier protein (ACP), as a cofactor, is also an essential part of FAS. We found that ten unigenes encoded ACP. From Fig 5, we can see that almost all the unigenes encoding FAS were up-regulated in the de novo synthesis of 16:0-ACP and 18:0-ACP in seeds compared to the leaves. Only two unigenes were down-regulated, which meant that the up-regulation of these unigenes would increase the fatty acid content in seeds. After this, the 16:0-ACP and 18:0-ACP are catalyzed by acyl-ACP desaturase (AAD, EC: 1.14.19.2) to form 16:1-ACP and 18:1-ACP [55], or 18:0-ACP is desaturated by fatty acid desaturase (FAD, EC: 1.14.19.-) to form 18:2/3-ACP. Finally, under the control of acyl-ACP thioesterase (FAT, EC: 3.1.2.14 3.1.2.-) and palmitoyl-CoA hydrolase (PCH, EC: 3.1.2.2), free fatty acids are released from the acyl carrier protein (ACP). Three unigenes that encoded AAD were all up-regulated, which suggested that B. napus tends to produce unsaturated fatty acid in the seeds. Four unigenes that encoded FATB were identified, of which three unigenes were down-regulated to form 16:0 palmitic acid and 18:0 stearic acid. Two unigenes that encoded FATA were all up-regulated to form 18:1oleic acid, which was the most common fatty acid in B. napus seeds (59.14%).
Table 5. Enzymes/protein related to FA biosynthesis and metabolism identified by annotation of the B. napus unigenes.
| Symbol | Enzymes/Protein | EC Number | Number of unigenes | Up/Down(numbers of unigenes) |
|---|---|---|---|---|
| Fatty acid biosynthesis | ||||
| ACP | Acyl carrier protein | 10 | 3up5down | |
| accB/bccp | acetyl-CoA carboxylase biotin carboxyl carrier protein | 4 | 3up1down | |
| ACC | Acetyl-CoA carboxylase 1/Biotin carboxylase | EC:6.4.1.2 | 3 | 2up |
| accA/accD | Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha/beta | EC:6.4.1.2 | 3 | 1up1down |
| MAT | Malonyl-CoA-acyl carrier protein transacylase | EC:2.3.1.39 | 2 | 1up |
| KAS III | 3-oxoacyl (ketoacyl)-[acyl-carrier-protein] synthase III | EC:2.3.1.180 | 3 | 1up |
| KAS II | 3-oxoacyl-[acyl-carrier-protein] synthase II | EC:2.3.1.179 | 6 | 5up1down |
| KAS I | 3-oxoacyl-[acyl-carrier-protein] synthase I | EC:2.3.1.41 | 1 | 1up |
| KAR | 3-oxoacyl-[acyl-carrier-protein] reductase | EC:1.1.1.100 | 2 | 2up |
| HAD | hydroxyacyl-acyl-ACP dehydratase | EC:4.2.1.- | 5 | 4up1down |
| EAR | Enoyl-[acyl-carrier-protein] reductase [NADH] | EC:1.3.1.9 | 1 | 1up |
| FATA | Oleoyl-acyl carrier protein thioesterase | EC:3.1.2.14 3.1.2.- | 2 | 2up |
| FATB | Palmitoyl/stearoyl-acyl carrier protein thioesterase | EC:3.1.2.14 3.1.2.- | 4 | 3down |
| Fatty acid elongation | ||||
| KCS1 | 3-ketoacyl-CoA synthase | EC:2.3.1.- | 21 | 3up9down |
| KCR2(KR) | Ketoacyl-CoA Reductase | EC:1.1.1.- | 3 | 1down |
| HACD(PSH1) | Hydroxyacyl-CoA Dehydratase | EC:4.2.1.- | 7 | 3up |
| ECR | enoyl-CoA reductase | EC:1.3.1.38 | 2 | 2up |
| PPT | Palmitoyl-protein thioesterase 1 | EC:3.1.2.22 | 6 | 3up1down |
| PCH | palmitoyl-CoA hydrolase | EC:3.1.2.2 | 1 | |
| Fatty acid desaturation | ||||
| AAD | Acyl-ACP desaturase | EC:1.14.19.2 | 3 | 3up |
| FAD2 | Omega-6 fatty acid desaturase, endoplasmic reticulum | EC:1.14.19.- | 1 | 1up |
| FAD3 | Omega-3 fatty acid desaturase, endoplasmic reticulum | EC:1.14.19.- | 1 | 1up |
| FAD5 | Palmitoyl-monogalactosyldiacylglycerol delta(δ)-7 desaturase, chloroplastic | EC:1.14.19.- | 4 | 1down |
| FAD6 | Omega-6 fatty acid desaturase, chloroplastic | EC:1.14.19.- | 2 | 2down |
| FAD7 | Omega-3 fatty acid desaturase, chloroplastic | EC:1.14.19.- | 6 | 1up2down |
| FAD8 | Omega-3 fatty acid desaturase, chloroplastic | EC:1.14.19.- | 2 | 2down |
| Fatty acid metabolism | ||||
| ACAT | acetyl-CoA acyltransferase | EC:2.3.1.16 | 1 | 1up |
| AACT | Acetyl-CoA C-acetyltransferase | EC:2.3.1.9 | 4 | 2up |
| ACAD | Acyl-CoA dehydrogenase | EC:1.3.99.3 | 3 | 1up1down |
| ACOX | acyl-coenzyme A oxidase | EC:1.3.3.6 | 11 | 1up5down |
| LACS | long-chain acyl-CoA synthetase | EC:6.2.1.3 | 10 | 3up6down |
| ADH | Alcohol dehydrogenase | EC:1.1.1.1 | 18 | 5up5down |
| ALDH | Aldehyde dehydrogenase | EC:1.2.1.3 | 23 | 10up6down |
| MFP2 | enoyl-CoA hydratase/3-hydroxybutyryl-CoA dehydrogenase 2 | EC:4.2.1.17 1.1.1.35 1.1.1.21 | 12 | 6up3down |
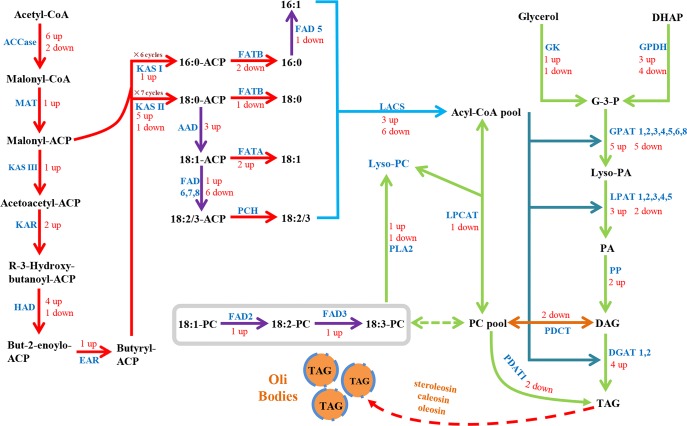
Fig 5. Overview of de novo fatty acid (FA) and triacylglycerol (TAG) biosynthesis pathways.
Indentified enzymes include: ACCase, aacetyl-CoA carboxylase carboxyl transferase (EC:6.4.1.2); MAT, Malonyl-CoA-ACP transacylase (EC:2.3.1.39); KAS, 3- oxoacyl ACP synthase (KASI, EC: 2.3.1.41; KASII, EC: 2.3.1.179; KAS III, EC: 2.3.1.180); KAR, 3-oxoacyl ACP reductase (EC:1.1.1.100); HAD, 3R-hydroxymyristoyl ACP dehydrase (EC:4.2.1.-); EAR, enoyl-ACP reductase I (EC:1.3.1.9); FATA/B, fatty acyl-ACP thioesterase A/B (EC:3.1.2.14 3.1.2.-); AAD, acyl-ACP desaturase (EC:1.14.19.2); PCH, palmitoyl-CoA hydrolase (EC:3.1.2.2); LACS, long-chain acyl-CoA synthetase (EC:6.2.1.3); FAD2/6, D12(ω6)-Desaturase (EC:1.14.19.-); FAD3/7/8, D15(ω3)-Desaturase (EC:1.14.19.-); GK, glycerol kinase (EC:2.7.1.30); ATS1/GPAT, glycerol-3-phosphate acyltransferase (EC:2.3.1.15); LPAT, lysophosphatidyl acyltransferase (EC:2.3.1.51); PP, phosphatidate phosphatase (EC:3.1.3.4); DGAT1, diacylglycerol O-acyltransferase 1 (EC:2.3.1.20); PDAT1, phospholipid: diacylglycerol acyltransferase 1 (EC:2.3.1.158); LPCAT, lysophosphatidylcholine acyltransferase (EC:2.3.1.23 2.3.1.67); PLA2, Phospholipase A2 (EC:3.1.1.4). Numbers indicated the numbers of each enzyme, and “up” meant up-regulation comparing 25DAP seeds with leaves, and “down” meant down-regulation. Lipid substrates are abbreviated: 16:0, palmitic acid; 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid.
In addition, ten unigenes that encoded long-chain acyl-CoA synthetases (LACS, EC: 6.2.1.3), which catalyze the esterification of free fatty acids to CoA upon arrival in the cytoplasm [56], and 15 unigenes that encoded acyl CoA binding protein (ACBP), which binds medium and long-chain acyl-CoA esters with a very high affinity and might function as an intracellular carrier of acyl-CoA esters [57], were also identified. Fig 5 and S5 Table showed that although three unigenes encoding LACS were up-regulated and six were down-regulated, eight unigenes encoding ACBP were up-regulated and three were down-regulated in seeds compared to leaves. This result meant that ACBP might play a critical role in improving oil content in the seeds rather than LACS.
ACCase is a crucial enzyme in de novo fatty acid synthesis, and its overexpression could alter the fatty acid composition of seeds and increase the fatty acid content, which would lead to an increased oleic acid content and seeds yield [58, 59]. The transcriptional level of the unigenes encoding ACCase in our transcriptome data was consistent with the reported results. Six unigenes were up-regulated and only two were down-regulated in seeds compared to leaves. The next key enzymes/proteins in fatty acid synthesis are ACP co-factor and the KAS enzymes in FAS. Research on Brassica juncea revealed that the functional expression of an ACP from Azospirillum brasilense could improve the content of 18:1 and 18:2 in seeds, and enhanced the ratio of monounsaturated (C18:1)/saturated fatty acids and linoleic (C18:2)/linolenic (C18:3) acid. It also reduced erucic acid (C22:1) levels [60]. In our transcriptome, three unigenes that encoded ACP were up-regulated and five were down-regulated. The results suggested that these three unigenes might be very important in the composition and content of fatty acids. We identified three KAS types in plastids (KAS I, KAS II, KAS III). During the first turn of the cycle, the condensation reaction was catalyzed by KAS III which condensed acetyl-CoA with malonyl-ACP to form acetoacetyl-ACP. For the next six turns of the cycle, KAS I catalyzed the condensation reaction to form 16:0-ACP. Finally, KAS II catalyzed 16:0-ACP to elongate to 18:0-ACP. Overexpression of KAS III induced an increase in the levels of 16:0 in tobacco, but reduced the rate of lipid synthesis [61]. Likewise, the suppression of KAS II led to an increase in 16:0 accumulation (53%), but there were deformities in some of the transgenic offspring [62]. Changes to KAS I caused a mutant that had a different polar lipid composition, disrupted embryo development and reduced fatty acid levels (~33.6% of the wild type) in its seeds [63], which suggested that KAS I was also very important to fatty acid synthesis. Unigenes that encoded KAS were almost all up-regulated in seeds compared to leaves in our transcriptome, which indicated that KAS was crucial to the change seen in the quality and content of fatty acids in B. napus seeds.
Genes related to TAG and OB biosynthesis
We identified 43 unigenes that encoded seven enzymes involved in the suggested pathway for TAG biosynthesis (Table 6, Fig 5) [3, 64]. Three unigenes that encoded glycerol kinase (GK, EC: 2.7.1.30) and twelve unigenes that encoded glycerol-3-phosphate dehydrogenase (GPDH, EC: 1.1.1.8 1.1.5.3) were identified. They catalyzed the glycerol to glycerol-3-phosphate (G-3-P) step, an initial substrate in the TAG pathway. Then 11 unigenes that encoded the key enzyme of TAG biosynthesis, glycerol-3-phosphate acyltransferase (GPAT, EC: 2.3.1.15; one for GPAT1, two for GPAT2, two for GPAT3, one for GPAT4, two for GPAT5, two for GPAT6 and one for GPAT8), were identified. These enzymes catalyzed the first acylation of G-3-P at the sn-1 position to form lysophosphatidic acid (Lyso-PA). The second acylation was catalyzed by 1-acyl-sn-glycerol-3-phosphate acyltransferase (LPAT, EC: 2.3.1.51; four for LPAT1, one for LPAT2, one for LPAT3 and one for LPAT4), to form phosphatidic acid (PA) at the sn-2 position of G-3-P. Among these 34 unigenes, 12 unigenes were up-regulated and 12 unigenes were down-regulated in seeds compared to leaves, which showed that they were important in both seeds and leaves. After the second acylation, the dephosphorylation of the resultant PA was catalyzed by phosphatidate phosphatase (PP, EC: 3.1.3.4), a key regulator of lipid homeostasis, to form diacylglycerol (DAG). Two unigenes that encoded PP were all up-regulated, which would increase the TAG substrate levels. The final acylation reaction was that several kinds of enzymes catalyzed DAG to TAG. Three kinds of enzymes, differing in their acyl donor, were identified. One was diacylglycerol O-acyltransferase (DGAT, EC: 2.3.1.20), encoded by five unigenes (two for DGAT1 and three for DGAT2), which catalyzed DAG at the sn-3 position using a fatty acyl-CoA molecule. Four unigenes were up-regulated among the five unigenes, which indicated that DGAT was a crucial TAG synthesis enzyme in seeds. The second enzyme was phospholipid diacylglycerol acyltransferase1 (PDAT1, EC 2.3.1.43), encoded by two unigenes, which catalyzed DAG using PC as the acyl donor. But the synthesis of DAG by PDAT1 was dependent on lysophosphatidylcholine acyltransferase (LPCAT, EC: 2.3.1.23) activity to regenerate PC from lyso-PC [65]. It was interesting that one unigene that encoded LPCAT and two unigenes that encoded PDAT1 were down-regulated in seeds compared to leaves in B. napus according to our transcriptome data, which suggested that the two enzymes might play more roles in TAG synthesis in leaves than seeds. The last enzyme was diacylglycerol cholinephosphotransferase (PDCT, EC: 2.7.8.2), encoded by three unigenes, which catalyzed the transfer of the phosphocholine head-group from PC to DAG, leading to an increase in the desaturation of fatty acids to DAG and subsequently to TAG [66]. Two unigenes were down-regulated, which indicated that PDCT and PDAT1 played important roles in TAG synthesis in B. napus leaves. Previous studies also demonstrated that ectopic expression of DGAT, a key enzyme regulating the rate of the Kennedy pathway, could improve the oil content in Arabidopsis, soybean and maize seeds [67–69]. In addition, phospholipase A2 (PLA2, EC: 3.1.1.4), encoded by two unigenes, was identified and might be involved in membrane lipid synthesis associated with PDAT1 and LPCAT, such as PC to TAG biosynthesis.
Table 6. Enzymes related to TAG biosynthesis and metabolism identified by annotation of the B. napus unigenes.
| Symbol | Enzymes/Protein | EC Number | Number of unigenes | Up/Down(numbers of unigenes) |
|---|---|---|---|---|
| TAG biosynthesis | ||||
| GK | Glycerol kinase | EC:2.7.1.30 | 3 | 1up1down |
| GPDH | Glycerol-3-phosphate dehydrogenase | EC:1.1.1.8 1.1.5.3 | 12 | 3up4down |
| GPAT1 | Glycerol-3-phosphate acyltransferase 1 | EC:2.3.1.15 | 1 | 1up |
| GPAT2 | Glycerol-3-phosphate acyltransferase 2 | EC:2.3.1.15 | 2 | 2down |
| GPAT3 | Glycerol-3-phosphate acyltransferase 3 | EC:2.3.1.15 | 2 | 2up |
| GPAT4 | Glycerol-3-phosphate 2-O-acyltransferase 4 | EC:2.3.1.15 | 1 | 1down |
| GPAT5 | Glycerol-3-phosphate acyltransferase 5 | EC:2.3.1.15 | 2 | 1up |
| GPAT6 | Glycerol-3-phosphate acyltransferase 6 | EC:2.3.1.15 | 2 | 1up1down |
| GPAT8 | Glycerol-3-phosphate acyltransferase 8 | EC:2.3.1.15 | 1 | 1down |
| LPAT1 | 1-acyl-sn-glycerol-3-phosphate acyltransferase 1, chloroplastic | EC:2.3.1.51 | 4 | 1down |
| LPAT2 | 1-acyl-sn-glycerol-3-phosphate acyltransferase 2 | EC:2.3.1.51 | 1 | 1up |
| LPAT3 | 1-acyl-sn-glycerol-3-phosphate acyltransferase 3 | EC:2.3.1.51 | 1 | 1up |
| LPAT4 | 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 | EC:2.3.1.51 | 1 | 1down |
| LPAT5 | 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 | EC:2.3.1.51 | 1 | 1up |
| PP | Phosphatidate phosphatase | EC:3.1.3.4 | 2 | 2up |
| DGAT1 | Diacylglycerol O-acyltransferase 1 | EC:2.3.1.20 | 2 | 1up |
| DGAT2 | Diacylglycerol O-acyltransferase 2 | EC:2.3.1.20 | 3 | 3up |
| PDAT1 | Phospholipid:diacylglycerol acyltransferase 1 | EC:2.3.1.158 | 2 | 2down |
| ACBP | Acyl-CoA-binding protein | 15 | 8up3down | |
| Acyl editing | ||||
| PLA2 | Phospholipase A2 | EC:3.1.1.4 | 2 | 1up1down |
| LPCAT | lysophosphatidylcholine acyltransferase | EC:2.3.1.23 2.3.1.67 | 1 | 1down |
| PDCT | Diacylglycerol cholinephosphotransferase | EC 2.7.8.2 | 3 | 2down |
| TAG metabolism | ||||
| TAGL | Triacylglycerol lipase | EC:3.1.1.3 | 12 | 3up4down |
| MAGL | Monoacylglycerol lipase | EC:3.1- | 3 | 1up1down |
Once synthesized, the TAG molecules can be stored in the form of an OB surrounded by a membrane composed of a layer of phospholipids embedded with several proteins, such as oleosin, caleosin and steroleosin, in mature seeds [53, 70]. We identified 16 unigenes that encoded oleosin, three encoding caleosin and two encoding steroleosin (Table 7). Olesosin, which contains a hydrophilic oil body-binding domain flanked by two amphipathic domains, helps stabilize OBs by increasing space bit resistance and charge repulsion, which prevent the fusion of OBs [53, 71]. Caleosin was not only involved in the synthesis and metabolism of OBs, but also plays a role in plant drought tolerance and TAG mobilization during germination, possibly by facilitating interactions with vacuoles [71–73]. Steroleosin, in addition to being an oil body-anchoring domain, might represent a class of dehydrogenases/reductases that may play a role in signal transduction by various sterols [74]. Among the 21 unigenes encoding oil body proteins, only two unigenes were down-regulated (one for olesion and one for caleosin). Among the 19 up-regulated unigenes in seeds, some unigenes were not detected in leaves at the transcriptional level (Table 7). This demonstrated that oleosin, caleosin and steroleosin play crucial roles in the synthesis of OBs in B. napus seeds, which will help future functional studies of B. napus.
Table 7. Unigenes annotated as oleosin, caleosin and steroleosin and the expression in 25DAP seeds and leaves.
| Oil body protein | Unigenes ID | Leaves | Seeds | FDR | Log2FC | Regulated |
|---|---|---|---|---|---|---|
| Oleosin | c1584.graph_c0 | |||||
| c23541.graph_c0 | 6.312222768 | 3.656048324 | 0.594927299 | -0.739596381 | down | |
| c24038.graph_c0 | 0 | 372.5670435 | 2.28E-08 | 9.015085824 | up | |
| c24532.graph_c0 | 0 | 725.9146663 | 2.86E-11 | 11.44138334 | up | |
| c28274.graph_c0 | 0 | 7990.140177 | 1.11E-16 | 15.82084718 | up | |
| c29002.graph_c0 | 0 | 2.385682868 | 0.033975201 | 3.61024562 | up | |
| c29397.graph_c0 | 0 | 14.47071134 | 1.74E-05 | 6.580151515 | up | |
| c32762.graph_c0 | 0.533087123 | 109.6065151 | 6.50E-07 | 7.311413204 | up | |
| c32777.graph_c0 | 0.536044935 | 4314.157298 | 2.40E-13 | 12.67767571 | up | |
| c33249.graph_c0 | 2.651180655 | 7784.451408 | 7.38E-12 | 11.42973502 | up | |
| c33254.graph_c0 | 0 | 1281.10938 | 3.30E-13 | 13.06117547 | up | |
| c33463.graph_c0 | 0 | 42.25251576 | 3.73E-07 | 7.99515731 | up | |
| c35049.graph_c0 | 0 | 124.6277195 | 3.14E-09 | 9.736084183 | up | |
| c37562.graph_c0 | 1.046391746 | 18864.00137 | 6.66E-15 | 13.95741831 | up | |
| c49494.graph_c0 | ||||||
| c52336.graph_c0 | ||||||
| caleosin | c33604.graph_c0 | 101.6787801 | 58.06664131 | 0.833270798 | -0.793341799 | down |
| c34268.graph_c0 | 0 | 1935.17207 | 1.15E-14 | 14.27874692 | up | |
| c46025.graph_c0 | 0 | 103.2154874 | 1.64E-07 | 8.296078219 | up | |
| steroleosin | c37523.graph_c0 | 0 | 270.6438792 | 4.88E-11 | 11.24821215 | up |
| c40308.graph_c0 | 0 | 464.4814328 | 2.25E-12 | 12.36463432 | up |
Genes related to fatty acid desaturation
Fatty acid desaturation contains two steps. the first step is the formation of monounsaturated fatty acids from saturated fatty acids in plastids, which is catalyzed by AAD [75]. The second step is the formation of unsaturated bonds on the monounsaturated fatty acids at specifically defined positions (△12, △15 or △16), which is catalyzed by enzymes located on the membranes of the endoplasmic reticulum and chloroplast [76], including △12(w6)-desaturase (FAD2 and FAD6, EC: 1.14.19.-), which desaturates oleic acid (18:1) to form linoleic acid (18:2), and △15(w3)-desaturase (FAD3, FAD7 and FAD8, EC: 1.14.19.-), which further desaturates linoleic acid (18:2) to form α-linolenic acid (18:3). Besides these desaturations, we also identified palmitoyl-monogalactosyldiacylglycerol delta(δ)-7 desaturase (FAD5, EC: 1.14.19.-), which affects the accumulation of 16:3Δ7,10,13 by catalyzing 16:0 MGDG to form 16:1 MGDG at position (△7) in leaves [3, 77, 78]. We found that three unigenes encoded AAD, one encoded FAD2, one encoded FAD3, four encoded FAD5, two encoded FAD6, six encoded FAD7 and two encoded FAD8 (Table 5). The unigenes that encoded AAD, FAD2 and FAD3 were up-regulated, which showed that these unigenes might play an important role in oleic acid (18:1) biosynthesis in seeds, for the unigenes encoded FAD2 and FAD3 were identified in endoplasmic reticulum. Six unigenes out of the seven that encoded FAD6, 7 and 8, and one unigene that encoded FAD5 were down-regulated, which could explain the high PG, MGDG, DGDG and SQDG contents in leaves for these unigenes were identified in chloroplastic, which are produced by the synthesis pathways for prokaryotic galactolipid, sulfolipid and phospholipid [3]. These results will help us to further explore lipid synthesis by leaves.
Genes related to the catabolism pathways for TAGs and fatty acids
The long-chain, insoluble TAGs are hydrolyzed in two steps. First, the TAGs are catalyzed by triacylglycerol lipase (TAGL, EC: 3.1.1.3) to hydrolyze the ester bonds that link fatty acyl chains to the glycerol backbone by releasing free fatty acids from DAG and TAG. The last ester bond is hydrolyzed by monoacylglycerol lipase (MAGL, EC: 3.1.-) [3]. Twelve unigenes that encoded TAGL and three encoding MAGL were identified in the B. napus transcriptome. We found that there were four down-regulated and three up-regulated unigenes for TAGL, and one down-regulated and one up-regulated unigene for MAGL (Table 6), which showed that these unigenes in both leaves and seeds were crucial for lipid degradation. The second step in TAG catabolism is the catabolism of fatty acids to form acetyl-CoA, which is further broken down by oxidation or other metabolic pathways [79]. According to the KEGG pathway assignment and annotation of the unigenes in the transcriptome, 82 unigenes that encoded eight kinds of enzymes related to fatty acid catabolism were identified; three key enzymes were acyl-CoA oxidase (ACOX, EC: 1.3.3.6), enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (MFP2, EC: 4.2.1.17 1.1.1.35 1.1.1.211) and acetyl-CoA acyltransferase (ACAT, EC: 2.3.1.16), which were encoded by 11, 12 and one unigenes, respectively (Table 5). The acetyl-CoA generated by fatty acid catabolism is used to produce energy for the cell via the citrate cycle or participates in TAG biosynthesis.
TAG and fatty acid catabolism proceeds in an opposite direction to their synthesis. So, the way to increase the accumulation of lipids may be to suppress the catabolism of TAG and fatty acids, which would improve the quality of B. napus.
Detection of TFs involved in lipid synthesis
Many TFs were involved in the synthesis and deposition of seed oil, such as LEC1, LEC2, ABI3, WRI1 and FUS3 (http://lipidlibrary.aocs.org/plantbio/transfactors/index.htm) [27, 28]. In this study, we identified that 3387 unigenes annotated with 1122 independent Arabidopsis TFs coding sequences belonged to 49 known TF families [47]. We found that the largest number of unigenes (667) was annotated to the Trihelix family, followed by the C2H2 family (456) (S6 Table). We identified 27 unigenes that encoded 11 TFs that are involved in oil biosynthesis according to research by Fobert (Table 8). These 11 TFs were ABI3, LEC1, WRI1, ADOF1, EMF2, AP2, LEC2, FUS3, GL2, HSI2-L1 and HSI2, and they might play a more important role in the synthesis of seed oil than in leaf oil. However, there were no unigenes that showed homology to L1L, PKL, FIE or SWN, which indicated that these TFs did not have much of a role in oil synthesis. To further understand the function of these 11 TFs, we analyzed the expression of the unigenes encoding these TFs (Table 8). We found that nearly all the unigenes were up-regulated in seeds compared to leaves, except for one ADOF1 unigene and one AP2 unigene. Among the up-regulated unigenes, the unigenes encoding ABI3, LEC1, FUS3 and GL2 in leaves had no expression at the transcription level, which revealed that these TFs played an important role in seed oil synthesis, but were probably not involved in oil synthesis in leaves. WRI1 was much more highly expressed in seeds than in leaves, which suggested that it had an extremely important role in oil synthesis, which was consistent with the function of WRI1 during fatty acid biosynthesis and photosynthesis, where it regulates the expression of GT1-element and/or GCC-box containing genes [80]. We also performed a wide expression analysis of all the transcription factor families (Table 9). Among these transcription factor families, ABI3VP1, AtRKD, CPP, E2F-DP, GRF, JUMONJI, MYB-related, PHD and REM may play an important role in lipid biosynthesis by seeds because the up-regulated unigenes in these transcription factor families made up a larger percentage (over 90% in all expressed unigenes) than the down-regulated unigenes (Table 9). This result showed that the unigenes in these TF families might be involved in or even contribute to the oil synthesis in seeds, which would lay a foundation for further research on transcription factor regulation during lipid biosynthesis. In summary, these analyses could provide further information about the regulation mechanism underlying TFs’ roles in oil synthesis.
Table 8. The relative expression of unigenes identified as the TF involved in oil synthesis.
| TF name | Unigenes ID | Leaves | Seeds | FDR | Log2FC | Regulated |
|---|---|---|---|---|---|---|
| ABI3(Abscisic Acid Insensitive 3) | c42986.graph_c0 | 0 | 53.901553 | 3.05E-10 | 10.58245 | up |
| LEC1(Leafy cotyledon1) | c33792.graph_c0 | 0 | 22.168763 | 2.17E-06 | 7.3483149 | up |
| WRI1(Wrinkled1) | c40435.graph_c0 | 1.1940946 | 73.933088 | 3.01E-05 | 5.8758729 | up |
| ADOF1(Arabidopsis Dof Zinc Finger Protein 1) | c34995.graph_c0 | 75.485511 | 24.245782 | 0.5465201 | -1.62022 | down |
| c22022.graph_c0 | ||||||
| EMF2(Embryonic Flower 2) | c23937.graph_c0 | 0.2626646 | 3.7354577 | 0.0396252 | 3.1316306 | up |
| c43162.graph_c0 | 6.0134657 | 19.844284 | 0.4507925 | 1.725307 | up | |
| AP2(APETALA2) | c42473.graph_c0 | 11.026932 | 13.492089 | 0.7413012 | 0.3010018 | up |
| c2509.graph_c0 | 3.7956765 | 0.3490075 | 0.0485946 | -2.682913 | down | |
| LEC2(Leafy cotyledon2) | c57560.graph_c0 | |||||
| c55216.graph_c0 | ||||||
| FUS3(Fusca3) | c33720.graph_c2 | 0 | 10.742026 | 0.0004825 | 5.3306493 | up |
| c38470.graph_c0 | 0 | 30.22454 | 4.49E-08 | 8.767464 | up | |
| GL2(GLABRA2) | c26728.graph_c0 | 0 | 7.737113 | 0.0016967 | 4.8439302 | up |
| c40770.graph_c0 | 0 | 6.9014965 | 0.0001735 | 5.7197442 | up | |
| c40770.graph_c1 | 0 | 7.5332505 | 9.66E-05 | 5.9406611 | up | |
| HSI2-L1/HSL1(HSI2-Like 1) | c32344.graph_c0 | |||||
| c45245.graph_c0 | 0.9631632 | 6.6234755 | 0.0779525 | 2.7608182 | up | |
| c45808.graph_c0 | 1.2847384 | 9.3659689 | 0.0664691 | 2.8455825 | up | |
| c43966.graph_c0 | 7.6220108 | 8.9851268 | 0.7515839 | 0.2482162 | up | |
| HSI2(High-Level Expression of Sugar-Inducible Gene 2) | c27173.graph_c0 | |||||
| c28819.graph_c0 | 0.1216735 | 4.2177701 | 0.0239492 | 3.5552057 | up | |
| c32124.graph_c0 | 0.3471751 | 10.723232 | 0.0031922 | 4.2128966 | up | |
| c45955.graph_c0 | 2.114279 | 12.57168 | 0.1269483 | 2.5626317 | up | |
| c28424.graph_c0 | 0.9865357 | 4.1651317 | 0.3120835 | 1.7648368 | up | |
| c27807.graph_c0 | 0 | 5.1606096 | 0.0083859 | 4.2048549 | up | |
| c41698.graph_c0 | 0.2267668 | 3.8850453 | 0.0053936 | 3.8479633 | up |
Table 9. The expression distribution of unigenes in transcript factor families in B.napus.
| TF families | Numbers of expressed unigenes | Numbers of up-reuglated unigenes against leaves | Numbers of down-reuglated unigenes against leaves | Percentage of up-regulated unigenes against leaves | Percentage of down-regulated unigenes against leaves |
|---|---|---|---|---|---|
| ABI3VP1 | 18 | 17 | 94.44% | 5.56% | |
| Alfin-like | 7 | 5 | 2 | 71.43% | 28.57% |
| AP2-EREBP | 88 | 38 | 50 | 43.18% | 56.82% |
| ARF | 31 | 23 | 8 | 74.19% | 25.81% |
| ARID | 4 | 3 | 1 | 75.00% | 25.00% |
| ARR-B | 22 | 13 | 9 | 59.09% | 40.91% |
| AtRKD | 1 | 1 | 0 | 100.00% | 0.00% |
| BBR/BPC | 7 | 3 | 4 | 42.86% | 57.14% |
| bHLH | 178 | 75 | 103 | 42.13% | 57.87% |
| bZIP | 60 | 34 | 26 | 56.67% | 43.33% |
| BZR | 9 | 4 | 5 | 44.44% | 55.56% |
| C2C2-CO-like | 30 | 6 | 24 | 20.00% | 80.00% |
| C2C2-Dof | 25 | 14 | 11 | 56.00% | 44.00% |
| C2C2-Gata | 22 | 10 | 12 | 45.45% | 54.55% |
| C2C2-YABBY | 4 | 3 | 1 | 75.00% | 25.00% |
| C2H2 | 379 | 222 | 157 | 58.58% | 41.42% |
| C3H | 261 | 141 | 120 | 54.02% | 45.98% |
| CAMTA | 27 | 17 | 10 | 62.96% | 37.04% |
| CCAAT-HAP2 | 6 | 4 | 2 | 66.67% | 33.33% |
| CCAAT-HAP3 | 5 | 2 | 3 | 40.00% | 60.00% |
| CCAAT-HAP5 | 8 | 7 | 1 | 87.50% | 12.50% |
| CPP | 5 | 5 | 0 | 100.00% | 0.00% |
| E2F-DP | 7 | 7 | 0 | 100.00% | 0.00% |
| EIL | 6 | 3 | 3 | 50.00% | 50.00% |
| G2-like | 35 | 21 | 14 | 60.00% | 40.00% |
| GeBP | 12 | 7 | 5 | 58.33% | 41.67% |
| GRAS | 33 | 12 | 21 | 36.36% | 63.64% |
| GRF | 10 | 10 | 0 | 100.00% | 0.00% |
| Homeobox | 87 | 58 | 29 | 66.67% | 33.33% |
| HRT | 1 | 0 | 1 | 0.00% | 100.00% |
| HSF | 28 | 15 | 13 | 53.57% | 46.43% |
| JUMONJI | 4 | 4 | 0 | 100.00% | 0.00% |
| MADS | 34 | 19 | 15 | 55.88% | 44.12% |
| MYB | 49 | 28 | 21 | 57.14% | 42.86% |
| MYB-related | 2 | 2 | 0 | 100.00% | 0.00% |
| NAC | 75 | 46 | 29 | 61.33% | 38.67% |
| NLP | 8 | 5 | 3 | 62.50% | 37.50% |
| Orphan | 3 | 0 | 3 | 0.00% | 100.00% |
| PHD | 19 | 19 | 0 | 100.00% | 0.00% |
| RAV | 3 | 0 | 3 | 0.00% | 100.00% |
| REM | 27 | 26 | 1 | 96.30% | 3.70% |
| SBP | 9 | 6 | 3 | 66.67% | 33.33% |
| TCP | 49 | 21 | 28 | 42.86% | 57.14% |
| Trihelix | 518 | 266 | 252 | 51.35% | 48.65% |
| TUB | 9 | 6 | 3 | 66.67% | 33.33% |
| VOZ | 4 | 3 | 1 | 75.00% | 25.00% |
| Whirly | 2 | 0 | 2 | 0.00% | 100.00% |
| WRKY | 182 | 113 | 69 | 62.09% | 37.91% |
| ZF-HD | 18 | 12 | 6 | 66.67% | 33.33% |
| CCAAT-DR1 | 0 | 0 | 0 | 0 | 0 |
Real-time PCR analysis of selected TFs
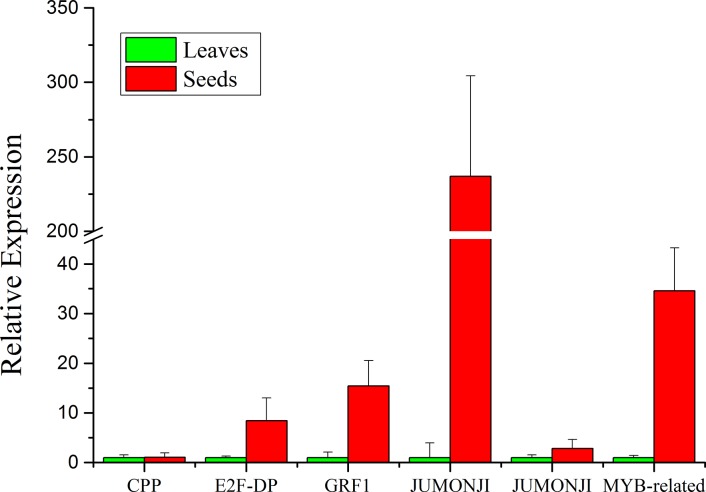
To confirm the expression difference of identified transcript factors in seeds and leaves, nine unigenes were selected for qRT-PCR analysis (Fig 6). Among these nine unigenes, three unigenes encoding transcript factors LEC1, HSI2 and REM16 were detected no expression in leaves, and only showed expression in seeds, indicating that these three transcript factors played more important roles in seeds. The other unigenes for transcript factors CPP, E2F-DP, GRF1, JUMONJI and MYB-related were up-regulated in seeds, and the most increased was the unigene JUMONJI, followed by MYB-related, GRF, E2F-DP, JUMONJI and CPP. Because the main metabolisms in seeds are fatty acids and lipids synthesis metabolism, so the high expression level of these unigenes for transcript factors might be involved in the fatty acids and lipids synthesis in seeds. The results of qRT-PCR analysis confirmed the transcriptome data. These TFs might provide the new clue for understanding the mechanism of fatty acids and lipids biosynthesis in seeds.
Fig 6. Relative expression of selected unigenes for transcript factors between leaves and developing seeds.
Conclusion
Although the sequencing of the whole B. napus genome has been finished [38], which could provide huge genomic information for scientific research, the regulation of lipids synthesis between leaves and seeds in B. napus was still unclear. This study has revealed how the genes involved in the biosynthesis and metabolism of lipids were regulated by analyzing the B. napus seeds and leaves transcriptome. We found 47216 unigenes. Information about these unigenes will aid future genomic gene expression assay research and can serve as a reference transcriptome for future B. napus experiments. We identified the unigenes that encoded key enzymes and TFs that were involved in the metabolic pathways for fatty acids, and TAG biosynthesis and metabolism. We also found some genes, TFs and proteins that played extremely important roles in the accumulation of fatty acids and lipids when we compared the seeds with the leaves, such as ACCase, HAD, KASII, PP, DGAT1,2, ABI3, LEC1, WRI1, FUS3, oleosin, caleosin and steroleosin. These results will offer molecular guidance to further, more targeted experiments on B. napus. We identified some new TFs that might promote the lipid biosynthesis metabolic pathway in seeds by the transcriptome and qRT-PCR analysis, such as JUMONJI, MYB-related, GRF, E2F-DP, CPP and REM16. The gene expression regulation analysis revealed the cause of the different lipid contents between seeds and leaves and how they have evolved different functions. This study has provided insights into the molecular mechanism underlying lipid biosynthesis, and has laid the foundations for further improvements to seeds lipids through genomics research.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
The unigenes information was deposited in the Sequence Read Archive (SRA) database in NCBI (Accession number, SRR1916242).
Funding Statement
This work was funded by the National Natural Science Foundation of China (http://www.nsfc.gov.cn/) 31271760, 31471527 to XL Tan, 31071672 to Z Wang, and the The Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions (http://202.119.175.131/pages/index.aspx) to XL Tan, Z Wang and ZY Zhang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, et al. Update of the LIPID MAPS comprehensive classification system for lipids. Journal of lipid research. 2009;50 Suppl:S9–14. 10.1194/jlr.R800095-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Subramaniam S, Fahy E, Gupta S, Sud M, Byrnes RW, Cotter D, et al. Bioinformatics and systems biology of the lipidome. Chemical reviews. 2011;111(10):6452–90. 10.1021/cr200295k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, et al. Acyl-lipid metabolism. The Arabidopsis book / American Society of Plant Biologists. 2013;11:e0161 10.1199/tab.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dormann P, Benning C. Galactolipids rule in seed plants. Trends in plant science. 2002;7(3):112–8. . [DOI] [PubMed] [Google Scholar]
- 5. Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature. 2001;411(6840):909–17. 10.1038/35082000 . [DOI] [PubMed] [Google Scholar]
- 6. Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature. 2005;438(7070):1040–4. 10.1038/nature04224 . [DOI] [PubMed] [Google Scholar]
- 7. Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Lipids in photosystem II: interactions with protein and cofactors. Biochimica et biophysica acta. 2007;1767(6):509–19. 10.1016/j.bbabio.2006.12.009 . [DOI] [PubMed] [Google Scholar]
- 8. Vu HS, Shiva S, Roth MR, Tamura P, Zheng L, Li M, et al. Lipid changes after leaf wounding in Arabidopsis thaliana: expanded lipidomic data form the basis for lipid co-occurrence analysis. The Plant journal: for cell and molecular biology. 2014;80(4):728–43. 10.1111/tpj.12659 . [DOI] [PubMed] [Google Scholar]
- 9. Tzen J, Cao Y, Laurent P, Ratnayake C, Huang A. Lipids, Proteins, and Structure of Seed Oil Bodies from Diverse Species. Plant physiology. 1993;101(1):267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y, Beisson F, Pollard M, Ohlrogge J. Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry. 2006;67(9):904–15. 10.1016/j.phytochem.2006.02.015 . [DOI] [PubMed] [Google Scholar]
- 11. Miquel M, Browse J. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. The Journal of biological chemistry. 1992;267(3):1502–9. . [PubMed] [Google Scholar]
- 12. Chen X, Chen G, Truksa M, Snyder CL, Shah S, Weselake RJ. Glycerol-3-phosphate acyltransferase 4 is essential for the normal development of reproductive organs and the embryo in Brassica napus. Journal of experimental botany. 2014;65(15):4201–15. 10.1093/jxb/eru199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gómez-Campo C, Prakash S. 2 Origin and domestication. Developments in plant genetics and breeding. 1999;4:33–58. [Google Scholar]
- 14. Prakash S, Wu X-M, Bhat S. History, Evolution, and Domestication. Plant breeding reviews. 2011;35:19. [Google Scholar]
- 15. Xiao G, Zhang ZQ, Yin CF, Liu RY, Wu XM, Tan TL, et al. Characterization of the promoter and 5'-UTR intron of oleic acid desaturase (FAD2) gene in Brassica napus. Gene. 2014;545(1):45–55. 10.1016/j.gene.2014.05.008 . [DOI] [PubMed] [Google Scholar]
- 16. Hu X, Sullivan-Gilbert M, Gupta M, Thompson SA. Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus L.). TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2006;113(3):497–507. 10.1007/s00122-006-0315-1 . [DOI] [PubMed] [Google Scholar]
- 17. Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG. hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant physiology. 2002;129(4):1723–31. 10.1104/pp.006353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konishi T, Shinohara K, Yamada K, Sasaki Y. Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant & cell physiology. 1996;37(2):117–22. . [DOI] [PubMed] [Google Scholar]
- 19. Brown AP, Affleck V, Fawcett T, Slabas AR. Tandem affinity purification tagging of fatty acid biosynthetic enzymes in Synechocystis sp. PCC6803 and Arabidopsis thaliana. Journal of experimental botany. 2006;57(7):1563–71. 10.1093/jxb/erj150 . [DOI] [PubMed] [Google Scholar]
- 20. Yurchenko O, Singer SD, Nykiforuk CL, Gidda S, Mullen RT, Moloney MM, et al. Production of a Brassica napus Low-Molecular Mass Acyl-Coenzyme A-Binding Protein in Arabidopsis Alters the Acyl-Coenzyme A Pool and Acyl Composition of Oil in Seeds. Plant physiology. 2014;165(2):550–60. 10.1104/pp.114.238071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knutzon DS, Hayes TR, Wyrick A, Xiong H, Maelor Davies H, Voelker TA. Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels. Plant physiology. 1999;120(3):739–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor DC, Barton DL, Giblin EM, MacKenzie SL, Van Den Berg C, McVetty P. Microsomal Lyso-Phosphatidic Acid Acyltransferase from a Brassica oleracea Cultivar Incorporates Erucic Acid into the sn-2 Position of Seed Triacylglycerols. Plant physiology. 1995;109(2):409–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends in biochemical sciences. 2006;31(12):694–9. 10.1016/j.tibs.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reue K, Brindley DN. Thematic Review Series: Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. Journal of lipid research. 2008;49(12):2493–503. 10.1194/jlr.R800019-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV Jr. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. Journal of lipid research. 2008;49(11):2283–301. 10.1194/jlr.R800018-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad I, Sharma AK, Daniell H, Kumar S. Altered lipid composition and enhanced lipid production in green microalga by introduction of brassica diacylglycerol acyltransferase 2. Plant biotechnology journal. 2014. 10.1111/pbi.12278 . [DOI] [PMC free article] [PubMed]
- 27. Baud S, Lepiniec L. Physiological and developmental regulation of seed oil production. Progress in lipid research. 2010;49(3):235–49. 10.1016/j.plipres.2010.01.001 . [DOI] [PubMed] [Google Scholar]
- 28. Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. The Plant journal: for cell and molecular biology. 2008;54(4):608–20. 10.1111/j.1365-313X.2008.03461.x . [DOI] [PubMed] [Google Scholar]
- 29. Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Molecular biology and evolution. 2009;26(12):2731–44. 10.1093/molbev/msp188 . [DOI] [PubMed] [Google Scholar]
- 30. Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, et al. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae How introns influence and enhance eukaryotic gene expression Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Nucleic acids research. 2012;40(20):10084–97. Epub 2012/09/12 2003/04/26 1997/12/24. 10.1093/nar/gks804 10.1016/s0968-0004(03)00052-5 10.1105/tpc.9.11.1963 PubMed Central PMCID: PMCPMC3488244 PMC157050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Metzker ML. Sequencing technologies—the next generation. Nature reviews Genetics. 2010;11(1):31–46. 10.1038/nrg2626 . [DOI] [PubMed] [Google Scholar]
- 32. Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, et al. BRAD, the genetics and genomics database for Brassica plants. BMC plant biology. 2011;11:136 10.1186/1471-2229-11-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genetics. 2011;43(10):1035–9. 10.1038/ng.919 . [DOI] [PubMed] [Google Scholar]
- 34. Beulé T, Camps C, Debiesse S, Tranchant C, Dussert S, Sabau X, et al. Transcriptome analysis reveals differentially expressed genes associated with the mantled homeotic flowering abnormality in oil palm (Elaeis guineensis). Tree Genetics & Genomes. 2011;7(1):169–82. 10.1007/s11295-010-0323-9 [DOI] [Google Scholar]
- 35. Guimaraes PM, Brasileiro AC, Morgante CV, Martins AC, Pappas G, Silva OB Jr., et al. Global transcriptome analysis of two wild relatives of peanut under drought and fungi infection. BMC genomics. 2012;13:387 10.1186/1471-2164-13-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H, Dong Y, Yang J, Liu X, Wang Y, Yao N, et al. De novo transcriptome of safflower and the identification of putative genes for oleosin and the biosynthesis of flavonoids. PloS one. 2012;7(2):e30987 10.1371/journal.pone.0030987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trick M, Long Y, Meng J, Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant biotechnology journal. 2009;7(4):334–46. 10.1111/j.1467-7652.2008.00396.x . [DOI] [PubMed] [Google Scholar]
- 38. Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–3. 10.1126/science.1253435 . [DOI] [PubMed] [Google Scholar]
- 39. Natarajan P, Parani M. De novo assembly and transcriptome analysis of five major tissues of Jatropha curcas L. using GS FLX titanium platform of 454 pyrosequencing. BMC genomics. 2011;12:191 10.1186/1471-2164-12-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y, Huang Z, Ao Y, Li W, Zhang Z. Transcriptome analysis of yellow horn (Xanthoceras sorbifolia Bunge): a potential oil-rich seed tree for biodiesel in China. PloS one. 2013;8(9):e74441 10.1371/journal.pone.0074441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larson TR, Graham IA. Technical Advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. The Plant journal: for cell and molecular biology. 2001;25(1):115–25. . [DOI] [PubMed] [Google Scholar]
- 42. Larson TR, Edgell T, Byrne J, Dehesh K, Graham IA. Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. The Plant journal: for cell and molecular biology. 2002;32(4):519–27. . [DOI] [PubMed] [Google Scholar]
- 43. Niewitetzki O, Tillmann P, Becker HC, Mollers C. A new near-infrared reflectance spectroscopy method for high-throughput analysis of oleic acid and linolenic acid content of single seeds in oilseed rape (Brassica napus L.). J Agric Food Chem. 2010;58(1):94–100. 10.1021/jf9028199 . [DOI] [PubMed] [Google Scholar]
- 44. Kumar S, Chauhan JS, Kumar A. Screening for erucic acid and glucosinolate content in rapeseed-mustard seeds using near infrared reflectance spectroscopy. Journal of food science and technology. 2010;47(6):690–2. 10.1007/s13197-010-0120-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29(7):644–52. http://www.nature.com/nbt/journal/v29/n7/abs/nbt.1883.html#supplementary-information. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5(7):621–8. 10.1038/nmeth.1226 . [DOI] [PubMed] [Google Scholar]
- 47. Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant physiology. 2006;140(3):818–29. 10.1104/pp.105.072280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z, Chen Y, Fang H, Shi H, Chen K, Zhang Z, et al. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Molecular genetics and genomics: MGG. 2014;289(5):1023–35. 10.1007/s00438-014-0853-1 . [DOI] [PubMed] [Google Scholar]
- 49. Hou R, Bao Z, Wang S, Su H, Li Y, Du H, et al. Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PloS one. 2011;6(6):e21560 10.1371/journal.pone.0021560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25(1):25–9. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic acids research. 2008;36(Database issue):D480–4. 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hills MJ. Control of storage-product synthesis in seeds. Current opinion in plant biology. 2004;7(3):302–8. 10.1016/j.pbi.2004.03.003 . [DOI] [PubMed] [Google Scholar]
- 53. Huang AHC. Oil Bodies and Oleosins in Seeds. Annual review of plant physiology and plant molecular biology. 1992;43(1):177–200. 10.1146/annurev.pp.43.060192.001141 [DOI] [Google Scholar]
- 54. Voelker T, Kinney AJ. VARIATIONS IN THE BIOSYNTHESIS OF SEED-STORAGE LIPIDS. Annual review of plant physiology and plant molecular biology. 2001;52(1):335–61. 10.1146/annurev.arplant.52.1.335 . [DOI] [PubMed] [Google Scholar]
- 55. Rismani-Yazdi H, Haznedaroglu BZ, Bibby K, Peccia J. Transcriptome sequencing and annotation of the microalgae Dunaliella tertiolecta: pathway description and gene discovery for production of next-generation biofuels. BMC genomics. 2011;12:148 10.1186/1471-2164-12-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. The Biochemical journal. 1997;323 (Pt 1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xiao S, Chye ML. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Progress in lipid research. 2011;50(2):141–51. 10.1016/j.plipres.2010.11.002 . [DOI] [PubMed] [Google Scholar]
- 58. Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J. Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant physiology. 1997;113(1):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Madoka Y, Tomizawa K, Mizoi J, Nishida I, Nagano Y, Sasaki Y. Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant & cell physiology. 2002;43(12):1518–25. . [DOI] [PubMed] [Google Scholar]
- 60. Jha JK, Sinha S, Maiti MK, Basu A, Mukhopadhyay UK, Sen SK. Functional expression of an acyl carrier protein (ACP) from Azospirillum brasilense alters fatty acid profiles in Escherichia coli and Brassica juncea. Plant physiology and biochemistry: PPB / Societe francaise de physiologie vegetale. 2007;45(6–7):490–500. 10.1016/j.plaphy.2007.03.001 . [DOI] [PubMed] [Google Scholar]
- 61. Dehesh K, Tai H, Edwards P, Byrne J, Jaworski JG. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant physiology. 2001;125(2):1103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pidkowich MS, Nguyen HT, Heilmann I, Ischebeck T, Shanklin J. Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc Natl Acad Sci U S A. 2007;104(11):4742–7. 10.1073/pnas.0611141104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu GZ, Xue HW. Arabidopsis beta-ketoacyl-[acyl carrier protein] synthase i is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. The Plant cell. 2010;22(11):3726–44. 10.1105/tpc.110.075564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bates PD, Stymne S, Ohlrogge J. Biochemical pathways in seed oil synthesis. Current opinion in plant biology. 2013;16(3):358–64. 10.1016/j.pbi.2013.02.015 . [DOI] [PubMed] [Google Scholar]
- 65. Xu J, Carlsson AS, Francis T, Zhang M, Hoffman T, Giblin ME, et al. Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC plant biology. 2012;12:4 10.1186/1471-2229-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lu C, Xin Z, Ren Z, Miquel M, Browse J. An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci U S A. 2009;106(44):18837–42. 10.1073/pnas.0908848106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lardizabal K, Effertz R, Levering C, Mai J, Pedroso MC, Jury T, et al. Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant physiology. 2008;148(1):89–96. 10.1104/pp.108.123042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, et al. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant physiology. 2001;126(2):861–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, Li J, et al. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nature genetics. 2008;40(3):367–72. 10.1038/ng.85 . [DOI] [PubMed] [Google Scholar]
- 70. Shimada TL, Hara-Nishimura I. Oil-body-membrane proteins and their physiological functions in plants. Biological & pharmaceutical bulletin. 2010;33(3):360–3. . [DOI] [PubMed] [Google Scholar]
- 71. Frandsen GI, Mundy J, Tzen JT. Oil bodies and their associated proteins, oleosin and caleosin. Physiologia plantarum. 2001;112(3):301–7. . [DOI] [PubMed] [Google Scholar]
- 72. Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, et al. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant molecular biology. 2000;44(4):463–76. . [DOI] [PubMed] [Google Scholar]
- 73. Poxleitner M, Rogers SW, Lacey Samuels A, Browse J, Rogers JC. A role for caleosin in degradation of oil-body storage lipid during seed germination. The Plant journal: for cell and molecular biology. 2006;47(6):917–33. 10.1111/j.1365-313X.2006.02845.x . [DOI] [PubMed] [Google Scholar]
- 74. Lin LJ, Tai SS, Peng CC, Tzen JT. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant physiology. 2002;128(4):1200–11. 10.1104/pp.010928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Murata N, Wada H, Gombos Z. Modes of Fatty-Acid Desaturation in Cyanobacteria. Plant and Cell Physiology. 1992;33(7):933–41. [Google Scholar]
- 76. Tasaka Y, Gombos Z, Nishiyama Y, Mohanty P, Ohba T, Ohki K, et al. Targeted mutagenesis of acyl-lipid desaturases in Synechocystis: evidence for the important roles of polyunsaturated membrane lipids in growth, respiration and photosynthesis. The EMBO journal. 1996;15(23):6416–25. [PMC free article] [PubMed] [Google Scholar]
- 77. Heilmann I, Pidkowich MS, Girke T, Shanklin J. Switching desaturase enzyme specificity by alternate subcellular targeting. Proc Natl Acad Sci U S A. 2004;101(28):10266–71. 10.1073/pnas.0402200101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heilmann I, Mekhedov S, King B, Browse J, Shanklin J. Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant physiology. 2004;136(4):4237–45. 10.1104/pp.104.052951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jaworski JG, Clough RC, Barnum SR. A Cerulenin Insensitive Short Chain 3-Ketoacyl-Acyl Carrier Protein Synthase in Spinacia oleracea Leaves. Plant physiology. 1989;90(1):41–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu XL, Liu ZH, Hu ZH, Huang RZ. BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. Journal of integrative plant biology. 2014;56(6):582–93. 10.1111/jipb.12158 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The unigenes information was deposited in the Sequence Read Archive (SRA) database in NCBI (Accession number, SRR1916242).