Abstract
Adverse life events can induce two kinds of memory with opposite valence, dependent on timing: “negative” memories for stimuli preceding them and “positive” memories for stimuli experienced at the moment of “relief.” Such punishment memory and relief memory are found in insects, rats, and man. For example, fruit flies (Drosophila melanogaster) avoid an odor after odor-shock training (“forward conditioning” of the odor), whereas after shock-odor training (“backward conditioning” of the odor) they approach it. Do these timing-dependent associative processes share molecular determinants? We focus on the role of Synapsin, a conserved presynaptic phosphoprotein regulating the balance between the reserve pool and the readily releasable pool of synaptic vesicles. We find that a lack of Synapsin leaves task-relevant sensory and motor faculties unaffected. In contrast, both punishment memory and relief memory scores are reduced. These defects reflect a true lessening of associative memory strength, as distortions in nonassociative processing (e.g., susceptibility to handling, adaptation, habituation, sensitization), discrimination ability, and changes in the time course of coincidence detection can be ruled out as alternative explanations. Reductions in punishment- and relief-memory strength are also observed upon an RNAi-mediated knock-down of Synapsin, and are rescued both by acutely restoring Synapsin and by locally restoring it in the mushroom bodies of mutant flies. Thus, both punishment memory and relief memory require the Synapsin protein and in this sense share genetic and molecular determinants. We note that corresponding molecular commonalities between punishment memory and relief memory in humans would constrain pharmacological attempts to selectively interfere with excessive associative punishment memories, e.g., after traumatic experiences.
Keywords: Drosophila, memory, pain, punishment, relief, Synapsin
Introduction
Painful, traumatic experiences can have a molding influence on behavior. Current research is focused on the “negative” memories that such experiences induce: stimuli experienced before a painful event become predictors of danger and will be avoided when encountered again. While in principle adaptive, danger predictions can also contribute to maladaptive behavior and undesired psychological states (e.g., stress, anxiety, panic). Under such circumstances, any means to counteract these effects is welcome. We therefore extend the focus toward “backward conditioning,” that is to memories related to stimuli perceived after a painful event (Moscovitch and LoLordo, 1968; Solomon and Corbit, 1974; Plotkin and Oakley, 1975; Heth, 1976; Wagner, 1981; Wagner and Larew, 1985; Gerber et al., 2014; Navratilova and Porreca, 2014).
In fruit flies, odor-shock training (“forward conditioning” of the odor) leads to conditioned avoidance of the odor during subsequent tests, whereas shock-odor training (backward conditioning of the odor) leads to conditioned approach (Tanimoto et al., 2004). Corresponding effects are seen in humans, rabbits, rats, and in the appetitive domain in bees (references in preceding paragraph as well as Hellstern et al., 1998; Andreatta et al., 2010, 2012; Franklin et al., 2013a, b; Felsenberg et al., 2014; Mohammadi et al., 2014). Such timing-dependent valence reversal makes sense, as after odor-shock training the odor predicts punishment, whereas shock-odor training associates the odor with relief from shock. We therefore refer to these processes as punishment learning and relief learning, respectively (Fig. 1A).
Figure 1.
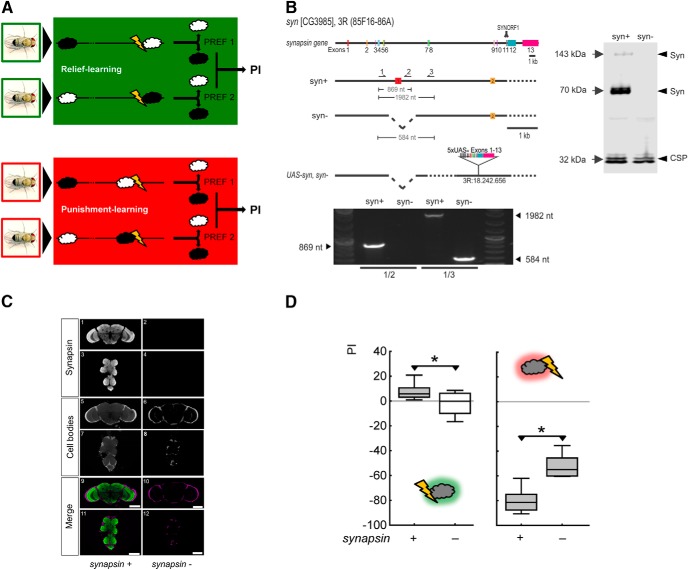
Mutant flies lacking Synapsin are impaired in both relief memory and punishment memory. A, Schematic of the experimental paradigm for relief learning and punishment learning. For relief learning, an odor is presented upon the cessation of shock, whereas for punishment learning an odor is presented before the onset of shock. In both cases, a control odor is presented temporally far removed from shock. During the test, the flies can choose between the relief-trained versus the control odor, and between the punishment-trained versus the control odor, respectively. Swapping the chemical identity of the odors in reciprocally trained flies allows a PI to be calculated from the difference in preference between these reciprocally trained flies. The PI thus measures associative memory and runs no risk of confounding differences in innate preference for either odor, or in nonassociative memory components. For further details see Materials and Methods. B, Upper left, Genomic organization of the Drosophila synapsin gene. Syn contains 13 exons and spans a 13.7 kb genomic region in the 3R (85F16–86A) cytological interval [flybase (www.flybase.org); the second of the 14 exons previously suggested on the basis of syn-cDNA sequences (Klagges et al., 1996) apparently represents a splice artifact (B. Klagges and E. Buchner, personal communication)]. Accordingly, the coding region for the epitope LFGGMEVCGL that is recognized by the monoclonal antibody SYNORF1 is encoded by exon 11. The syn97CS strain (labeled synapsin− for simplicity) carries a 1.4 kb deletion spanning parts of the regulatory region and the first exon of the synapsin gene. The arrows indicate the binding sites for the PCR primers upstream (1), within (2), and downstream (3) of the deletion. The effector strain UAS-syn, synapsin− contains the syn-cDNA plus 156 base pairs upstream (indicated by * between UAS and Exons 1–13). Lower left, In a single-fly PCR approach, primer combination 1/2 yields an 869 nt product in synapsin+ wild-type but not in synapsin− mutants, whereas primer pair 1/3 yields a 1982 nt product in synapsin+ and a 584 nt product in synapsin−. Right, Western blot from material obtained from three adult fly heads stained for Synapsin and for CSP as a loading control. The single band at ∼143 kDa and the double band at ∼70 kDa, where Synapsin isoforms are expected (Klagges et al., 1996), are found in synapsin+ but not in synapsin− flies. C, Synapsin immunoreactivity is absent in whole-mount preparations of synapsin− mutant flies. In the bottom row (Merge), anti-Synapsin staining is shown in green, and cell-body counterstaining with propidium iodide in magenta, displayed as frontal optical sections (0.9 μm) of synapsin+ (left column) and synapsin− (right column) brain and thoracic nervous system. Scale bar, 100 μm. D, Left, Shows that relief memory is intact in synapsin+ wild-type flies, but is abolished in mutant flies lacking Synapsin (synapsin−). Right, Shows that punishment memory is impaired, but is not abolished, in synapsin− mutant flies. *p < 0.05 for the between-genotype comparison within an experiment; a gray shading of the boxes indicates p < 0.05/2 in comparisons of either genotype to chance levels (zero) within one experiment. PI, indicating the difference in preference between reciprocally trained flies, and thus learned approach (positive scores) and learned avoidance (negative scores), respectively. The middle line of the box plots represents the median, the box boundaries the 25 and 75% quartiles, and the whiskers the 10 and 90% quantiles, respectively.
For punishment learning, reasonably clear working hypotheses exist (see Discussion), but much less is known about relief learning, in any experimental system. In Drosophila, relief memory is less strong than punishment memory, requires six rather than just one training trial, and is strongest when using relatively mild shock intensities (Yarali et al., 2008). These parametric requirements were the likely reason why relief learning was overlooked in classical studies (Quinn et al., 1974; Tully and Quinn, 1985). Whenever the current study compares relief memory and punishment memory, six trials and the same shock intensity are used in either case.
Given that relatively little is known about the mechanisms of relief memory, we decided to focus on an evolutionarily conserved process of synaptic plasticity and memory. We chose the Synapsin protein as a study case (coded by the synapsin gene: CG3985), a ubiquitously and abundantly expressed, and evolutionarily conserved, presynaptic phosphoprotein (Klagges et al., 1996; Nuwal et al., 2011; Diegelmann et al., 2013a). It can tether reserve-pool vesicles to the cytoskeleton and, during learning, regulate their recruitment to the readily releasable pool in a phosphorylation-dependent way. Thus, more synaptic vesicles are made eligible for release upon subsequent activation of the cell during retrieval. In this sense, Synapsin is thought to function during learning in a way that determines memory strength. It contributes to the regulation of synaptic output and various forms of memory throughout the animal kingdom (Silva et al., 1996; Hilfiker et al., 1999; Garcia et al., 2004; Gitler et al., 2004; Südhof, 2004; Greco et al., 2013; Diegelmann et al., 2013a; Sadanandappa et al., 2013; Vasin et al., 2014). Regarding associative processing in Drosophila, Synapsin null mutants show reduced scores in one-trial strong-shock punishment memory, reduced spatial memory in the heat-box paradigm and, as larvae, reduced odor-sugar memory (Godenschwege et al., 2004; Michels et al., 2005, 2011; Knapek et al., 2010; Walkinshaw et al., 2015). We reasoned that Synapsin is worthy of study for its role in relief memory and/or punishment memory because if it were specifically required for either form of memory it would provide a target to tip the balance between punishment memory and relief memory by systemic manipulation. Given the conserved function of Synapsin, this may offer translation potential to, e.g., selectively erase the punishment-memory component but not the relief-memory component related to a traumatic episode. On the other hand, showing a common requirement of Synapsin for both relief memory and punishment memory would caution against such approaches.
Materials and Methods
Genotypes and rearing of flies.
To compare flies with versus without Synapsin, we compared the deletion mutant syn97CS (Fig. 1) to a Canton-S (CS) wild-type strain. The syn97CS strain had undergone 13 outcrossing steps to this very CS wild-type strain to yield effectively identical genetic backgrounds as described previously (Godenschwege et al., 2004; Michels et al., 2005, 2011). To ensure a stably identical genetic background over the course of this project, 13 further outcrossing steps were undertaken. For simplicity, these strains are referred to as synapsin− for the deletion mutant and synapsin+ for the wild-type throughout this study.
We used the following parental driver and effector strains (in addition to the mentioned status, all strains are homozygous w1118): elav-Gal4 [X] (strain c155 from Lin and Goodman, 1994); mb247-Gal4 [III], synapsin−, which was generated by using mb247-Gal4 (Schulz et al., 1996), recombined into the synapsin− mutant background; UAS-syn [III], synapsin−, which was generated on the basis of Löhr et al. (2002); tub-Gal80ts [II]; UAS-syn [III], synapsin− (Michels et al., 2011); and UAS-RNAi-syn [III] (Michels et al., 2011).
For experiments we used the F1 progeny of the following crosses. RNAi: To knock down Synapsin, elav-Gal4 [X] females were crossed to UAS-RNAi-syn [III] males, yielding double-heterozygous elav-Gal4/+;; UAS-RNAi-syn/+ flies. As an effector control, females without any transgene were crossed to males carrying UAS-RNAi-syn, yielding single-heterozygous UAS-RNAi-syn/+. As a driver control, elav-Gal4 females were crossed to males without any transgene, yielding single-heterozygous elav-Gal4/+. Rescue: To rescue Synapsin, female mb247-Gal4 [III], synapsin− flies were crossed to male UAS-syn [III], synapsin−, yielding double-heterozygous mb247-Gal4/UAS-syn flies in the homozygous synapsin− mutant background. As a driver control, female mb247-Gal4, synapsin− were crossed to male synapsin−, yielding single-heterozygous mb247-Gal4/+ in the homozygous synapsin− mutant background. As an effector control, synapsin− females were crossed to male UAS-syn, synapsin− to yield single-heterozygous UAS-syn/+ in the homozygous synapsin− mutant background. Induced rescue: To restrict expression of Synapsin to the adult stage we made use of the temperature-inducible transgene tub-Gal80ts. We crossed female mb247-Gal4 [III], synapsin− to male tub-Gal80ts [II]; UAS-syn [III], synapsin−, yielding triple-heterozygous tub-Gal80ts/+; mb247-Gal4/UAS-syn in the homozygous synapsin− mutant background. As an effector control we used the offspring of female synapsin− and male tub-Gal80ts; UAS-syn, synapsin− to yield double-heterozygous tub-Gal80ts/+; UAS-syn/+, in the homozygous synapsin− mutant background. As a driver control, mb247-Gal4, synapsin− females were crossed to male synapsin− to yield heterozygous mb247-Gal4/+ in the homozygous synapsin− mutant background.
We note that preliminary experiments using elav-Gal4 as a driver strain did not result in a rescue of associative memory (data not shown); this is in accordance with the lack of rescue observed when using this driver to restore rutabaga function (Zars et al., 2000).
All flies were kept in mass culture at 25°C, 60–70% humidity, and on a 16/8 h light/dark cycle. Unless mentioned otherwise, 1- to 5-d-old flies were collected and kept at 18°C until the following experimental day. Experiments were performed at 22–25°C and 75–85% relative humidity. For the induced rescue experiment, flies were raised at 18°C and, after collection, either shifted to 30°C for 4 d to allow inactivation of Gal80ts and thus expression of Synapsin (Induced), or retained at 18°C for 4 d (Control).
Throughout, we used flies in groups of ∼100, handled in a tube system based on Tully and Quinn (1985) and modified to allow the handling of four groups of flies in parallel. Training was performed in dim red light to allow sight for the experimenter (but not for the flies); tests were run in darkness. The electric shock was applied via an electrifiable grid, covering the inner side of the training tubes. A vacuum pump ensured removal of odor-saturated air. As odorants, 80 μl benzaldehyde (BA; CAS number 100-52-7; Fluka) or 110 μl limonene (LM; CAS number 5989-27-5; Sigma-Aldrich) were applied in Teflon containers of 5 mm or 7 mm diameter, respectively.
Learning experiments and behavioral controls.
For punishment learning and for relief learning, flies received six training trials. In the following example, BA is the to-be-learned odor and LM is the control odor. At time 0:00, flies were loaded into the experimental setup, which took ∼1 min. After an additional accommodation period of 3 min, LM was presented for 15 s. Then, for punishment learning, BA was presented from 7:15 to 7:30. At 7:30, the electric shock was delivered. Thus, for punishment learning, the interstimulus interval (ISI) between the onset of the shock and the onset of BA was −15 s. The shock consisted of six pulses of 100 V, each 1.2 s long and followed by the next pulse after an onset–onset interval of 5 s. At 12:00, the flies were transferred back to food vials for 16 min until the next trial started.
For relief learning, all parameters were identical, except that BA was presented from 8:20 to 8:35, leading to a shock-offset to odor-onset interval of 25 s, which corresponds to the optimal delay in this paradigm (Tanimoto et al., 2004; Yarali et al., 2008; Yarali et al., 2009b).
In half of the cases, BA served as the to-be-learned odor and LM as the control, while these assignments were swapped in the other half.
For the experiment describing the ISI functions in synapsin+ wild-type and synapsin− mutant flies, the timing of odor and shock was as indicated in the Results section.
Once training was completed, a 16 min break was given before the animals were again loaded into the setup for the memory test. After an accommodation period of 5 min, the animals were transferred to the choice point of a T-maze, where they could choose between BA and LM. After 2 min, the arms of the maze were closed and the number of animals (denoted as # in the following equation) within each arm was counted. The relative preference between BA and LM (PREF) was then calculated as follows:
The difference in PREF scores between the two reciprocally trained sets of flies was then calculated to obtain an index of associative memory performance (PI) that ranges between −100 and 100, and indicates conditioned approach by positive PIs and conditioned avoidance by negative PIs (Tully and Quinn (1985); Gerber and Stocker (2007) feature discussions of why these PIs yield measures of associative memory, cleared of nonassociative effects):
All procedures were the same for the one-odor version of the tasks, except that LM was omitted.
Behavioral controls.
To test for shock avoidance, the flies were loaded into the experimental setup. After an accommodation period of 2 min, the red light was switched off and flies were transferred to the choice point where they could enter either arm of the maze; 10 s later, shock was applied in one arm of the maze as specified above. Ten seconds after the onset of the last shock pulse, the maze was closed and the flies were collected and counted. A preference index for the shock (PREFSHOCK) was calculated to provide negative values for avoidance of the electrified arm:
To assess olfactory behavior, the flies were loaded into the experimental setup. After an accommodation period of 4 min, the red light was switched off and the flies were brought to the choice point of the T-maze and allowed to choose between a blank arm with air only and the other arm scented with odor (either BA or LM); after 2 min, the maze was closed and the flies were collected and counted. A preference index (PREFODOR) was calculated as follows:
Please note that, ever since their introduction as a control procedure in Drosophila, these tests for olfactory behavior do not involve a choice between the two odors; otherwise a failure in the ability to detect the odors in the mutant could not be distinguished from an indifference between the two odors in the wild-type.
For the odor-exposure and shock-exposure controls, the flies were handled as in normal training, except that we omitted either the shock (in the case of the odor-exposure control) or the odors (in the shock-exposure control). Then, the preference toward BA and LM was measured as described in the preceding paragraph (Eqs. 4a,b).
Statistical analyses of behavioral data.
Nonparametric statistics was used throughout. Kruskal–Wallis or Mann–Whitney U tests were used to compare multiple or two groups of flies, respectively. To test for differences from zero, we used the one-sample sign tests. The significance level was p < 0.05. For multiple comparisons within a dataset, p levels were adjusted by a Bonferroni correction (p < 0.05 divided by the number of comparisons), a conservative approach to maintain the experiment-wide error-rate of <5%. Data were plotted as box plots, representing the median as the middle line, the 25 and 75% quantiles as boundaries of the box, and the 10 and 90% quantiles as whiskers.
Single fly PCR.
PCRs were performed according to Gloor et al. (1993), using material from individual flies. The primer binding sites were upstream (primer 1: 5′-AGAAAATTTGGCTTGCATGG-3′), within (primer 2: 5′-CGGGGTCTCAGTTTTGTTG-3′), or downstream (primer 3: 5′-CCTCTACTTTTGGCTGCCTG-3′) of the deletion (Fig. 1).
Immunohistochemistry and Western blotting.
For whole-mount immunohistochemistry, brains were dissected in Ringer's solution and fixed for 2 h in 4% formaldehyde with PBS containing 0.3% Triton X-100 (PBST) as the solvent. Samples were blocked in 3% normal goat serum (Jackson ImmunoResearch) and subsequently incubated overnight with the mouse monoclonal anti-Synapsin antibody SYNORF1 3C11 [Developmental Studies Hybridoma Bank (DSHB); diluted 1:20 in PBST; Klagges et al., 1996; Godenschwege et al., 2004]. The sample was then incubated overnight with an Alexa 488-coupled goat anti-mouse Ig (diluted 1:200 in PBST; Invitrogen) to detect the primary antibody. All incubation steps were followed by multiple PBST washes. Incubations with antibodies were done at 4°C; all other steps were performed at room temperature. The brains were mounted in Vectashield mounting medium (Vector Laboratories), containing propidium iodide for counterstaining of DNA and hence of cell nuclei. Preparations were examined under a confocal microscope.
For Western blots, three adult heads per lane were homogenized in 10 μl Laemmli buffer. The sample was heated to 70°C for 5 min and centrifuged for 2 min before electrophoresis. Proteins were separated by 12.5% SDS-PAGE in a Multigel chamber (100 mA, 2 h; Peqlab) and transferred to a nitrocellulose membrane (Kyhse-Andersen, 1984). The membrane was blocked overnight (5% milk powder in 1 × TBST). Immunoreactions were successively performed with two mouse monoclonal antibodies: SYNORF1 for Synapsin detection (Klagges et al., 1996; dilution 1:100) and ab49 (Zinsmaier et al., 1990, 1994; DSHB; dilution 1:133) for detection of the Cysteine String Protein (CSP; Arnold et al., 2004) as a loading control (incubation time: 1.5 h). The membrane was then incubated with the secondary antibody (goat anti-mouse IgG-HRP coupled, 1:3700; 1 h). The incubation steps were followed by multiple washing procedures (1 × TBST). Visualization was achieved with the ECL Western blot detection reagents (GE Healthcare Life Sciences).
Analysis of Synapsin phosphorylation by LC-MS/MS.
Brains of experimentally naive adult Drosophila were dissected in Ringer's solution containing halt protease and phosphatase inhibitor cocktail, EDTA-free (1:100; Thermo Fisher Scientific) to prevent enzymatic protein degradation. Samples were then stored at −80°C in a liquid-free manner. Five adult brains were resolubilized in 50 μl of water containing 8 m freshly deionized urea. Tissue and cell destruction was achieved by means of a microglass potter and pulsed sonification on ice for 1 h. Biological membranes were destroyed and membrane proteins solubilized by the addition of 200 μl of 50 mm NH4HCO3 buffer, pH 8.0, containing 2 mm dithiothreitol and 1% (w/v) RapiGest SF surfactant (Waters) to obtain a final concentration of 2 m urea and 0.75% RapiGest. After gently shaking at room temperature for 60 min, 10 mm methyl methane thiosulfonic acid was added for an additional 1 h for thiomethylation of previously reduced cysteins. Limited proteolysis was started by adding 1 μg trypsin (Promega; Trypsin Gold) followed by incubation at room temperature for 12 h. After complete digestion, RapiGest was hydrolyzed by adding trifluoroacetic acid (TFA) to a final concentration of 1% and incubation for 1 h, at room temperature. Precipitated RapiGest fragments were spun down at 10 000 × g for 10 min and the supernatant was applied to an Empore Universal Resin SPE column (3M), equilibrated with 2 ml methanol and subsequently washed with 0.1% TFA. Resin-bound peptides were washed with 5 ml of 0.1% TFA and eluted twice with 0.5 ml of 70% acetonitrile (ACN), 0.1% TFA. Eluates were pooled and dried in a vacuum evaporator centrifuge (Savant; Thermo Fisher Scientific). Proteome analysis was performed on a hybrid dual-pressure linear ion trap/orbitrap mass spectrometer (LTQ Orbitrap Velos Pro; Thermo Scientific) equipped with an EASY-nLC ultra HPLC (Thermo Scientific). Samples were resolubilized in 50 μl of 0.1% TFA and 2% ACN and divided in five 10 μl aliquots as technical replicates, corresponding to a sample amount of one adult brain per injection, on a 75 μm i.d., 25 cm PepMap C18-column, packed with 2 μm resin (Dionex). Separation was achieved by applying a gradient from 2% ACN to 35% ACN in 0.1% formic acid over a 220 min gradient at a flow rate of 300 nl/min. The LTQ Orbitrap Velos Pro MS used exclusively CID fragmentation with wideband activation (pseudo-MS3 for neutral losses of phosphate residues) when acquiring MS/MS spectra. The spectra acquisition consisted of an orbitrap full MS (FTMS) scan followed by up to 15 LTQ MS/MS experiments (TOP15) on the most abundant ions detected in the full MS scan. Essential MS settings were as follows: FTMS (resolution 60,000; m/z range 400–2000) and MS/MS (Linear Trap; minimum signal threshold 500; wideband isolation; dynamic exclusion time setting 30 s; singly charged ions were excluded from selection). Normalized collision energy was set to 35%, and activation time to 10 ms.
Raw data processing, protein identification, and phosphopeptide assignment of the high-resolution orbitrap data were performed by PEAKS Studio 7.0 (Bioinformatics Solutions). FDR was set to <1%. Phosphosites were accepted as confident for p < 0.005 (modified t test, included in PEAKS Studio 7.0). The prediction of putative kinases responsible for the motif-specific phosphorylations was performed using the NetPhosK 1.0 Server (Blom et al., 2004).
Results
Synapsin is a phosphoprotein
Given that Drosophila Synapsin has been described as a phosphoprotein (Nuwal et al., 2011), we first sought to replicate and potentially extend the description of which sites of Synapsin are indeed phosphorylated. We found by mass spectrometry (LC-MS/MS) that in experimentally naive, synapsin+ wild-type flies the Synapsin protein features 28 phosphorylated sites and that these sites encompass consensus motifs for 11 kinases (Fig. 2, Table 1). We further note that proteins corresponding to both the edited and the nonedited version of Synapsin (Diegelmann et al., 2006) were found to be expressed (Fig. 2, Table 1).
Figure 2.
Phosphorylation sites of Synapsin and abundance of edited and nonedited Synapsin. LC-MS/MS analysis of experimentally naive, wild-type fly brains to map phosphorylated sites across the Synapsin protein (see also Table 1). Eighty-five LC-MS/MS runs were performed, consisting of a combination of 17 biological and five technical replicates each (coverage of the longest Synapsin protein isoform of 97%). This identified 28 phosphorylated sites of Synapsin: one at tyrosine, 10 at threonine, and 17 at serine. Twenty-three phosphorylated sites were identified for the first time, while five had been reported before (Zhai et al., 2008; Nuwal et al., 2011). Of the seven phosphorylated sites both reported by Nuwal et al. (2011) and covered by the present data, we can confirm three, while we found four of these sites to be nonphosphorylated. Both the edited and the nonedited forms of the Synapsin protein were found. That is, the pre-mRNA of the synapsin transcript is modified from the N-terminal motif RRFS (nonedited) to RGFS (edited) such that the PKA consensus motif RRFS is compromised (Diegelmann et al., 2006). In n = 85 LC-MS/MS runs the nonedited protein motif (RRFS) was found 54 times (p = 1.71E-07) and the edited protein motif (RGFS) was found 22 times (p = 9.44E-13). A phosphorylation at the motifs' serine was reliably detected only once (sic) for the edited, but not at all for the nonedited protein motif (Table 1; S22/S6). The workflow optimized sensitivity for proteome and phosphorylation site analysis of sample amounts corresponding to only a single brain. Therefore, a separation of isoforms before MS was not warranted, such that discrimination between isoforms is not possible. Given a 97% coverage, however, it is possible to ascertain the longest isoform (isoform D, E2QCY9_DROME; www.uniprot.org); this D isoform emerges from transcription starting at the first start codon and read through at the first stop codon (Klagges et al., 1996; Jungreis et al., 2011). A shorter isoform based on transcription from the second start codon and thus lacking 16 aa at the N terminus (Q24546_DROME; www.uniprot.org) was confirmed previously (Nuwal et al., 2011). Blue bars below the sequence indicate peptide-spectra matches (PSMs) identified by LC-MS/MS and the PEAKS de novo sequencing algorithm. The red “P” boxes indicate phosphorylation (p < 0.005). Note the S1003 → G mutation (white “G” box).
Table 1.
Phosphosites of the Synapsin protein determined by LC-MS/MS
| Amino acid | Phosphosite in Synapsin isoform D E2QCY9_DROME 1041 AS | Phosphosite in Synapsin Q24546_DROME 1025 ASc | Frequency of detection (n = 85) | P value | Predicted kinases (NetPhosK-Score) | Lit. |
|---|---|---|---|---|---|---|
| Sa | 22 | 6 | 1 | 6.31E-07 | PKA, 0.85 | |
| RSK, 0.65 | ||||||
| T | 86 | 70 | 40 | 7.94E-04 | p38MAPK, 0.53 | |
| T | 89 | 73 | 43 | 1.26E-03 | ||
| S | 99 | 83 | 6 | 2.00E-03 | PKA, 0.66 | |
| PKG, 0.59 | ||||||
| T | 102 | 86 | 0 | c.a.n.pb | PKC, 0.79 | |
| cdc2, 0.50 | Nuwal et al., 2011 | |||||
| cdk5, 0.75 | ||||||
| S | 103 | 87 | 4 | 2.51E-03 | p38MAPK, 0.58 | |
| GSK3, 0.53 | Nuwal et al., 2011 | |||||
| S | 107 | 91 | 38 | 2.00E-03 | ||
| S | 117 | 101 | 4 | 2.00E-06 | RSK, 0.57 | |
| S | 128 | 112 | 25 | 1.58E-03 | cdc2, 0.50 | |
| S | 136 | 120 | 11 | 2.00E-03 | ||
| T | 138 | 122 | 37 | 1.58E-03 | PKC, 0.76 | |
| T | 195 | 179 | 7 | 1.58E-03 | ||
| T | 200 | 184 | 11 | 1.26E-03 | ||
| Y | 204 | 188 | 8 | 1.26E-03 | ||
| T | 208 | 192 | 4 | 1.58E-03 | PKC, 0.71 | |
| cdk5, 0.65 | ||||||
| S | 480 | 464 | 81 | 5.01E-10 | RSK, 0.55 | |
| GSK3, 0.51 | Nuwal et al., 2011 | |||||
| T | 482 | 466 | 26 | 2.51E-09 | CKII, 0.52 | Nuwal et al., 2011 |
| cdk5, 0.62 | ||||||
| T | 489 | 473 | 1 | 1.58E-04 | p38MAPK, 0.53 | |
| GSK3, 0.51 | ||||||
| S | 512 | 496 | 24 | 1.00E-03 | ||
| S | 522 | 506 | 2 | 1.26E-08 | PKC, 0.79 | |
| PKA, 0.52 | ||||||
| S | 523 | 507 | 4 | 5.01E-10 | RSK, 0.59 | |
| S | 525 | 509 | 2 | 1.26E-08 | Zhai et al., 2008 | |
| S | 526 | 510 | 18 | 7.94E-12 | PKA, 0.67 | |
| CKI, 0.60 | Zhai et al., 2008 | |||||
| S | 549 | 533 | 29 | 1.00E-11 | PKA, 0.81 | |
| DNAPK, 0.61 | ||||||
| RSK, 0.60 | ||||||
| T | 551 | 535 | 56 | 7.94E-14 | PKC, 0.78 | |
| S | 554 | 538 | 0 | c.a.n.pb | Nuwal et al., 2011 | |
| S | 725 | 709 | 26 | 1.58E-03 | ||
| T | 728 | 712 | 17 | 3.16E-04 | PKC, 0.51 | |
| S | 752 | 736 | 5 | 5.01E-04 | ||
| S | 952 | 936 | 3 | 3.98E-07 | CKI, 0.50 | |
| S | 977 | 961 | 0 | c.a.n.pb | cdc2, 0.50 | Nuwal et al., 2011 |
| Y | 998 | 982 | 0 | c.a.n.pb | Nuwal et al., 2011 |
aOnly in the edited version of the protein was a phosphorylation at this site detected.
bc.a.n.p, covered as nonphosphorylated (peptides containing this motive were found as nonphosphorylated only).
cFor convenience we added amino acid numbering as reported previously (Zhai et al., 2008; Nuwal et al., 2011) using the matrix (Q24546_DROME; http://www.uniprot.org).
Synapsin− mutant flies have defects in both relief memory and punishment memory
Mutant flies lacking Synapsin (synapsin−; Fig. 1B,C) are deficient in both relief memory and punishment memory scores (Fig. 1D). That is, after shock-odor training, the associative performance indices for the synapsin+ wild-type strain are higher than for the synapsin− mutant, indicating reduced relief memory in the synapsin− mutant (U = 72.0, p < 0.05; N = 16, 16). The synapsin+ wild-type flies show the expectedly small, yet significant relief-memory scores (p < 0.05/2) while the performance indices for synapsin− mutant flies are not different from zero (p > 0.05/2). We conclude that relief memory is abolished in Synapsin null mutant flies.
After odor-shock training, synapsin− mutant flies show less negative performance indices, and thus less punishment memory, than synapsin+ wild-type flies (U = 3.0, p < 0.05; N = 12, 12). As both genotypes show significantly negative performance indices (p < 0.05/2 in both cases), we conclude that punishment memory is impaired, but not abolished, in Synapsin null mutant flies.
Is relief-memory and punishment-memory strength indeed lessened?
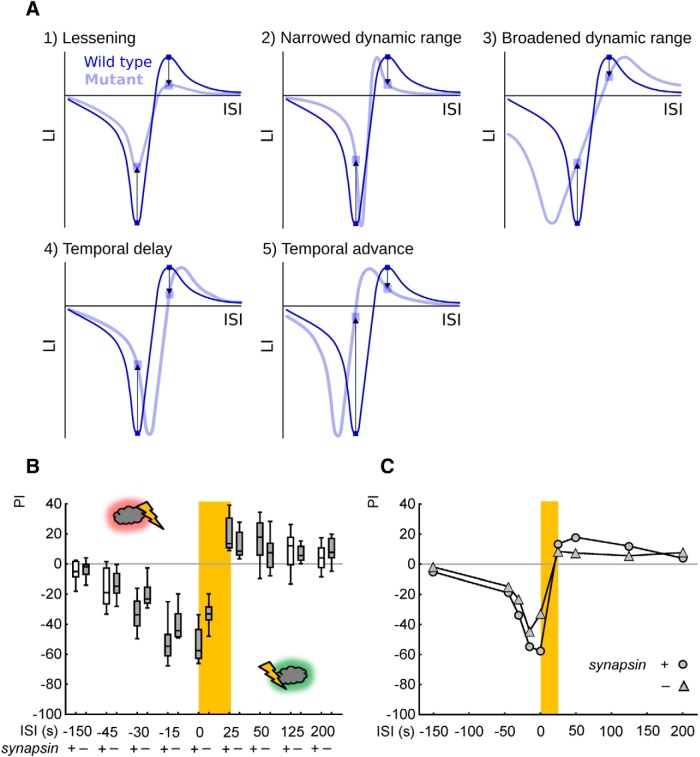
Given the known dependence of conditioned valence on the timing between odor and shock (the ISI; Gerber et al., 2014), there are actually five scenarios as to how a lack of Synapsin could lead to defects in both relief memory and punishment memory. That is, suppose only one ISI each was chosen for relief training and punishment training (Fig. 3, vertical arrows). Under such conditions all scenarios, but the one in Figure 3A1, predict that the respective maximum of the relief-memory and punishment-memory scores is actually unchanged. In other words, a reduction in the associative memory score using a single given ISI in any given mutant is not sufficient to argue for a lessening in strength of the underlying associative processes, which would show by a consistent lessening of associative memory scores and thus a dampening of scores along the y-axis of the ISI function (Fig. 3A1). Therefore, we decided to compare the full ISI function between synapsin+ wild-type and synapsin− mutant flies.
Figure 3.
Memory strength is lessened, rather than features of coincidence detection modulated, in mutant flies lacking Synapsin. A, When using only one ISI for relief learning and one ISI for punishment learning, less strong scores in both relief memory and punishment memory could be a result of a lessening in strength of the associative memory (A1), a narrowed dynamic range of associative coincidence detection (A2), a broadened dynamic range (A3), a temporal delay (A4) or a temporal advance (A5), if the ISIs happened to be chosen as indicated by the arrows. We therefore decided to compare the full ISI function between synapsin+ and synapsin− mutant flies. B, Associative performance indices of wild-type synapsin+ (respective left plots) and the mutant synapsin− flies (respective right plots) for the indicated ISIs. For statistics, see body text. A gray shading of the boxes indicates p < 0.05/10 and p < 0.05/8, respectively, in comparisons to chance levels (zero). Other details as in Figure 1. C, The median PIs from B are plotted across the ISIs. The consistent lessening of scores throughout the ISI function resembles scenario A1, suggesting a lessening of associative memory in the mutant synapsin− flies.
As shown in Figure 3, B and C, we find that after punishment learning the performance indices are consistently less negative in synapsin− mutant flies than in the synapsin+ wild-type (U = 63, 43, 30, 31, 26; p > 0.05 for the ISIs of −150, − 45 s, and p < 0.05 for the ISIs of −30, − 15, 0 s; N = 10–12). Likewise, after relief learning the performance indices are consistently less positive in synapsin− mutant than synapsin+ wild-type flies (U = 42, 316, 59, 182; p > 0.05 for the ISIs of 25, 125, 200 s and p < 0.05 for the ISI of 50 s; N = 12–30). In other words, after both punishment-memory and relief-learning memory strength is lessened, corresponding to the scenario in Figure 3A1. This argues that a lack of Synapsin entails a lessening in strength of associative memory rather than an alteration in the temporal properties of coincidence detection (Fig. 3A2–A5).
Task-relevant sensory and motor faculties are intact in synapsin− mutant flies
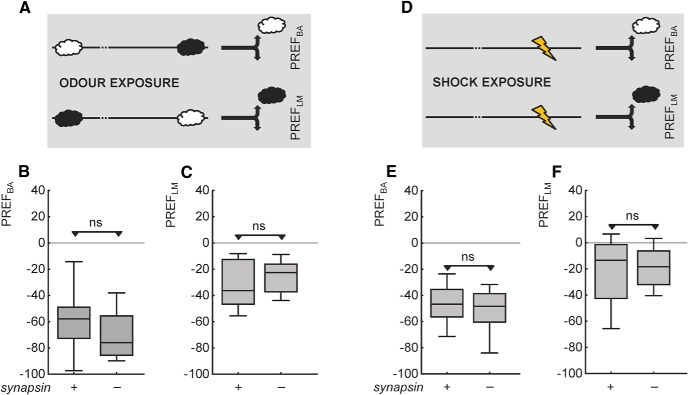
To ascertain whether the lessening in strength of both relief memory and punishment memory in the synapsin− mutant is actually secondary to some sort of sensory or motor impairment, we test whether behavior toward the to-be-associated stimuli is impaired. This is found not to be the case. There is no between-genotype difference in shock avoidance (Fig. 4A; U = 104, p > 0.05; N = 16, 16). Likewise, synapsin+ wild-type and synapsin− mutant flies do not differ in their behavior toward BA or LM (Fig. 4B,C; U tests: U = 116, 158, p > 0.05 in both cases; N = 19, 16, 20, 16). We conclude that avoidance of the shock as well as of both the odors in question is indistinguishable between genotypes, in experimentally naive animals.
Figure 4.
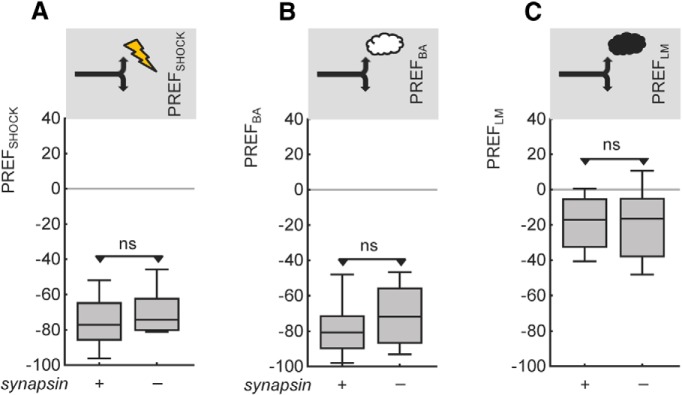
Behavior toward the to-be-associated stimuli is normal in experimentally naive mutant flies lacking Synapsin. Avoidance of the shock (A) and of the odors (B: BA; C: LM) is not different between experimentally naive flies of the two genotypes. ns: p > 0.05. Other details as in Figure 1.
These kinds of control procedures have been state of the art since the first studies of associative odor learning in flies (Quinn et al., 1974; Dudai et al., 1976; Tully and Quinn, 1985). However, testing olfactory behavior in experimentally naive animals only shows that at the beginning of the learning experiment the mutants are normal in sensorimotor ability; whether these faculties are still intact at the moment of the test remains unclear (Préat, 1998; Michels et al., 2005; see discussion in Gerber and Stocker, 2007; Knapek et al., 2010). For example, the handling during the experiment, exposure to odors, and/or exposure to the shocks during training can nonassociatively alter olfactory behavior (Préat, 1998; Boyle and Cobb, 2005; Colomb et al., 2007; Knapek et al., 2010; Sadanandappa et al., 2013). If the mutant differed from the wild-type in its susceptibility to these effects, the mutant but not the wild-type could be distorted in its olfactory behavior at the moment of test, and this could be mistaken as an associative memory phenotype. Indeed, for isoamyl acetate and 1-butanol, synapsin+ wild-type and synapsin− mutant flies differ in olfactory behavior after such exposure (H. Tanimoto, personal communication). We therefore run two kinds of “sham training” control: animals are handled just as in normal training, but either the shock is omitted (odor exposure, Fig. 5A) or the odors are omitted (shock exposure, Fig. 5D). After this kind of treatment, we test whether synapsin+ wild-type and synapsin− mutant flies differ in their behavior toward either odor. We do not find any between-genotype differences in these tests (Fig. 5B,C,E,F; U = 146, 163, 113, 120, p > 0.05 in all cases; N = 16–21). We conclude that also at the moment of test, those sensory and motor faculties that are required to show learned odor behavior are not defective in the synapsin− mutant.
Figure 5.
Olfactory behavior is normal in mutant flies lacking Synapsin also after training-like stimulus exposure. Genotypes do not differ in olfactory behavior after either odor exposure (A; B: BA, C: LM) or shock exposure (D; E: BA, F: LM). Other details as in Figure 4.
Do the synapsin− mutant defects reflect defects in odor discrimination?
Both the control procedures for olfactory behavior, the testing of experimentally naive animals (Fig. 4B,C), and the testing of animals after sham training (Fig. 5) feature a test situation where the flies chose between an odor arm versus a blank arm of the T-maze. However, both relief memory and punishment memory are assessed in a choice situation between the trained odor and the control odor (Fig. 1A). The rationale, throughout the literature, for not running the control procedures with two odors as well is that if both odors were (approximately) equally salient, one might obtain choice scores of approximately zero in both wild-type and mutant flies, but possibly for different reasons: the wild-type flies may be effectively indifferent between the two odors, while the mutants may be anosmic. However, intact performance in one-odor control procedures still allows the argument that the synapsin− mutant defect in two-odor memory tests may not actually be due to defects in memory, but to an inability of the synapsin− mutant flies to tell the two odors apart. If this is the case, deficits should not be observed in nondiscriminatory tasks, i.e., if one-odor versions of the task are used (Fig. 6A). However, also in these nondiscriminatory tasks, deficits in relief memory and punishment memory are found (Fig. 6B; U tests: U = 67, 21, p < 0.05 in both cases; N = 16, 16, 16, 16).
Figure 6.
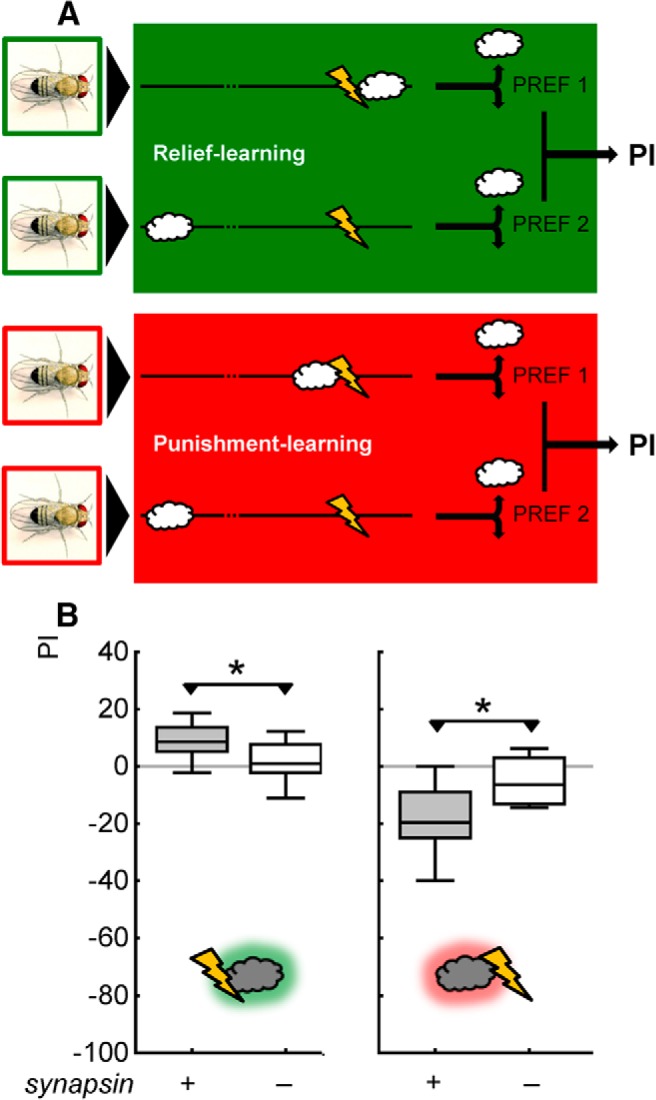
Mutants lacking Synapsin are defective also in nondiscriminatory relief- and punishment-learning tasks. A, Schematic of the one-odor versions of the relief- and punishment-learning tasks. The procedure is as in the two-odor version of the paradigm (Fig. 1A; see Materials and Methods), except that one odor is omitted. That is, for one-odor relief learning, the odor (BA) is presented upon the cessation of shock, while for punishment learning the odor is presented before the onset of shock. In both cases, a second experimental group receives unpaired presentations of odor and punishment. The difference in odor preference between paired- and unpaired-trained groups indicates associative memory, and is quantified as PI. B, Also in nondiscriminatory, one-odor versions of the paradigm, relief memory and punishment memory are strongly impaired in mutant flies lacking Synapsin (synapsin−). Other details as in Figure 1D.
We conclude that the defects of the synapsin− mutant flies after relief learning and punishment learning reflect defects in associative memory and are unrelated to odor discrimination ability.
RNAi-mediated knock-down phenocopies the relief memory and punishment memory defect
We next test whether the defect of the synapsin− mutant flies in relief memory and punishment memory is indeed due to the lack of the Synapsin protein, or, despite our outcrossing efforts, to residual differences in genetic background or to side effects of the deletion. To this end, we use an RNAi approach, combining a UAS-RNAi-syn effector strain with the broad neuronal driver elav-Gal4 (knockdown).
A reduction in Synapsin levels (Fig. 7A) leads to reduced relief memory in the knock-down flies compared with both genetic controls (Fig. 7B; U = 19, 38, p < 0.05/3 in both cases; N = 12, 15, 14; a test across all genotypes yields: p < 0.05, H = 12, df = 2). Both genetic controls show small yet significant relief-memory scores (p < 0.05/3 in both cases), while the scores of knock-down flies are not significantly different from zero (p > 0.05/3). We conclude that relief memory is intact in control flies but abolished in knock-down flies.
Figure 7.
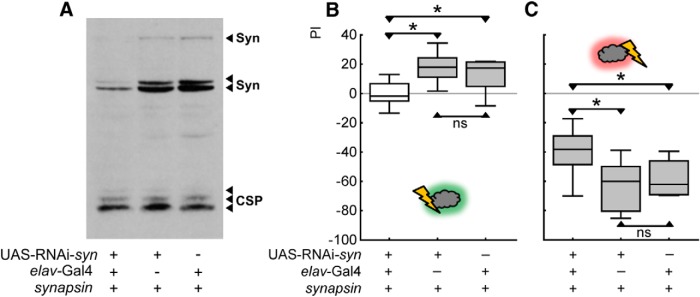
RNAi-mediated knockdown of Synapsin impairs both relief memory and punishment memory. A, Western blot of material obtained from three heads stained for Synapsin and for CSP as a loading control. The blot is loaded with double heterozygous elav-Gal4;; UAS-RNAi-syn flies to the left (knockdown), UAS-RNAi-syn heterozygous flies in the middle (effector control), and elav-Gal4 heterozygote flies to the right (driver control). In the knock-down flies, a reduction of all Synapsin isoforms is apparent. B, Relief memory is abolished in knock-down flies compared with controls. C, Punishment memory is reduced in knock-down flies compared with controls. *p < 0.05/3 and ns: p > 0.05/3 are used for pairwise comparisons. Gray shading of the boxes indicates significance from chance (zero) at p < 0.05/3.
Knock-down flies also show reduced punishment memory compared with both genetic control flies (Fig. 7C; U = 47, 48, p < 0.05/3 in both cases; N = 13, 19, 15; a test across all genotypes yields: p < 0.05, H = 9, df = 2). All three genotypes show significant levels of punishment-memory (p < 0.05/3 in all three cases). We conclude that punishment memory is reduced, but is not abolished, in flies with reduced levels of Synapsin.
Given that two independent means of reducing Synapsin levels (i.e., the deletion mutant and the RNAi-mediated knockdown) yield concordant memory defects, we conclude that it is the effect upon Synapsin levels, rather than their respectively different potential genetic background or off-target effects, which is responsible for the defects in relief memory and punishment memory.
Locally restoring Synapsin restores relief memory and punishment memory
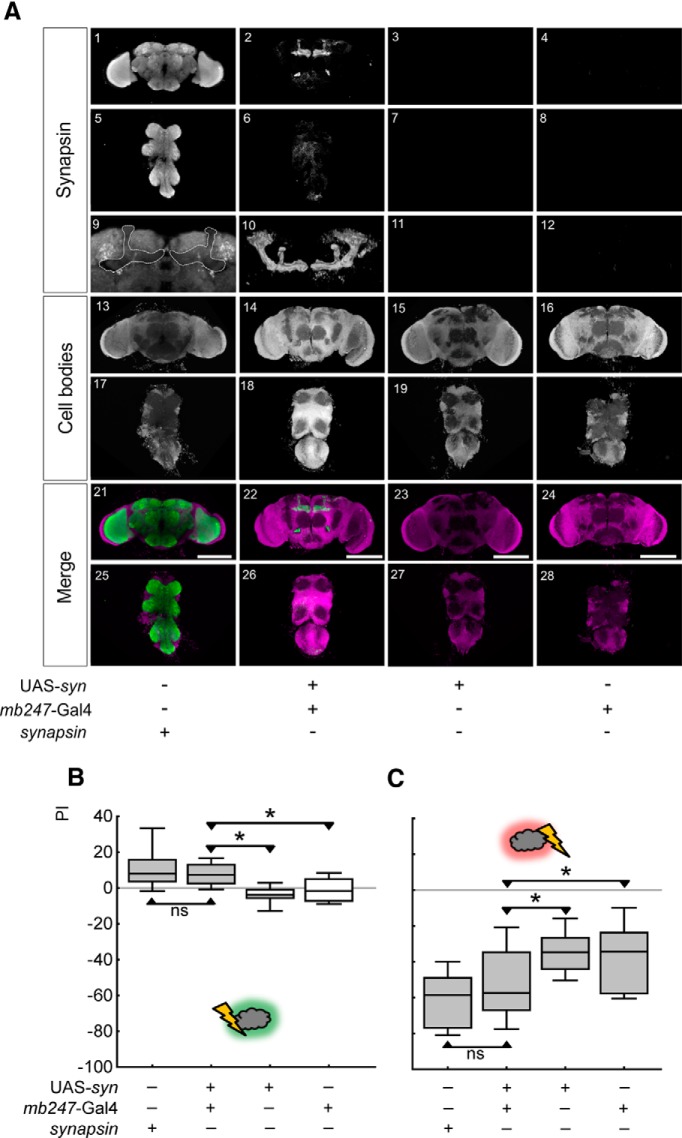
To further scrutinize the role of the Synapsin protein, we test whether restoring Synapsin expression locally in the mushroom bodies, using mb247-Gal4 as a driver and UAS-syn as an effector in the synapsin− mutant background, can rescue the mutant defect in relief memory and/or punishment memory.
The expression of Synapsin in the rescue flies is indeed restricted to the mushroom bodies (Fig. 8A). Upon such local expression, we find that both relief memory and punishment memory are fully rescued: after relief learning, rescue flies perform better than both control genotypes (Fig. 8B; U = 26, 48, p < 0.05/3 in both cases; N = 17, 15, 15, 15), and actually do as well as synapsin+ wild-type flies (U = 115, p > 0.05/3; N = 15, 17; a test across all genotypes yields: p < 0.05, H = 22, df = 3). We conclude that restoring Synapsin locally in the mushroom body fully restores the synapsin− mutant defect in relief memory.
Figure 8.
Locally restoring Synapsin restores relief memory and punishment memory. A, Expression pattern of Synapsin in flies of the indicated genotypes. In the bottom row (Merge), anti-Synapsin staining in brains and thoracic nervous systems is shown in green, and cell-body counterstaining with propidium iodide is shown in magenta, from 0.9 μm frontal optical sections of the indicated genotypes. In A9–12, the mushroom body regions from A1–4 are shown at higher magnification. In A9 the mushroom bodies are indicated by the stippled line. In A10 the expression of Synapsin using the mb247-Gal4 driver is shown as a 3D display. Scale bars: 100 μm. B, Relief memory of synapsin− mutant flies is fully restored upon rescue expression of Synapsin using the mb247-Gal4 driver. C, Punishment memory, too, is fully restored upon locally expressing Synapsin. *p < 0.05/3 and ns: p > 0.05/3 are used for pairwise comparisons. Gray shading of the boxes indicates significance from chance (zero) at p < 0.05/4.
Also after punishment learning, rescue flies perform better than either kind of control flies (Fig. 8C; U = 150, 185, p < 0.05/3 in both cases; N = 26, 26, 26, 26). Indeed, their punishment-memory scores are as high as those of synapsin+ wild-type flies (U = 290, p > 0.05/3; N = 26, 26; a test across all genotypes yields: p < 0.05, H = 30, df = 3). We conclude that the synapsin− mutant defect in punishment memory is also fully restored upon restoring Synapsin locally in the mushroom body.
We conclude that restoring Synapsin in the set of cells covered by mb247-Gal4, covering the mushroom body α, β, and γ lobes, and with faint if any background expression (Aso et al., 2009; Fig. 8A), is sufficient to restore the defects in both relief memory and punishment memory, which ensue upon a lack of Synapsin.
Acutely and locally restoring Synapsin restores relief memory and punishment memory
We next wanted to see whether Synapsin functions during embryonic, larval, and pupal development, or rather acutely during adulthood. Using tub-Gal80ts we combined the expression of Gal80ts, a temperature-dependent inhibitor of Gal4, together with mb247-Gal4 as a driver and UAS-syn as an effector. Thus, the induction of Synapsin expression in the mushroom bodies can be achieved by raising the temperature acutely only during adulthood, which inactivates Gal80ts, releases the inhibition of Gal4, and allows Synapsin expression in the mb247-Gal4 pattern (compare Figs. 9A10, 10A10). As compared with constitutive expression of Synapsin, such induced expression is notably faint (compare Figs. 8A22, 10A22).
Figure 9.
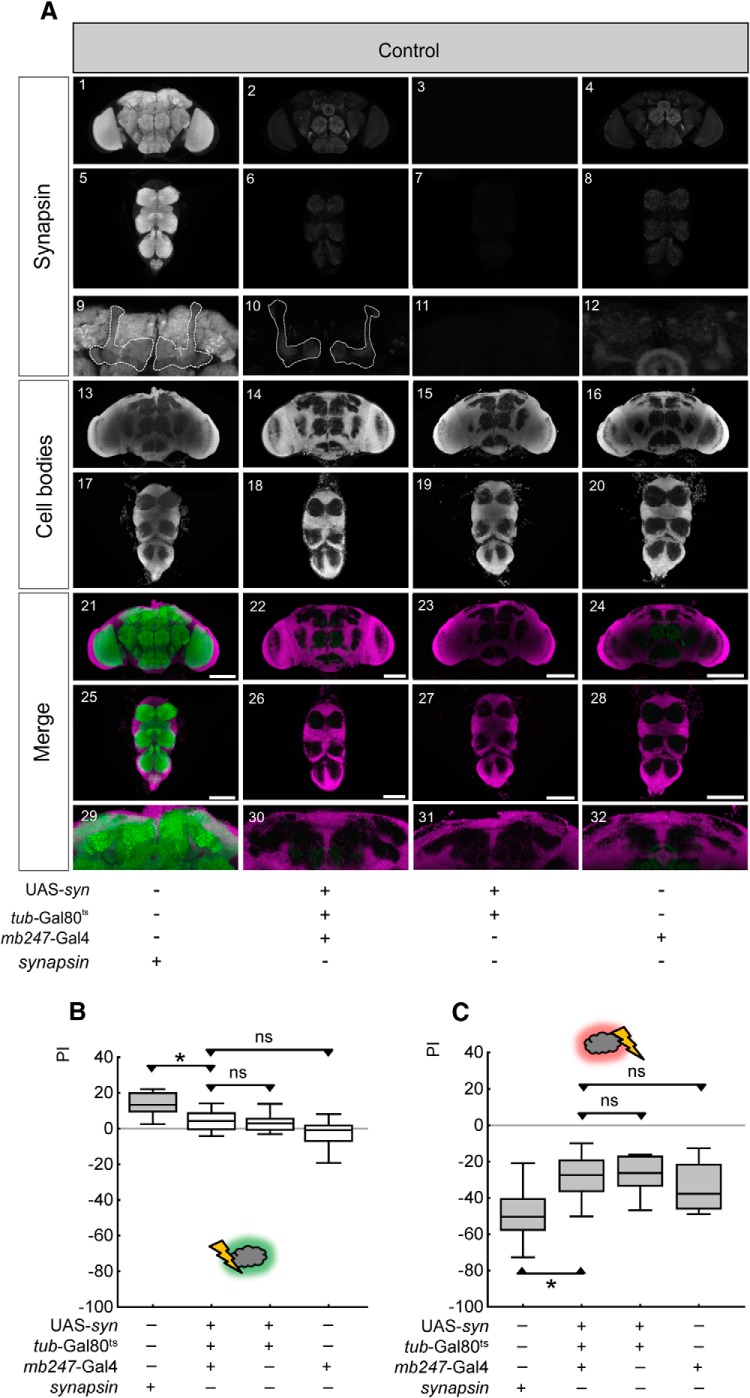
Relief memory and punishment memory remain impaired in control conditions without acute and local restoration of Synapsin. A, Expression pattern of Synapsin in uninduced control flies of the indicated genotypes. In the bottom rows (Merge), anti-Synapsin staining in brains and thoracic nervous systems is shown in green, while cell-body counterstaining with propidium iodide is shown in magenta. The mushroom body region of A1–4 is shown at higher magnification in A9–12. The stippled line in A9 and A10 indicates the mushroom body neuropil. The mushroom body region of A21–24 is shown at higher magnification in A29–32. Note the absence of anti-Synapsin staining in A10 and A30. Scale bars: 100 μm. B, C, In uninduced control conditions, relief memory (B) and punishment memory (C) remain abolished in the experimental genotype. *p < 0.05/3 and ns: p > 0.05/3 are used for pairwise comparisons. Gray shading of the boxes indicates significance from chance (zero) at p < 0.05/4.
Figure 10.
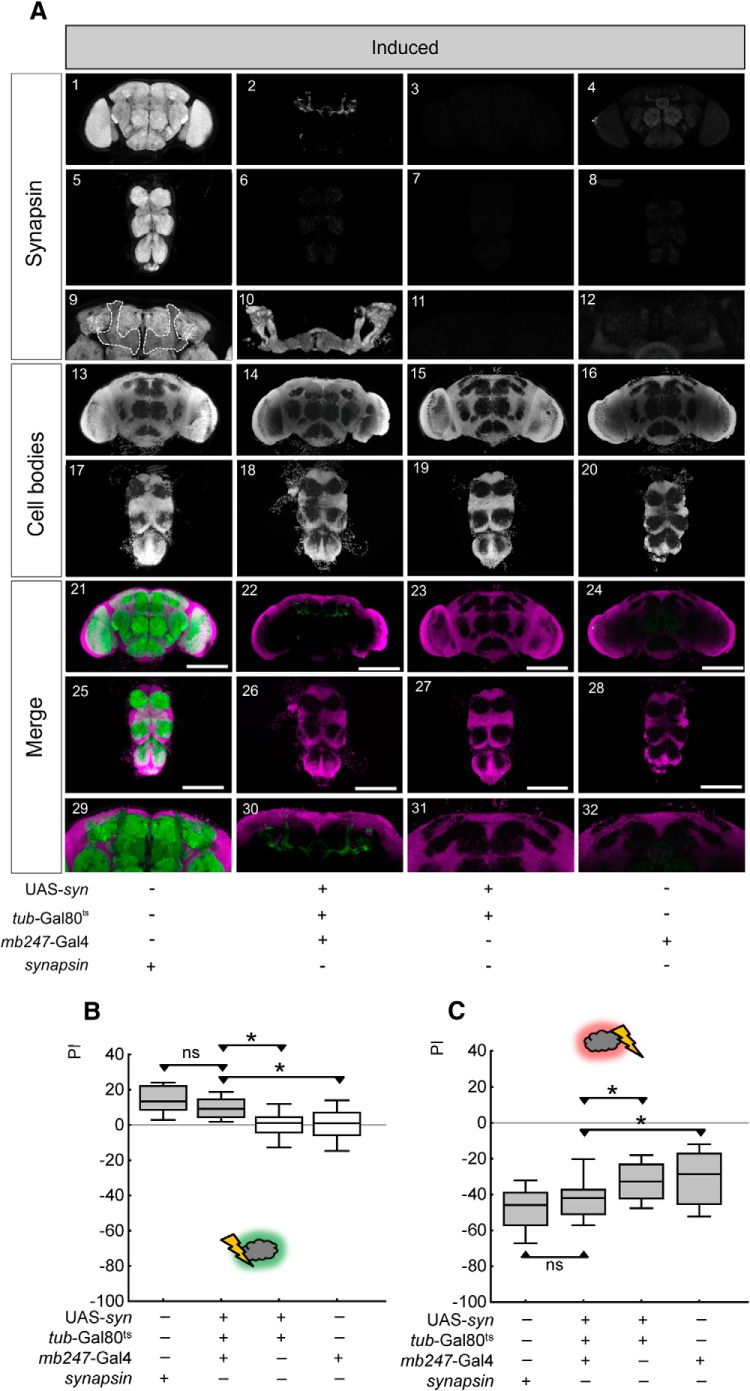
Acutely and locally inducing Synapsin expression restores relief memory and punishment memory. A, Expression pattern of Synapsin in induced rescue flies of the indicated genotypes. In the bottom row (Merge), anti-Synapsin staining in brains and thoracic nervous systems is shown in green, while cell-body counterstaining with propidium iodide is shown in magenta. The mushroom body region of A1–4 is shown at higher magnification in A9–12. The stippled line in A9 indicates the mushroom body neuropil. In A10 the expression of Synapsin induced by the mb247-Gal4 driver is shown as a 3D display. The mushroom body region of A21–24 is shown at higher magnification in A29–32. Scale bars: 100 μm. B, Relief memory of synapsin− mutant flies is restored upon acutely induced local expression of Synapsin using the mb247-Gal4 driver in combination with Gal80ts. C, Punishment memory, too, is fully restored upon acutely and locally expressing Synapsin. *p < 0.05/3 and ns: p > 0.05/3 are used for pairwise comparisons. Gray shading of the boxes indicates significance from chance (zero) at p < 0.05/4.
In control conditions without induction, relief-memory scores in the experimental genotype are less than in the synapsin+ wild-type flies (Fig. 9B; U = 46, p < 0.05/3, N = 21, 21), and not different from effector control and driver control (U = 116, 68, p > 0.05/3, N = 21, 21). Acute induction of Synapsin expression restores relief memory to levels indistinguishable from those observed in synapsin+ wild-type flies (Fig. 10B; U = 151, p > 0.05/3, N = 16, 16). Indeed, relief-memory scores upon acute Synapsin expression are higher than in effector- and in driver-control flies (U = 99, 116, p < 0.05/3; N = 16, 16).
Likewise, after punishment learning acute Synapsin expression supports a full rescue compared with synapsin+ wild-type flies (Fig. 10C; U = 231, p > 0.05/3, N = 25, 22); these flies indeed show stronger punishment-memory scores than both the effector control and driver control (U = 135, 117, p < 0.05/3 in both cases, N = 22, 19). Without induction, punishment memory remains at levels less than in synapsin+ wild-type flies (Fig. 9C; U = 38; p < 0.05/3, N = 15, 12) and indistinguishable from effector- and driver-control flies (U = 69, 51, p > 0.05/3 in both cases, N = 13, 12).
We conclude that restoring Synapsin at fairly low levels, acutely during adulthood, and locally in the mushroom bodies, restores the synapsin− mutant defects in both relief memory and punishment memory.
Discussion
We report Synapsin as a determinant for memory strength in a behavioral assay of timing-dependent associative plasticity (Figs. 1, 3, 6–9). The defects upon a lack of Synapsin do not reflect any task-relevant sensorimotor impairment, as manifest either in naive responsiveness to odors or shock (Fig. 4) or in responsiveness to the odors after sham-training procedures (Fig. 5). Also, the outcrossing regimen for the mutant, the RNAi-phenocopy of the mutant phenotype, and the spatially and temporally specific rescue make it highly unlikely that effects of genetic background or off-target effects of our genetic manipulations impact these results.
The “fingerprint” of Synapsin dysfunction is distinct from what has been found for the w1118 mutation, which shifts the overall balance between relief memory and punishment memory toward generally more negative valence (Yarali et al., 2009b). That is, in the w1118 mutants any event including an electric shock establishes a more negative net effect in memory. Fittingly, Nakamura et al. (1999) reported a relation of genetic variance in the human ABCG1 gene, the homolog of the white gene of Drosophila, to the susceptibility for panic and mood disorders.
We find that punishment memory is partially but not completely abolished both in Synapsin null mutant flies and upon an RNAi-mediated knockdown of Synapsin. The observation that this defect is partial is in agreement with what Godenschwege et al. (2004) and Knapek et al. (2010) reported for the Synapsin null mutant, namely a 25–30% decrement in punishment-memory scores. A somewhat stronger yet still partial defect (50%) was also seen after larval odor-sugar learning (Michels et al., 2005, 2011). Interestingly, in wild-type flies an application of cold shock between training and test induces a partial amnesia of punishment memory, and the residual amnesia-resistant memory does not require Synapsin (Knapek et al., 2010). Given that relief memory is fully abolished upon loss of function of Synapsin, one would thus reason that cold shock should fully abolish relief memory–and intriguingly this is what Diegelmann et al. (2013b) have found. The emerging scenario is that punishment learning establishes two short-term memory components: one that is Synapsin dependent and amnesia sensitive, and one that is Synapsin independent and amnesia resistant. In contrast, relief learning establishes only Synapsin-dependent and amnesia-sensitive short-term memory. This implies that if in a natural succession of events a traumatic experience were to induce both relief memory and punishment memory (for the stimuli experienced, respectively, after and before the trauma), an amnesic cold-shock treatment would leave only a punishment memory.
Our observation that restoring Synapsin in the Synapsin null mutant fully restores both relief- and punishment-memory scores (Fig. 8) demonstrates that it is the absence of the Synapsin protein that causes the defects in associative memory, rather than other effects of the deletion or of differences in genetic background that remained despite our extensive outcrossing regimen. The current study is thus the first actually to prove a role of the Synapsin protein in the associative memory of adult Drosophila.
The rescue of associative memory was obtained by restoring Synapsin both in a temporally specific way, acutely only during adulthood (Fig. 10), and in a spatially specific way, in the mushroom body (Figs. 8, 10). Notably, this was the case for both relief memory and punishment memory. This prompts the question of how the fly brain is organized on the cellular and/or molecular level to establish memories that support opposite behaviors (i.e., conditioned avoidance vs approach). For such a discussion, a brief sketch of the current working hypothesis on punishment learning is warranted (for review, see Heisenberg, 2003; Gerber et al., 2004; Davis, 2005; Margulies et al., 2005; Keene and Waddell, 2007; Kahsai and Zars, 2011; Gerber et al., 2014; see also the seminal recent work of Aso et al., 2014a, b).
A given odor activates a specific combination of olfactory sensory neurons, according to the ligand profile of the expressed receptor protein. Sensory neurons expressing a given receptor converge at typically a single glomerulus in the antennal lobe; within these glomeruli they provide output to olfactory projection neurons. The pattern of activated projection neurons is additionally shaped by lateral connections between glomeruli. The olfactory projection neurons further connect to two target areas, the mushroom body and the lateral horn. Thus, dependent on receptor expression, ligand profile, and connectivity within this system, odors can be coded combinatorially across these ascending pathways. Both mushroom body and lateral horn then connect to premotor circuitry.
Processing of the electric shock is less well understood. What is clear is that shock activates a subset of dopaminergic neurons mediating an internal punishment signal. These neurons provide input (mb-input neurons) to a sufficient number of mushroom body neurons to cover the full olfactory stimulus space (Ito et al., 1998; Schwaerzel et al., 2003; Riemensperger et al., 2005; Kim et al., 2007; Aso et al., 2009; Claridge-Chang et al., 2009; Mao and Davis, 2009; Aso et al., 2010, 2012; Burke et al., 2012; Pech et al., 2013; Das et al., 2014; Galili et al., 2014; larval Drosophila: Schroll et al., 2006; Selcho et al., 2009).
Thus, only in an odor-specific set of mushroom body neurons does a coincidence of odor- and shock-induced activity take place, resulting in a modulation of connection between these mushroom body neurons and their postsynaptic partners, the mushroom body output neurons (mb-output neurons). The AC-cAMP-PKA signaling cascade is one of the necessary processes for detecting and enacting this coincidence (Zars et al., 2000; Thum et al., 2007; Blum et al., 2009; Tomchik and Davis, 2009; Boto et al., 2014; see Gervasi et al., 2010 for appetitive learning). Arguably Synapsin is one of the relevant targets of this cascade in the context of associative learning (Michels et al., 2011). If after training the learned odor is encountered again, activity in the mb-output neurons—because of their modified input from the mushroom body neurons—is altered such that learned odor avoidance can take place (Séjourné et al., 2011; Menzel, 2012). Nontrained odors do not support conditioned avoidance, unless sufficiently similar in quality and/or intensity to the trained one (Yarali et al., 2009a; Niewalda et al., 2011; Campbell et al., 2013; Barth et al., 2014).
The mushroom body is under intense study with respect to the organization of punishment learning versus reward learning. The emerging picture is that punishment learning and reward learning may engage different sets of mushroom body neurons as well as different input and output neurons, as if the mushroom body was internally multiplexed according to valence (mb: Perisse et al., 2013; Boto et al., 2014; mb-input: Liu et al., 2012; mb-output: Séjourné et al., 2011; Plaçais et al., 2013; Aso et al., 2014a, b). With respect to the mb-input neurons, those dopaminergic neurons that are required for punishment learning, defined, for example, by the TH-Gal4 driver, are dispensable for relief-memory (Yarali and Gerber, 2010). It is not known which TH-Gal4-negative mb-input neurons and which mb-output neurons are participating in relief learning. Whether different subsets of the mushroom body neurons harbor the memory trace after relief learning and punishment learning likewise remains to be determined.
Alternatively, relief memory and punishment memory may be dissociated at the level not of the Synapsin protein as such, but at the level of its phosphorylation pattern. Given the 28 phosphorylated sites of Synapsin, targeted by up to 11 different kinases (Fig. 2, Table 1; Michels et al., 2011; Nuwal et al., 2011; Sadanandappa et al., 2013; Diegelmann et al., 2013a), it is conceivable that different kinases and/or phosphorylation sites of the Synapsin protein could be used during relief learning and punishment learning. Likewise, the mRNA editing observed for one of the phosphorylation sites of Synapsin (Diegelmann et al., 2006) could be selectively involved in relief learning and punishment learning, in particular as the proteins corresponding to both the edited and the nonedited version are indeed expressed (Fig. 2, Table 1).
To summarize, the current results regarding Synapsin point to shared genetic and molecular determinants for relief memory and punishment memory in Drosophila. Given our shared evolutionary heritage in general and the conserved role of Synapsin in mammalian associative memory in particular, this may caution against systemic pharmacological approaches for reducing excessively strong punishment memory in humans, for example, after traumatic experiences. This is because such traumatic experiences may induce both relief memory and punishment memory. If the target of the pharmacological treatment affects both memories, a systemic drug treatment will unwittingly reduce relief memory as well and may thus have a net detrimental effect (for a more detailed discussion see Gerber et al., 2014). For example, in the case of Synapsin, a pharmacological erasure of Synapsin-dependent memory would abolish relief memory but would leave punishment memory partially intact (see also the discussion of cold-shock amnesia above). In this sense, and with due caveats in mind, the current study can help in avoiding adverse effects of medical treatment after traumatic experiences.
Footnotes
This study received institutional support from the Leibniz Institut für Neurobiologie, the Wissenschaftsgemeinschaft Gottfried Wilhelm Leibniz, the State of Sachsen-Anhalt, the Center for Behavioral Brain Sciences Magdeburg, and the Otto von Guericke Universität Magdeburg, as well as grant support from the Deutsche Forschungsgemeinschaft (SFB 779 Motivated behavior). Support, comments, and discussions with E. Buchner, M. Heisenberg, T. Saumweber, A. Yarali, and the members of our research group are gratefully acknowledged, as are the comments by the reviewers. We thank K. Gerber, K. Tschirner, and C. Tauber for their help with outcrossing of flies, Western blotting, and histology; K. Öchsner, H. Kaderschabek, R. Blumenstein, F. Unterstab, and M. Dombach for workshop support; Evren Pamir for help with Figure 3A; and R. Glasgow for language editing.
The authors declare no competing interests.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License Creative Commons Attribution 4.0 International, which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Andreatta M, Mühlberger A, Yarali A, Gerber B, Pauli P. A rift between implicit and explicit conditioned valence in human pain relief learning. Proc Biol Sci. 2010;277:2411–2416. doi: 10.1098/rspb.2010.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M, Fendt M, Mühlberger A, Wieser MJ, Imobersteg S, Yarali A, Gerber B, Pauli P. Onset and offset of aversive events establish distinct memories requiring fear and reward networks. Learn Mem. 2012;19:518–526. doi: 10.1101/lm.026864.112. [DOI] [PubMed] [Google Scholar]
- Arnold C, Reisch N, Leibold C, Becker S, Prüfert K, Sautter K, Palm D, Jatzke S, Buchner S, Buchner E. Structure–function analysis of the cysteine string protein in Drosophila: cysteine string, linker and C terminus. J Exp Biol. 2004;207:1323–1334. doi: 10.1242/jeb.00898. [DOI] [PubMed] [Google Scholar]
- Aso Y, Grübel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H. The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet. 2009;23:156–172. doi: 10.1080/01677060802471718. [DOI] [PubMed] [Google Scholar]
- Aso Y, Siwanowicz I, Bräcker L, Ito K, Kitamoto T, Tanimoto H. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20:1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Herb A, Ogueta M, Siwanowicz I, Templier T, Friedrich AB, Ito K, Scholz H, Tanimoto H. Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet. 2012;8:e1002768. doi: 10.1371/journal.pgen.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, Rubin GM. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014a;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais PY, Robie AA, Yamagata N, Schnaitmann C, Rowell WJ, Johnston RM, Ngo TT, Chen N, Korff W, Nitabach MN, Heberlein U, Preat T, Branson KM, Tanimoto H, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife. 2014b;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth J, Dipt S, Pech U, Hermann M, Riemensperger T, Fiala A. Differential associative training enhances olfactory acuity in Drosophila melanogaster. J Neurosci. 2014;34:1819–1837. doi: 10.1523/JNEUROSCI.2598-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Blum AL, Li W, Cressy M, Dubnau J. Short- and long-term memory in Drosophila require cAMP signaling in distinct neuron types. Curr Biol. 2009;19:1341–1350. doi: 10.1016/j.cub.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto T, Louis T, Jindachomthong K, Jalink K, Tomchik SM. Dopaminergic modulation of cAMP drives nonlinear plasticity across the Drosophila mushroom body lobes. Curr Biol. 2014;24:822–831. doi: 10.1016/j.cub.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle J, Cobb M. Olfactory coding in Drosophila larvae investigated by cross-adaptation. J Exp Biol. 2005;208:3483–3491. doi: 10.1242/jeb.01810. [DOI] [PubMed] [Google Scholar]
- Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, Waddell S. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492:433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RA, Honegger KS, Qin H, Li W, Demir E, Turner GC. Imaging a population code for odor identity in the Drosophila mushroom body. J Neurosci. 2013;33:10568–10581. doi: 10.1523/JNEUROSCI.0682-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li H, Hirsh J, Miesenböck G. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139:405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomb J, Grillenzoni N, Stocker RF, Ramaekers A. Complex behavioural changes after odour exposure in Drosophila larvae. Anim Behav. 2007;73:587–594. doi: 10.1016/j.anbehav.2006.04.016. [DOI] [Google Scholar]
- Das G, Klappenbach M, Vrontou E, Perisse E, Clark CM, Burke CJ, Waddell S. Drosophila learn opposing components of a compound food stimulus. Curr Biol. 2014;24:1723–1730. doi: 10.1016/j.cub.2014.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Diegelmann S, Nieratschker V, Werner U, Hoppe J, Zars T, Buchner E. The conserved protein kinase-A target motif in synapsin of Drosophila is effectively modified by pre mRNA editing. BMC Neurosci. 2006;7:76. doi: 10.1186/1471-2202-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diegelmann S, Klagges B, Michels B, Schleyer M, Gerber B. Maggot learning and Synapsin function. J Exp Biol. 2013a;216:939–951. doi: 10.1242/jeb.076208. [DOI] [PubMed] [Google Scholar]
- Diegelmann S, Preuschoff S, Appel M, Niewalda T, Gerber B, Yarali A. Memory decay and susceptibility to amnesia dissociate punishment-from relief-learning. Biol Lett. 2013b;9:20121171. doi: 10.1098/rsbl.2012.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y, Jan YN, Byers D, Quinn WG, Benzer S. dunce, a mutant of Drosophila deficient in learning. Proc Natl Acad Sci U S A. 1976;73:1684–1688. doi: 10.1073/pnas.73.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenberg J, Plath JA, Lorang S, Morgenstern L, Eisenhardt D. Short- and long-term memories formed upon backward conditioning in honeybees (Apis mellifera) Learn Mem. 2014;21:37–45. doi: 10.1101/lm.031765.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin JC, Lee KM, Hanna EK, Prinstein MJ. Feeling worse to feel better: pain-offset relief simultaneously stimulates positive affect and reduces negative affect. Psychol Sci. 2013a;24:521–529. doi: 10.1177/0956797612458805. [DOI] [PubMed] [Google Scholar]
- Franklin JC, Puzia ME, Lee KM, Lee GE, Hanna EK, Spring VL, Prinstein MJ. The nature of pain offset relief in nonsuicidal self-injury: a laboratory study. Clin Psychol Sci. 2013b;1:110–119. doi: 10.1177/2167702612474440. [DOI] [Google Scholar]
- Galili DS, Dylla KV, Lüdke A, Friedrich AB, Yamagata N, Wong JY, Ho CH, Szyszka P, Tanimoto H. Converging circuits mediate temperature and shock aversive olfactory conditioning in Drosophila. Curr Biol. 2014;24:1712–1722. doi: 10.1016/j.cub.2014.06.062. [DOI] [PubMed] [Google Scholar]
- Garcia CC, Blair HJ, Seager M, Coulthard A, Tennant S, Buddles M, Curtis A, Goodship JA. Identification of a mutation in synapsin I, a synaptic vesicle protein, in a family with epilepsy. J Med Genet. 2004;41:183–186. doi: 10.1136/jmg.2003.013680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B, Stocker RF. The Drosophila larva as a model for studying chemosensation and chemosensory learning: a review. Chem Senses. 2007;32:65–89. doi: 10.1093/chemse/bjl030. [DOI] [PubMed] [Google Scholar]
- Gerber B, Tanimoto H, Heisenberg M. An engram found? Evaluating the evidence from fruit flies. Curr Opin Neurobiol. 2004;14:737–744. doi: 10.1016/j.conb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Gerber B, Yarali A, Diegelmann S, Wotjak CT, Pauli P, Fendt M. Pain-relief learning in flies, rats, and man: basic research and applied perspectives. Learn Mem. 2014;21:232–252. doi: 10.1101/lm.032995.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasi N, Tchénio P, Preat T. PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron. 2010;65:516–529. doi: 10.1016/j.neuron.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Gitler D, Takagishi Y, Feng J, Ren Y, Rodriguiz RM, Wetsel WC, Greengard P, Augustine GJ. Different presynaptic roles of synapsins at excitatory and inhibitory synapses. J Neurosci. 2004;24:11368–11380. doi: 10.1523/JNEUROSCI.3795-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR. Type I repressors of P element mobility. Genetics. 1993;135:81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godenschwege TA, Reisch D, Diegelmann S, Eberle K, Funk N, Heisenberg M, Hoppe V, Hoppe J, Klagges BR, Martin JR, Nikitina EA, Putz G, Reifegerste R, Reisch N, Rister J, Schaupp M, Scholz H, Schwärzel M, Werner U, Zars TD, et al. Flies lacking all synapsins are unexpectedly healthy but are impaired in complex behaviour. Eur J Neurosci. 2004;20:611–622. doi: 10.1111/j.1460-9568.2004.03527.x. [DOI] [PubMed] [Google Scholar]
- Greco B, Managò F, Tucci V, Kao HT, Valtorta F, Benfenati F. Autism-related behavioral abnormalities in synapsin knockout mice. Behav Brain Res. 2013;251:65–74. doi: 10.1016/j.bbr.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Hellstern F, Malaka R, Hammer M. Backward inhibitory learning in honeybees: a behavioral analysis of reinforcement processing. Learn Mem. 1998;4:429–444. doi: 10.1101/lm.4.5.429. [DOI] [PubMed] [Google Scholar]
- Heth CD. Simultaneous and backward fear conditioning as a function of number of CS–UCS pairings. J Exp Psychol Anim Behav Process. 1976;2:117–129. doi: 10.1037/0097-7403.2.2.117. [DOI] [PubMed] [Google Scholar]
- Hilfiker S, Pieribone VA, Czernik AJ, Kao HT, Augustine GJ, Greengard P. Synapsins as regulators of neurotransmitter release. Philos Trans R Soc Lond B Biol Sci. 1999;354:269–279. doi: 10.1098/rstb.1999.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Suzuki K, Estes P, Ramaswami M, Yamamoto D, Strausfeld NJ. The organization of extrinsic neurons and their implications in the functional roles of the mushroom bodies in Drosophila melanogaster Meigen. Learn Mem. 1998;5:52–77. [PMC free article] [PubMed] [Google Scholar]
- Jungreis I, Lin MF, Spokony R, Chan CS, Negre N, Victorsen A, White KP, Kellis M. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011;21:2096–2113. doi: 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahsai L, Zars T. Learning and memory in Drosophila: behavior, genetics, and neural systems. Int Rev Neurobiol. 2011;99:139–167. doi: 10.1016/B978-0-12-387003-2.00006-9. [DOI] [PubMed] [Google Scholar]
- Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- Kim YC, Lee HG, Han KA. D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci. 2007;27:7640–7647. doi: 10.1523/JNEUROSCI.1167-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagges BR, Heimbeck G, Godenschwege TA, Hofbauer A, Pflugfelder GO, Reifegerste R, Reisch D, Schaupp M, Buchner S, Buchner E. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J Neurosci. 1996;16:3154–3165. doi: 10.1523/JNEUROSCI.16-10-03154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapek S, Gerber B, Tanimoto H. Synapsin is selectively required for anesthesia-sensitive memory. Learn Mem. 2010;17:76–79. doi: 10.1101/lm.1661810. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022X(84)90040-X. [DOI] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Liu C, Plaçais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, Tanimoto H. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488:512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- Löhr R, Godenschwege T, Buchner E, Prokop A. Compartmentalization of central neurons in Drosophila: a new strategy of mosaic analysis reveals localization of presynaptic sites to specific segments of neurites. J Neurosci. 2002;22:10357–10367. doi: 10.1523/JNEUROSCI.22-23-10357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr Biol. 2005;15:R700–R713. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R. The honeybee as a model for understanding the basis of cognition. Nat Rev Neurosci. 2012;13:758–768. doi: 10.1038/nrn3357. [DOI] [PubMed] [Google Scholar]
- Michels B, Diegelmann S, Tanimoto H, Schwenkert I, Buchner E, Gerber B. A role for Synapsin in associative learning: the Drosophila larva as a study case. Learn Mem. 2005;12:224–231. doi: 10.1101/lm.92805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels B, Chen YC, Saumweber T, Mishra D, Tanimoto H, Schmid B, Engmann O, Gerber B. Cellular site and molecular mode of synapsin action in associative learning. Learn Mem. 2011;18:332–344. doi: 10.1101/lm.2101411. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Bergado-Acosta JR, Fendt M. Relief learning is distinguished from safety learning by the requirement of the nucleus accumbens. Behav Brain Res. 2014;272:40–45. doi: 10.1016/j.bbr.2014.06.053. [DOI] [PubMed] [Google Scholar]
- Moscovitch A, LoLordo VM. Role of safety in the pavlovian backward fear conditioning procedure. J Comp Physiol Psychol. 1968;66:673–678. doi: 10.1037/h0026548. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ueno S, Sano A, Tanabe H. Polymorphisms of the human homologue of the Drosophila white gene are associated with mood and panic disorders. Mol Psychiatry. 1999;4:155–162. doi: 10.1038/sj.mp.4000515. [DOI] [PubMed] [Google Scholar]
- Navratilova E, Porreca F. Reward and motivation in pain and pain relief. Nat Neurosci. 2014;17:1304–1312. doi: 10.1038/nn.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewalda T, Völler T, Eschbach C, Ehmer J, Chou WC, Timme M, Fiala A, Gerber B. A combined perceptual, physico-chemical, and imaging approach to ‘odour-distances’ suggests a categorizing function of the Drosophila antennal lobe. PLoS One. 2011;6:e24300. doi: 10.1371/journal.pone.0024300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwal T, Heo S, Lubec G, Buchner E. Mass spectrometric analysis of synapsins in Drosophila melanogaster and identification of novel phosphorylation sites. J Proteome Res. 2011;10:541–550. doi: 10.1021/pr100746s. [DOI] [PubMed] [Google Scholar]
- Pech U, Pooryasin A, Birman S, Fiala A. Localization of the contacts between Kenyon cells and aminergic neurons in the Drosophila melanogaster brain using SplitGFP reconstitution. J Comp Neurol. 2013;521:3992–4026. doi: 10.1002/cne.23388. [DOI] [PubMed] [Google Scholar]
- Perisse E, Yin Y, Lin AC, Lin S, Huetteroth W, Waddell S. Different kenyon cell populations drive learned approach and avoidance in Drosophila. Neuron. 2013;79:945–956. doi: 10.1016/j.neuron.2013.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaçais PY, Trannoy S, Friedrich AB, Tanimoto H, Preat T. Two pairs of mushroom body efferent neurons are required for appetitive long-term memory retrieval in Drosophila. Cell Rep. 2013;5:769–780. doi: 10.1016/j.celrep.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Plotkin HC, Oakley DA. Backward conditioning in the rabbit (Oryctolagus cuniculus) J Comp Physiol Psychol. 1975;88:586–590. doi: 10.1037/h0076426. [DOI] [PubMed] [Google Scholar]
- Préat T. Decreased odor avoidance after electric shock in Drosophila mutants biases learning and memory tests. J Neurosci. 1998;18:8534–8538. doi: 10.1523/JNEUROSCI.18-20-08534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn WG, Harris WA, Benzer S. Conditioned behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1974;71:708–712. doi: 10.1073/pnas.71.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T, Völler T, Stock P, Buchner E, Fiala A. Punishment prediction by dopaminergic neurons in Drosophila. Curr Biol. 2005;15:1953–1960. doi: 10.1016/j.cub.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Sadanandappa MK, Blanco Redondo B, Michels B, Rodrigues V, Gerber B, VijayRaghavan K, Buchner E, Ramaswami M. Synapsin function in GABA-ergic interneurons is required for short-term olfactory habituation. J Neurosci. 2013;33:16576–16585. doi: 10.1523/JNEUROSCI.3142-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Völler T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, Fiala A. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Schulz RA, Chromey C, Lu MF, Zhao B, Olson EN. Expression of the D-MEF2 transcription in the Drosophila brain suggests a role in neuronal cell differentiation. Oncogene. 1996;12:1827–1831. [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séjourné J, Plaçais PY, Aso Y, Siwanowicz I, Trannoy S, Thoma V, Tedjakumala SR, Rubin GM, Tchénio P, Ito K, Isabel G, Tanimoto H, Preat T. Mushroom body efferent neurons responsible for aversive olfactory memory retrieval in Drosophila. Nat Neurosci. 2011;14:903–910. doi: 10.1038/nn.2846. [DOI] [PubMed] [Google Scholar]
- Selcho M, Pauls D, Han KA, Stocker RF, Thum AS. The role of dopamine in Drosophila larval classical olfactory conditioning. PLoS One. 2009;4:e5897. doi: 10.1371/journal.pone.0005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Rosahl TW, Chapman PF, Marowitz Z, Friedman E, Frankland PW, Cestari V, Cioffi D, Südhof TC, Bourtchuladze R. Impaired learning in mice with abnormal short-lived plasticity. Curr Biol. 1996;6:1509–1518. doi: 10.1016/S0960-9822(96)00756-7. [DOI] [PubMed] [Google Scholar]
- Solomon RL, Corbit JD. An opponent-process theory of motivation. I. Temporal dynamics of affect. Psychol Rev. 1974;81:119–145. doi: 10.1037/h0036128. [DOI] [PubMed] [Google Scholar]
- Südhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Tanimoto H, Heisenberg M, Gerber B. Experimental psychology: event timing turns punishment to reward. Nature. 2004;430:983. doi: 10.1038/430983a. [DOI] [PubMed] [Google Scholar]
- Thum AS, Jenett A, Ito K, Heisenberg M, Tanimoto H. Multiple memory traces for olfactory reward learning in Drosophila. J Neurosci. 2007;27:11132–11138. doi: 10.1523/JNEUROSCI.2712-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik SM, Davis RL. Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron. 2009;64:510–521. doi: 10.1016/j.neuron.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Vasin A, Zueva L, Torrez C, Volfson D, Littleton JT, Bykhovskaia M. Synapsin regulates activity-dependent outgrowth of synaptic boutons at the Drosophila neuromuscular junction. J Neurosci. 2014;34:10554–10563. doi: 10.1523/JNEUROSCI.5074-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AR. SOP: a model of automatic memory processing in animal behavior. In: Spear NE, Miller RR, editors. Information processing in animals: memory mechanisms. Hillsdale, NJ: Erlbaum; 1981. pp. 5–47. [Google Scholar]
- Wagner AR, Larew MB. Opponent processes and pavlovian inhibition. In: Miller RR, Spear NE, editors. Information processing in animals: conditioned inhibition. Hillsdale, NJ: Erlbaum; 1985. pp. 233–265. [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, Davis RL. Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics. 2015;199:1173–1182. doi: 10.1534/genetics.114.173575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarali A, Gerber B. A neurogenetic dissociation between punishment-, reward-, and relief-learning in Drosophila. Front Behav Neurosci. 2010;4:189. doi: 10.3389/fnbeh.2010.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarali A, Niewalda T, Chen Y, Tanimoto H, Duerrnagel S, Gerber B. ‘Pain relief’ learning in fruit flies. Anim Behav. 2008;76:1173–1185. doi: 10.1016/j.anbehav.2008.05.025. [DOI] [Google Scholar]
- Yarali A, Ehser S, Hapil FZ, Huang J, Gerber B. Odour intensity learning in fruit flies. Proc Biol Sci. 2009a;276:3413–3420. doi: 10.1098/rspb.2009.0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarali A, Krischke M, Michels B, Saumweber T, Mueller MJ, Gerber B. Genetic distortion of the balance between punishment and relief learning in Drosophila. J Neurogenet. 2009b;23:235–247. doi: 10.1080/01677060802441372. [DOI] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier KE, Hofbauer A, Heimbeck G, Pflugfelder GO, Buchner S, Buchner E. A cysteine-string protein is expressed in retina and brain of Drosophila. J Neurogenet. 1990;7:15–29. doi: 10.3109/01677069009084150. [DOI] [PubMed] [Google Scholar]
- Zinsmaier KE, Eberle KK, Buchner E, Walter N, Benzer S. Paralysis and early death in cysteine string protein mutants of Drosophila. Science. 1994;263:977–980. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]