Abstract
Controllable release is particularly important for the delivery of small interfering RNA (siRNA), as siRNAs have a high susceptibility to enzymatic degradation if release is premature, yet lack silencing activity if they remain inaccessible within the cytoplasm. To overcome these hurdles, novel and tailorable mPEG-b-poly(5-(3-(amino)propoxy)-2-nitrobenzyl methacrylate) (mPEG-b-P(APNBMA)n) diblock copolymers containing light-sensitive o-nitrobenzyl moieties and pendant amines are employed to provide both efficient siRNA binding, via electrostatic and hydrophobic interactions, as well as triggered charge reversal and nucleic acid release. In particular, siRNA/mPEG-b-P(APNBMA)23.6 polyplexes show minimal aggregation in physiological salt and serum, and enhanced resistance to polyanion-induced unpackaging compared to polyethylenimine preparations. Cellular delivery of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes reveal greater than 80% cellular transfection, as well as rapid and widespread cytoplasmic distribution. Additionally, UV irradiation indicates ~70% reduction in targeted gene expression following siRNA/mPEG-b-P(APNBMA)23.6 polyplex treatment, as compared to 0% reduction in polyplex treated cells without UV irradiation, and only ~30% reduction for Lipofectamine treated cells. The results herein highlight the potential of these light-sensitive copolymers with a well-defined on/off switch for applications including cellular patterning for guided cell growth and extension, and cellular microarrays for exploring protein and drug interactions that require enhanced spatiotemporal control of gene activation.
Keywords: siRNA, RNA interference, polyplexes, diblock copolymer, photo-responsive
1. Introduction
Small interfering RNA (siRNA) has emerged as a promising tool for modulating gene expression and offers significant opportunities for the treatment of a range of acquired and hereditary diseases.[1] However, the clinical application of siRNAs has been limited by their high susceptibility to enzymatic hydrolysis, rapid clearance from systemic circulation, limited cellular uptake, and inability to efficiently traffic to the cytoplasm of cells.[2] In particular, the difficulty in controlling binding vs. release of siRNAs is a key factor limiting translation of siRNA nanostructures.[3] For example, in a well-known study, Davis and coworkers highlighted the importance of siRNA binding stability in the development of siRNA/cyclodextrin-containing polymer (siRNA/CDP) polyplexes for the treatment of Ewing’s sarcoma.[4] When introduced into systemic circulation in mice, the siRNA/CDP polyplexes transiently accumulated in the glomerular basement membrane, and disassembled due to interactions with heparan sulfate polyanions. Other studies have also reported rapid polyanion- or serum-induced disassembly in siRNA polyplexes,[5] emphasizing the critical need for carriers that provide stable binding within the polyanion-rich extracellular environment. At the same time, multiple reports also indicate that intracellular unbinding and siRNA release are essential to maximize gene silencing activity following delivery to the cytoplasm.[6]
Additionally, inefficient packaging of siRNA has proven to be a key challenge in designing siRNA carriers. Specifically, the short anionic chain length and rod-like solution conformation of siRNA limits its electrostatic interactions with cationic carriers and makes siRNA significantly more difficult to package than DNA.[7] Accordingly, several efforts have focused on improving the stability of siRNA encapsulation via structural adaptation of the siRNA cargo,[8] such as self-crosslinked siRNA multimers[8a–c] and sponge-like RNAi superstructures that are able to maintain highly stable RNA prior to cellular processing.[8d, e] Alternative approaches to improve the stability of siRNA polyplexes have focused on the development of modified polymers with enhanced siRNA binding capacity.[9] Among these approaches, the most commonly explored are efforts to tune siRNA binding association by modifying the polymer molecular weight and/or charge density. Multiple examples show that increasing the polymer molecular weight, in particular, can lead to significant enhancements in nucleic acid encapsulation.[10] In one such example, Howard and coworkers explored the ability to enhance siRNA binding by tailoring the molecular weight and percentage of deacetylated primary amines (degree of deacetylation) in siRNA/chitosan polyplexes.[10c] In this work, the authors demonstrated efficient complexation with increasing molecular weight for all tested degrees of deacetylation when using chitosan molecules with molecular weights greater than ~65 kDa, and identified aggregates greater than 500 nm for lower molecular weight preparations. Hydrophobic modification of polycations has also been explored to improve the binding affinity in gene delivery carriers by providing cooperative binding interactions,[9a, 11] and hydrophobic modification also enhances serum stability[12] and increases adsorption to the cell membrane.[13] However, with all such approaches, the ability to sufficiently enhance siRNA binding affinity for extracellular stabilization has been hindered by the need for siRNA liberation prior to assembly of the RNA-induced silencing complex in the cytoplasm.[6a, b] For example, in the use of linear polyethylenimine (PEI) for siRNA delivery, Shim et al. reported limited siRNA unpackaging and low gene silencing efficiency despite efficient cellular uptake and cytoplasmic localization of these structures,[6b] and other studies have reported similar effects.[5a, 14] As such, a delivery strategy that balances the requirements of strong binding stability during circulation and reduced binding stability in the cytosol is particularly attractive.
The need to control siRNA binding vs. release has motivated the development of stimuli responsive carriers whose interactions with siRNA depend upon intracellular or externally-applied cues.[6c, 15] For example, Kwon and coworkers demonstrated efficient gene silencing using acid-degradable ketalized linear PEI (KL-PEI).[6b, c] Intravenous injection of PEGylated siRNA/KL-PEI polyplexes targeting green fluorescent protein (GFP) produced significantly reduced GFP mRNA and protein levels in solid tumors as well as whole blood of mice. Stimuli-responsive carriers with sensitivity to external triggers (e.g. light, magnetic fields, or ultrasound) offer increased flexibility in timing and localization of nucleic acid release, which further motivates their application in gene transfer systems requiring activation of genes in defined cellular subpopulations, including efforts to engineer complex tissue architectures.[16] Photo-responsive materials are particularly attractive due to the versatility in photo-responsive chemistries and spatial resolution afforded by light. In fact, delivery vehicles containing the o-nitrobenzyl (o-NB) linker undergo structural changes in response to ultraviolet (UV) and near-infrared (NIR) irradiation and have enabled light-triggered release of siRNA.[17] For example, Li et al. recently demonstrated cationic amphiphilic macromolecules with a single photolabile NB linker between a long hydrophobic alkyl tail and a cationic/hydrophilic head for siRNA complexation.[18] These amphiphiles assembled in aqueous solution into micelles and bound siRNA within the cationic corona. While UV cleavage of surface-associated siRNA resulted in reduced luciferase expression following in vitro delivery to MDA-MB-231 (human breast cancer) cells, the amphiphiles did not demonstrate the enhanced binding stability necessary for continued siRNA association in the absence of a light trigger. These studies highlight the tremendous potential of photo-responsive carriers for controlling gene expression, and motivate their use to directly activate siRNA release for delivery. However, further enhancements are needed to incorporate strong binding affinity in concert with activated siRNA release and subsequent gene knockdown.
We recently demonstrated the design and synthesis of mPEG-b-poly(5-(3-(amino)propoxy)-2-nitrobenzyl methacrylate (mPEG-b-P(APNBMA)n) copolymers with tunable molecular weight, low dispersity, and o-NB-linked pendant ammonium cations to enable light-triggered polymer cleavage and charge reversal (to carboxylate anions) along the P(APNBMA)n backbone.[19] In that work, mPEG-b-P(APNBMA)7.9 facilitated efficient pDNA encapsulation, improved salt stability, and light-mediated pDNA release. Additionally, mPEG-b-P(APNBMA)n polyplexes did not compromise the viability of mouse embryonic fibroblast (NIH/3T3) cells, either with or without UV light irradiation.
In the present study, we assessed the feasibility of using mPEG-b-P(APNBMA)n copolymers containing hydrophobic and cationic binding units as carriers for siRNA. The inclusion of o-NB groups decoupled the binding vs. release of the siRNA payload and introduced the capacity for light-responsive initiation of gene silencing. We identified mPEG-b-P(APNBMA)n designs and polyplex assembly conditions to facilitate strong siRNA association and stability of these photocleavable structures. Furthermore, polyplex delivery to NIH/3T3 cells revealed high levels of cellular uptake, rapid and widespread cytoplasmic distribution of delivered siRNA polyplexes, and gene-specific and light-mediated siRNA release and silencing in response to UV irradiation, highlighting a unique approach for spatiotemporal control of gene expression.
2. Results and Discussion
2.1. siRNA binding studies
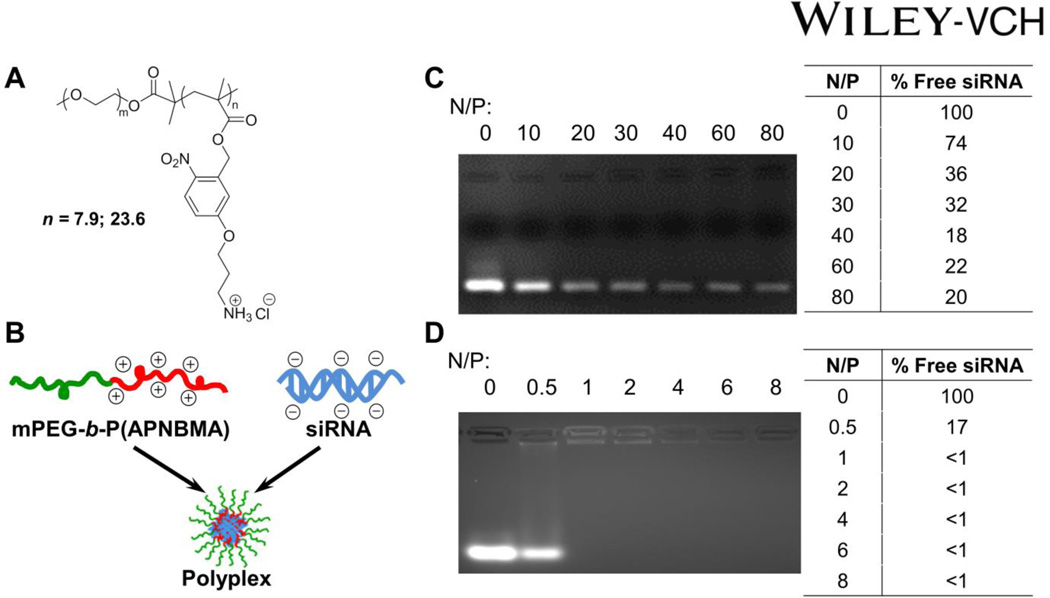
Efficient siRNA binding typically requires higher N/P ratios and/or higher molecular weight polycations as compared with DNA condensation.[3] Hence, we sought to test the siRNA binding capacity of mPEG-b-P(APNBMA)n (Mn = 7,900 g/mol, n = 7.9 or Mn = 13,100 g/mol, n = 23.6; Figure 1A) block copolymers as a function of P(APNBMA)n block length. mPEG-b-P(APNBMA)n was added to siRNA at various charge ratios to form siRNA/mPEG-b-P(APNBMA)n polyplexes (Figure 1B), and the resulting solutions were analyzed via gel electrophoresis. Previous studies utilizing mPEG-b-P(APNBMA)7.9 for pDNA delivery indicated efficient complexation at N/P ≥ 5; however, in the preparation of siRNA polyplexes, mPEG-b-P(APNBMA)7.9 was unable to efficiently complex siRNA (Fig. 1C) based on the minimal changes that were noted in ethidium bromide fluorescence upon polymer addition, as well as the persistence of a free siRNA band for N/P ratios of 10 – 80. Specifically, while there was an initial reduction in ethidium bromide fluorescence following mPEG-b-P(APNBMA)7.9 addition at N/P = 10 and small changes in fluorescence at N/P = 20 and 40, polymer addition above an N/P ratio of 40 did not induce additional polyplex formation. Several research groups have identified similar reductions in siRNA complexation efficiency with carriers that exhibited high affinity for DNA.[10c, 20]
Figure 1.
(A) Chemical structure of mPEG-b-P(APNBMA)n polymers. (B) Cartoon schematic of siRNA/mPEG-b-P(APNBMA)n polyplex formation. Representative siRNA mobility assay of (C) siRNA/mPEG-b-P(APNBMA)7.9 and (D) siRNA/mPEG-b-P(APNBMA)23.6 polyplexes in 4 wt% agarose gels stained with ethidium bromide. Lane 1 of both gels is siRNA alone (N/P = 0), while the remaining lanes show siRNA complexed with polymer at varied N/P ratios. Tabulated values indicate percentages of free siRNA based on ImageJ analysis of the free siRNA band.
In contrast, mPEG-b-P(APNBMA)23.6 provided sufficiently strong interactions to complex siRNA at low N/P ratios (Figure 1D). Upon addition of mPEG-b-P(APNBMA)23.6 at N/P 0.5, there was an initial ~83% reduction in ethidium bromide fluorescence. Further addition of the polymer (N/P ≥ 1) fully inhibited migration and low levels of fluorescence became visible in the wells. The fluorescence in the wells decreased with continued polymer addition, and disappeared completely above an N/P of 4, suggesting that mPEG-b-P(APNBMA)23.6 effectively complexed siRNA. Several studies have indicated effective condensation of siRNA using PEG-amine methacrylate block copolymers, which display similar architectures to mPEG-b-P(APNBMA).[21] For example, Duvall and coworkers reported siRNA complexation at N/P ≥ 1 for PEG-b-PDMAEMA (Mn = 17,035 g/mol) by using electrophoresis.[9a] Additionally, Rice and coworkers previously demonstrated that the stability of polyplex association could be enhanced through the incorporation of hydrophobic acridine groups into polycations.[11c] Given the high siRNA binding affinity of mPEG-b-P(APNBMA)23.6, it is possible that the cationic and hydrophobic components in the APNBMA block may cooperatively condense and stabilize the siRNA/mPEG-b-P(APNBMA)23.6 structures.
2.2. Physical characterization of polyplexes formed with mPEG-b-P(APNBMA)23.6
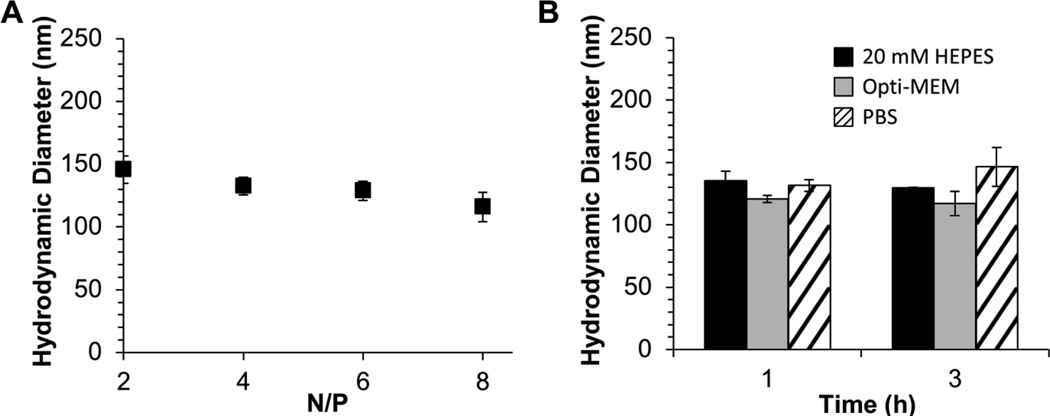
Based on the increased binding efficiency of the 23.6 repeat unit copolymer, this polymer was used for subsequent investigations. The hydrodynamic diameters (DH) of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes were measured using dynamic light scattering (DLS) (Figure 2A) to determine whether these structures would be suitable for cellular uptake, as well as to determine whether DH decreased as polymer binding affinity (and N/P ratio) increased. Polyplexes were formed at varied charge ratios ranging from N/P = 2 to 8 based on the electrophoretic complexation data. The values of polyplex DH for all polyplex formulations were below 150 nm, significantly lower than the size limit that has been suggested for endocytic uptake (< 200 nm).[22] Furthermore, the sizes of the siRNA/mPEG-b-P(APNBMA)23.6 polyplexes were relatively constant as a function of N/P. These findings indicated a high binding affinity between siRNA and the longer polycation, consistent with electrophoresis data. Furthermore, the efficient siRNA complexation combined with the small DH at N/P ≥ 4 revealed an enhanced siRNA binding capacity compared to other PEG-polycation block copolymers used for siRNA delivery.[23]
Figure 2.
Polyplex characterization using DLS. (A) DH of polyplexes determined at various N/P ratios. (B) Representative DH of polyplexes prepared at N/P = 4 following a 1 h or a 3 h incubation in 20 mM HEPES (pH = 6; black), Opti-MEM (gray), or PBS containing 150 mM NaCl (diagonal lines). Each data point represents the mean ± standard deviation for a total of three separately prepared samples.
Stealth coatings such as PEG are well known to promote colloidal stability and resist non-specific protein adsorption and opsonization in a number of gene delivery structures.[24] Thus, we tested if the incorporation of the PEG block provided nonfouling characteristics in the presence of physiological salt solutions and serum. Polyplexes were incubated in phosphate buffered saline (PBS, 150 mM NaCl) or Opti-MEM (transfection medium containing protein supplements). Subsequently, the average DH values of the polyplexes were measured by DLS to determine whether the polyplexes exhibited salt- or serum-induced changes in size as compared with polyplexes that were incubated in formulation buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, pH 6.0) (Figure 2B). The polyplex sizes remained constant at ~120 nm following 1 h and 3 h incubations in Opti-MEM at 23 °C. Incubation in PBS over a 3 h period resulted in a slight increase in polyplex size from ~130 nm at 1 h to ~145 nm following a 3 h incubation. In contrast, Zhao et al. reported siRNA/PEI (25 kDa) polyplexes greater than 1 µm in diameter following a 1 h incubation in physiological saline (150 mM NaCl).[25] In the presented studies, all polyplexes retained small sizes that were below the 200 nm threshold for endocytic uptake.[26] Based on the relative stability of the polyplex structures and the minimal increase in DH, these studies suggested that the PEG block formed a protective corona following polyplex formation to resist both salt-induced aggregation and protein adsorption.
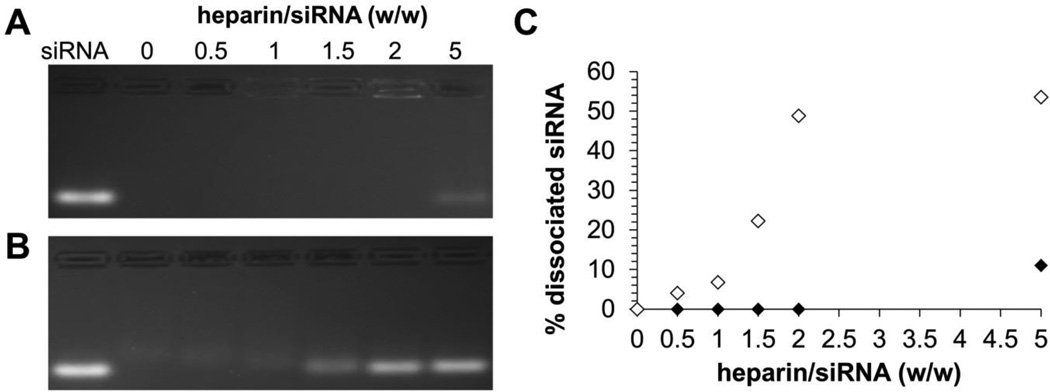
Polyanionic glycosaminoglycans (GAGs) such as heparin are major components of the extracellular matrix and have been shown to displace nucleic acids from their cationic carriers in numerous studies.[5a, 27] Thus, to evaluate the stability of mPEG-b-P(APNBMA)23.6 polyplexes against competing polyanions, we tested the susceptibility of these polyplexes to heparin displacement as compared with siRNA/PEI polyplexes. siRNA/PEI polyplexes were prepared at N/P = 6 to ensure effective binding of siRNA, and the mPEG-b-P(APNBMA)23.6 and PEI polyplexes were incubated with increasing heparin concentrations. Polyplex dissociation and siRNA release were monitored by electrophoresis (Figure 3). Gel mobility assays indicated greatly enhanced heparin stability for mPEG-b-P(APNBMA)23.6 structures, which continued to associate with siRNA and exclude ethidium bromide fluorescence in the wells of the gel for heparin/siRNA (w/w) ratios ≤ 1.5 (Figure 3A). An increase in the heparin/siRNA ratio to 2 resulted in polyplex loosening and demonstrated low levels of fluorescence in the well of the gel. Further increases of the heparin/siRNA ratio to 5 resulted in ~10% siRNA dissociation from the mPEG-b-P(APNBMA)23.6 structures (Figure 3C). In contrast, PEI preparations indicated siRNA displacement for heparin/siRNA (w/w) ratio ≥ 1.5 (Figure 3B) and ~50% siRNA dissociated for heparin/siRNA (w/w) ratio of 5. A number of studies have identified enhanced polyplex stability through the incorporation of hydrophobic groups.[28] For example, Duvall and coworkers demonstrated increased stability against heparin-mediated disassembly with increased butyl methacrylate (BMA) content in the use of PEG-b-P(DMAEMA-co-BMA) block copolymers for siRNA delivery.[9a] We hypothesize that hydrophobic groups in the cationic block may provide additional binding interactions to stabilize the siRNA polyplexes and provide increased stability against polyanion-mediated dissociation.
Figure 3.
Heparin displacement of (A) siRNA/mPEG-b-P(APNBMA)23.6 polyplexes (N/P = 4), and (B) siRNA/PEI polyplexes (N/P = 6) incubated in the presence of increasing amounts of heparin. (C) Quantification of free siRNA in siRNA/mPEG-b-P(APNBMA)23.6 polyplexes (black) and siRNA/PEI polyplexes (white) based on gel image analysis using ImageJ.
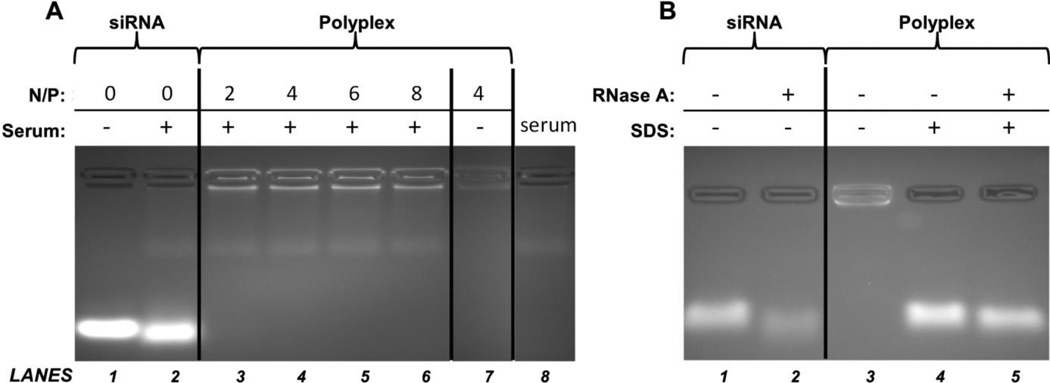
The interactions between serum proteins and polyplexes also can lead to polyplex disassembly and subsequent degradation of siRNA by serum nucleases. Hence, electrophoresis and serum/nuclease stability assays were performed to assess the structure of the serum-exposed mPEG-b-P(APNBMA)23.6 polyplexes (Figure 4A). Gel mobility shift assays on free siRNA treated with whole mouse serum produced a modest broadening of the siRNA band due to degradation (Figure 4A, Lane 2) compared to untreated siRNA (Lane 1). In contrast, no free siRNA band was visible for all serum-treated polyplex samples (Lanes 3 – 7). The increased background fluorescence in serum treated samples was attributed to interactions between ethidium bromide and serum components, as similar fluorescence occurred in the serum control (Lane 8). These data indicated that the siRNA was not displaced from the polyplex following incubation in serum.
Figure 4.
Gel mobility shift and nuclease stability analyses of siRNA polyplexes. (A) Stability of free siRNA and siRNA/mPEG-b-P(APNBMA)23.6 polyplexes against serum-mediated disassembly. Free siRNA and siRNA/mPEG-b-P(APNBMA)23.6 polyplexes prepared at N/P = 2, 4, 6, and 8 were analyzed in the presence (+) or absence (−) of serum. (B) Stability of free siRNA and siRNA polyplexes (N/P = 4) against RNase A-mediated degradation. Free siRNA and siRNA/mPEG-b-P(APNBMA)23.6 polyplexes were treated with RNase A (+) or RNase A-free (−) buffers, the nuclease reactions were terminated, and the polyplexes were either disassembled via SDS incubation (+) or left intact (−).
siRNA/mPEG-b-P(APNBMA)23.6 polyplexes were exposed to RNase A to investigate the susceptibility of encapsulated siRNA to nuclease-mediated degradation (Figure 4B). The treatment of free siRNA with RNase A resulted in increased electrophoretic mobility of the siRNA, broadening of the siRNA band, and ~65% reduction in ethidium bromide fluorescence intensity (Lane 2). In contrast, siRNA released from RNase A-treated mPEG-b-P(APNBMA)23.6 polyplexes (Lane 5) indicated an ~14% reduction in ethidium bromide fluorescence intensity compared to siRNA released from polyplexes without nuclease treatment (Lane 4). These combined serum and nuclease stability results indicated that the binding affinity of the mPEG-b-P(APNBMA)23.6 for siRNA was sufficiently strong to prevent serum-mediated polyplex disassembly and provide enhanced resistance to nuclease degradation.
2.3. UV-induced unpackaging of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes
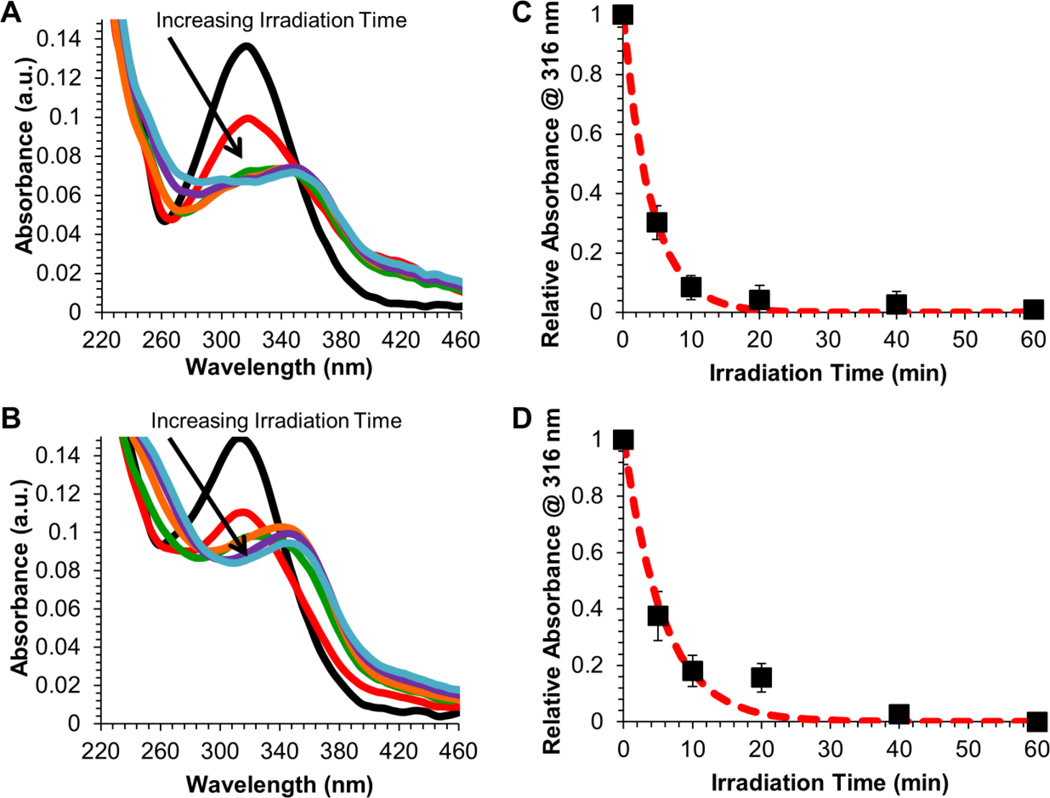
Given the efficient packaging and enhanced salt and serum stability of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes, UV irradiation studies were performed to assess the efficacy of light-triggered siRNA release. Free mPEG-b-P(APNBMA)23.6 block copolymer as well as siRNA/mPEG-b-P(APNBMA)23.6 polyplexes were irradiated with 365 nm light, and UV/Vis spectroscopy was used to monitor changes in the characteristic absorbance of the o-NB ester at 316 nm to determine the extent of photocleavage (Figure 5).[19]
Figure 5.
UV absorbance spectral changes of (A) mPEG-b-P(APNBMA)23.6 and (B) siRNA/mPEG-b-P(APNBMA)23.6 polyplexes following exposure to UV irradiation (365 nm, 200 W/m2) for 0 (black), 5 (red), 10 (orange), 20 (green), 40 (blue), or 60 (purple) min. Normalized absorbance (filled squares) at 316 nm for (C) mPEG-b-P(APNBMA)23.6 and (D) siRNA/mPEG-b-P(APNBMA)23.6 polyplexes as a function of UV irradiation time. The log of the normalized intensity, for which the red dashed line indicates an exponential decay fit to the normalized intensity. Error bars represent the standard deviation determined from the mean of three independent absorbance measurements.
The free polymer absorbance spectra displayed dramatic decreases in absorbance at 316 nm for UV exposure times up to 10 min, and smaller changes at longer UV exposure times (Figure 5A). Irradiation of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes yielded similar results under the same conditions, with a large decrease in absorbance after 10 min of irradiation (Figure 5B). This behavior was consistent with the typical cleavage behavior of o-NB-containing polymers, which disassemble in response to irradiation with UV light to form a carboxylic acid and nitrosobenzaldehyde.[29] The spectral shifts also were similar to those previously noted for mPEG-b-P(APNBMA)n in pDNA polyplexes,[19] and the decrease in absorbance as a function of irradiation time followed an exponential decay for both the polymer and siRNA polyplex preparations. In particular, both the free polymer and polyplex spectra suggested complete cleavage of the o-NB group, based on the change in absorbance at 316 nm.
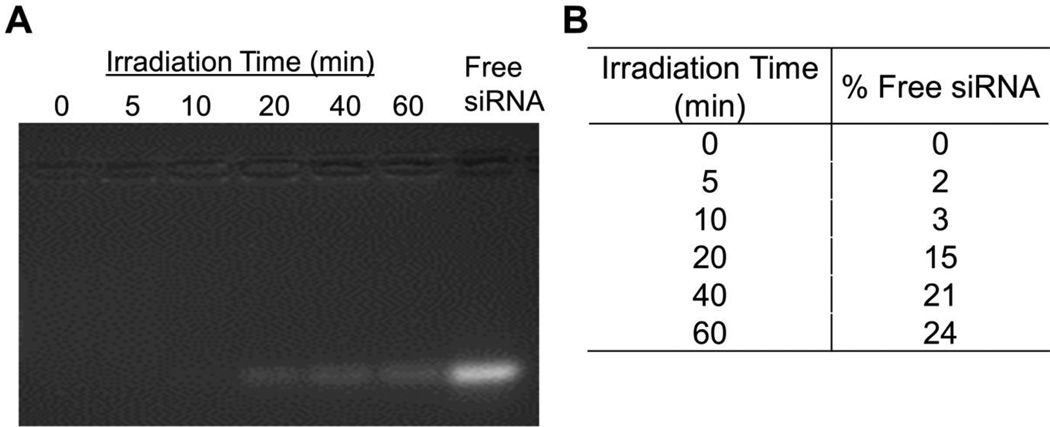
The irradiated polyplexes were collected and analyzed via electrophoresis to determine the extent of light-induced siRNA release (Figure 6A). When the polyplexes were irradiated for short periods of time (≤ 10 min irradiation), siRNA migration was limited, but after ≥ 20 min of exposure, significant amounts of free siRNA migrated down the gel. After 60 min of irradiation, additional siRNA migrated on the gel, yet release remained incomplete. Specifically, quantification of the free siRNA band intensities by ImageJ analysis revealed that ~15% of the siRNA was released after a 20 min irradiation, and the levels of released siRNA increased up to ~24% as irradiation times increased (Figure 6B).
Figure 6.
(A) Electrophoretic gel migration patterns of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes after exposure to UV irradiation (365 nm, 200 W/m2) for varying periods of time. Free siRNA is shown for comparison. (B) Quantification of ethidium bromide fluorescence in the siRNA band using ImageJ analysis software.
These studies highlight a novel release property that has not been previously demonstrated in other light-sensitive nucleic acid binding polymers.[30] Specifically, this system provides robust polyplex stability against polyanion- or serum-mediated disassembly, along with light-mediated release of a significant fraction of siRNA following light exposure. Light-induced structural changes remove the hydrophobic and cationic moieties responsible for siRNA binding and elicit a charge reversal along the mPEG-b-P(APNBMA)23.6 polymer backbone, which may account for the enhanced dissociation of the siRNA structures in this work as compared with other approaches. Meanwhile, it is possible that the apparent lack of complete release following irradiation was an artifact in quantification caused by the continued association of cleaved polymer fragments with the siRNA. The continued association of these fragments could reduce ethidium bromide binding, especially given that these fragments contained phenyl rings that might prohibit the intercalation of the ethidium bromide molecules. In previous work, we noted that irradiation of pDNA/mPEG-b-P(APNBMA)7.9 polyplexes also produced incomplete release of pDNA, although in that case, the released pDNA exhibited reduced mobility.[19] These distinct behaviors may be caused by differences in siRNA vs. pDNA association with polycations, as siRNA/polycation interactions are weaker;[3] hence, it is reasonable that the siRNA would exhibit enhanced dissociation from both the polymer backbone and polymer fragments in comparison to pDNA-based polyplexes.
2.4. Cellular uptake of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes
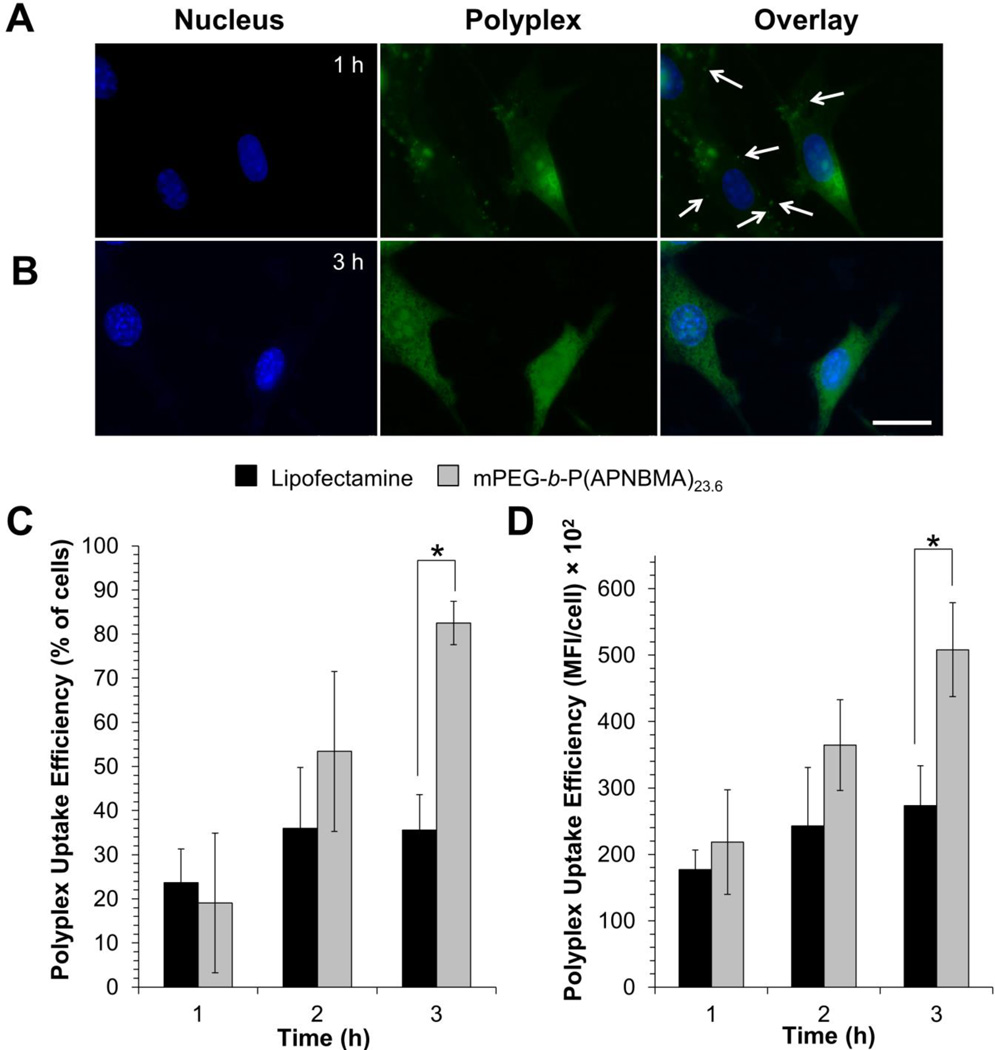
Given that mPEG-b-P(APNBMA)23.6 formed compact, salt- and serum-stable polyplexes that released siRNA upon light exposure, the polymer’s ability to mediate cellular uptake in NIH/3T3 cells was tested. Polyplexes were formulated with siRNA that was labeled with YOYO-1 iodide, visualized using fluorescence microscopy, and quantified via flow cytometry. Fluorescence microscopy showed rapid uptake of the labeled polyplexes, as indicated by the appearance of large quantities of intracellular YOYO-1-labeled structures after 1 h (Figure 7A). The punctate appearance in many of these structures suggested that the polyplexes were entrapped within endomembrane vesicular compartments within the cells, consistent with previous studies investigating the trafficking of siRNA polyplexes[6b] and other nanostructures. Notably, at slightly longer periods of polyplex exposure, a diffuse polyplex staining pattern was apparent. After 3 h, there was a clear increase in intracellular YOYO-1 iodide, as well as a loss of the punctate structures and a shift to a uniformly diffuse intracellular distribution (Figure 7B). Although the staining pattern alone cannot confirm cytoplasmic localization of these polyplexes, the endosomal escape/cytoplasmic release of at least some of the polyplexes is a likely explanation;[31] however, further experiments would be needed to verify this phenomenon.”
Figure 7.
Representative fluorescence microscopy images showing the internalization of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes (N/P = 4) following a (A) 1 h, and (B) 3 h transfection. siRNA was labeled with YOYO-1 iodide (green); the nucleus was stained with Hoechst (blue). Arrows indicate punctate polyplex structures, and the scale bar represents 25 µm. Quantification of YOYO-1-labeled siRNA/Lipofectamine RNAiMax (black) and siRNA/mPEG-b-P(APNBMA)23.6 (light gray) assemblies assemblies using flow cytometry: (C) uptake as a percentage of cells that internalized lipoplexes or polyplexes, and (D) the mean fluorescence intensity (MFI, arbitrary units) of the cells that had internalized lipoplexes or polyplexes. Each data point represents the mean ± standard deviation for a total of three separately prepared samples. An asterisk (*) indicates a statistically significant difference at a given time point between the mPEG-b-P(APNBMA)23.6 polyplexes and the Lipofectamine lipoplexes (p < 0.001).
Flow cytometry was used to quantify the efficiency of cellular uptake and the amount of intracellular YOYO-1 iodide-labeled siRNA polyplexes (Figures 7C and 7D). Following 1 h of polyplex exposure, the fraction of cells that had internalized siRNA/mPEG-b-P(APNBMA)23.6 polyplexes was comparable to the fraction that had internalized Lipofectamine preparations. Notably, the uptake efficiency of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes continued to increase more significantly than that of siRNA/Lipofectamine assemblies, whose uptake efficiency remained constant after 2 h (Figure 7C) at 36 ± 8% of cells with internalized siRNA/Lipofectamine. Previous studies have indicated comparable levels of cellular uptake under similar experimental conditions using siRNA/Lipofectamine assemblies in NIH/3T3 cells. For example, Kang et al. reported ~45% uptake following 4 h transfection using a 9 nM siRNA/Lipofectamine solution.[32] The average intracellular concentration of siRNA also increased up to 3 h post-transfection for the mPEG-b-P(APNBMA)23.6 polyplexes but remained constant for siRNA/Lipofectamine assemblies (Figure 7D). PDMAEMA-containing block copolymers have similarly indicated near 100% cellular uptake in the preparation of siRNA delivery structures.[9a] Several studies have indicated that enhanced cellular uptake of gene delivery polyplexes could be due to excess polycations that remain free in solution after polyplex formation.[33]
2.5. NIH/3T3 cell viability
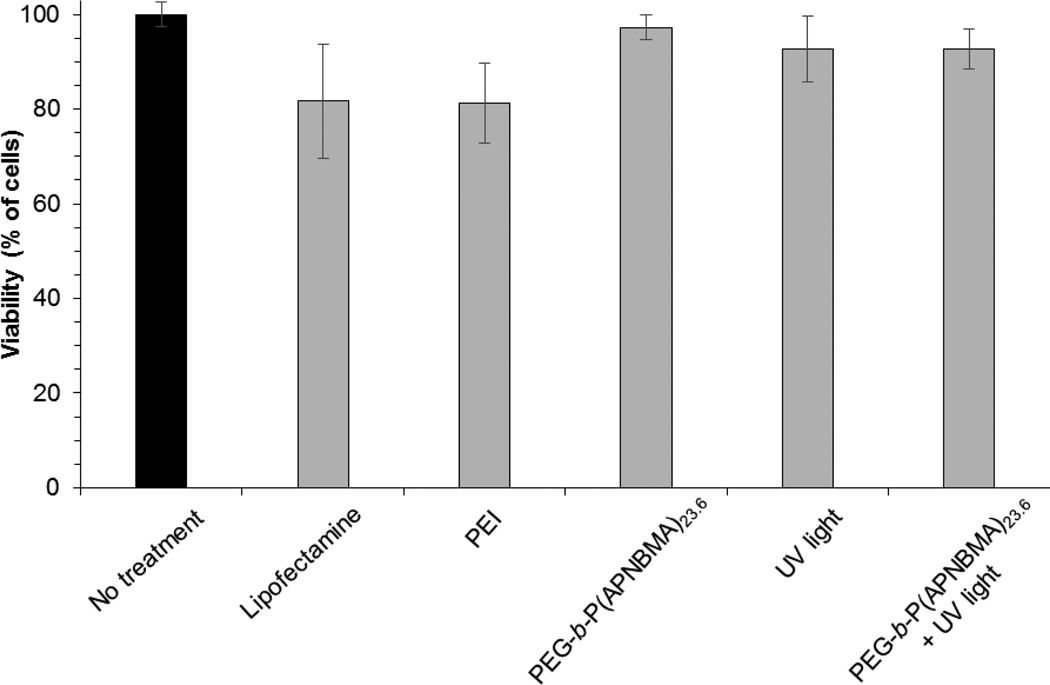
Numerous studies have indicated significant cellular toxicity when using cationic polymers due to non-specific interactions with the cell surface and subsequent membrane disruption.[34] However, as previously demonstrated, mPEG-b-P(APNBMA)n copolymers exhibited significantly reduced cytotoxicity as compared to PEI, and pDNA/mPEG-b-P(APNBMA)7.9 polyplexes exhibited minimal cytotoxicity both with and without UV irradiation.[19] Thus, we verified the expected lack of cytotoxicity in cell populations transfected by siRNA polyplexes under similar conditions using the Alamar Blue (AB) cell survival assay. AB-based quantification revealed greater than 90% cell survival in all samples, and no additive/synergistic effects were found in cells that were treated with UV light in combination with siRNA/mPEG-b-P(APNBMA)23.6 polyplexes vs. UV light alone (Figure 8). As anticipated, these results indicated similar biocompatibility of the siRNA assemblies as noted in pDNA/mPEG-b-P(APNBMA)7.9 polyplexes, suggesting their potential for further in vitro and in vivo delivery applications. Additionally, cells treated with siRNA/PEI (N/P = 6) and siRNA/Lipofectamine indicated 81% and 82% viability, respectively, demonstrated improved biocompatibility of mPEG-b-P(APNBMA)n polymers.
Figure 8.
Cell viabilities 48 h after polyplex treatment, UV exposure, or the combination of polyplexes and UV exposure as measured by the AB assay. Percent viabilities are relative to untreated cells. Each data point represents the mean ± standard deviation for a total of at least three separately prepared and analyzed samples. The untreated control sample is shown in black. PEI polyplexes were prepared at N/P = 6.
2.6. Gene silencing capacity of photocleavable polyplexes
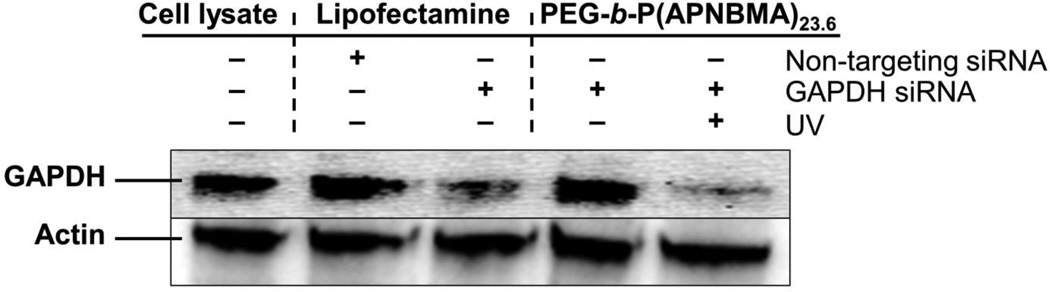
Based on the finding of triggered siRNA release in the characterization of polyplexes in cell-free studies, we also sought to test the on/off binding and silencing capacity of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes in NIH/3T3 cells using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a target (Figure 9). Cells were transfected for 3 h and then irradiated for 20 min, as the fluorescence microscopy analyses had indicated the robust cytoplasmic distribution of the polyplexes within the 3 h time frame (Figure 7B). Cells transfected with anti-GAPDH siRNA/mPEG-b-P(APNBMA)23.6 polyplexes exhibited a significantly higher level of GAPDH silencing following UV irradiation than cells treated with Lipofectamine assemblies, and moreover, these irradiated cells also exhibited enhanced silencing as compared to siRNA/mPEG-b-P(APNBMA)23.6 treated cells in the absence of UV irradiation. Specifically, quantification of the integrated optical density of GAPDH bands obtained through Western blot analysis indicated a 65 ± 4 % (n = 4) reduction in GAPDH levels for cells transfected with siRNA/mPEG-b-P(APNBMA)23.6 polyplexes and UV light, but no reduction in GAPDH levels in cells transfected with the siRNA/mPEG-b-P(APNBMA)23.6 polyplexes in the absence of UV light, and only a ~30% reduction in GAPDH levels in cells transfected with Lipofectamine. The silencing behavior was specific to GAPDH, as delivery of non-targeting siRNA (ON-TARGETplus non-targeting siRNA) only minimally affected GAPDH levels.
Figure 9.
Representative western blot analysis of NIH/3T3 cell extracts collected 48 h post-transfection (n = 4). Cells were treated with GAPDH targeted siRNA/mPEG-b-P(APNBMA)23.6 polyplexes with (+) or without (−) UV exposure (20 min), or with Lipofectamine lipoplexes containing either non-targeting or GAPDH targeting siRNAs.
These results highlight the unique capability of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes to maintain stable association within cells in the absence of light. Additionally, ~70% reduction in GAPDH levels indicated an enhanced gene silencing capacity compared to several other stimuli-responsive siRNA carriers, which report ~50% protein silencing in response to changes in enzyme concentration and pH.[35] Based on the combined siRNA release and gene silencing studies, it is worth noting that complete siRNA release from the irradiated polyplexes does not appear to be necessary for gene silencing, as only 15% free siRNA (based on siRNA release in gel electrophoresis following 20 min of UV-irradiation) was required to obtain this silencing effect. The studies presented in this work highlight the potential of mPEG-b-P(APNBMA)n polymers to direct siRNA delivery and further reveal an enhanced silencing capacity of mPEG-b-P(APNBMA)23.6 compared to a number of light-responsive siRNA delivery carriers.[17b, 18]
3. Conclusions
In summary, we demonstrated a novel photo-responsive system for localized cytoplasmic delivery and light-activated release of siRNA. mPEG-b-P(APNBMA)23.6 provided enhanced siRNA binding, to form salt-stable copolymer-based polyplexes that resisted aggregation following incubation in PBS, as compared to substantial growth for siRNA/PEI polyplexes (to > 1 µm in diameter).[25] Serum exposure of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes indicated tight association, in contrast to poly(L-lysine)-based polyplexes that showed > 90% siRNA dissociation following 1 h of serum incubation.[5b] Delivery of mPEG-b-P(APNBMA)23.6 polyplexes to NIH/3T3 cells facilitated > 80% cellular uptake compared to 40% for Lipofectamine lipoplexes, and cytoplasmic release of siRNA-containing polyplexes within 3 h of polyplex exposure. UV-irradiation of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes cleaved the o-NB and facilitated charge reversal on the polymer backbone and siRNA release. Furthermore, gene silencing experiments revealed the exquisite specificity and utility of this delivery approach. Specifically, by stimulating the photo-responsive polyplexes with UV light, we achieved ~70% reduction in protein levels, as compared to no changes in protein levels for samples treated with siRNA/mPEG-b-P(APNBMA)23.6 polyplexes without irradiation, and only 30% reduction in Lipofectamine treated cells. We anticipate that the capacity for efficient cytoplasmic release and distribution, as well as the user-controlled release mechanism allowing for on/off control of siRNA binding and activity, will introduce significant versatility into the delivery platform relevant to a range of biological applications in developmental biology, regenerative medicine, and drug screening.
4. Experimental Section
Materials
Photocleavable mPEG-b-P(APNBMA)n (Mn = 7,900 g/mol, n = 7.9; Mn = 13,100 g/mol, n = 23.6) polymers were grown off of a 5 kDa mPEG-Br macroinitiator via atom-transfer radical polymerization as previously described.[19] ON-TARGETplus siRNA (anti-GAPDH) as well as a non-targeting siRNA sequence were purchased from ThermoFisher Scientific (Pittsburgh, PA). Lipofectamine RNAiMax, and the bis-intercalating dye, YOYO-1 iodide were purchased from Life Technologies (Grand Island, NY). RNase A and 25 kDa branched PEI were obtained from Sigma-Aldrich (St. Louis, MO). A bicinchoninic acid (BCA) protein assay kit and bovine serum albumin (BSA) were obtained from Pierce (Rockford, IL). Dulbecco’s modification of Eagle’s medium (DMEM) and PBS (150 mM NaCl) solutions were purchased from Corning Life Sciences - Mediatech Inc. (Manassas, VA). The ECL Plus Western Blotting Detection Kit was obtained from GE Healthcare Bioscience (Buckinghamshire, UK). Antibodies for Western blotting were purchased from AbCam (Cambridge, CA). Other Western blotting reagents were obtained from Pierce Biotechnology (Rockford, IL, USA). All other cell culture reagents were purchased from Fisher Scientific (Pittsburgh, PA).
siRNA polyplex formation and characterization
Polyplexes were formed using mixtures of the ON-TARGETplus siRNA and mPEG-b-P(APNBMA)n polymers. Prior to complexation, siRNA solutions were prepared at 40 µg/mL in 20 mM HEPES buffer, pH 6.0. Polyplexes were formed by the addition of polymer solution to an equal volume of siRNA solution followed by gentle vortexing. Polymer solutions contained polymer over the range of concentrations appropriate to form polyplexes at the desired polymer nitrogen to siRNA phosphate (N/P) ratios. Polyplexes were incubated at room temperature for 10 min prior to further analyses. Polyplex formation was analyzed using agarose gel electrophoresis. For electrophoresis, 25 µL of polyplex solution was added to 5 µL of loading solution (3/7 (v/v) glycerol/water). Subsequently, the polyplex solution was added to the wells of a 4.0 wt% agarose gel containing 0.5 µg/mL of ethidium bromide. Gels were run at 100 V for 1 h and subsequently imaged using a Bio-Rad Gel Doc XR (Hercules, CA). Polyplexes (75 µL of solution) were mixed with 200 µL HEPES buffer, PBS, or Opti-MEM, and subsequently incubated at 23 °C for either 1 h or 3 h prior to particle size determination. A CNI Optoelectronics Co., Ltd. (Changchun, China) 532 nm, 427.6 mW laser, coupled with a Brookhaven Instruments Corporation (Holtsville, NY) BI-200SM goniometer equipped with an inline 532 nm filter from Intor, Inc. (Socorro, NM), was used for DLS analyses. The intensity auto-correlation function was recorded at an angle of 90° and analyzed using a quadratic cumulant fit. All light scattering experiments were performed at 25 °C.
Stability of mPEG-b-P(APNBMA)23.6 polyplexes against heparin
Polyplexes containing 1 µg of siRNA were formed as described using either mPEG-b-P(APNBMA)23.6 or PEI and the polyplexes subsequently were incubated with heparin solutions for 30 min at 37 °C; the heparin solutions were used over the reported range of heparin/siRNA (w/w) ratios. Then, samples were analyzed by electrophoresis as described herein.
Serum stability of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes
The serum stability of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes was monitored by two complementary approaches. Whole mouse serum was used to investigate the stability against serum-mediated disassembly, and RNase A treatment was used to monitor siRNA degradation following nuclease exposure. For serum stability analysis, free siRNA (1 µg) or mPEG-b-P(APNBMA)23.6 polyplexes containing 1 µg of siRNA were incubated in 25 µL of whole mouse serum for 2 h. The serum nuclease activity was terminated by adding 20 µL of a buffer comprised of 0.16 M ethylene diamine tetraacetic acid (EDTA), 0.67 M NaOH, and 0.16 M NaCl, and the solutions were placed on ice for 10 min prior to analysis by agarose gel electrophoresis.
RNase stability of siRNA/mPEG-b-P(APNBMA)23.6 polyplexes
To monitor polyplex resistance to enzymatic degradation, RNase A was added to prepared polyplex samples containing 1 µg of siRNA, resulting in solution concentrations of 5 µg/mL siRNA and 0.5 µg/ml RNase A. These solutions were incubated for 5 min at 37 °C, and then the RNase activity was terminated by adding 8 µL termination buffer. siRNA release was initiated by adding 2 µL of 10 mg/mL sodium dodecyl sulfate (SDS) solution and incubating the resulting polyplex-SDS mixture for 5 min. The extent of siRNA degradation was subsequently analyzed by agarose gel electrophoresis.
Photocleavage and siRNA release
Photocleavage studies were performed using 300 µL of either the siRNA polyplex solution or free mPEG-b-P(APNBMA)n solution. These samples were loaded into a chamber that was prepared by sealing two glass slides with a rubber gasket. The chamber was irradiated with 365 nm light at 200 W/m2 (Omnicure S2000, Lumen Dynamics, Mississauga, Ontario, Canada) for 0 min, 5 min, 10 min, 20 min, 40 min, or 60 min. After irradiation, samples were removed from the chamber and collected for UV-visible (UV-Vis) absorbance measurements and agarose gel electrophoresis analyses. Absorbance measurements were performed using a Thermo Scientific NanoDrop 1000 Spectrophotometer (Waltham, MA).
Cell culture
Mouse embryonic fibroblast (NIH/3T3) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured according to ATCC protocols at 37 °C and 5 vol% CO2 in DMEM supplemented with 10 vol% fetal bovine serum (FBS) and 1 vol% penicillin-streptomycin.
Cell transfection
For transfections with mPEG-b-P(APNBMA)n polymers polyplexes were formed as described. Lipofectamine RNAiMax assemblies were formed according to the manufacturer’s protocols. Cells were seeded into 6-well plates at a density of 15,000 cells/cm2 and allowed to adhere for 24 h. Growth medium was subsequently replaced with Opti-MEM and polyplex solutions or Lipofectamine RNAiMax solutions containing 3.0 pmol siRNA/cm2 were added dropwise to the cells at 24 h post-seeding. After a 3 h incubation with the transfection reagents, the cells were washed with PBS and complete growth medium was added to each well. For transfections with UV treatment, cells were allowed to recover for 30 min in complete growth medium following the 3 h transfection. Subsequently, the medium was replaced with Opti-MEM and the samples were placed on a hot plate set to 37 °C and irradiated for 20 min at 365 nm (I0 = 200 W/m2). Cells then were washed with PBS and supplemented with complete growth medium for the remainder of the culture period.
Cellular uptake analyses
Polyplex internalization was monitored by two complementary approaches. Polyplex distributions were visualized using cell staining analyses, and flow cytometry was used as a quantitative measure of uptake. For both approaches, siRNA was labeled for detection by adding 1.5 eq of YOYO-1 iodide and incubating the labeling mixture at 37 °C for 1 h prior to polyplex formation. To prepare cells for uptake analyses, cells were synchronized by serum starvation. Cells were plated at a density of 2,000 cells/cm2 and incubated overnight. For synchronization, the medium was removed and replaced with fresh DMEM containing 0.75 vol% FBS and 1 vol% antibiotics, according to standard procedures.[36] The cells were grown for an additional 48 h, and then the cells were released from starvation by replacing the low serum medium with fully supplemented growth medium containing 10 vol% serum and 1 vol% antibiotics.
Transfections were performed 12 h after synchronization, using polyplex suspensions containing 3.0 pmol siRNA/cm2. Transfections were halted after 1 h, 2 h, or 3 h of polyplex exposure by removing the transfection media, rinsing the cells with PBS, washing with 10 µg/mL heparin solutions to remove extracellular polyplexes,[37] and washing again with PBS. For cell staining, cells were fixed with 4% paraformaldehyde in PBS for 15 min, permeabilized with 0.2 vol% Tween-20 in PBS (0.2% PBSt), and blocked with 3% BSA in 0.2% PBSt. Cells were stained with 4 µg/mL Hoechst dye solution and stored at 4 °C prior to imaging. For flow cytometry analyses, cells were collected for analysis by standard trypsin-mediated collection protocols. Subsequently, cells were resuspended in PBS, filtered through a 35 µm nylon mesh to remove aggregates, and stored at 4 °C until analysis. Polyplex uptake efficiency was calculated as the percentage of cells containing polyplexes. Specifically, scatter plots from untransfected cells were gated for autofluorescence and the percentage of cells with fluorescence levels above the autofluorescence threshold was determined by comparison with the gate position. A total of 10,000 cells were analyzed for each sample.
Cell toxicity
Polyplex and UV toxicity were evaluated in NIH/3T3 cells using the AB assay according to the manufacturer’s protocols. Cells were seeded into 6-well plates at a density of 15,000 cells/cm2 and allowed to adhere for 24 h prior to toxicity analyses. The cells were rinsed with PBS, transfected with polyplex solutions, and UV treated as described above. Subsequently, the cells were rinsed with PBS and the medium was replaced with complete growth medium. After 48 h, AB was added directly into the culture medium to a final concentration of 10 vol%, and the AB-containing solutions were incubated with the cells for an additional 6 h at 37 °C, 5 vol% CO2. AB fluorescence was measured using a GloMax-multi detection system plate reader (Promega, Madison, WI). To determine the baseline fluorescence, AB was added to media without cells.
Western blot analysis
siRNA-mediated gene silencing was analyzed by standard Western blotting analyses. After a 48 h transfection, protein was extracted from NIH/3T3 cells by incubating the cells with a lysis solution composed of 0.5 vol% Triton X-100, 0.5% sodium deoxycholate, 150 mM NaCl, 5 mM Tris-HCl (pH 7.4), 5 mM EDTA, and 1× Halt Protease and Phosphatase Inhibitor cocktail. For each sample, total protein content was measured using a BCA Protein Assay Reagent Kit. Equal amounts of cellular protein from each cell extract were subjected to 4–20% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the separated proteins were subsequently transferred onto a poly(vinylidene fluoride) membrane. The membrane was blocked in 5 vol% BSA in Tris-HCl buffered saline (50 mM Tris-HCl, pH 7.4, 150 mM NaCl) containing 0.1 vol% Tween 20 (TBSt) for 1 h at room temperature, and then the membrane was incubated with an anti-GAPDH rabbit monoclonal IgG primary antibody (0.4 µg/mL in TBSt) overnight at 4 °C. Subsequently, the membrane was incubated with a solution of goat anti-rabbit polyclonal IgG antibody conjugated to horseradish peroxidase (HRP) (0.04 µg/mL in TBSt) for 1 h at room temperature. Target proteins were visualized with the ECL Plus chemiluminescent substrate. Subsequently, the membrane was stripped for 15 min using Restore Western Blot stripping buffer and probed for actin using anti-actin rabbit polyclonal IgG (1 µg/mL in TBSt) and a goat anti-rabbit polyclonal IgG HRP conjugated secondary antibody. The chemiluminescence signal was quantified with the aid of ImageJ software.
Acknowledgements
The authors thank the National Institute of General Medical Sciences of the National Institutes of Health (NIH) for financial support through an Institutional Development Award (IDeA) under grant number P20GM103541. The statements herein do not reflect the views of the NIH. We acknowledge the Delaware Biotechnology Institute (DBI) and Delaware Economic Development Office (DEDO) for financial support through the Bioscience Center for Advanced Technology (Bioscience CAT) award (12A00448). We also thank the Center for Molecular and Engineering Thermodynamics at the University of Delaware for providing access to the DLS instrument.
Contributor Information
Abbygail A. Foster, Department of Chemical and Biomolecular Engineering, Newark, DE 19716, USA
Chad T. Greco, Department of Chemical and Biomolecular Engineering, Newark, DE 19716, USA
Matthew D. Green, Department of Chemical and Biomolecular Engineering, Newark, DE 19716, USA
Thomas H. Epps, III, Email: thepps@udel.edu, Department of Chemical and Biomolecular Engineering, Newark, DE 19716, USA.
Millicent O. Sullivan, Email: msullivan@udel.edu, Department of Chemical and Biomolecular Engineering, Newark, DE 19716, USA.
References
- 1.a) Jacque JM, Triques K, Stevenson M. Nature. 2002;418:435. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dorsett Y, Tuschl T. Nat. Rev. Drug. Discov. 2004;3:318. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]; c) Behlke MA. Mol. Ther. 2006;13:644. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Nat. Rev. Drug. Discov. 2007;6:443. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Castanotto D, Rossi JJ. Nature. 2009;457:426. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Proc. Natl. Acad. Sci. USA. 2007;104:15549. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK, Kornbrust DJ, Davis ME. Proc. Natl. Acad. Sci. USA. 2007;104:5715. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gary DJ, Puri N, Won YY. J. Control. Release. 2007;121:64. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Zuckerman JE, Choi CH, Han H, Davis ME. Proc. Natl. Acad. Sci. USA. 2012;109:3137. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Bioconjugate Chem. 2006;17:1209. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]; b) Buyens K, Meyer M, Wagner E, Demeester J, De Smedt SC, Sanders NN. J. Control. Release. 2010;141:38. doi: 10.1016/j.jconrel.2009.08.026. [DOI] [PubMed] [Google Scholar]; c) Vader P, van der Aa LJ, Engbersen JF, Storm G, Schiffelers RM. Pharm. Res. 2012;29:352. doi: 10.1007/s11095-011-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. J. Control. Release. 2008;130:57. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]; b) Shim MS, Kwon YJ. Bioconjugate Chem. 2009;20:488. doi: 10.1021/bc800436v. [DOI] [PubMed] [Google Scholar]; c) Shim MS, Chang S, Kwon YJ. Biomater. Sci. 2014;2:35. doi: 10.1039/c3bm60187k. [DOI] [PubMed] [Google Scholar]; d) Liao ZX, Ho YC, Chen HL, Peng SF, Hsiao CW, Sung HW. Biomaterials. 2010;31:8780. doi: 10.1016/j.biomaterials.2010.07.086. [DOI] [PubMed] [Google Scholar]
- 7.Wagner E. Acc. Chem. Res. 2012;45:1005. doi: 10.1021/ar2002232. [DOI] [PubMed] [Google Scholar]
- 8.a) Jeong JH, Mok H, Oh YK, Park TG. Bioconjugate Chem. 2009;20:5. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]; b) Mok H, Lee SH, Park JW, Park TG. Nat. Mater. 2010;9:272. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]; c) Lee SH, Mok H, Jo S, Hong CA, Park TG. Biomaterials. 2011;32:2359. doi: 10.1016/j.biomaterials.2010.11.062. [DOI] [PubMed] [Google Scholar]; d) Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Nat. Mater. 2012;11:316. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Shopsowitz KE, Roh YH, Deng ZJ, Morton SW, Hammond PT. Small. 2014;10:1623. doi: 10.1002/smll.201302676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. ACS Nano. 2013;7:8870. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu Z, Zhang Z, Zhou C, Jiao Y. Prog. Polym. Sci. 2010;35:1144. [Google Scholar]
- 10.a) Johnson RN, Chu DS, Shi J, Schellinger JG, Carlson PM, Pun SH. J. Control. Release. 2011;155:303. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kang HC, Bae YH. Biomaterials. 2011;32:4914. doi: 10.1016/j.biomaterials.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Liu X, Howard KA, Dong M, Andersen MO, Rahbek UL, Johnsen MG, Hansen OC, Besenbacher F, Kjems J. Biomaterials. 2007;28:1280. doi: 10.1016/j.biomaterials.2006.11.004. [DOI] [PubMed] [Google Scholar]; d) Kang HC, Kang HJ, Bae YH. Biomaterials. 2011;32:1193. doi: 10.1016/j.biomaterials.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Mann A, Thakur G, Shukla V, Singh AK, Khanduri R, Naik R, Jiang Y, Kalra N, Dwarakanath BS, Langel U, Ganguli M. Mol. Pharm. 2011;8:1729. doi: 10.1021/mp2000814. [DOI] [PubMed] [Google Scholar]; f) Godbey WT, Wu KK, Mikos AG. J. Biomed. Mater. Res. 1999;45:268. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 11.a) Kuhn PS, Levin Y, Barbosa MC. Physica A: Statistical Mechanics and its Applications. 1999;274:8. [Google Scholar]; b) Philipp A, Zhao X, Tarcha P, Wagner E, Zintchenko A. Bioconjugate Chem. 2009;20:2055. doi: 10.1021/bc9001536. [DOI] [PubMed] [Google Scholar]; c) Kizzire K, Khargharia S, Rice KG. Gene Ther. 2013;20:407. doi: 10.1038/gt.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Nguyen HK, Lemieux P, Vinogradov SV, Gebhart CL, Guerin N, Paradis G, Bronich TK, Alakhov VY, Kabanov AV. Gene Ther. 2000;7:126. doi: 10.1038/sj.gt.3301052. [DOI] [PubMed] [Google Scholar]; b) Kuo JH. Biotechnol. Appl. Biochem. 2003;37:267. doi: 10.1042/BA20020123. [DOI] [PubMed] [Google Scholar]
- 13.a) Incani V, Tunis E, Clements BA, Olson C, Kucharski C, Lavasanifar A, Uludag H. J. Biomed. Mater. Res. A. 2007;81:493. doi: 10.1002/jbm.a.31249. [DOI] [PubMed] [Google Scholar]; b) Wong SY, Pelet JM, Putnam D. Progress Polym. Sci. 2007;32:799. [Google Scholar]
- 14.Han L, Tang C, Yin C. Biomaterials. 2013;34:5317. doi: 10.1016/j.biomaterials.2013.03.060. [DOI] [PubMed] [Google Scholar]
- 15.a) Lin YL, Jiang G, Birrell LK, El-Sayed ME. Biomaterials. 2010;31:7150. doi: 10.1016/j.biomaterials.2010.05.048. [DOI] [PubMed] [Google Scholar]; b) Won YW, Yoon SM, Lee KM, Kim YH. Mol. Ther. 2011;19:372. doi: 10.1038/mt.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Kelley EG, Albert JN, Sullivan MO, Epps TH., III Chem. Soc. Rev. 2013;42:7057. doi: 10.1039/c3cs35512h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Nucleic Acids Res. 2008;36:559. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S. Nat Mater. 2009;8:736. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ziauddin J, Sabatini DM. Nature. 2001;411:107. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]; e) Aytar BS, Muller JP, Kondo Y, Abbott NL, Lynn DM. ACS Appl. Mater. Interfaces. 2013;5:8283. doi: 10.1021/am402594z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Soliman M, Allen S, Davies MC, Alexander C. Chem. Commun. 2010;46:5421. doi: 10.1039/c0cc00794c. [DOI] [PubMed] [Google Scholar]; b) Huschka R, Barhoumi A, Liu Q, Roth JA, Ji L, Halas NJ. ACS Nano. 2012;6:7681. doi: 10.1021/nn301135w. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Braun GB, Pallaoro A, Wu G, Missirlis D, Zasadzinski JA, Tirrell M, Reich NO. ACS Nano. 2009;3:2007. doi: 10.1021/nn900469q. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Wang H, Sun C, Du J, Wang J. RSC Adv. 2014;4:1961. [Google Scholar]
- 19.Green MD, Foster AA, Greco CT, Roy R, Lehr RM, Epps TH, III, Sullivan MO. Polym. Chem. 2014;5:5535. doi: 10.1039/C4PY00638K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura Y, Kogure K, Futaki S, Harashima H. J. Control. Release. 2007;119:360. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 21.a) Gary DJ, Lee H, Sharma R, Lee JS, Kim Y, Cui ZY, Jia D, Bowman VD, Chipman PR, Wan L, Zou Y, Mao G, Park K, Herbert BS, Konieczny SF, Won YY. ACS Nano. 2011;5:3493. doi: 10.1021/nn102540y. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Deshpande MC, Garnett MC, Vamvakaki M, Bailey L, Armes SP, Stolnik S. J. Control. Release. 2002;81:185. doi: 10.1016/s0168-3659(02)00052-4. [DOI] [PubMed] [Google Scholar]
- 22.Elsabahy M, Wooley KL. Chem. Soc. Rev. 2012;41:2545. doi: 10.1039/c2cs15327k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a) Sun TM, Du JZ, Yan LF, Mao HQ, Wang J. Biomaterials. 2008;29:4348. doi: 10.1016/j.biomaterials.2008.07.036. [DOI] [PubMed] [Google Scholar]; b) Li J, Cheng D, Yin T, Chen W, Lin Y, Chen J, Li R, Shuai X. Nanoscale. 2014;6:1732. doi: 10.1039/c3nr05024f. [DOI] [PubMed] [Google Scholar]
- 24.a) Harris JM, Chess RB. Nat. Rev. Drug. Discov. 2003;2:214. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]; b) Owens DE, III, Peppas NA. Int. J. Pharm. 2006;307:93. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]; c) Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. Gene Ther. 1999;6:595. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]; d) Peracchia MT, Harnisch S, Pinto-Alphandary H, Gulik A, Dedieu JC, Desmaele D, d'Angelo J, Muller RH, Couvreur P. Biomaterials. 1999;20:1269. doi: 10.1016/s0142-9612(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Qin Y, Liang Y, Zou H, Peng X, Huang H, Lu M, Feng M. Biomacromolecules. 2013;14:1777. doi: 10.1021/bm400177q. [DOI] [PubMed] [Google Scholar]
- 26.Grayson AC, Doody AM, Putnam D. Pharm. Res. 2006;23:1868. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 27.Ruponen M, Ronkko S, Honkakoski P, Pelkonen J, Tammi M, Urtti A. J. Biol. Chem. 2001;276:33875. doi: 10.1074/jbc.M011553200. [DOI] [PubMed] [Google Scholar]
- 28.a) Alshamsan A, Haddadi A, Incani V, Samuel J, Lavasanifar A, Uludag H. Mol. Pharm. 2009;6:121. doi: 10.1021/mp8000815. [DOI] [PubMed] [Google Scholar]; b) Takahashi T, Kono K, Itoh T, Emi N, Takagishi T. Bioconjugate Chem. 2003;14:764. doi: 10.1021/bc025663f. [DOI] [PubMed] [Google Scholar]
- 29.a) Il'ichev YV, Schworer MA, Wirz J. J. Am. Chem. Soc. 2004;126:4581. doi: 10.1021/ja039071z. [DOI] [PubMed] [Google Scholar]; b) Zhao H, Sterner ES, Coughlin EB, Theato P. Macromolecules. 2012;45:1723. [Google Scholar]
- 30.a) Handwerger RG, Diamond SL. Bioconjugate Chem. 2007;18:717. doi: 10.1021/bc060280t. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Feng L, Yang X, Shi X, Tan X, Peng R, Wang J, Liu Z. Small. 2013;9:1989. doi: 10.1002/smll.201202538. [DOI] [PubMed] [Google Scholar]
- 31.Kim SH, Jeong JH, Kim TI, Kim SW, Bull DA. Mol. Pharm. 2009;6:718. doi: 10.1021/mp800161e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang H, Fisher MH, Xu D, Miyamoto YJ, Marchand A, Van Aerschot A, Herdewijn P, Juliano RL. Nucleic Acids Research. 2004;32:4411. doi: 10.1093/nar/gkh775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Thibault M, Astolfi M, Tran-Khanh N, Lavertu M, Darras V, Merzouki A, Buschmann MD. Biomaterials. 2011;32:4639. doi: 10.1016/j.biomaterials.2011.03.010. [DOI] [PubMed] [Google Scholar]; b) Xu T, Liu W, Wang S, Shao Z. Int. J. Nanomed. 2014;9:3231. doi: 10.2147/IJN.S64308. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yue Y, Jin F, Deng R, Cai J, Chen Y, Lin MC, Kung HF, Wu C. J. Control. Release. 2011;155:67. doi: 10.1016/j.jconrel.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 34.a) Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. Mol. Ther. 2005;11:990. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]; b) Xue HY, Liu S, Wong HL. Nanomedicine (Lond) 2014;9:295. doi: 10.2217/nnm.13.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.a) Yingyuad P, Mevel M, Prata C, Kontogiorgis C, Thanou M, Miller AD. J. RNAi Gene Silencing. 2014;10:490. [PMC free article] [PubMed] [Google Scholar]; b) Hong BJ, Chipre AJ, Nguyen ST. J. Am. Chem. Soc. 2013;135:17655. doi: 10.1021/ja404491r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markovits J, Pommier Y, Kerrigan D, Covey JM, Tilchen EJ, Kohn KW. Cancer Res. 1987;47:2050. [PubMed] [Google Scholar]
- 37.a) Reilly MJ, Larsen JD, Sullivan MO. Mol. Pharm. 2012;9:1031. doi: 10.1021/mp200372s. [DOI] [PubMed] [Google Scholar]; b) Reilly MJ, Larsen JD, Sullivan MO. Mol. Pharm. 2012;9:1280. doi: 10.1021/mp200583d. [DOI] [PubMed] [Google Scholar]